Abstract
Skeletal muscle tissue shows an extraordinary cellular plasticity, but the underlying molecular mechanisms are still poorly understood. Here, we use a combination of experimental and computational approaches to unravel the complex transcriptional network of muscle cell plasticity centered on the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), a regulatory nexus in endurance training adaptation. By integrating data on genome-wide binding of PGC-1α and gene expression upon PGC-1α overexpression with comprehensive computational prediction of transcription factor binding sites (TFBSs), we uncover a hitherto-underestimated number of transcription factor partners involved in mediating PGC-1α action. In particular, principal component analysis of TFBSs at PGC-1α binding regions predicts that, besides the well-known role of the estrogen-related receptor α (ERRα), the activator protein 1 complex (AP-1) plays a major role in regulating the PGC-1α-controlled gene program of the hypoxia response. Our findings thus reveal the complex transcriptional network of muscle cell plasticity controlled by PGC-1α.
INTRODUCTION
A sedentary lifestyle can lead to an imbalance between energy intake and expenditure and favors the development of a number of chronic diseases like obesity and type 2 diabetes. Regular exercise, on the other hand, is an effective way to reduce the risk for these lifestyle-related pathologies (1). The health benefits of exercise are at least in part induced by changes in skeletal muscle tissue. Muscle cells exhibit a high plasticity and thus a remarkably complex adaptation to increased contractile activity. For example, endurance training induces mitochondrial biogenesis, increases capillary density, and improves insulin sensitivity (1, 2). To achieve such a complex plastic response, a number of different signaling pathways are activated in an exercising muscle, for example, p38 mitogen-activated protein kinase (MAPK)-mediated protein phosphorylation events, increased intracellular calcium levels, or the activation of the metabolic sensors AMP-dependent protein kinase (AMPK) and sirtuin-1 (SIRT1) (3). While the temporal coordination of the numerous inputs is not clear, all of the major signaling pathways converge on the peroxisome proliferator-activated receptor (PPAR) γ coactivator 1α (PGC-1α) to either induce Ppargc1a gene expression, promote posttranslational modifications of the PGC-1α protein, or do both (4, 5). Upon activation, PGC-1α mediates the muscular adaptations to endurance exercise by coactivating various transcription factors (TFs) involved in the regulation of diverse biological programs such as mitochondrial biogenesis, angiogenesis, reactive oxygen species (ROS) detoxification, or glucose uptake (3). Accordingly, transgenic (TG) expression of PGC-1α in mouse skeletal muscle at physiological levels not only induces mitochondrial biogenesis but also drives a fiber-type conversion toward a more oxidative, slow-twitch phenotype (6), while muscle-specific Ppargc1a-knockout animals exhibit several symptoms of pathological inactivity (7, 8).
Coregulators are part of multicomponent regulatory protein complexes that are well suited to translate external stimuli into changes in promoter and enhancer activities by combining various enzymatic activities to modulate histones and chromatin structure and recruit other TFs (9). Thus, dynamic assembly of distinct coregulator complexes enables the integration of many different signaling pathways, leading to a coordinated and specific regulation of entire biological programs by multiple TFs (10, 11). For example, PGC-1α not only recruits histone acetylases (12), the TRAP/DRIP/Mediator (13), and the SWI/SNF protein complexes (14) but also binds to and coactivates a myriad of different transcription factors, even though a systematic inventory of TF binding partners has not been compiled yet (15). Thus, the specific control exerted by the PGC-1α-dependent transcriptional network might provide an explanation for the dynamic and coordinated muscle adaptation to exercise. Since PGC-1α in skeletal muscle not only confers a trained phenotype but also ameliorates several different muscle diseases (16), the unraveling of the PGC-1α-controlled transcriptional network in skeletal muscle would be of great interest to identify putative therapeutic targets within this pathway.
Therefore, we aimed at obtaining a global picture of the coregulatory activity of PGC-1α in skeletal muscle cells. More precisely, by combining data on the genome-wide binding locations of PGC-1α and the gene expression profiles in response to PGC-1α overexpression with comprehensive computational prediction of transcription factor binding site (TFBS) occurrence, we sought to unveil the biological processes that are regulated by PGC-1α, to identify the transcription factors that partner with PGC-1α, and to determine the mechanistic details of PGC-1α-regulated transcription. We not only mapped the locations on the DNA where PGC-1α was bound but also delineated the target genes whose expression is either directly or indirectly affected by PGC-1α and identified novel putative transcription factor partners that mediated PGC-1α's action. In particular, our results strongly suggest that the activator protein 1 (AP-1) complex is a major regulatory partner of PGC-1α, with AP-1 and PGC-1α together regulating the hypoxic response gene program in muscle cells in vitro and in vivo.
MATERIALS AND METHODS
Cell culture and small interfering RNA (siRNA) transfection.
C2C12 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. To obtain myotubes, the C2C12 myoblasts were allowed to reach 90% confluence and the medium was changed to DMEM supplemented with 2% horse serum (differentiation medium) for 72 h.
The siRNAs for the knockdown of NFE2L2, FOS, JUN, ATF3, NFYC, ZFP143, GTF2I, the nontargeting siRNA pool, and the DharmaFECT1 transfection reagent were purchased from Dharmacon (Fisher Scientific), and the siRNA transfection was performed according to the Thermo Scientific DharmaFECT transfection reagent siRNA transfection protocol. Briefly, after 3 days of differentiation, the respective siRNAs (50 nM final concentration) were added to the medium. Twenty-four hours after siRNA transfection, the cells were infected with adenovirus (AV) expressing either PGC-1α (AV-PGC-1α) or the green fluorescent protein (AV-GFP). Then, 48 h after adenoviral infection, the cells were harvested.
Differentiated C2C12 cells were infected with AV expressing short hairpin RNA (shRNA) for estrogen-related receptor α (AV-shERRα) (kindly provided by Anastasia Kralli, Scripps Research Institute, La Jolla, CA) to knock down and inactivate ERRα or shGFP as a control. The infected cells were kept in culture for 4 days. Afterwards, cells were infected with the AV-Flag-PGC-1α or AV-GFP and kept in culture for two additional days. As a supplement to the previously infected AV shERRα cells, 2 μM ERRα inverse agonist XCT-790 was added. To the remaining cells, 0.02% dimethyl sulfoxide (DMSO) as a vehicle was added to the differentiated medium. All the experiments have been performed in biological triplicates. For RNA isolation, TRIzol was used according to the TRIzol reagent RNA isolation protocol (Invitrogen). Three conditions were used for further analysis: AV-shGFP plus AV-GFP plus vehicle, AV-shGFP plus AV-Flag-PGC-1α plus vehicle, and AV-shERRα plus AV-Flag-PGC-1α plus 2 μM XCT-790.
ChIP and ChIP sequencing (ChIP-Seq).
