Abstract
The development of the craniofacial muscles requires reciprocal interactions with surrounding craniofacial tissues that originate from cranial neural crest cells (CNCCs). However, the molecular mechanism involved in the tissue-tissue interactions between CNCCs and muscle progenitors during craniofacial muscle development is largely unknown. In the current study, we address how CNCCs regulate the development of the tongue and other craniofacial muscles using Wnt1-Cre; Alk5fl/fl mice, in which loss of Alk5 in CNCCs results in severely disrupted muscle formation. We found that Bmp4 is responsible for reduced proliferation of the myogenic progenitor cells in Wnt1-Cre; Alk5fl/fl mice during early myogenesis. In addition, Fgf4 and Fgf6 ligands were reduced in Wnt1-Cre; Alk5fl/fl mice and are critical for differentiation of the myogenic cells. Addition of Bmp4 or Fgf ligands rescues the proliferation and differentiation defects in the craniofacial muscles of Alk5 mutant mice in vitro. Taken together, our results indicate that CNCCs play critical roles in controlling craniofacial myogenic proliferation and differentiation through tissue-tissue interactions.
INTRODUCTION
The craniofacial musculature consists of the eye, masticatory, facial, tongue, and other head muscles. The development of the craniofacial musculature is distinct from that of the trunk in terms of the origin of the muscles and the genetic programs underlying myogenesis (1). Lineage-tracing approaches have indicated that most of the craniofacial muscles originate from cranial paraxial mesoderm, whereas tongue muscles originate from muscle precursors that migrate from the occipital somites and eye muscles originate from mixed populations of paraxial mesoderm and prechordal mesoderm (2, 3). Irrespective of this heterogeneity, the migrating myogenic precursors undergo proliferation and differentiation at designated sites, where they interact with surrounding tissues during the two-phase process of myogenesis. During primary myogenesis, myogenic precursors proliferate and generate primary myotubes; this occurs in mice during embryonic days 10.5 (E10.5) to E12.5. Secondary myofibers are created by the fusion of fetal myoblasts and preexisting primary myofibers or between primary myofibers at E14.5 to E17.5 (4).
Myogenic cells do not appear to have intrinsic muscle-patterning information but gain this information from interactions with surrounding tissues, such as tendons (5). Supporting tissues in the tongue bud primarily originate from cranial neural crest cells (CNCCs), which belong to a migratory multipotent population that gives rise to bones, connective tissues, nerves, glial cells, smooth muscle cells, dentin, and tendons in the craniofacial region (6, 7). The migration, specification, and survival of CNCCs play significant roles in craniofacial morphogenesis. The role of neural crest cells in myogenesis has been investigated in both trunk and craniofacial myogenesis. Neural crest cells induce myogenesis in somite dermomyotomes by secreting Notch ligands that transiently activate Notch signaling in myogenic progenitors (8). Previous studies using CNC ablation approaches have demonstrated that tissue-tissue interactions between CNCCs and craniofacial myogenic populations play a role in craniofacial myogenesis (9–12). Myogenic cells from muscleless chn mutant zebrafish were able to form normal branchial muscles after being grafted into wild-type hosts, suggesting that CNCCs play an instructive role in muscle formation (11). Taken together, these studies indicate that CNCCs control muscle patterning or differentiation; however, the underlying molecular and cellular mechanisms of the CNCC-myogenic interactions remain to be elucidated.
Transforming growth factor β (TGF-β) signaling in both myogenic precursors and CNCCs is important for tongue myogenesis. Specifically, our previous study has shown that loss of Smad4 in Myf5-positive muscle precursors caused defective muscle differentiation and fusion during tongue development without affecting cell migration or survival (13). We have also shown that loss of Tgfbr2 in CNCCs results in microglossia due to defects in myogenic cell proliferation and differentiation via tissue-tissue interactions (12). However, these muscle defects were not detectable at early myogenic stages, during which CNCCs guide migrating myogenic precursors for muscle growth and patterning. The signaling cascade downstream of TGF-β that controls the early primary myogenesis of tongue muscles is still poorly understood.
In this study, we investigated three different groups of craniofacial muscles, namely, the tongue, eye, and masticatory muscles, to study the molecular mechanism of tissue-tissue interactions between CNCCs and myogenic precursors. We show that the early formation of craniofacial muscles is severely affected in Wnt1-Cre; Alk5fl/fl mice. We found that the Alk5-mediated TGF-β signaling pathway in CNCCs regulates the expression of Bmp and Fgf genes during craniofacial primary myogenesis. Exogenous Bmp4 and Fgfs can rescue proliferation and differentiation defects in primary cell culture in vitro. Loss of Alk5 receptors in CNCCs also results in impaired tendon development and decreased Scleraxis expression, suggesting the Scleraxis expression in CNCCs is downstream of Alk5-mediated TGF-β signaling.
MATERIALS AND METHODS
Generation of Wnt1-Cre; Alk5fl/fl mice.
Wnt1-Cre transgenic mice have been described previously (7). We crossed Wnt1-Cre; Alk5fl/+ mice with Alk5fl/fl mice to generate Wnt1-Cre; Alk5fl/fl mice. Genotyping was carried out using PCR primers as previously described (14). Mice expressing ZsGreen Cre reporter were obtained from Jackson Laboratory. Animal usage was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Southern California.
Histological analysis and immunostaining.
Hematoxylin and eosin (H&E) and immunofluorescence staining were performed following standard procedures. The following antibodies were used for immunostaining: mouse anti-myosin heavy chain (anti-MHC) (DSHB), mouse anti-MyoD1 (Abcam), rabbit anti-phospho-histone H3 (Santa Cruz Biotechnology), rabbit anti-active caspase-3 (Abcam), rabbit anti-phospho-Smad1/5/8 (Cell Signaling), mouse anti-Pax7 (DSHB), rabbit antidesmin (Abcam), and mouse antimyogenin (Abcam). Following MHC immunostaining, immunofluorescence images were acquired after analyzing 10 fields from each condition. The results were assessed for statistical significance using Student's t test.
In situ hybridization.
In situ hybridization was performed following standard procedures. Digoxigenin-labeled antisense probes were generated from mouse cDNA clones that were kindly provided by several laboratories: Bmp4 (Malcolm Snead, University of Southern California), Pitx2 (Marina Campione, Albert Einstein College of Medicine), Fgf4 and Fgf6 (Pascal Maire, Institute Cochin, France), and Scleraxis (Eric N. Olson, University of Texas Southwestern Medical Center).
Quantitative RT-PCR.
The mRNA levels of Bmp4, Fgf4, Fgf6, and Scleraxis were analyzed by quantitative real-time reverse transcription (RT)-PCR (Bio-Rad iCycler system). Tongue primordium was dissected at E11.5, E12.5, and E13.5, and total RNA was subsequently extracted. The mRNAs were reverse transcribed into cDNAs using RNeasy Mini and QuantiTect reverse transcription kits (Qiagen), followed by real-time PCR with specific primers. Gene-specific primer sequences were obtained from the Primer Bank (15). Real-time PCR was performed using SYBR Super Mix kits (Bio-Rad). Values were normalized against GAPDH (glyceraldehyde-3-phosphate dehydrogenase) using the 2ΔΔCT method (16). Global gene expression analysis was performed as previously described (17). Data are shown as means and standard deviations (SD).
Cell culture.
Tongue primordium, eye, and masticatory muscle tissues were dissected from E12.5 or E13.5 embryos and cut into small pieces. The tissue blocks were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum at 37°C as previously described (12). The cultures were treated with recombinant mouse Bmp4 (15 ng/ml; R&D Systems) for 2 days for the proliferation assays. The cultures were switched to differentiation medium supplemented with 2% horse serum for 1 week for the differentiation assays. Where indicated, recombinant mouse Fgf4 or Fgf6 (10 ng/ml; R&D Systems) or recombinant mouse Bmp4 (15 ng/ml; R&D Systems) was added to the medium. The medium was changed every other day.
Tongue organ culture.
Timed-pregnant Wnt1-Cre; Alk5fl/fl mice were sacrificed at E11.5. The tongues were dissected and cultured in serum-free medium as previously described (12). Where indicated, the tongues were treated with Affi-Gel blue agarose beads (Bio-Rad) containing Bmp4 for 24 h in culture.
