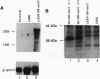
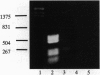
Abstract
Chronic myelogenous leukemia evolves in two clinically distinct stages: a chronic and a blast crisis phase. The molecular changes associated with chronic phase to blast crisis transition are largely unknown. We have identified a cDNA clone, DR-nm23, differentially expressed in a blast-crisis cDNA library, which has approximately 70% sequence similarity to the putative metastatic suppressor genes, nm23-H1 and nm23-H2. The deduced amino acid sequence similarity to the proteins encoded by these two latter genes is approximately 65% and includes domains and amino acid residues (the leucine zipper-like and the RGD domain, a serine and a histidine residue in the NH2- and in the COOH-terminal portion of the protein, respectively) postulated to be important for nm23 function. DR-nm23 mRNA is preferentially expressed at early stages of myeloid differentiation of highly purified CD34+ cells. Its constitutive expression in the myeloid precursor 32Dc13 cell line, which is growth-factor dependent for both proliferation and differentiation, results in inhibition of granulocytic differentiation induced by granulocyte colony-stimulating factor and causes apoptotic cell death. These results are consistent with a role for DR-nm23 in normal hematopoiesis and raise the possibility that its overexpression contributes to differentiation arrest, a feature of blastic transformation in chronic myelogenous leukemia.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedi A., Zehnbauer B. A., Barber J. P., Sharkis S. J., Jones R. J. Inhibition of apoptosis by BCR-ABL in chronic myeloid leukemia. Blood. 1994 Apr 15;83(8):2038–2044. [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Daley G. Q., Van Etten R. A., Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990 Feb 16;247(4944):824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gishizky M. L., Witte O. N. Initiation of deregulated growth of multipotent progenitor cells by bcr-abl in vitro. Science. 1992 May 8;256(5058):836–839. doi: 10.1126/science.1375394. [DOI] [PubMed] [Google Scholar]
- Gong J., Traganos F., Darzynkiewicz Z. A selective procedure for DNA extraction from apoptotic cells applicable for gel electrophoresis and flow cytometry. Anal Biochem. 1994 May 1;218(2):314–319. doi: 10.1006/abio.1994.1184. [DOI] [PubMed] [Google Scholar]
- Greenberger J. S., Sakakeeny M. A., Humphries R. K., Eaves C. J., Eckner R. J. Demonstration of permanent factor-dependent multipotential (erythroid/neutrophil/basophil) hematopoietic progenitor cell lines. Proc Natl Acad Sci U S A. 1983 May;80(10):2931–2935. doi: 10.1073/pnas.80.10.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groffen J., Stephenson J. R., Heisterkamp N., de Klein A., Bartram C. R., Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984 Jan;36(1):93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hailat N., Keim D. R., Melhem R. F., Zhu X. X., Eckerskorn C., Brodeur G. M., Reynolds C. P., Seeger R. C., Lottspeich F., Strahler J. R. High levels of p19/nm23 protein in neuroblastoma are associated with advanced stage disease and with N-myc gene amplification. J Clin Invest. 1991 Jul;88(1):341–345. doi: 10.1172/JCI115299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa M., Rukstalis D. B., Tanabe T., Chodak G. W. High levels of nm23 expression are related to cell proliferation in human prostate cancer. Cancer Res. 1994 Mar 1;54(5):1313–1318. [PubMed] [Google Scholar]
- Kantarjian H. M., Deisseroth A., Kurzrock R., Estrov Z., Talpaz M. Chronic myelogenous leukemia: a concise update. Blood. 1993 Aug 1;82(3):691–703. [PubMed] [Google Scholar]
- Keim D., Hailat N., Melhem R., Zhu X. X., Lascu I., Veron M., Strahler J., Hanash S. M. Proliferation-related expression of p19/nm23 nucleoside diphosphate kinase. J Clin Invest. 1992 Mar;89(3):919–924. doi: 10.1172/JCI115672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneuville P., Heisterkamp N., Groffen J. Expression of the chronic myelogenous leukemia-associated p210bcr/abl oncoprotein in a murine IL-3 dependent myeloid cell line. Oncogene. 1991 Feb;6(2):275–282. [PubMed] [Google Scholar]