Chromatin immunoprecipitation (ChIP) was performed according to the Agilent Mammalian ChIP-on-chip protocol, version 10.0. For each immunoprecipitation, approximately 1 × 108 C2C12 cells were differentiated into myotubes and infected with AV-Flag-PGC-1α. For cross-linking protein complexes to DNA-binding elements, the cells were incubated in a 1% formaldehyde solution for 10 min, followed by the addition of glycine to a final concentration of 125 mM to quench the effect of the formaldehyde. The cells were rinsed in 1× phosphate-buffered saline (PBS), harvested in ice-cold 1× PBS using a silicone scraper, and pelleted by centrifugation. The pelleted cells were either used immediately or flash frozen and stored for later. The cells were then lysed at 4°C using two lysis buffers containing 0.5% NP-40–0.25% Triton X-100 and 0.1% sodium deoxycholate–0.5% N-lauroylsarcosine, respectively. The chromatin was then sheared by sonication to obtain DNA fragments of about 100 to 600 bp in length. Fifty microliters of the sonicated lysate was saved as input DNA. The immunoprecipitation was performed overnight at 4°C using magnetic beads (protein G Dynabeads; Invitrogen), which were previously coated with monoclonal antibodies like the monoclonal anti-Flag M2 antibody (Sigma) for the ChIP of PGC-1α or with the monoclonal anti-c-Fos (9F6) rabbit antibody (catalog no. 2250; Cell Signaling) for the ChIP of FOS. The beads carrying the precipitate were washed five times for the c-Fos antibody and six times for the Flag antibody with radioimmunoprecipitation assay (RIPA) buffer and once with Tris-EDTA (TE) that contained 50 mM NaCl to eliminate unspecific binding of DNA to the beads. For elution, the beads were resuspended in elution buffer containing 1% SDS, placed in a 65°C water bath for 15 min, and vortexed every 2 min. To reverse the cross-links, the samples were incubated at 65°C overnight. The following day, the RNA and the cellular proteins were digested using RNase A and proteinase K. The DNA was precipitated, and the success of the chromatin immunoprecipitation was validated by semiquantitative real-time PCR. The ChIP experiments were performed in triplicates. The ChIP of PGC-1α was further used for sequencing. The ChIP-Seq experiment on overexpressed PGC-1α in C2C12 cells was performed in biological duplicates. At the joint Quantitative Genomics core facility of the University of Basel and the Department of Biosystems Science and Engineering (D-BSSE) of the ETH Zurich in Basel, Switzerland, DNA libraries were prepared using the standard Illumina ChIP-Seq protocol, as described by the manufacturer, and the immunoprecipitated samples were sequenced on the Genome Analyzer II. In order to keep only high-quality data, the sequenced reads were filtered based on the quality score of each read and its alignments. Reads were retained when the Phred score was ≥20, the read length was ≥25 bp, and the number of wrongly called nucleotides (Ns) was ≤2. Those reads that passed the filter (6,711,717 for the first immunoprecipitated sample [IP], 36,580,431 for the second IP, 17,899,074 for the first whole-cell extract [WCE], and 35,525,221 for the second WCE) were aligned with the mouse genome (UCSC mm9 assembly), using Bowtie version 0.12.7 (17) with parameters –best –strata -a –m 100. The number of aligned reads equaled 5,699,648 for the first IP sample, 16,053,370 for the first WCE, 21,448,059 for the second IP, and 32,244,584 for the second WCE.
Identification of bound regions.
To identify regions that were significantly enriched in the ChIP, we passed a 200-bp-long sliding window along the genome, sliding by 25 bp between consecutive windows, and estimated the fraction of all ChIP reads (fIP) that fall within the window, as well as the fraction fWCE of reads from the whole-cell extract that fall in the same window (which we estimate from a 2,000-bp-long window centered on the same genomic location). A Z score quantifying the enrichment in the ChIP of each window was computed as
where σ2IP and σ2WCE are the variances of the IP and WCE read frequencies, respectively, which are given by
respectively.
The enrichments were reproducible across biological replicates. Using only the first sequencing data set, we called peaks at a Z cutoff of 4.5; we then compared these with the Z scores from the corresponding regions of the second data set, and the Pearson correlation coefficient was found to be 0.778. Similarly, we called peaks at a Z cutoff of 4.5 using only the second sequencing data set; when we compared these peaks with the Z scores of the corresponding regions from the first data set, the Pearson correlation coefficient was found to be 0.782.
To obtain a final set of binding peaks, we combined the reads from the two biological replicates, computing the Z score of each window as
We conservatively considered all windows with a Z score larger than 4.5 to be significantly enriched (false discovery rate [FDR], 0.6%). The final binding peaks were obtained by merging consecutive windows that all passed the cutoff and by considering the “peak” to correspond to the top-scoring window, i.e., corresponding to the summit of the ChIP-Seq signal. To determine the PGC-1α distribution genome-wide, peaks were annotated according to their closest Mus musculus RefSeq transcripts. We defined peaks as “intronic” (peak center lying inside an intron), “exonic” (peak center lying inside an exon), “upstream of TSS” (peak center lying between kb −10 and 0 relative to the closest transcription start site [TSS]), “downstream of TES” (peak center lying between kb 0 and +10 relative to the closest transcription end site [TES]), or “intergenic” (peak center located farther than 10 kb from the nearest transcript). Moreover, we computed the ratio between observed and expected peak location distributions, obtained by generating 100 peak sets composed of 7,512 random peaks each.
Motif finding and TFBS overrepresentation.
The binding peak regions were aligned with orthologous regions from 6 other mammalian species—human (hg18), rhesus macaque (rheMac2), dog (canFam2), horse (equCab1), cow (bosTau3), and opossum (monDom4)—using T-Coffee (18). A collection of 190 mammalian regulatory motifs (position weight matrices [WMs]) representing the binding specificities of approximate 350 mouse TFs (in many cases, sequence specificities of multiple closely related TFs were represented with the same WM) were downloaded from the SwissRegulon website (19). TFBSs for all known motifs were predicted using the MotEvo algorithm (20) on the alignments of all 7,512 peak sequences. Only binding sites with a posterior probability of ≥0.1 were considered for the further steps of the analysis. In order to create a background set of regions to assess the overrepresentation of binding sites within our regions, we created randomized alignments by shuffling the multiple alignment columns, maintaining both the gap patterns and the conservation patterns of the original alignments. TFBSs were predicted on the shuffled alignments using the same MotEvo settings as those for the original peak alignments. Overrepresentation of motifs in the PGC-1α binding peaks was calculated by comparing total predicted TFBS occurrence within binding peaks with the predicted TFBS occurrence in the shuffled alignments. We evaluated the enrichment of TFBSs for each motif x by collecting the sum nx of the posterior probabilities of its predicted sites in the peak alignments as well as the corresponding sum n′x in the shuffled alignments and computed a Z score:
where Lx and L′x are the total lengths of the original and shuffled alignments, respectively, while fx and f′x are given by the equations nx × lx = fx × Lx and n′x × lx = f′x × L′x, with lx being the length of motif x.
PCA of TFBS occurrence in binding peaks.
The input matrix N for the principal component analysis (PCA) contained the total number of predicted binding sites Npm in each of the 7,512 binding peaks p (rows) for each of the 190 mammalian regulatory motifs m (columns). After mean centering the columns of this matrix, Ñpm = Npm − 〈Nm〉, i.e., subtracting the average site count for each motif, singular value decomposition (SVD) was used to factorize this matrix: Ñ = U · S · VT, where U is a P × M matrix whose columns are the left singular vectors of Ñ; S is an M × M diagonal matrix containing the singular values, and VT (the transpose of V) is an M × M matrix whose rows are the right singular vectors, with P the number of peaks and M the number of motifs. The SVD was performed using the “svd” package of the “R” programming language.
Gene expression arrays.
Whole-gene expression after 48 h of transfection with adenovirus was measured in C2C12 cells with Affymetrix GeneChip Mouse Gene 1.0 ST microarrays at the Life Science Training core facility of the University of Basel. Raw probe intensities were corrected for background and unspecific binding using the Bioconductor package “affy” (21). Subsequently, probes were classified as expressed or nonexpressed by using the “Mclust” R package (22), and after removal of nonexpressed probes, the intensity values were quantile normalized across all samples. Using mapping of the probes to the UCSC collection of mouse mRNAs, probes were then associated with a comprehensive collection of mouse promoters available from the SwissRegulon database (19). The log2 expression level of a given promoter was calculated as the weighted average of the expression levels of all probes associated with it. Log2 expression levels were then compared between overexpressed PGC-1α and the control GFP sample; for each promoter, the change in expression level across the two conditions was measured by log2 fold change (log2FC), computed as the difference between the mean of the log2 values in PGC-1α and the mean of the log2 values in GFP. The significance of the expression change was assessed by a Z score, which was computed as
where n = 3 is the number of replicate samples, ĒPGC1α is the mean log2 expression across the PGC-1α samples, ĒGFP is the mean log2 expression across the GFP samples, and σ2PGC1α and σ2GFP are the variances of log2 expression levels across the replicates for the PGC-1α and control samples, respectively. Promoters were considered significantly upregulated when log2FC was ≥1 and Z was ≥3 and significantly downregulated when log2FC was ≤−1 and Z was ≤−3.
