Abstract
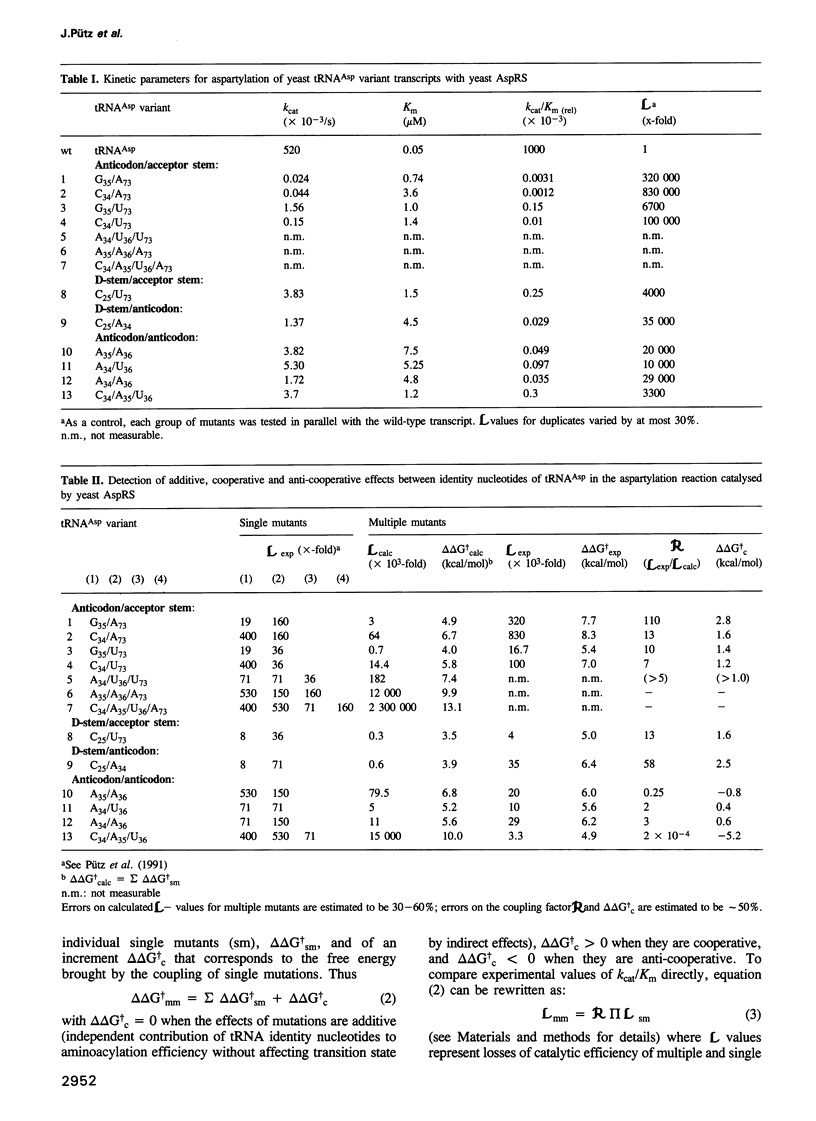
We have investigated the functional relationship between nucleotides in yeast tRNAAsp that are important for aspartylation by yeast aspartyl-tRNA synthetase. Transcripts of tRNAAsp with two or more mutations at identity positions G73, G34, U35, C36 and base pair G10-U25 have been prepared and the steady-state kinetics of their aspartylation were measured. Multiple mutations affect the catalytic activities of the synthetase mainly at the level of the catalytic constant, kcat. Kinetic data were expressed as free energy variation at transition state of these multiple mutants and comparison of experimental values with those calculated from results on single mutants defined three types of relationships between the identity nucleotides of this tRNA. Nucleotides located far apart in the three-dimensional structure of the tRNA act cooperatively whereas nucleotides of the anticodon triplet act either additively or anti-cooperatively. These results are related to the specific interactions of functional groups on identity nucleotides with amino acids in the protein as revealed by the crystal structure of the tRNAAsp/aspartyl-tRNA synthetase complex. These relationships between identity nucleotides may play an important role in the biological function of tRNAs.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackers G. K., Smith F. R. Effects of site-specific amino acid modification on protein interactions and biological function. Annu Rev Biochem. 1985;54:597–629. doi: 10.1146/annurev.bi.54.070185.003121. [DOI] [PubMed] [Google Scholar]
- Bare L. A., Uhlenbeck O. C. Specific substitution into the anticodon loop of yeast tyrosine transfer RNA. Biochemistry. 1986 Sep 23;25(19):5825–5830. doi: 10.1021/bi00367a072. [DOI] [PubMed] [Google Scholar]
- Bonnet J., Ebel J. P. Interpretation of incomplete reactions in tRNA aminoacylation. Aminoacylation of yeast tRNA Val II with yeast valyl-tRNA synthetase. Eur J Biochem. 1972 Dec 4;31(2):335–344. doi: 10.1111/j.1432-1033.1972.tb02538.x. [DOI] [PubMed] [Google Scholar]
- Carter P. J., Winter G., Wilkinson A. J., Fersht A. R. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell. 1984 Oct;38(3):835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Dietrich A., Kern D., Bonnet J., Giegé R., Ebel J. P. Interpretation of tRNA-mischarging kinetics. Eur J Biochem. 1976 Nov 1;70(1):147–158. doi: 10.1111/j.1432-1033.1976.tb10965.x. [DOI] [PubMed] [Google Scholar]
- Dreher T. W., Tsai C. H., Florentz C., Giegé R. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyl-tRNA synthetase determined by three anticodon loop nucleotides. Biochemistry. 1992 Sep 29;31(38):9183–9189. doi: 10.1021/bi00153a010. [DOI] [PubMed] [Google Scholar]