FACS.
At E12.5, tongue primary cells were isolated as described above. Fluorescence-activated cell sorting (FACS) analyses were performed as reported previously (12).
Microarray data accession numbers.
Gene expression profiling data from the mandibular arch and tongue bud from Alk5 mutant mouse models have been deposited in GEO under accession numbers GSE52357 and GSE52358, respectively.
RESULTS
Loss of the gene for TGF-β type 1 receptor (Alk5) in CNCCs results in a reduction of craniofacial muscles and microglossia.
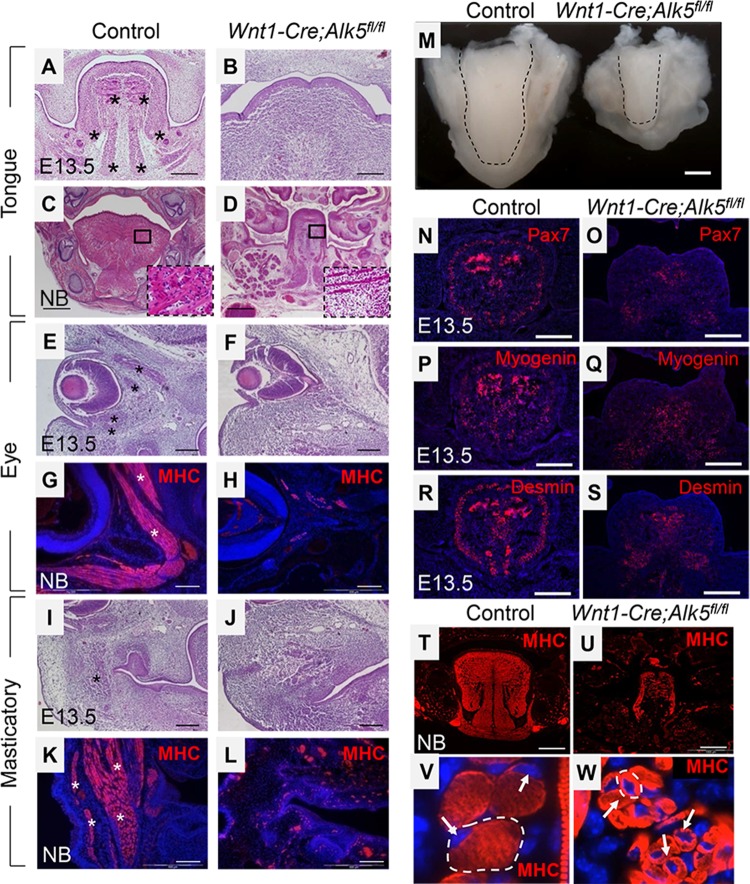
Mice lacking TGF-β type 1 receptor in CNCCs (Wnt1-Cre; Alk5fl/fl mice) exhibit craniofacial defects, including facial and palatal clefting, hemorrhage, mandibular hypoplasia, delayed tooth development, and microglossia (14, 18). In order to investigate the severity of the craniofacial muscle defects in Wnt1-Cre; Alk5fl/fl mice, we first examined the size and shape of tongues at E13.5 and the newborn stage. At E13.5, the sizes of control and Wnt1-Cre; Alk5fl/fl tongues were comparable, but the morphology of Wnt1-Cre; Alk5fl/fl tongues was abnormally wide and flat compared to the control tongues (data not shown). Histological analysis also confirmed that Wnt1-Cre; Alk5fl/fl tongues have strikingly abnormal morphology and reduced muscle mass at E13.5 (Fig. 1A and B). At the newborn stage, Wnt1-Cre; Alk5fl/fl mice exhibit microglossia with reduced tongue muscle formation (Fig. 1C, D, and M). The size of the myofibers in Wnt1-Cre; Alk5fl/fl tongues was also reduced compared to controls (Fig. 1T to W).
FIG 1.
Craniofacial muscle defects in the tongue, eye, and masticatory regions of Wnt1-Cre; Alk5fl/fl mice, including microglossia. (A to D) Hematoxylin and eosin staining of sections of tongues from E13.5 and newborn (NB) control and Wnt1-Cre; Alk5fl/fl mice. The boxed areas in panels C and D are shown magnified in the insets. (E to L) Hematoxylin and eosin staining and MHC immunostaining of the eye (E to H) and masticatory (I to L) regions from E13.5 and newborn control and Wnt1-Cre; Alk5fl/fl mice. The asterisks indicate muscle fibers. (M) Oral views of tongues from newborn control and Wnt1-Cre; Alk5fl/fl mice. The dashed lines indicate the outline of the tongue. (N to S) Immunostaining of the tongue from E13.5 control and Wnt1-Cre; Alk5fl/fl mice with myogenic markers Pax7, myogenin, and desmin. (T to W) MHC immunostaining of cross sections of tongues from newborn control and Wnt1-Cre; Alk5fl/fl mice. The arrows indicate nuclei that are centrally located in muscle fibers of Wnt1-Cre; Alk5fl/fl mice versus peripherally located in control mice. The dashed lines outline single myofibers. DAPI was used to counterstain nuclei (blue). Scale bars, 200 μm (A, B, E to L, and N to S), 500 μm (C, D, T, and U), and 1 mm (M).
Although most myoblasts were incorporated into myofibers in newborn Wnt1-Cre; Alk5fl/fl mice, the myofibers were dystrophic and immature. During muscle maturation, primary myofibers thicken by fusing with adjacent myoblasts or myofibers, resulting in peripherally located nuclei beneath the plasma membrane. In contrast, nuclei in Wnt1-Cre; Alk5fl/fl tongue myofibers were centrally located, indicating immature myofibers (Fig. 1V and W). We also found that myofibers of intrinsic and extrinsic tongue muscles in E13.5 Wnt1-Cre; Alk5fl/fl mice were disorganized compared to control tongues (Fig. 1C and D). We have found that loss of Alk5 signaling in CNCCs does not affect commitment of fetal myogenic cells based on Pax7, myogenin, and desmin expression (Fig. 1N to S). Thus, loss of Alk5 signaling in CNCCs affects the terminal differentiation of myogenic cells. Our results suggest that CNCCs exert control over tongue muscle formation, differentiation, maturation, and patterning. Furthermore, the muscle defects were not restricted to the tongue organ. Total muscle mass was also dramatically reduced in the eye and masticatory muscles of Wnt1-Cre; Alk5fl/fl mice, indicating that CNCCs play a crucial role in the formation of multiple craniofacial muscles (Fig. 1E to L).
Cell proliferation is significantly reduced but apoptosis is unaffected in myogenic cells of Wnt1-Cre; Alk5fl/fl mice.
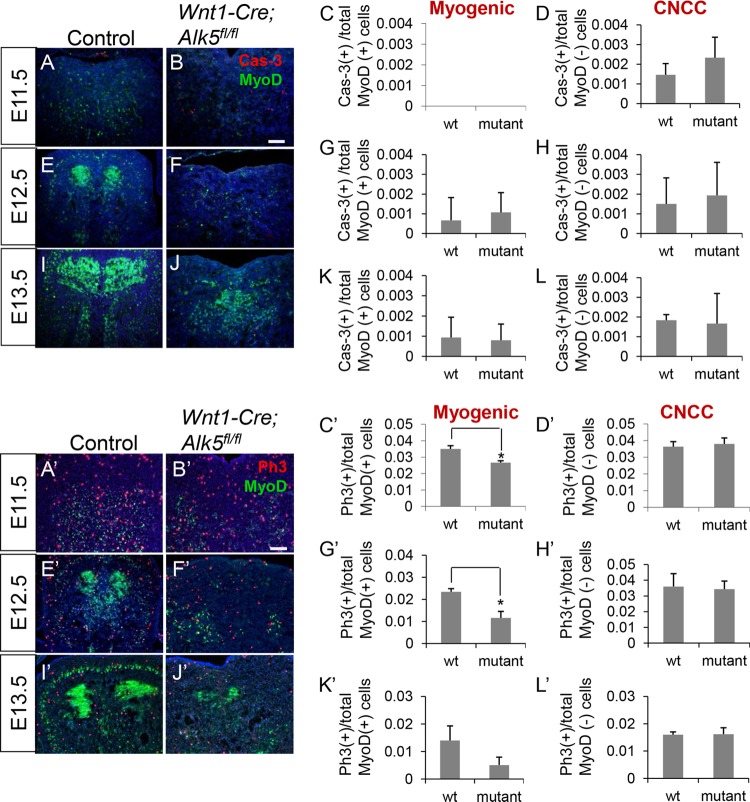
Previous reports have demonstrated that apoptosis is increased in the branchial arches at E10.5 in Wnt1-Cre; Alk5fl/fl mice (18). We hypothesized that reduced craniofacial muscle mass in Wnt1-Cre; Alk5fl/fl mice might be due to increased myogenic cell apoptosis, particularly since the migration of myogenic precursors was not affected. We analyzed tissue sections of E11.5-to-E13.5 embryos for apoptosis by coimmunostaining with anti-active caspase-3 and anti-MyoD, standard apoptosis and myogenic markers, respectively. We previously showed that CNCCs occupy the entire tongue primordium, surrounding myogenic precursors, by LacZ staining of Wnt1-Cre; R26R and Myf5-Cre; R26R mice at E11.5 (13). At E13.5, Wnt1-Cre; green fluorescent protein (GFP)-positive CNCCs and MyoD-positive cells also exhibit close spatial association in the craniofacial regions, indicating that MyoD-negative cells are mostly CNCCs (data not shown). Thus, myogenic and CNCC apoptosis indices were calculated by determining the number of caspase-3-positive cells out of the total number of MyoD-positive or MyoD-negative cells.