Peaks were assigned to promoters by proximity. To assign each peak to a promoter, we calculated the distance from the center of the peak to the center of neighboring promoters; whenever the peak was closer than 10 kb from at least one promoter, it was assigned to the nearest promoter and, thus, to its associated gene.
GO enrichment analysis.
Gene identifiers (IDs) were extracted from differentially regulated promoters and divided into four groups: upregulated promoters with an assigned binding peak, upregulated promoters without an assigned binding peak, downregulated promoters with an assigned peak, and downregulated promoters without an assigned peak. These four gene sets were used as input for the functional analysis tool FatiGO (23) to identify significantly overrepresented gene ontology (GO) categories compared to all Mus musculus genes. Only GO terms having an FDR-adjusted P value of ≤0.05 were considered significant.
Motif activity at direct and indirect targets of PGC-1α.
To integrate the information from the PGC-1α binding peaks, we extended motif activity response analysis (MARA) (24) to model the direct and indirect regulatory effects of PGC-1α. Given the input expression data and the computationally predicted binding sites, MARA infers, for each of 190 regulatory motifs m, the activity Ams of the motif in each sample s when the motif occurs outside a region of PGC-1α and the activities A*ms of the motifs when they occur within a PGC-1α binding peak. That is, changes in the motif activities Ams upon overexpression of PGC-1α indicate indirect regulatory effects of PGC-1α on each motif m, whereas changes in the motif activities A*ms reflect direct regulatory effects of PGC-1α as mediated by each motif m. For each promoter p that was not associated with any PGC-1α binding peak (which we designate indirect targets), we modeled its log-expression in sample s, eps, in terms of the predicted number of TFBSs Npm that occur in the proximal promoter region (running from −500 to +500 relative to the TSS) for each regulatory motif m. That is, MARA assumes the linear model
where cp is the basal expression of promoter p, c̃s is a sample-dependent normalization constant, and Ams is the regulatory activity of motif m in sample s, which is inferred by the model. Formally, Ams quantifies the amount by which the expression of promoter p in sample s would be reduced if a binding site for motif m were to be deleted from the promoter.
For each “direct target” promoter p that has an associated PGC-1α binding peak, which we defined as promoters with a peak within 1 kb or with a peak within 100 kb that was highly conserved according to the PhastCons score of the region (25), we model its expression in terms of the predicted TFBSs in the binding peak, i.e.,
where N*pm is the number of predicted TFBSs for motif m in the peak associated with promoter p and A*ms is the motif activity of regulator m in sample s when this motif occurs in the context of PGC-1α binding. That is, the inferred motif activities Ams quantify the activities of regulatory motifs when they occur independently of PGC-1α binding, and the motif activities A*ms quantify the activities of motifs when they occur in a PGC-1α binding peak, i.e., the latter activities reflect direct effects of a PGC-1α while the former activities reflect indirect effects.
MARA predicts activities for 190 different mammalian regulatory motifs, associated with roughly 350 mouse TFs. Besides motif activities, MARA also calculates error bars δms for each motif m in each sample s. Using these, MARA calculates, for each motif m, an overall significance measure for the variation in motif activities across the samples analogous to a Z statistic:
For each motif, we calculate both a Z score zm associated with its indirect activity changes and a Z score z*m associated with its direct activity changes. MARA also ranks the confidence on predicted target promoters of each motif by a Bayesian procedure that quantifies the contribution of that factor to explaining the promoter's expression variation by a chi-square value (for details, see reference 24). The parameters used for motif stratification were (i) the Z score z*m for direct activity changes; (ii) the Z score zm for indirect motif activity changes; (iii) the Z score z̄*m for direct motif activity changes, computed by averaging the sample replicates; and (iv) the Z score z̄m for indirect motif activity changes, computed by averaging the sample replicates. The latter two measures were used to show which in direction the motif activity changes when overexpressing PGC-1α with respect to the control condition. All motifs m for which either the direct or indirect motif activities were changing significantly (z ≥ 2) were subsequently selected.
De novo motif finding.
PhyloGibbs (26) was used to identify de novo motifs across the 200 top enriched PGC-1α peaks. The parameters used were -D 1 -z 1 -y 200 -m 10, corresponding to searching on multiple alignments for a single motif of length 10 with a total of 200 sites. The resulting motif was scanned for similarity to the other known motifs from our data set using STAMP (27), with the following settings: Pearson correlation coefficient for column comparison metric, Smith-Waterman for the alignment method, and penalty of 0.5 and 0.25 for gap opening and gap extension, respectively.
Real-time PCR and target gene validation.
Putative target genes of distinct transcription factor–PGC-1α combinations were chosen according to three criteria: first, positive transcriptional regulation by PGC-1α by more than 2-fold; second, presence of a PGC-1α binding peak within a 10-kb distance from the TSS; and third, prediction of targeting by MARA with a positive chi-square score. The sequences of the primers used in real-time PCR experiments are depicted in Table S1 in the supplemental material. Relative mRNA was quantified by quantitative PCR (qPCR) on a StepOnePlus system (Applied Biosystems) using Power SYBR green PCR master mix (Applied Biosystems).
The values are presented as the mean ± standard error of the mean (SEM). Student's t test was performed, and a P value of <0.05 was considered significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Animals.
Mice were housed in a conventional facility with a 12-h night/12-h day cycle with free access to chow diet pellet and water. For the experiments, 22- to 23-week-old skeletal muscle-specific HSA-PGC-1α knockout (MKO) male mice and 8-week-old PGC-1α muscle-specific transgenic (TG) male mice were used as previously described (6–8). All experiments were performed according to the criteria outlined for the care and use of laboratory animals and with approval of the veterinary office of the Basel canton and the Swiss authorities.
Treadmill running.
Treadmill running was performed with the TG mice on the Columbus Instruments motorized treadmill with an electric shock grid. The mice were acclimatized to the treadmill and then allowed to run till exhaustion. The running protocol is as follows: 10 m/min for 5 min with an increase of 2 m/min every 5 min until 26 m/min and an inclination of 5 degrees. The speed of 26 m/min was kept until exhaustion of the mice (7, 28, 29). Mice were killed and tissues were collected 3 h after exercise.
RNA isolation of muscle tissue.
Gastrocnemius and quadriceps were used to isolate RNA by TRIzol according to the TRIzol reagent RNA isolation protocol (Invitrogen).
Microarray data accession number.
The Gene Expression Omnibus (GEO) accession number for the ChIP-Seq and gene expression array data reported in this paper is GSE51191.
RESULTS
Broad recruitment of PGC-1α to the mouse genome.