- Ebel J. P., Giegé R., Bonnet J., Kern D., Befort N., Bollack C., Fasiolo F., Gangloff J., Dirheimer G. Factors determining the specificity of the tRNA aminoacylation reaction. Non-absolute specificity of tRNA-aminoacyl-tRNA synthetase recognition and particular importance of the maximal velocity. Biochimie. 1973 May;55(5):547–557. doi: 10.1016/s0300-9084(73)80415-8. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry. 1987 Dec 15;26(25):8031–8037. doi: 10.1021/bi00399a001. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Knill-Jones J., Lowe D. M., Wilkinson A. J., Blow D. M., Brick P., Carter P., Waye M. M., Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985 Mar 21;314(6008):235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Florentz C., Dreher T. W., Rudinger J., Giege R. Specific valylation identity of turnip yellow mosaic virus RNA by yeast valyl-tRNA synthetase is directed by the anticodon in a kinetic rather than affinity-based discrimination. Eur J Biochem. 1991 Jan 1;195(1):229–234. doi: 10.1111/j.1432-1033.1991.tb15698.x. [DOI] [PubMed] [Google Scholar]
- Florentz C., Giegé R. Contact areas of the turnip yellow mosaic virus tRNA-like structure interacting with yeast valyl-tRNA synthetase. J Mol Biol. 1986 Sep 5;191(1):117–130. doi: 10.1016/0022-2836(86)90427-4. [DOI] [PubMed] [Google Scholar]
- Gangloff J., Keith G., Ebel J. P., Dirheimer G. Structure of aspartate-tRNA from brewer's yeast. Nat New Biol. 1971 Mar 24;230(12):125–126. doi: 10.1038/newbio230125a0. [DOI] [PubMed] [Google Scholar]
- Garcia A., Giege R. Footprinting evidence for close contacts of the yeast tRNA(Asp) anticodon region with aspartyl-tRNA synthetase. Biochem Biophys Res Commun. 1992 Jul 31;186(2):956–962. doi: 10.1016/0006-291x(92)90839-d. [DOI] [PubMed] [Google Scholar]
- Garcia A., Giegé R., Behr J. P. New photoactivatable structural and affinity probes of RNAs: specific features and applications for mapping of spermine binding sites in yeast tRNA(Asp) and interaction of this tRNA with yeast aspartyl-tRNA synthetase. Nucleic Acids Res. 1990 Jan 11;18(1):89–95. doi: 10.1093/nar/18.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Kern D., Ebel J. P. Incorrect aminoacylations catalysed by E. coli valyl-tRNA synthetase. Biochimie. 1972;54(10):1245–1255. doi: 10.1016/s0300-9084(72)80065-8. [DOI] [PubMed] [Google Scholar]
- Giegé R., Puglisi J. D., Florentz C. tRNA structure and aminoacylation efficiency. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- Lesser D. R., Kurpiewski M. R., Jen-Jacobson L. The energetic basis of specificity in the Eco RI endonuclease--DNA interaction. Science. 1990 Nov 9;250(4982):776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- Lorber B., Kern D., Dietrich A., Gangloff J., Ebel J. P., Giegé R. Large scale purification and structural properties of yeast aspartyl-tRNA synthetase. Biochem Biophys Res Commun. 1983 Nov 30;117(1):259–267. doi: 10.1016/0006-291x(83)91569-3. [DOI] [PubMed] [Google Scholar]
- Mans R. M., Pleij C. W., Bosch L. tRNA-like structures. Structure, function and evolutionary significance. Eur J Biochem. 1991 Oct 15;201(2):303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- Mildvan A. S., Weber D. J., Kuliopulos A. Quantitative interpretations of double mutations of enzymes. Arch Biochem Biophys. 1992 May 1;294(2):327–340. doi: 10.1016/0003-9861(92)90692-p. [DOI] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K., Schimmel P. Functional contacts of a transfer RNA synthetase with 2'-hydroxyl groups in the RNA minor groove. Nature. 