Although apoptosis of CNCCs was increased in the eye and masticatory areas in Wnt1-Cre; Alk5fl/fl mice at E11.5, we did not detect a significant increase in apoptosis in myogenic precursors (data not shown). Moreover, cell apoptosis was not significantly increased in either myogenic cells or CNCCs of Wnt1-Cre; Alk5fl/fl tongues (Fig. 2A to L). Thus, our data indicate that reduced muscle formation in Wnt1-Cre; Alk5fl/fl mice is not due to increased myogenic cell death in the tongue.
FIG 2.
Myogenic cell proliferation is significantly reduced but apoptosis is unaffected in tongues of Wnt1-Cre; Alk5fl/fl mice. (A, B, E, F, I, and J) Double immunostaining of caspase-3 (Cas-3) (red) and MyoD (green) in sections of the tongue region of E11.5, E12.5, and E13.5 control and Wnt1-Cre; Alk5fl/fl mice. (C, D, G, H, K, and L) Quantification of apoptosis in MyoD-positive myogenic cells and MyoD-negative CNCCs from control (wild-type [wt]) and Wnt1-Cre; Alk5fl/fl (mutant) mice. (A′, B′, E′, F′, I′, and J′) Double immunostaining of phospho-histone 3 (Ph3; red) and MyoD (green) in tongue sections from E11.5, E12.5 and E13.5 control and Wnt1-Cre; Alk5fl/fl mice. (C′, D′, G′, H′, K′, and L′) Quantification of proliferation in MyoD-positive myogenic cells and MyoD-negative CNCCs from control (wt) and Wnt1-Cre; Alk5fl/fl (mutant) mice. Scale bars, 100 μm. *, P < 0.05; n = 3. The error bars indicate standard deviation.
Next, we investigated whether proliferation of myogenic precursor cells is altered in Wnt1-Cre; Alk5fl/fl mice, resulting in decreased craniofacial muscle mass. We performed cell proliferation assays using tissue sections from E11.5, E12.5, and E13.5 Wnt1-Cre; Alk5fl/fl and control embryos (Fig. 2A′ to L′). We found that CNCC proliferation in tongues of Wnt1-Cre; Alk5fl/fl mice was indistinguishable from that in control mice (Fig. 2D′, H′, and L′), consistent with a previous report that mesenchymal cell proliferation in the branchial arches of Wnt1-Cre; Alk5fl/fl mice was unaffected (18). In contrast, myogenic cell proliferation was significantly reduced in the tongue (Fig. 2C′, G′, and K′) and in other craniofacial muscles (data not shown) of Wnt1-Cre; Alk5fl/fl mice at E11.5, E12.5, and E13.5. Thus, proliferation of tongue, eye, and masticatory myogenic cells during craniofacial development requires Alk5-mediated TGF-β signaling in CNCCs.
Downregulation of Bmp4 and Fgfs in the tongues of Wnt1-Cre; Alk5fl/fl mice.
To identify candidate genes that might be responsible for the muscle defects in Wnt1-Cre; Alk5fl/fl tongues, we performed microarray assays using tongue bud tissues from E11.5 and E13.5 Wnt1-Cre; Alk5fl/fl and control mice. We chose E11.5 and E13.5 because they represent key stages of early tongue myogenesis. Between E11.5 and E12.5, myogenic precursors migrate to the tongue bud and have direct contact with CNCCs, after which the myogenic population expands. By E13.5, myoblasts initiate terminal differentiation to generate multinucleate myofibers. We focused on downregulated genes based on the hypothesis that the reduction of a signaling molecule(s) from CNCCs results in the defects in myogenic cell proliferation and differentiation. We identified genes with greater than 1.5-fold changes and a P value of less than 0.05, indicating a significant difference. Based on preliminary phenotype analyses, we identified genes associated with cell proliferation, muscle differentiation, and tendon formation, as listed in Table 1 (GSE52357) and Table 2 (GSE52358). There was no overlap between the lists of downregulated genes at E11.5 and E13.5, suggesting that different signaling pathways govern the proliferation and differentiation processes.
TABLE 1.
Downregulated genes related to muscle development in tongue buds from E11.5 Wnt1-Cre; Alk5fl/fl and control mice
| Gene name | Gene symbol | Fold changea | P valueb |
|---|---|---|---|
| Six1 | Sine oculis-related homeobox 1 | −1.88 | 0.008809 |
| Eya1 | Eyes absent 1 homolog | −1.69 | 0.016867 |
| Eya4 | Eyes absent 4 homolog | −2.17 | 0.021839 |
| Lef1 | Lymphoid enhancer binding factor 1 | −1.80 | 0.042653 |
| Pax3 | Paired box gene 3 | −4.01 | 0.001484 |
| Pitx1 | Paired-like homeodomain transcription factor 1 | −2.14 | 0.001238 |
| Bmp4 | Bone morphogenetic protein 4 | −2.23 | 0.021048 |
| Fst | Follistatin | −3.14 | 0.030522 |
Fold change > 1.5.
P value < 0.05.
TABLE 2.
Downregulated genes in tongues from E13.5 Wnt1-Cre;Alk5fl/fl and control mice
| Gene function and name | Gene symbol | Fold changea | P valueb |
|---|---|---|---|
| Muscle differentiation | |||
| Tcap | Titin-cap | −1.61 | 0.003541 |
| Myl3 | Myosin, light polypeptide 3 | −1.56 | 0.006601 |
| Cav3 | Caveolin3 | −2.00 | 0.014199 |
| Tnnc1 | Troponin C, cardiac/slow skeletal | −1.64 | 0.021082 |
| Tnnc2 | Troponin C2, fast | −1.97 | 0.000987 |
| Tnni1 | Troponin I, skeletal, slow 1 | ||
| Tnni2 | Troponin I, skeletal, fast 2 | −1.94 | 0.000210 |
| Tnnt2 | Troponin T2, cardiac | −1.82 | 0.000902 |
| Tnnt3 | Troponin T3, skeletal, fast | −2.35 | 0.000898 |
| Casq1 | Calsequestrin 1 | −1.63 | 0.031895 |
| Chrna1 | Cholinergic receptor, nicotinic, alpha polypeptide 1 | −1.72 | 0.002628 |
| Acta1 | Actin, alpha 1 | −2.04 | 0.009624 |
| Tmod1 | Tropomodulin 1 | −1.57 | 0.010005 |
| Myf6 | Myogenic factor 6 | −2.41 | 0.044509 |
| Fgf4 | Fibroblast growth factor 4 | −1.85 | 0.003976 |
| Fgf6 | Fibroblast growth factor 6 | −2.26 | 0.000587 |
| Connective tissue and tendon formation | |||
| Scx | Scleraxis | −2.40 | 4.01E−05 |
| Thbs4 | Thrombospondin 4 | −3.24 | 2.23E−05 |
| Col8a1 | Collagen | −2.78 | 5.85E−06 |
| Col19a1 | Collagen | −2.48 | 0.002539 |
| Col6a1 | Collagen | −1.89 | 0.011672 |
| Cstl5 | Follistatin-like 5 | −1.86 | 0.000436 |
| Cbln5 | Fibulin 5 | −1.90 | 0.003835 |
| Tgfb3 | Transforming growth factor β3 | −1.58 | 0.005932 |
Fold change > 1.5.