- Leone A., Flatow U., King C. R., Sandeen M. A., Margulies I. M., Liotta L. A., Steeg P. S. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell. 1991 Apr 5;65(1):25–35. doi: 10.1016/0092-8674(91)90404-m. [DOI] [PubMed] [Google Scholar]
- Ma X. P., Calabretta B. DNA binding and transactivation activity of A-myb, a c-myb-related gene. Cancer Res. 1994 Dec 15;54(24):6512–6516. [PubMed] [Google Scholar]
- MacDonald N. J., De la Rosa A., Benedict M. A., Freije J. M., Krutsch H., Steeg P. S. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J Biol Chem. 1993 Dec 5;268(34):25780–25789. [PubMed] [Google Scholar]
- Mandai M., Konishi I., Koshiyama M., Mori T., Arao S., Tashiro H., Okamura H., Nomura H., Hiai H., Fukumoto M. Expression of metastasis-related nm23-H1 and nm23-H2 genes in ovarian carcinomas: correlation with clinicopathology, EGFR, c-erbB-2, and c-erbB-3 genes, and sex steroid receptor expression. Cancer Res. 1994 Apr 1;54(7):1825–1830. [PubMed] [Google Scholar]
- Miller A. D., Rosman G. J. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989 Oct;7(9):980-2, 984-6, 989-90. [PMC free article] [PubMed] [Google Scholar]
- Morishita K., Parganas E., Matsugi T., Ihle J. N. Expression of the Evi-1 zinc finger gene in 32Dc13 myeloid cells blocks granulocytic differentiation in response to granulocyte colony-stimulating factor. Mol Cell Biol. 1992 Jan;12(1):183–189. doi: 10.1128/mcb.12.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamori S., Ishikawa O., Ohhigashi H., Kameyama M., Furukawa H., Sasaki Y., Inaji H., Higashiyama M., Imaoka S., Iwanaga T. Expression of nucleoside diphosphate kinase/nm23 gene product in human pancreatic cancer: an association with lymph node metastasis and tumor invasion. Clin Exp Metastasis. 1993 Mar;11(2):151–158. doi: 10.1007/BF00114973. [DOI] [PubMed] [Google Scholar]
- Okabe-Kado J., Kasukabe T., Honma Y., Hayashi M., Henzel W. J., Hozumi M. Identity of a differentiation inhibiting factor for mouse myeloid leukemia cells with NM23/nucleoside diphosphate kinase. Biochem Biophys Res Commun. 1992 Feb 14;182(3):987–994. doi: 10.1016/0006-291x(92)91829-f. [DOI] [PubMed] [Google Scholar]
- Rosengard A. M., Krutzsch H. C., Shearn A., Biggs J. R., Barker E., Margulies I. M., King C. R., Liotta L. A., Steeg P. S. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989 Nov 9;342(6246):177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- Rovera G., Valtieri M., Mavilio F., Reddy E. P. Effect of Abelson murine leukemia virus on granulocytic differentiation and interleukin-3 dependence of a murine progenitor cell line. Oncogene. 1987 Mar;1(1):29–35. [PubMed] [Google Scholar]
- Rowley J. D. Identification of the constant chromosome regions involved in human hematologic malignant disease. Science. 1982 May 14;216(4547):749–751. doi: 10.1126/science.7079737. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E., Pierschbacher M. D. New perspectives in cell adhesion: RGD and integrins. Science. 1987 Oct 23;238(4826):491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- Sposi N. M., Zon L. I., Carè A., Valtieri M., Testa U., Gabbianelli M., Mariani G., Bottero L., Mather C., Orkin S. H. Cell cycle-dependent initiation and lineage-dependent abrogation of GATA-1 expression in pure differentiating hematopoietic progenitors. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6353–6357. doi: 10.1073/pnas.89.14.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl J. A., Leone A., Rosengard A. M., Porter L., King C. R., Steeg P. S. Identification of a second human nm23 gene, nm23-H2. Cancer Res. 1991 Jan 1;51(1):445–449. [PubMed] [Google Scholar]
- Yamashiro S., Urano T., Shiku H., Furukawa K. Alteration of nm23 gene expression during the induced differentiation of human leukemia cell lines. Oncogene. 1994 Sep;9(9):2461–2468. [PubMed] [Google Scholar]