1992 Jun 11;357(6378):513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Grosjean H., Ebel J. P., Florentz C., Giegé R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990 Apr 19;344(6268):787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J. P., Florentz C., Giegé R. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990 Oct;72(10):735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Bergdoll M., Dumas P., Vlassov V. V., Westhof E., Ebel J. P., Giegé R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J Mol Biol. 1985 Aug 5;184(3):455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Rudinger J., Puglisi J. D., Pütz J., Schatz D., Eckstein F., Florentz C., Giegé R. Determinant nucleotides of yeast tRNA(Asp) interact directly with aspartyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5882–5886. doi: 10.1073/pnas.89.13.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Behlen L. S., DiRenzo A. B., Uhlenbeck O. C. Recognition of yeast tRNA(Phe) by its cognate yeast phenylalanyl-tRNA synthetase: an analysis of specificity. Biochemistry. 1992 May 5;31(17):4161–4167. doi: 10.1021/bi00132a002. [DOI] [PubMed] [Google Scholar]
- Sarai A., Takeda Y. Lambda repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6513–6517. doi: 10.1073/pnas.86.17.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Parameters for the molecular recognition of transfer RNAs. Biochemistry. 1989 Apr 4;28(7):2747–2759. doi: 10.1021/bi00433a001. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. The anticodon contains a major element of the identity of arginine transfer RNAs. Science. 1989 Dec 22;246(4937):1595–1597. doi: 10.1126/science.2688091. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Senger B., Despons L., Walter P., Fasiolo F. The anticodon triplet is not sufficient to confer methionine acceptance to a transfer RNA. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10768–10771. doi: 10.1073/pnas.89.22.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söll D. The accuracy of aminoacylation--ensuring the fidelity of the genetic code. Experientia. 1990 Dec 1;46(11-12):1089–1096. doi: 10.1007/BF01936918. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Sarai A., Rivera V. M. Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proc Natl Acad Sci U S A. 1989 Jan;86(2):439–443. doi: 10.1073/pnas.86.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Tsai C. H., Dreher T. W. Second-site suppressor mutations assist in studying the function of the 3' noncoding region of turnip yellow mosaic virus RNA. J Virol. 1992 Sep;66(9):5190–5199. doi: 10.1128/jvi.66.9.5190-5199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A. Additivity of mutational effects in proteins. Biochemistry. 1990 Sep 18;29(37):8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- Wells T. N., Fersht A. R. Use of binding energy in catalysis analyzed by mutagenesis of the tyrosyl-tRNA synthetase. Biochemistry. 1986 Apr 22;25(8):1881–1886. doi: 10.1021/bi00356a007. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson A. J., Fersht A. R., Blow D. M., Winter G. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry. 1983 Jul 19;22(15):3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Chastain M., Puglisi J. D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991 Dec;11(6):764–769. [PubMed] [Google Scholar]