P value < 0.05.
In the tongue bud tissue of Wnt1-Cre; Alk5fl/fl mice, only a small number of genes were downregulated at E11.5 (Table 1), including transcription factor genes Six1, Eya1, Eya4, and Pax3, which are expressed in myogenic precursors during early myogenesis (19). Follistatin, which has been reported to be expressed during muscle growth (20), was also downregulated. In E13.5 tongue samples, we identified downregulated genes that are involved in aspects of muscle differentiation, such as myofiber assembly and muscle contraction, including Troponin C, Troponin I, Troponin T, Alpha 1 actin, and Myosin light chain (Table 2). Myogenic factor 6 (Myf6), whose product is also known as MRF4 or herculin, was also downregulated. Mrf4 encodes one of the myogenic regulatory factors (MRFs) that regulate fusion of myoblasts and differentiation (21). Alteration of these genes may explain the myogenic differentiation defects in Wnt1-Cre; Alk5fl/fl mice. In addition, we found that connective-tissue- and tendon-specific genes were downregulated, among them collagen genes, thrombospondin 4, and Scleraxis. Scleraxis-positive cells are tendon precursors, and CNCCs give rise to tendons, as well as craniofacial connective tissues. Scleraxis downregulation in Wnt1-Cre; Alk5fl/fl tongues may be relevant to the phenotypes in craniofacial muscle patterning and organization, because tendon precursors develop in close spatial proximity to myogenic precursors in order to connect muscles to bones.
In search of possible downstream regulators of Alk5-mediated TGF-β signaling, we found that Bmp4 transcripts were downregulated in Wnt1-Cre; Alk5fl/fl mice at E11.5 (Table 1). BMP signaling regulates the expansion of fetal myogenic progenitors during skeletal muscle development (22). We also found that Fgf4 and Fgf6 were downregulated in E13.5 tongue buds of Wnt1-Cre; Alk5fl/fl mice (Table 2). We previously showed that Fgf6 regulates myofiber fusion and differentiation in the tongue (13). Therefore, we decided to test whether Bmp4, Fgf4, and Fgf6 were responsible for the muscle defects in Wnt1-Cre; Alk5fl/fl mice.
Altered Bmp4 expression and Smad1/5/8 activation in Wnt1-Cre; Alk5fl/fl mice.
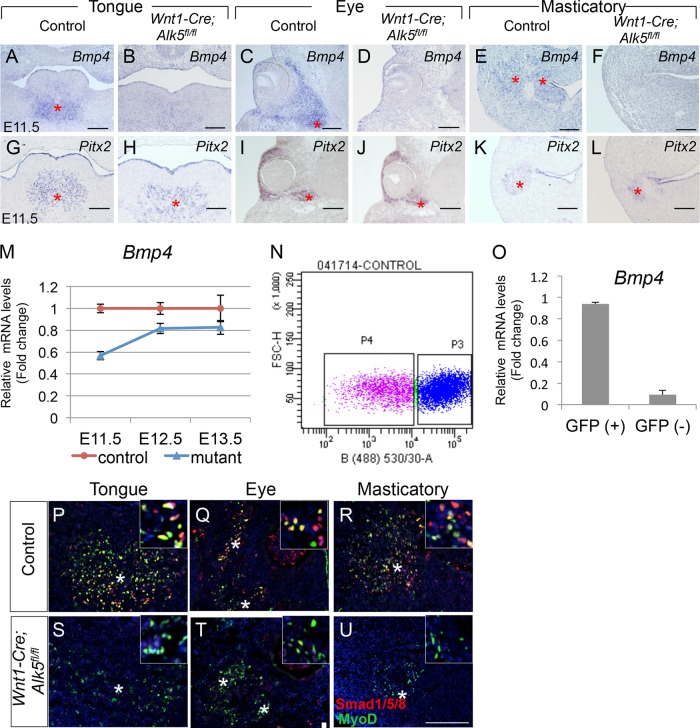
BMP signaling is required for bone and cartilage formation, but it also plays a role in muscle cell survival and differentiation. Recent studies have shown that BMP4 activates fetal and adult myogenic cell proliferation, in addition to inhibiting myogenic differentiation (22, 23). We performed real-time RT-PCR and found that Bmp4 mRNA expression was significantly reduced in the tongues of Wnt1-Cre; Alk5fl/fl mice at E11.5 and E12.5 (Fig. 3M). Next, we examined the expression patterns of Bmp4 and Pitx2 in the tongue bud, eye, and masticatory regions by in situ hybridization. Bmp4 expression was widespread in the mesenchyme and epithelium of the developing tongue and other craniofacial regions at E11.5 (Fig. 3A, C, and E). Pitx2, which labels cranial myogenic precursors (24), was detectable in a region adjacent to that of Bmp4-positive mesenchymal cells (Fig. 3G, I, and K), suggesting that Bmp4-expressing CNCCs have a close spatial relationship with myogenic precursor cells. In Wnt1-Cre; Alk5fl/fl mice, however, Bmp4 expression was dramatically reduced in the mesenchyme, whereas Pitx2-positive myogenic precursors were still detectable in all the craniofacial muscle anlagen (Fig. 3B, D, F, H, J, and L). To determine whether Bmp4 is expressed in CNCCs, CNC-derived (GFP-positive) cells were separated from myogenic (GFP-negative) cells from tongue cultures of Wnt1-Cre; ZsGreen mice by flow cytometric (FACS) analysis, followed by real-time RT-PCR (Fig. 3N and O). We found that Bmp4 expression was significantly reduced in the myogenic population compared to the CNC-derived GFP-positive population (Fig. 3O).
FIG 3.
Bmp4 expression in the mesenchyme and Smad1/5/8 activation in craniofacial muscles are reduced in Wnt1-Cre; Alk5fl/fl mice. (A to L) In situ hybridization of Bmp4 and Pitx2 in the tongue, eye, and masticatory regions of E11.5 control and Wnt1-Cre; Alk5fl/fl mice. The asterisks indicate expression of Bmp4 in control mice (A, C, and E) and migrated myogenic precursors (G to L). (M) Real-time RT-PCR of Bmp4 in E11.5, E12.5, and E13.5 control and Wnt1-Cre; Alk5fl/fl (mutant) mice. (N) Flow cytometry plot of GFP-positive cells (CNC derived; P3) and GFP-negative cells (myogenic cells; P4) from tongue cultures. (O) Real-time RT-PCR of Bmp4 expression in tongue CNC-derived cells [GFP (+)] and myogenic cells [GFP (–)] from control and Wnt1-Cre; ZsGreen mice. (P to U) Double immunostaining of phosphorylated Smad1/5/8 (red) and MyoD (green) in the tongue, eye, and masticatory regions of control and Wnt1-Cre; Alk5fl/fl mice. The asterisks indicate myogenic precursors. The insets show higher magnification. Scale bars, 200 μm (A to L) and 100 μm (P to U). The error bars indicate standard deviation.
To confirm that Bmp4 from CNCCs transduces Bmp signaling in myogenic cells, we examined the phosphorylation levels of Smad1/5/8, which are Bmp4 intracellular signaling mediators. Phosphorylation of Smad1/5/8 was detectable in approximately half of the myogenic precursors and was not detectable in the surrounding mesenchyme of the craniofacial area in E11.5 control mice (Fig. 3P to R). The phopho-Smad1/5/8-positive, MyoD-negative cells are most likely myogenic lineages that are not labeled by MyoD. In Wnt1-Cre; Alk5fl/fl mice, we failed to detect the phosphorylation of Smad1/5/8 in E11.5 myogenic precursors, indicating that Bmp4-controlled downstream Smad1/5/8 signaling was abrogated (Fig. 3S to U).