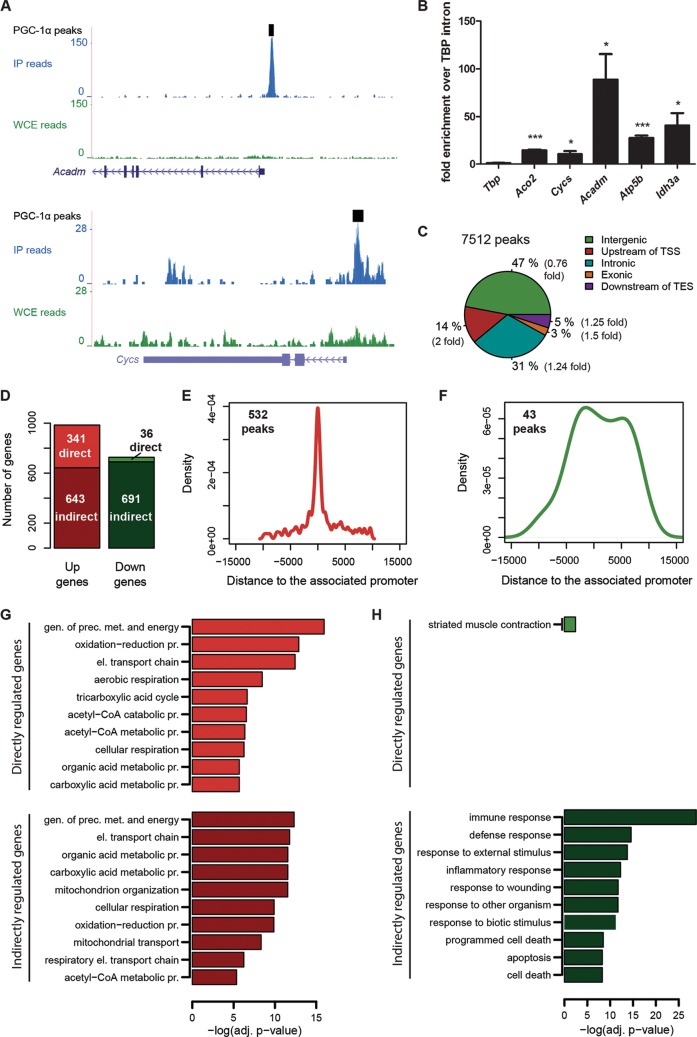
PGC-1α-dependent gene transcription has been studied in many different experimental contexts. In isolation, gene expression arrays, however, are unable to distinguish direct from indirect targets or to reveal the genomic sites where PGC-1α is recruited to enhancer and promoter elements, i.e., by coactivating TFs that directly bind to the DNA. Thus, we first performed chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) of PGC-1α in differentiated C2C12 mouse myotubes to identify the locations where PGC-1α is bound to the genome. To identify genomic regions that are significantly enriched in the ChIP, we slid a 200-bp window across the genome comparing the local ChIP read density with the read density from a background whole-cell extract sample. We selected all regions with a Z-statistic larger than 4.5 as significantly enriched (FDR, 0.6%) (see Fig. S1A in the supplemental material). Using this stringent cutoff, we identified 7,512 binding regions for PGC-1α via interaction with a TF genome-wide, which include binding regions in the promoters of known PGC-1α target genes (Fig. 1A), such as medium-chain-specific acyl coenzyme A (acyl-CoA) dehydrogenase (Acadm) and cytochrome c (Cycs) (30, 31). The enrichment of immunoprecipitated DNA fragments from the ChIP-Seq was validated for these and other PGC-1α target genes by semiquantitative real-time PCR (Fig. 1B). In absolute terms, the distribution of the ChIP-Seq peaks revealed that PGC-1α is mostly recruited at distal sites from the assigned targets and, to a lesser extent, to proximal regions of the gene or within an intronic sequence (Fig. 1C). However, compared to randomly selected DNA regions of equal size and number, PGC-1α binding peaks occur twice as often within 10 kb upstream of the transcription start site (TSS).
FIG 1.
Genome-wide DNA recruitment of PGC-1α in mouse muscle cells. (A) PGC-1α ChIP-Seq binding peaks (read densities) around the TSSs of the genes Acadm and Cycs obtained from the UCSC Genome Browser. (B) Real-time PCR validation of the ChIP enrichment measured at the promoter of a set of PGC-1α target genes. Bars represent fold enrichment over that of the Tbp intron; error bars represent SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Mapping ChIP-Seq PGC-1α peaks across the genome. Transcription start site (TSS) and transcription end site (TES) are relative to mm9 RefSeq transcripts. “Intergenic,” ≥10 kb from the nearest transcript; “Upstream of TSS,” kb −10 to 0 relative to the TSS; “Downstream of TES,” kb 0 to +10 relative to the TES. Numbers in parentheses indicate, for each category, the ratio between the percentage of PGC-1α peaks and the percentage of the same number of randomly distributed peaks. (D) Histogram illustrating the number of direct and indirect genes either up- or downregulated by overexpression of PGC-1α in muscle cells. Direct genes are those associated with promoters found within ±10 kb relative to the nearest peak. (E) Distribution of the distances of 532 peaks from their associated upregulated gene promoters. (F) Distribution of the distances of 43 peaks from their associated downregulated gene promoters. (G and H) Subset of the top significantly enriched GO Biological Process terms identified for directly and indirectly upregulated (G) and downregulated (H) PGC-1α target genes.
In parallel to the ChIP-Seq experiment, we furthermore analyzed gene expression patterns in differentiated muscle cells both under control conditions and under PGC-1α overexpression. Using a reference set of mouse promoters (19) and associating microarray probes with promoters by mapping to known transcripts, we found 1,566 promoters (corresponding to 984 genes) to be significantly upregulated (log2 fold change, ≥1; Z score, ≥3) and 1,165 promoters (corresponding to 727 genes) to be significantly downregulated (log2 fold change, ≤−1; Z score, ≤−3). Thus, similarly to previous reports, PGC-1α induced and repressed the transcription of almost the same number of genes, respectively, indicating that the physiological function of PGC-1α includes both the activation and the inhibition of substantial numbers of genes.
To combine the DNA-binding results from the ChIP-Seq with the data of the gene expression arrays, we then assigned ChIP-Seq peaks to the closest promoter (and the associated gene) within a maximum distance of 10 kb. In this way, about 30% of all peaks (2,295 of 7,512) could be associated with a target promoter. Inversely, for about 35% of all significantly upregulated genes (341 of 984), a PGC-1α binding peak is found within 10 kb of the promoter. Since some of the upregulated promoters may be regulated by more distal peaks, this is only a lower bound on the fraction of genes that are directly regulated. In stark contrast, only about 5% of all repressed genes harbor one or more PGC-1α DNA recruitment peaks in their vicinity (36 of 727), compared with 95% indirectly downregulated PGC-1α target genes (691 genes) (Fig. 1D). Moreover, the distribution of the distances between PGC-1α peaks and their associated promoters revealed a tight cluster of 532 peaks close to promoter regions for upregulated, direct PGC-1α target genes (Fig. 1E), whereas the distribution of the 43 peaks associated with downregulated genes was much wider, raising the possibility that the association of peaks with transcriptionally repressed genes was spurious (Fig. 1F). In summary, the strong enrichment of binding peaks near upregulated genes and the almost complete absence of binding peaks near downregulated genes suggest that direct regulation of transcription by PGC-1α is almost exclusively activating. We note that there is a large fraction of binding peaks (75%) that are associated with target genes that do not significantly alter their expression. These peaks may have been wrongly assigned, their functionality may be dependent on additional factors not active in these cells, or they may simply be spurious binding events that are not functional.
We next used this stratification of peaks and genes to study whether direct (i.e., with an associated binding peak) and indirect PGC-1α target genes exert different biological functions and identified gene ontology (GO) terms that were overrepresented in any of the four categories. First, we observed that the most significantly enriched functional categories for directly and indirectly upregulated genes were those related to mitochondria, oxidative phosphorylation, and energy production (Fig. 1G; see also Fig. S1B in the supplemental material). In contrast, GO analysis of indirectly downregulated PGC-1α target genes revealed a high prevalence of terms related to inflammation and immune response (Fig. 1H; see also Fig. S1C). Assuming that the assignment of peaks to repressed genes is not spurious, the few directly repressed PGC-1α targets exhibit an enrichment in functions related to muscle contraction, in particular for genes that are linked to contractile and metabolic properties of glycolytic, fast-twitch muscle fibers (Fig. 1H; see also Fig. S1D), as would be expected from the observed shift from glycolytic to oxidative fibers mediated by PGC-1α in muscle (6).
Modeling the direct and indirect gene regulatory effects of PGC-1α.