Addition of Bmp4 rescues the myogenic cell proliferation defect in Wnt1-Cre; Alk5fl/fl myogenic cells in vitro.
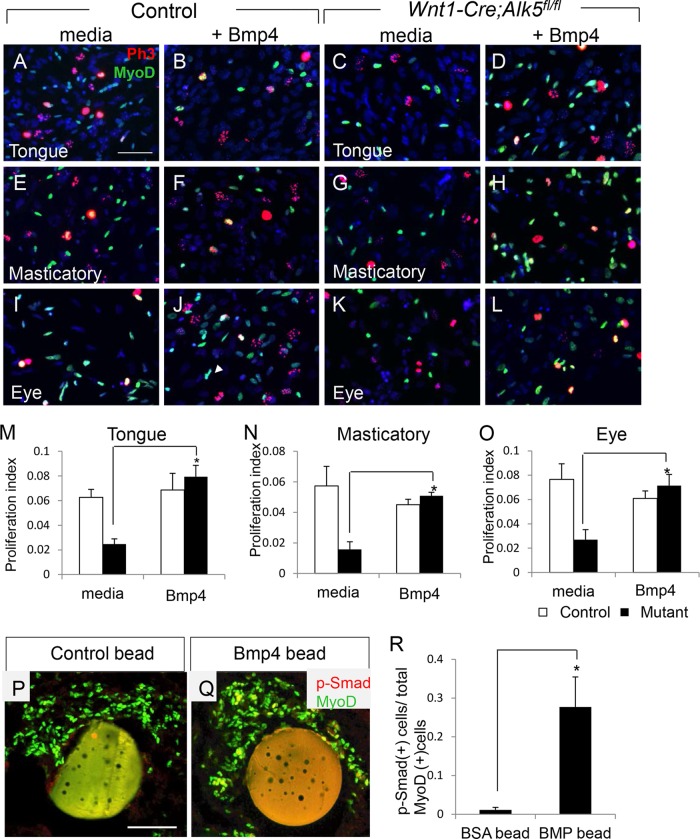
To examine the requirement for Bmp4 in myogenic cell proliferation in the craniofacial region, we added exogenous recombinant Bmp4 to in vitro cell cultures of embryonic fibroblasts isolated from eye, masticatory, and tongue tissues at E12.5. We chose embryonic stage E12.5 because myogenic proliferation defects occur at both E11.5 and E12.5, but the tongue primordium is more defined for microdissection at E12.5, helping to prevent contamination by mandibular tissues that may also be responsive to Bmp for osteogenesis. Myogenic cell proliferation was decreased 2-fold in Wnt1-Cre; Alk5fl/fl samples compared to control samples in medium (Fig. 4A, C, E, G, I, and K), consistent with our in vivo data (Fig. 2). The addition of Bmp4 to the culture medium rescued the reduction in cell proliferation in Wnt1-Cre; Alk5fl/fl samples (Fig. 4D, H, L, and M to O). We did not detect any effect on myogenic cell proliferation after addition of Fgf4 or Fgf6 (data not shown). Thus, our data indicate that Bmp4 expression in CNCCs is required for activating the proliferation of craniofacial myogenic cells during early myogenesis.
FIG 4.
Exogenous Bmp4 rescues the proliferation defect in Wnt1-Cre; Alk5fl/fl mice. (A to L) Double immunostaining of phospho-histone 3 (red) and MyoD (green) in cells from the tongue (A to D), masticatory (E to H), and eye muscle (I to L) regions of control and Wnt1-Cre; Alk5fl/fl mice that were untreated (media) or treated with exogenous Bmp4. (M to O) Quantification of proliferation in MyoD-positive myogenic cells from control or Wnt1-Cre; Alk5fl/fl (mutant) mice that were untreated (media) or treated with Bmp4. *, P < 0.05; n = 6. (P and Q) Coimmunostaining of MyoD and phospho-Smad1/5/8 in tongue explants from E11.5 Wnt1-Cre; Alk5fl/fl mice cultured with BSA control (P) or Bmp4 (Q) beads. (R) Quantification of phospho-Smad1/5/8-positive myogenic cells from the tongue explants treated with control (BSA) or Bmp4 (BMP) beads. Scale bars, 50 μm. The error bars indicate standard deviation.
In order to confirm that the rescue of proliferation by exogenous Bmp4 treatment correlates with restoration of phospho-Smad1/5/8 in Wnt1-Cre; Alk5fl/fl myogenic cells, we performed organ culture using E12.5 tongues with bovine serum albumin (BSA)-loaded (control) or Bmp4-loaded beads. As expected, we failed to detect phosphorylation of Smad1/5/8 in Wnt1-Cre; Alk5fl/fl tongues treated with control beads (Fig. 4P). However, the levels of phospho-Smad1/5/8 were restored in myogenic cells in Wnt1-Cre; Alk5fl/fl tongue explants treated with Bmp4-loaded beads (Fig. 4Q and R). Thus, exogenous Bmp4 can restore the activation of phospho-Smad1/5/8, which is critical for myogenic cell proliferation, and rescue the muscle cell proliferation defect in Wnt1-Cre; Alk5fl/fl mice.
Differentiation defects in Wnt1-Cre; Alk5fl/fl mice are due to decreased expression of Fgf4 and Fgf6 in the craniofacial muscles and can be rescued with exogenous Fgf proteins.
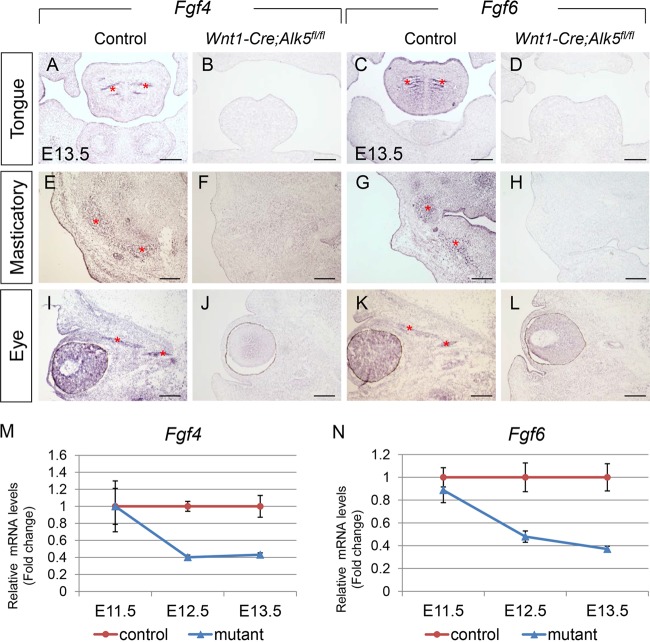
Based on our microarray data from E13.5 tongues, Fgf4 and Fgf6 are downregulated in Wnt1-Cre; Alk5fl/fl mice, suggesting a role for Fgfs in tongue myogenesis. We confirmed the microarray data using quantitative real-time RT-PCR (Fig. 5M and N). mRNA levels of Fgf4 and Fgf6 were reduced in Wnt1-Cre; Alk5fl/fl tongues to about 40% that of controls starting at E12.5, which corresponds to the time at which muscle differentiation begins. We also examined the expression patterns of Fgf4 and Fgf6 in the craniofacial region using in situ hybridization. We detected decreased expression of Fgf4 and Fgf6 in the tongue muscles, as well as in the ocular and masticatory muscles, of Wnt1-Cre; Alk5fl/fl mice compared to controls (Fig. 5A to L). Our results suggest that CNCC signals are either directly or indirectly required for Fgf expression in myogenic cells.
FIG 5.
Fgf4 and Fgf6 expression is reduced in craniofacial muscles of Wnt1-Cre; Alk5fl/fl mice. (A to L) In situ hybridization of Fgf4 and Fgf6 in the tongue, eye, and masticatory muscles of E13.5 control and Wnt1-Cre; Alk5fl/fl mice. The asterisks indicate muscle fibers. (M and N) Real-time RT-PCR of Fgf4 (M) and Fgf6 (N) in E11.5, E12.5, and E13.5 control and Wnt1-Cre; Alk5fl/fl (mutant) mice. Scale bars, 200 μm. The error bars indicate standard deviation.
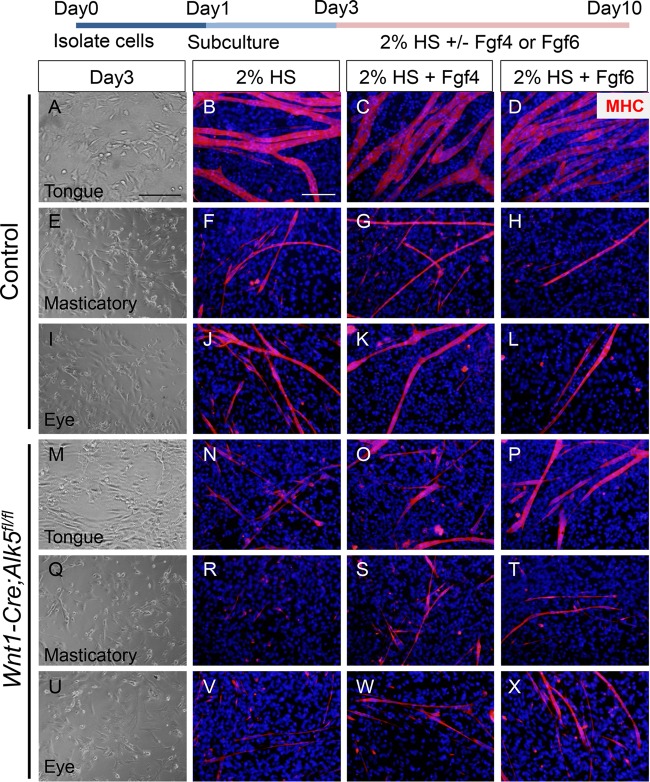
Fgf6 is expressed in developing tongue muscles and is responsible for muscle differentiation, especially muscle fusion (13). To evaluate the role of reduced expression of Fgf4 and Fgf6 in the muscle differentiation defect of Wnt1-Cre; Alk5fl/fl mice, we added exogenous Fgf4 and Fgf6 to an in vitro cell culture of mouse embryonic fibroblasts isolated from eye, masticatory, and tongue bud muscle tissues at E13.5. After culture in differentiation medium for a week, myofibers from Wnt1-Cre; Alk5fl/fl mice were poorly differentiated, as well as thinner and shorter than myofibers from control mice (Fig. 6B, F, J, N, R, and V). Strikingly, addition of Fgf4 or Fgf6 resulted in partial rescue of myofiber formation (Fig. 6C, D, G, H, K, L, O, P, S, T, W, and X). However, the addition of both Fgf4 and Fgf6 did not show any synergistic effect on myofiber formation in vitro, suggesting that other growth factors may also be involved in muscle differentiation (data not shown). Therefore, we conclude that the muscle differentiation defect in Wnt1-Cre; Alk5fl/fl mice is due to reduced Fgf4 and Fgf6 expression, which is downstream of Alk5-mediated TGF-β signaling in CNCCs.
FIG 6.
Exogenous Fgf4 and Fgf6 partially rescue myogenic differentiation in Wnt1-Cre; Alk5fl/fl mice. The diagram at the top depicts the experimental design. Primary cells from tongue, masticatory, and eye muscle tissues of control and Wnt1-Cre; Alk5fl/fl mice were isolated and cultured in growth medium. After 2 days, the primary cells were stimulated for differentiation in 2% horse serum medium (2% HS) with or without Fgf4 or Fgf6 for a week. (A to X) Light microscopy of cultures at day 3 (A, E, I, M, Q, and U) and MHC immunostaining of cultures at day 10 (B to D, F to H, J to L, N to P, R to T, and V to X). Scale bars, 100 μm (A, E, I, M, Q, and U) and 50 μm (B to D, F to H, J to L, N to P, R to T, and V to X).
We investigated whether Bmp4 expression during early myogenesis is responsible for the expression of Fgf4 and Fgf6 during later stages of myogenesis. To test this hypothesis, we cultured myogenic cells from Wnt1-Cre; Alk5fl/fl mice in the presence of Bmp4 and Fgf4/6 to determine if they had a synergistic effect in the rescue of differentiation of mutant myogenic cells. Surprisingly, Bmp4 inhibited myogenic cell differentiation of not only the Wnt1-Cre; Alk5fl/fl myogenic cells but also the control myogenic cells (data not shown). Previous studies have shown an inhibitory effect of Bmp, suggesting that Bmp inhibits premature muscle cell differentiation (25). Therefore, Bmp4 is not upstream of Fgf expression but inhibits muscle differentiation of myogenic cells during fetal myogenesis. In addition, we analyzed whether exogenous Bmp4 can rescue Fgf4 and Fgf6 expression in the Wnt1-Cre; Alk5fl/fl tongues by culturing Wnt1-Cre; Alk5fl/fl tongue buds for 24 h with control or Bmp4 beads and found that neither Fgf4 nor Fgf6 expression was rescued by exogenous Bmp4 (data not shown). Thus, the induction of Fgf4 and Fgf6 expression by CNCCs does not appear to be downstream of Bmp4.
CNC-derived tendon formation is compromised by loss of Alk5 signaling.
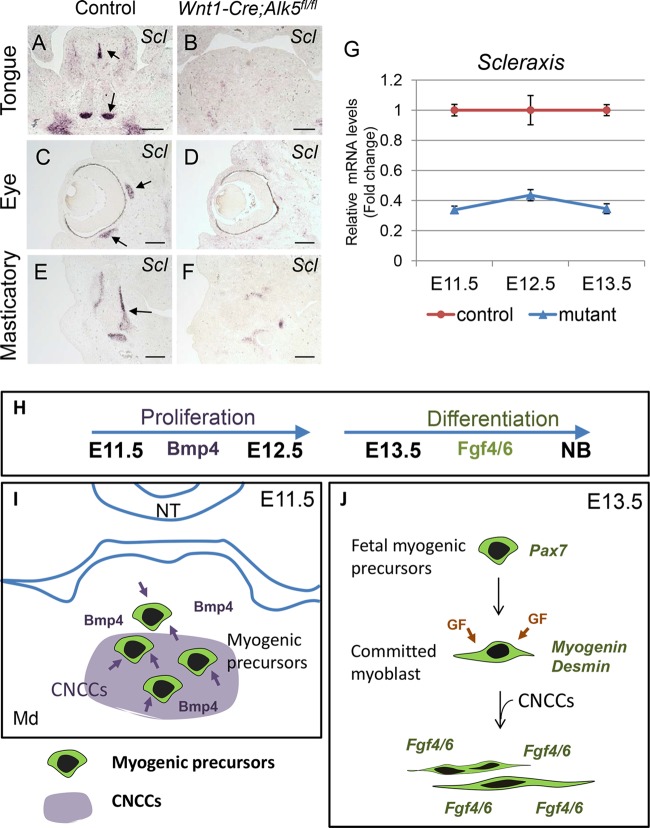
Muscle and tendon development involves reciprocal tissue-tissue interactions for the establishment of the musculoskeletal system (26). To determine if the induction and differentiation of tendon precursors are also disrupted in Wnt1-Cre; Alk5fl/fl mice, we analyzed the Scleraxis expression pattern in craniofacial tissue using in situ hybridization. Scleraxis is a bHLH transcription factor involved in the formation of tendons and connective tissues (27). At E13.5, the expression of Scleraxis was detectable and restricted to the musculotendinous junctions in craniofacial muscles, the lingual septum, and the tendons connecting the genioglossus in the tongue (Fig. 7A, C, and E). In contrast, Scleraxis expression in Wnt1-Cre; Alk5fl/fl mice was dramatically reduced overall at E13.5 (Fig. 7B, D, and F). Real-time RT-PCR confirmed that Scleraxis expression was reduced in Wnt1-Cre; Alk5fl/fl mice at E11.5, E12.5, and E13.5, indicating that Scleraxis expression is downstream of Alk5-mediated TGF-β signaling in CNCCs (Fig. 7G).
FIG 7.