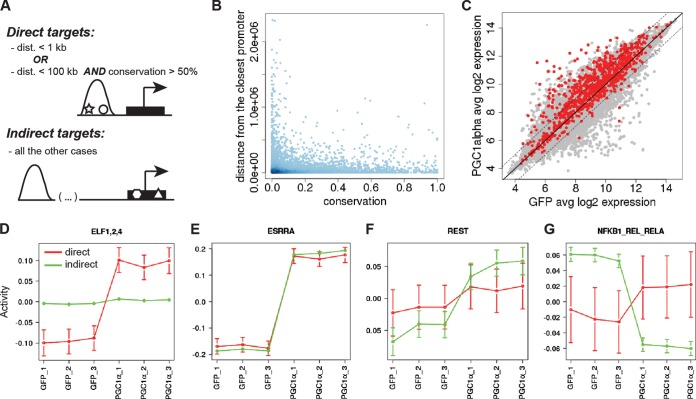
As a next step, we rigorously modeled the effects of PGC-1α on its target genes in terms of the occurrence of TFBSs for a large collection of mammalian regulatory motifs. We previously introduced a general framework, called motif activity response analysis (MARA) (24), for modeling the gene expression profiles as a linear function of the TFBSs occurring in the promoters and unknown regulatory “activities” of each of the regulators. As detailed in Materials and Methods, we here extended MARA to incorporate information from the PGC-1α ChIP-Seq data, with the aim of identifying which other TFs are involved in mediating both the direct and indirect regulatory effects of PGC-1α. Specifically, for all “direct target” promoters that were associated with a PGC-1α binding peak, we modeled the expression of the promoter in terms of the predicted TFBSs in the neighborhood of the binding peak, while for “indirect target” promoters, we modeled the promoter's expression in terms of the predicted TFBSs in the proximal promoter region, according to the conventional MARA approach (Fig. 2A and B).
FIG 2.
Four distinct mechanistic modes of action for gene expression regulated by PGC-1α and TF partners. (A) Classification of direct and indirect target genes in MARA (see Materials and Methods). (B) Distribution of peak distance from the closest promoter and PhastCons conservation score of the peak. (C) Distribution of log2 expression values for all mouse promoters. Expression values were averaged across the 3 GFP and the 3 PGC-1α samples. Direct targets are depicted in red; indirect targets are depicted in gray. (D to G) Activity plot of the motifs ELF1,2,4 (D), ESRRA (E), REST (F), and NFKB1_REL_RELA (G) as predicted by MARA (motif activity response analysis). Red, direct targets; green, indirect targets.
First, further supporting our analysis above, direct target promoters were almost exclusively upregulated and only in a few exceptional cases reached statistical significance for PGC-1α-repressed transcripts (Fig. 2C). Among the direct motif activities, the ESRRA position weight matrix was the top-ranking motif with a Z score of 6.04 (see Fig. S2 in the supplemental material). The corresponding TF estrogen-related receptor α (ERRα), an orphan nuclear receptor, has been extensively studied as a central binding partner for PGC-1α in the regulation of mitochondrial gene expression (30–32). To stratify the different motifs according to their predicted functions, we then divided all motifs into groups according to the behavior of both their direct and indirect activity changes. Strikingly, all motifs exhibited one of only four different motif activity patterns. First, 6 TFs (see Fig. S2) were predicted to positively regulate PGC-1α target genes only in the presence of PGC-1α (Fig. 2D). Second, we found 6 motifs (see Fig. S2) with significantly upregulated direct and indirect motif activities upon PGC-1α overexpression (Fig. 2E). To our surprise, ERRα was predicted to regulate PGC-1α target genes in this manner, even though in previous reports gene regulation by ERRα in the context of activated PGC-1α was suggested to be dependent on PGC-1α coactivation (30–32). Third, we found 13 motifs (see Fig. S2) that were predicted to regulate PGC-1α target genes but, however, only in the absence of PGC-1α (Fig. 2F). Fourth, there was a group of 28 motifs (see Fig. S2) that showed a significant decrease of indirect motif activity upon PGC-1α overexpression, but no significant change of their direct motif activity, including NF-κB (Fig. 2G), a central regulator of inflammation which is indirectly repressed by PGC-1α (33). Intriguingly, however, no motif was found that showed significant direct repression of target genes, reinforcing the hypothesis that PGC-1α-dependent gene repression is an indirect event.
Nuclear receptors and activator protein-1-like leucine zipper proteins are the main functional partners of PGC-1α in muscle cells.
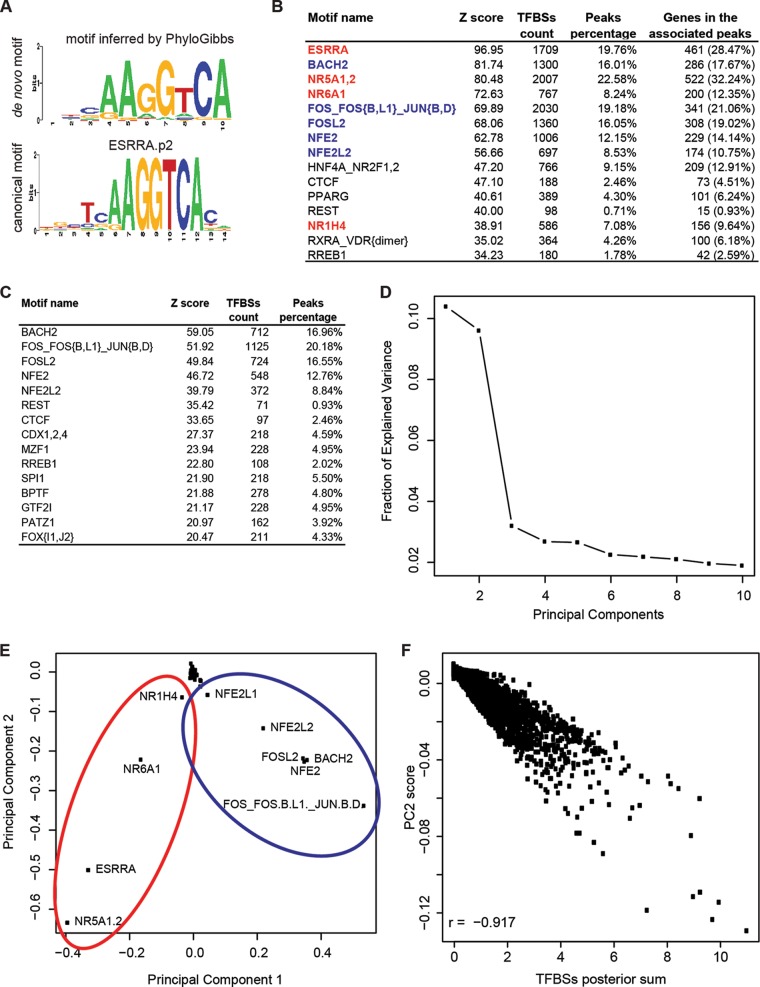
As a next step, we analyzed the occurrence of TF DNA-binding motifs in the PGC-1α peaks identified by ChIP-Seq. We first performed de novo motif prediction on the top 200 peaks, using PhyloGibbs (26). As shown in Fig. 3A, the motif that PhyloGibbs identified matches significantly (E value = 7.7834e−10, as calculated by STAMP [27]) the canonical ESRRA motif. In addition to the de novo prediction, we also used the same collection of 190 mammalian regulatory motifs used by MARA (19) to check which known TF DNA-binding motifs were significantly overrepresented in the PGC-1α peaks relative to a set of background regions. Many of the most significantly enriched motifs represent variations of nuclear receptor binding sequences that are based on the “AGT/GTCA” core hexamer and occur either alone or in direct, inverted, or everted repeats with variable spacing (Fig. 3B). Of these, the most significantly enriched motif was ESRRA, which is present in ∼20% of all peaks. Moreover, among all genes with at least one associated binding peak within 10 kb, ∼28% are associated with a peak containing a predicted ERRα site. Interestingly, besides the nuclear receptor motifs, we also found the DNA-binding element of the insulator protein CCCTC-binding factor (CTCF), and a set of highly similar DNA elements sharing the FOS-JUN-like recognition sequence “TGA(G/C)TCA” was bound by the TFs BACH2, FOS, FOSB, FOSL1, JUN, JUNB, JUND, FOSL2, NFE2, and NFE2L2 among the top 15 motifs enriched in PGC-1α peaks (Fig. 3B).