Tendon differentiation is compromised in the craniofacial region of Wnt1-Cre; Alk5fl/fl mice. (A to F) In situ hybridization of Scleraxis in the tongue (A and B), eye (C and D), and masticatory (E and F) regions of E13.5 control and Wnt1-Cre; Alk5fl/fl mice. The arrows indicate expression of Scleraxis in control mice. (G) Real-time RT-PCR of Scleraxis in the tongues of E11.5, E12.5, and E13.5 control and Wnt1-Cre; Alk5fl/fl (mutant) mice. Scale bars, 200 μm. (H to J) The Alk5-mediated TGF-β signaling pathway in CNCCs contributes to early and late myogenesis in the craniofacial region. Bmp4 from CNCCs induces proliferation of myogenic precursors during early myogenesis at E11.5 and E12.5. Later, TGF-β signaling in CNCCs induces the expression of Fgf4 and Fgf6 in myogenic cells, which is critical for myogenic differentiation of the craniofacial muscles. NT, neural tube; Md, mandible; GF, growth factors. The error bars indicate standard deviation.
DISCUSSION
TGF-β signaling in CNCCs in craniofacial myogenesis.
We have found that Bmp4 and Fgf4/6 act downstream of Alk5-mediated TGF-β signaling to control the proliferation of myogenic precursors and the initiation of muscle differentiation during early stages of craniofacial myogenesis. Noncanonical TGF-β signaling in CNCCs also plays a significant role in regulating muscle organization, proliferation, and differentiation during secondary myogenesis. We recently reported that craniofacial muscles, including the tongue, were also malformed in Wnt1-Cre; Tgfbr2fl/fl mice, and this phenotype was evident during secondary myogenesis (12, 28). In Wnt1-Cre; Tgfbr2fl/fl mice, noncanonical TGF-β signaling in the absence of TGF-β type 2 receptor (Tgfbr2) leads to misregulation of the Bmp5 and Fgf4/5/6 signaling pathways, affecting fetal myoblast cell proliferation and muscle differentiation. Thus, both canonical and noncanonical TGF-β signaling in CNCCs control the different stages of craniofacial myogenesis by activating alternative signaling mediators.
Bmp4 from CNCCs is critical for the proliferation of embryonic myogenic progenitors.
In this study, we have demonstrated that Bmp4 ligands are expressed in the tongue bud, eye, and masticatory regions in tissues adjacent to developing muscles. Bmps are members of the TGF-β superfamily and play a crucial role in cardiac and skeletal myogenesis (29). Bmp signaling from neural crest cells is involved in the inhibition of myogenic differentiation in somites. Previous studies have shown that Bmp4 signaling controls cell proliferation of Pax7-positive myogenic precursors (22). Bmp stimulates fetal muscle growth by increasing the number of satellite cells in chick limbs, but it is unclear whether Bmp4 also has a stimulatory effect on the proliferation of myogenic precursors during embryonic stages (22). Here, we have shown that Bmp4 is expressed in the mesenchyme that surrounds myogenic precursors and that Bmp4 induces proliferation during early embryonic myogenesis in the craniofacial region. CNCC expression of Bmp4 may promote expansion of myogenic precursors before they undergo terminal differentiation. Our finding that Bmp4 beads can restore Bmp signaling in Alk5 mutant tongue explants, as evidenced by the presence of phospho-Smad1/5/8, suggests that Bmp4 acts downstream of Alk5-mediated TGF-β signaling to control muscle formation.
Fgfs play an important role in primary muscle differentiation.
Members of the fibroblast growth factor (FGF) family control a variety of cellular events in limb development, with differential expression patterns (30). Moreover, skeletal muscle differentiation is regulated by the expression of MRFs that are induced by various factors, including FGFs (31, 32). Among the FGF family members, Fgf4, Fgf6, and the receptor Fgfr4 are expressed in the somite myotomes, in developing skeletal muscles in the limbs, and in the tongue (33–35). We found that Fgf4 and Fgf6 are also expressed in all developing muscles in the craniofacial region, including the tongue, eye, and masticatory muscles, with similar expression patterns. The expression of both Fgf4 and Fgf6 was abrogated in Wnt1-Cre; Alk5fl/fl mice, suggesting that their expression in craniofacial muscles is induced by CNCCs during early craniofacial myogenesis. We have previously shown that Fgf6 expression in the tongue muscles is important for muscle differentiation, especially muscle fusion in the tongue bud (13). In Wnt1-Cre; Alk5fl/fl mice, severe muscle differentiation defects are associated with the loss of Fgf4 and Fgf6. The muscle differentiation defects were rescued by exogenous Fgf4 and Fgf6. Different members of the FGF family that are expressed in different tissue compartments play distinct roles in muscle proliferation and differentiation. For example, in chick limb myogenesis, overexpression of mFgf4 (mouse Fgf4) inhibits terminal muscle differentiation (36). Our data show that Fgf4 and Fgf6 are crucial for muscle differentiation during craniofacial myogenesis in mice. However, the mechanism by which CNCCs induce Fgf4 and Fgf6 expression in the muscles to promote differentiation remains to be elucidated.
Role of CNCCs in muscle organization and differentiation.
Previous studies have shown that CNCCs instruct the patterning of surrounding tissues, including muscles (6, 37). We previously reported that TGF-β signaling-mediated scleraxis expression is required for tendon formation by CNCCs in developing tongue muscles (12). We found that a subpopulation of CNCCs are Scleraxis-positive mesenchymal cells and that this subpopulation does not precisely overlap the subset of Bmp4-expressing CNCCs in wild-type mice. The Scleraxis-expressing CNCCs differentiated into tendons in the craniofacial region. Disruption of Alk5 signaling in CNCCs reduced the population of Scleraxis-positive cells, resulting in defects in tendon formation, followed by aberrant muscle organization. Our data support previous lineage-tracing and quail chick chimera studies using myogenic precursors in the somites, which have shown that muscle-patterning information comes from extrinsic cues, not from myogenic cells themselves (19). Thus, CNCCs play multiple roles in myogenic proliferation and differentiation, as well as in muscle patterning.
Bmp and Fgf signaling in the differentiation of myofibers during primary myogenesis.
The timing of differentiation of myogenic precursors into primary myofibers is important for determining muscle type and organization. Our schematic diagram (Fig. 7H to J) summarizes Bmp and Fgf signaling in the early and later stages of craniofacial myogenesis. Migrated myogenic precursors expand their population in response to Bmp4 during E11.5 and E12.5 downstream of Alk5-mediated TGF-β signaling in CNCCs. Terminal muscle differentiation of myoblasts in the later myogenic stage is also dependent on Alk5-mediated TGF-β signaling in CNCCs mediated by Fgf4 and Fgf6. However, Bmp4 is not responsible for the expression of Fgf4 and Fgf6, and it actually inhibits muscle differentiation. Previous studies have shown that Bmp inhibits muscle differentiation by controlling cell cycle exit rather than promoting differentiation in adult satellite cells (23, 25). During embryonic myogenesis, Bmp inhibits muscle differentiation of myogenic precursors in the dermomyotome in zebrafish (38). It remains unclear how Bmp4 signaling is inhibited before the onset of primary myogenic differentiation in the craniofacial region, because the expression of Bmp4 inhibitors, including Noggin, Chordin, and Gremlin, was not significantly altered in our microarray data for E11.5 and E13.5 Wnt1-Cre; Alk5fl/fl mice.
Temporal and spatial regulation of craniofacial myogenesis by CNCCs.
Myogenesis is a multistep process during which myogenic precursors interact with surrounding tissues. After the migration of myogenic cells to the myogenic core in the first branchial arch, myogenic precursors are in direct contact with surrounding tissues that are mostly composed of CNCCs. Loss of Alk5 in CNCCs results in defects in myogenic proliferation and muscle organization and differentiation, suggesting that CNCCs control craniofacial myogenesis. In this study, we show that CNCCs play dual roles in early and late myogenesis via Bmp and Fgf signaling, which are involved in muscle proliferation and differentiation, respectively, as the schematic diagram depicts. This study will be crucial for understanding craniofacial muscle developmental defects and regeneration.
ACKNOWLEDGMENTS
We thank J. Mayo and B. Samuels for critical reading of the manuscript.
This study was supported by grants from the NIDCR, NIH (R01 DE014078 and R37 DE012711), to Yang Chai.
We declare that we have no competing financial interests.
Footnotes
Published ahead of print 9 June 2014
REFERENCES
- 1.Mootoosamy RC, Dietrich S. 2002. Distinct regulatory cascades for head and trunk myogenesis. Development 129:573–583 [DOI] [PubMed] [Google Scholar]
- 2.Noden DM, Trainor PA. 2005. Relations and interactions between cranial mesoderm and neural crest populations. J. Anat. 207:575–601. 10.1111/j.1469-7580.2005.00473.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harel I, Nathan E, Tirosh-Finkel L, Zigdon H, Guimaraes-Camboa N, Evans SM, Tzahor E. 2009. Distinct origins and genetic programs of head muscle satellite cells. Dev. Cell 16:822–832. 10.1016/j.devcel.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G. 2009. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 23:997–1013. 10.1101/gad.1769009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kardon G. 1998. Muscle and tendon morphogenesis in the avian hind limb. Development 125:4019–4032 [DOI] [PubMed] [Google Scholar]
- 6.Noden DM. 1983. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev. Biol. 96:144–165. 10.1016/0012-1606(83)90318-4 [DOI] [PubMed] [Google Scholar]
- 7.Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. 2000. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127:1671–1679 [DOI] [PubMed] [Google Scholar]
- 8.Rios AC, Serralbo O, Salgado D, Marcelle C. 2011. Neural crest regulates myogenesis through the transient activation of NOTCH. Nature 473:532–535. 10.1038/nature09970 [DOI] [PubMed] [Google Scholar]
- 9.Tzahor E, Kempf H, Mootoosamy RC, Poon AC, Abzhanov A, Tabin CJ, Dietrich S, Lassar AB. 2003. Antagonists of Wnt and BMP signaling promote the formation of vertebrate head muscle. Genes Dev. 17:3087–3099. 10.1101/gad.1154103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinon A, Lazar S, Marshall H, Buchmann-Moller S, Neufeld A, Elhanany-Tamir H, Taketo MM, Sommer L, Krumlauf R, Tzahor E. 2007. Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development 134:3065–3075. 10.1242/dev.002501 [DOI] [PubMed] [Google Scholar]
- 11.Schilling TF, Walker C, Kimmel CB. 1996. The chinless mutation and neural crest cell interactions in zebrafish jaw development. Development 122:1417–1426 [DOI] [PubMed] [Google Scholar]
- 12.Hosokawa R, Oka K, Yamaza T, Iwata J, Urata M, Xu X, Bringas P, Jr, Nonaka K, Chai Y. 2010. TGF-beta mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis. Dev. Biol. 341:186–195. 10.1016/j.ydbio.2010.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han D, Zhao H, Parada C, Hacia JG, Bringas P, Jr, Chai Y. 2012. A TGFbeta-Smad4-Fgf6 signaling cascade controls myogenic differentiation and myoblast fusion during tongue development. Development 139:1640–1650. 10.1242/dev.076653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao H, Oka K, Bringas P, Kaartinen V, Chai Y. 2008. TGF-beta type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev. Biol. 320:19–29. 10.1016/j.ydbio.2008.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Spandidos A, Wang H, Seed B. 2012. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40:D1144–D1149. 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 17.Iwata J, Tung L, Urata M, Hacia JG, Pelikan R, Suzuki A, Ramenzoni L, Chaudhry O, Parada C, Sanchez-Lara PA, Chai Y. 2012. Fibroblast growth factor 9 (FGF9)-pituitary homeobox 2 (PITX2) pathway mediates transforming growth factor beta (TGFbeta) signaling to regulate cell proliferation in palatal mesenchyme during mouse palatogenesis. J. Biol. Chem. 287:2353–2363. 10.1074/jbc.M111.280974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudas M, Kim J, Li WY, Nagy A, Larsson J, Karlsson S, Chai Y, Kaartinen V. 2006. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev. Biol. 296:298–314. 10.1016/j.ydbio.2006.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kardon G, Heanue TA, Tabin CJ. 2002. Pax3 and Dach2 positive regulation in the developing somite. Dev. Dyn. 224:350–355. 10.1002/dvdy.10107 [DOI] [PubMed] [Google Scholar]
- 20.Matzuk MM, Lu N, Vogel H, Sellheyer K, Roop DR, Bradley A. 1995. Multiple defects and perinatal death in mice deficient in follistatin. Nature 374:360–363. 10.1038/374360a0 [DOI] [PubMed] [Google Scholar]
- 21.Rhodes SJ, Konieczny SF. 1989. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 3:2050–2061. 10.1101/gad.3.12b.2050 [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Noulet F, Edom-Vovard F, Tozer S, Le Grand F, Duprez D. 2010. Bmp signaling at the tips of skeletal muscles regulates the number of fetal muscle progenitors and satellite cells during development. Dev. Cell 18:643–654. 10.1016/j.devcel.2010.02.008 [DOI] [PubMed] [Google Scholar]
- 23.Ono Y, Calhabeu F, Morgan JE, Katagiri T, Amthor H, Zammit PS. 2011. BMP signalling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell Death Differ. 18:222–234. 10.1038/cdd.2010.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih HP, Gross MK, Kioussi C. 2007. Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proc. Natl. Acad. Sci. U. S. A. 104:5907–5912. 10.1073/pnas.0701122104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrichs M, Wirsdoerfer F, Flohe SB, Schneider S, Wuelling M, Vortkamp A. 2011. BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC Cell Biol. 12:26. 10.1186/1471-2121-12-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweitzer R, Zelzer E, Volk T. 2010. Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development 137:2807–2817. 10.1242/dev.047498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. 2001. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128:3855–3866 [DOI] [PubMed] [Google Scholar]
- 28.Iwata JI, Suzuki A, Pelikan RC, Ho TV, Chai Y. 2013. Non-canonical transforming growth factor beta (TGFb) signaling in cranial neural crest cells causes tongue muscle developmental defects. J. Biol. Chem. 288:29760–29770. 10.1074/jbc.M113.493551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultheiss TM, Burch JB, Lassar AB. 1997. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 11:451–462. 10.1101/gad.11.4.451 [DOI] [PubMed] [Google Scholar]
- 30.Martin GR. 1998. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 12:1571–1586. 10.1101/gad.12.11.1571 [DOI] [PubMed] [Google Scholar]
- 31.Marics I, Padilla F, Guillemot JF, Scaal M, Marcelle C. 2002. FGFR4 signaling is a necessary step in limb muscle differentiation. Development 129:4559–4569 [DOI] [PubMed] [Google Scholar]
- 32.Hannon K, Kudla AJ, McAvoy MJ, Clase KL, Olwin BB. 1996. Differentially expressed fibroblast growth factors regulate skeletal muscle development through autocrine and paracrine mechanisms. J. Cell Biol. 132:1151–1159. 10.1083/jcb.132.6.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niswander L, Martin GR. 1992. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development 114:755–768 [DOI] [PubMed] [Google Scholar]
- 34.deLapeyriere O, Ollendorff V, Planche J, Ott MO, Pizette S, Coulier F, Birnbaum D. 1993. Expression of the Fgf6 gene is restricted to developing skeletal muscle in the mouse embryo. Development 118:601–611 [DOI] [PubMed] [Google Scholar]
- 35.Marcelle C, Wolf J, Bronner-Fraser M. 1995. The in vivo expression of the FGF receptor FREK mRNA in avian myoblasts suggests a role in muscle growth and differentiation. Dev. Biol. 172:100–114. 10.1006/dbio.1995.0008 [DOI] [PubMed] [Google Scholar]
- 36.Edom-Vovard F, Bonnin MA, Duprez D. 2001. Misexpression of Fgf-4 in the chick limb inhibits myogenesis by down-regulating Frek expression. Dev. Biol. 233:56–71. 10.1006/dbio.2001.0221 [DOI] [PubMed] [Google Scholar]
- 37.Trainor PA, Ariza-McNaughton L, Krumlauf R. 2002. Role of the isthmus and FGFs in resolving the paradox of neural crest plasticity and prepatterning. Science 295:1288–1291. 10.1126/science.1064540 [DOI] [PubMed] [Google Scholar]
- 38.Patterson SE, Bird NC, Devoto SH. 2010. BMP regulation of myogenesis in zebrafish. Dev. Dyn. 239:806–817. 10.1002/dvdy.22243 [DOI] [PMC free article] [PubMed] [Google Scholar]