FIG 3.
PCA reveals FOS-JUN-like leucine zippers as a new class of putative functional PGC-1α partners. (A) Sequence logo of the top position weight matrix discovered de novo by PhyloGibbs in the top 200 scoring peaks and of the corresponding canonical motif of ERRα as predicted by STAMP. (B) Top-scoring results of motif search performed on all 7,512 PGC-1α peaks with MotEvo. Motifs depicted in red and blue correspond to the clusters identified by PCA in panel D. (C) Top-scoring results of motif search performed on the 3,656 “non-ESRRA-like” peaks with MotEvo. (D) Fraction of explained variance of the top 10 PCA components. (E) PCA of the 7,512 PGC-1α peaks. Eigenmotif scores across principal component 1 (PC1) and principal component 2 (PC2) are shown. Red and blue ellipses highlight motif clusters, as identified by PC1, of nuclear hormone receptor-like zinc finger and FOS-JUN-like leucine zipper proteins, respectively. (F) Correlation between principal component 2 scores and binding site posterior sum for each peak relative to the top 10 PCA motifs. “r” refers to the Pearson correlation coefficient.
The identity of the exact nuclear receptor binding partner that is bound at each peak is difficult to deduce from DNA-binding motifs, since considerable promiscuity exists between receptors and DNA-binding elements in different configurations of hexameric repeats (34). Moreover, non-nuclear receptor-like TFs are less well studied in the context of PGC-1α-controlled gene expression. Thus, to identify which regulatory motifs are most overrepresented among peaks that do not contain nuclear receptor-like sites, we first manually grouped all of the motifs with a sequence logo very similar to that of ESRRA. Next, we discarded all peaks that had one or more predicted TFBSs for any of the motifs in this set. With the remaining 3,856 DNA sequences (51.33% of the peaks), we then again assessed the overrepresentation of each of the 190 mammalian regulatory motifs. In this analysis, “TGA(G/C)TCA” recognition elements, hence, FOS-JUN-like motifs, were the most significantly enriched among these peaks (Fig. 3C). This result suggests that PGC-1α peaks naturally fall into two classes: those containing ESRRA-like sites and those containing sites for FOS-JUN-like motifs.
We then constructed a matrix N, whose elements Npm contain the number of predicted TFBSs for each motif m in each peak region p. We then performed principal component analysis (PCA) on this site-count matrix to identify linear combinations of regulatory motifs that explain most of the variation in site-counts across the PGC-1α peaks. The first two components (out of 190 in total) clearly proved to be the most relevant ones, accounting for 10% and 9.6% of the total variation in our data set, respectively (Fig. 3D). Figure 3E shows the projection of all motifs on these first two principal components, with the names of the motifs with the largest projections indicated in the figure. Whereas most motifs have projections close to zero along the first component, there is one group of motifs with strong negative projections (ESRRA, NR1H4, NR5A1,2, and NR6A1) and one group of motifs with strong positive projections [BACH2, FOS_FOS(B,L1)_JUN(B,D), FOSL2, NFE2, NFE2L1, and NFE2L2]. These two sets of sites correspond precisely to the two classes of motifs identified above, confirming that the most significant variation in TFBSs across PGC-1α peaks is caused by the occurrence of either ESRRA-like motifs or FOS-JUN-like motifs. Most interestingly, these two clusters of motifs reflect structurally distinct classes of TFs; the negatively scoring eigenmotifs are characterized by binding of nuclear receptor-type zinc finger domains, while the eigenmotifs with a positive score correspond to activator protein 1 (AP-1)-like leucine zipper domains.
The second principal component corresponds to the strength of the binding signal for these 10 motifs, as confirmed by the robust negative correlation (r = −0.92) between the TFBS posterior sum per peak and the peak's projection along the second principal component (Fig. 3F).
Validation of top-scoring motifs reveals novel functional partners of PGC-1α.
Our analysis identified a number of so-far-uncharacterized TFs as potentially functional partners for PGC-1α-controlled gene expression in skeletal muscle cells. In order to experimentally validate some of these candidates, we sorted all TFs by a number of criteria, including TFBS overrepresentation in binding peaks, MARA activity upon PGC-1α overexpression, and the expression pattern of the TFs themselves. Table 1 shows the top 15 ranked TFs according to this selection. As expected, the well-known PGC-1α partner ERRα was identified as the most important factor. For our validation experiments, we chose the next two motifs [FOS_FOS(B,L1)_JUN(B,D) and ZNF143, which is also known as ZFP143] as well as three motifs from further down the list of the top 15 motifs (GTF2I, NFE2L2, and NFYC).
TABLE 1.
Global summary of all analyses performed on PGC-1α peaksf
| Motif name | PCAa | Overrepresentation inb: |
MARA activity Z scorec |
Log2 FC in expression arrayd | Absolute expression in PGC-1α samplee | Final ranking | ||
|---|---|---|---|---|---|---|---|---|
| All PGC-1α peaks | “Non ESRRA-like” peaks | Direct | Indirect | |||||
| ESRRA | Yes | 1 | 182 | 6.04 (14.78) | 15.49 (37.94) | 2.31 | 1,829.45 | 6 |
| FOS_FOS(B,L1)_JUN(B,D) | Yes | 5 | 2 | 0.88 (2.14) | 1.81 (−4.34) | 1.78 | 1,508.85 | 5 |
| ZNF143 | 27 | 28 | 2.48 (6.05) | 4.65 (9.68) | 0.38 | 384.36 | 5 | |
| BPTF | 21 | 12 | 1.38 (3.37) | 2.56 (−6.25) | −0.56 | 333.34 | 4 | |
| ESR1 | 17 | 50 | 2.33 (5.69) | 4.53 (11.04) | −0.47 | 232.42 | 4 | |
| FOSL2 | Yes | 6 | 3 | 0.88 (2.14) | 1.51 (3.65) | −0.98 | 717.09 | 4 |
| GTF2I | 34 | 13 | 2.09 (5.10) | 2.38 (−5.80) | −0.55 | 1,207.81 | 4 | |
| NFE2L2 | Yes | 8 | 5 | 0.57 (1.38) | 1.01 (−2.37) | −0.38 | 3,673.63 | 4 |
| NFY(A,B,C) | 96 | 116 | 2.37 (5.80) | 3.56 (7.62) | 1.07 | 2,409.48 | 4 | |
| NR5A1,2 | Yes | 3 | 188 | 3.53 (8.66) | 7.73 (17.00) | −0.08 | 80.97 | 4 |
| REST | 12 | 6 | 0.48 (1.15) | 2.41 (5.70) | −0.89 | 328.04 | 4 | |
| RREB1 | 15 | 10 | 1.56 (3.82) | 2.39 (−5.42) | 0.05 | 678.44 | 4 | |
| SP1 | 24 | 22 | 3.99 (9.76) | 0.61 (0.33) | −0.32 | 751.98 | 4 | |
| STAT2,4,6 | 29 | 23 | 0.35 (0.52) | 4.81 (−9.67) | −2.72 | 380.12 | 4 | |
| TLX1.3_NFIC (dimer) | 19 | 17 | 0.84 (−2.05) | 4.91 (−11.97) | −0.34 | 2,339.33 | 4 | |
Requirement for PCA: being among the top 10 motifs contributing most to PC1 and PC2.
Requirement for motif overrepresentation: being among the top 30 significant motifs; ranking position shown.
Requirement for MARA: having a Z score of ≥2.0. Numbers in parentheses show the difference between the PGC-1α state and the GFP state, representing the direction in which the motif activity changes following PGC-1α overexpression.
Requirement for the expression array (i): having a log2 fold change value of ≥1.0 (corresponding to 2-fold upregulation).
Requirement for the expression array (ii): having an absolute expression in the PGC-1α sample of ≥100.
The final score is the count of all analyses where a certain motif passed the defined cutoffs. The motifs chosen for validation and their corresponding values which satisfied the cutoffs are shown in bold.
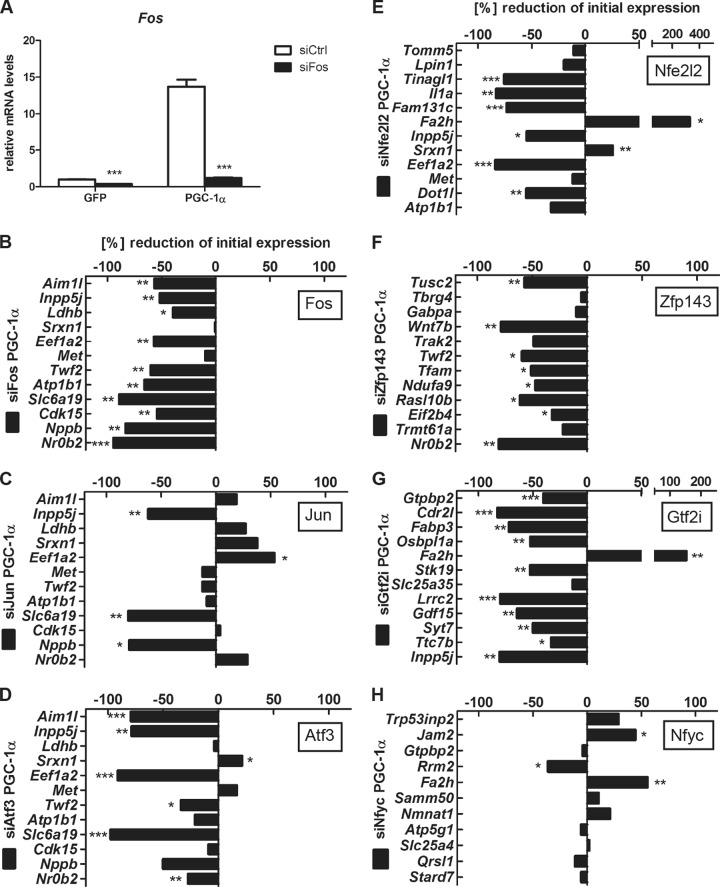
FOS, the most upregulated TF (log2 fold change = 1.78) among the TFs associated with the motif FOS_FOS(B,L1)_JUN(B,D), is a basic leucine zipper transcription factor known to heterodimerize with other leucine zipper proteins in order to form the AP-1 complex (35). The AP-1 complex furthermore contains JUN as well as ATF proteins. Thus, to dissect the function of the AP-1 protein complex, we also included JUN and ATF3, the most highly expressed isoforms of their respective protein families in muscle cells.
For each of these 7 TFs (ATF3, FOS, GTF2I, JUN, NFE2L2, NFYC, and ZFP143), we selected a dozen target genes based on the chi-square score of the MARA prediction, the presence of a PGC-1α binding peak with at least one predicted binding site for the factor of interest, and the presence of at least a 2-fold induction upon overexpression of PGC-1α. As summarized in Fig. 4 and Fig. S3 in the supplemental material, siRNA-based knockdown of all TFs resulted in a robust reduction of the target mRNAs from −40% to −75%. With the exception of NFYC and JUN, we found that the large majority of predicted target genes were downregulated upon knockdown of the factor, confirming our predictions (Fig. 4). The most consistent effects were observed for FOS and ZFP143 (all targets downregulated), followed by GTF2I (11 out of 12 downregulated) and NFE2L2 and ATF3 (10 out of 12 downregulated). Interestingly, distinct target genes of the AP-1 complex showed differential responsiveness to knockdown of the three AP-1 complex components FOS, JUN, and ATF3 (Fig. 4B, C, and D). Similarly, PGC-1α-mediated induction of a majority of the predicted target genes for NFE2L2 (Fig. 4E), ZFP143 (Fig. 4F), and GTF2I (Fig. 4G) was reduced upon knockdown of the respective TF compared to the expression in cells with overexpressed PGC-1α and a scrambled siRNA control. Surprisingly, only 1 of the 11 predicted target genes for NFYC that have been chosen for validation was significantly repressed by siRNA-induced reduction of this TF (Fig. 4H), suggesting that other TFs may be involved in mediating the regulatory effects of the NFYC regulatory motif.
FIG 4.
Validation of TFs associated with top-scoring motifs reveals novel functional PGC-1α partners. (A) siRNA-mediated knockdown efficiency for FOS. Bars represent fold induction over GFP/siCtrl value; error bars represent SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. See also Fig. S3 in the supplemental material. (B to H) Quantitative real-time PCR analysis of PGC-1α target genes whose associated peak contains at least one binding site for the motif FOS_FOS(B,L1)_JUN(B,D) (B to D), NFE2L2 (E), ZNF143, also known as ZFP143 (F), GTF2I (G), or NFY(A,B,C) (H). Bars represent % change compared to PGC-1α/siCtrl values. Error bars represent SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
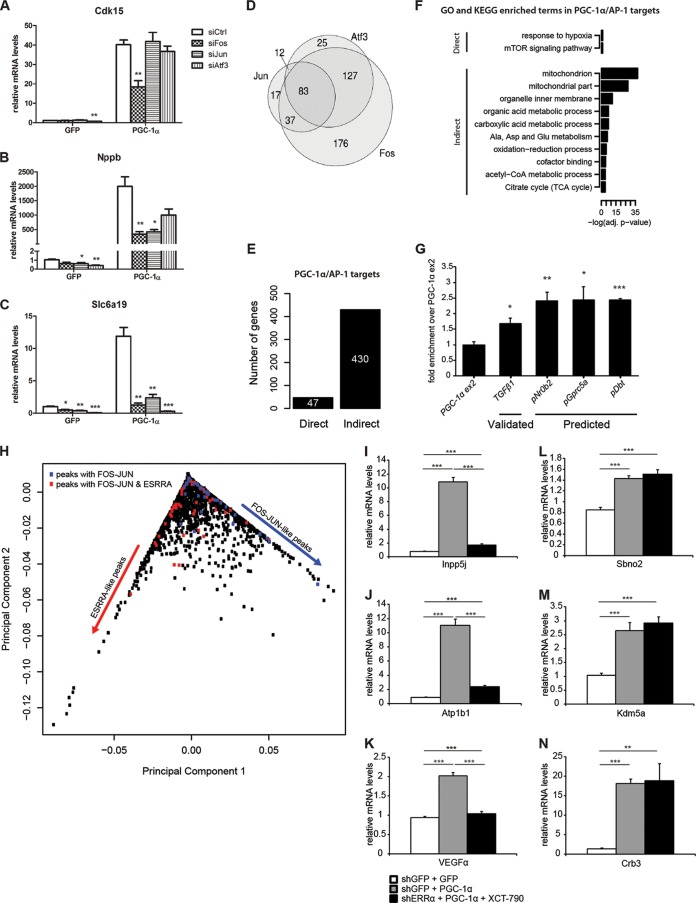
Functional interaction between PGC-1α and different compositions of the AP-1 protein complex.
Our targeted validation strategy revealed that PGC-1α target genes predicted to be regulated by the FOS-JUN-like motif react in distinct manners to siRNA-mediated knockdown of individual components of the AP-1 transcription factor protein complex. For example, some genes reacted only to reduction of FOS (Fig. 5A), while others were responsive to the knockdown of two (Fig. 5B) or even all three (Fig. 5C) AP-1 protein partners that we have tested using the siRNA-based approach. To further dissect the responsiveness of PGC-1α target genes to different AP-1 protein complexes, we performed global gene expression arrays upon knockdown of each of the three TF components of the AP-1 complex. Figure 5D depicts the number of genes that were induced by PGC-1α and that were, at the same time, downregulated by the siRNA knockdown of any of the three AP-1 complex members. Among a total of 477 genes, 89% responded to FOS knockdown, 52% to ATF3 knockdown, and 31% to JUN knockdown. Moreover, while 37% of all targets responded exclusively to FOS, the fraction of targets responding exclusively to either JUN or ATF3 was at most 5%. This analysis shows that, whereas different target genes respond differently to the knockdown of distinct AP-1 components, FOS is the dominant factor in determining AP-1 function under these conditions.
FIG 5.
PGC-1α controls the hypoxia gene program via a functional interaction with different configurations of the AP-1 protein complex. (A to C) Quantitative real-time PCR analysis of Cdk15 (A), Nppb (B), and Slc6a19 (C) mRNA levels in response to PGC-1α overexpression and either siFos, siJun, or siAtf3 knockdown. Data are normalized to mRNA levels in GFP adenovirus-infected cells. Error bars represent ±SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Venn diagram illustrating the overlap in number of genes upregulated by PGC-1α and downregulated by either FOS, JUN, or ATF3 knockdown. (E) Histogram illustrating the number of direct and indirect PGC-1α/AP-1 target genes. (F) Subset of the top significantly enriched Gene Ontology and KEGG terms identified for the two gene groups illustrated in panel E. TCA, tricarboxylic acid. (G) Quantitative real-time PCR validation of the ChIP enrichment of c-Fos measured at the gene TGFβ1 (validated) and at the promoters of Nr0b2, Gprc5a, and Dbt (predicted) target genes. Bars represent fold enrichment over PGC-1α exon 2 set as 1. Error bars represent SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (H) PCA of the 7,512 PGC-1α peaks. Eigenpeak scores across principal component 1 and principal component 2 are shown. Colored dots correspond to peaks associated to the 47 direct PGC-1α/AP-1 targets. Blue dots refer to genes associated with peaks containing only FOS-JUN TFBSs, while red dots refer to genes associated with peaks with FOS-JUN and ESRRA TFBSs, located either in the same peak or in distinct PGC-1α peaks. (I to K) Quantitative real-time PCR analysis of PGC-1α/AP-1 targets whose associated peaks contain an ESRRA binding site. The bars represent relative mRNA levels compared to AV-shGFP plus AV-GFP plus vehicle, which is set as 1. The error bars represent SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (L to N) Quantitative real-time PCR analysis of PGC-1α/AP-1 targets whose associated peaks (if any) do not contain an ESRRA binding site. The bars represent relative mRNA levels compared to AV-shGFP plus AV-GFP plus vehicle, which is set as 1. The error bars represent SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As shown in Fig. 3B, 341 genes were associated with a PGC-1α binding peak containing a predicted site for the FOS-JUN-like motif bound by the AP-1 complex. Of these genes, the expression of 55 was significantly induced by PGC-1α overexpression in muscle cells. In our siRNA-based validation experiment, we found that 47 out of these 55 PGC-1α-induced/AP-1 predicted targets were significantly downregulated by knockdown of the AP-1 complex components, and we called these genes “direct PGC-1α/AP-1 targets.” The remaining 430 genes out of 477 (Fig. 5D) were defined accordingly as “indirect PGC-1α/AP-1 targets” that lack a PGC-1α peak containing a FOS-JUN-like motif but still are regulated by PGC-1α and the AP-1 protein components (Fig. 5E). To reveal whether these gene categories exert distinct functions, GO and KEGG enrichment analyses were performed. Surprisingly, the 47 direct PGC-1α/AP-1 target genes showed a distinct and significant overrepresentation of the terms “response to hypoxia” (GO ID, 0001666; adjusted P value, 0.0247542) and “mTOR signaling pathway” (KEGG ID, mmu04150; adjusted P value, 0.030674) that were absent in the GO analysis of the remaining PGC-1α/AP-1 targets (Fig. 5F). Recruitment of FOS to the same regulatory regions as PGC-1α in the direct AP-1/PGC-1α target genes was subsequently validated by ChIP (Fig. 5G). These results suggest that AP-1, when interacting with PGC-1α, drives a synergic effect of response to hypoxia; on the other hand, when AP-1 and PGC-1α act separately and, furthermore, through downstream intermediate TFs, they regulate the expression of genes involved in mitochondrial organization and energy metabolism.
Intriguingly, several of the predicted AP-1/PGC-1α target genes are also under the control of PGC-1α working with other transcription factors. For example, the vascular endothelial growth factor (VEGF) or, based on the gene expression arrays, 8 OXPHOS genes seem likewise to be under the control of AP-1 as well as ERRα in the context of elevated PGC-1α in skeletal muscle (31, 36). We therefore assessed the predicted and experimental overlap of these two transcription factors in the regulation of AP-1/PGC-1α target genes. Interestingly, when the PCA of the PGC-1α peaks was stratified in terms of eigenpeaks, we observed two distinct groups of peaks associated with AP-1/PGC-1α target genes (Fig. 5H). First, some of these genes exclusively harbored peaks with FOS-JUN-like TFBSs, whereas the second group exhibited either peaks with both FOS-JUN- and ESRRA-like TFBSs or a combination of distinct peaks with either of these sites within 10 kb from their promoters (Fig. 5H). Next, we validated this prediction by investigating the change in expression of different AP-1/PGC-1α target genes in the context of reduced ERRα expression and function, elicited by a combination of shRNA-mediated knockdown and pharmacological treatment of muscle cells with the ERRα inverse agonist XCT-790 (31). In line with the PCA, two distinct groups of ERRα inhibition-sensitive (Fig. 5I to K) and -insensitive (Fig. 5L to N) AP-1/PGC-1α target genes were found.
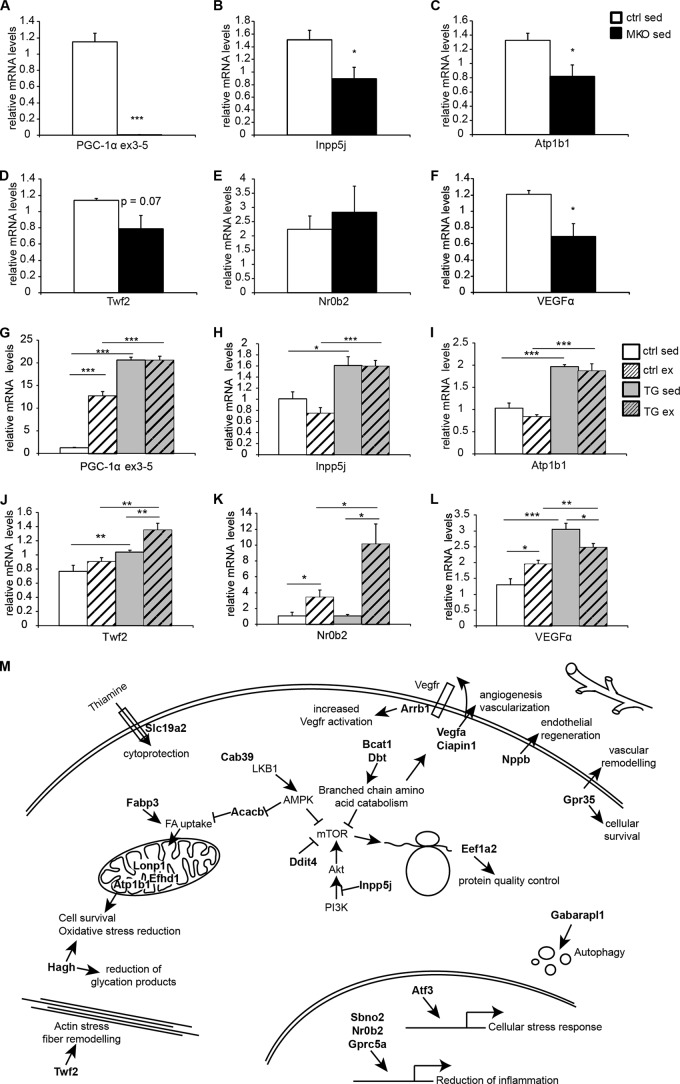
Finally, since all of the experiments were performed in differentiated myotubes in culture, we assessed whether similar gene expression changes of the direct AP-1/PGC-1α targets involved in hypoxic gene regulation are also observed in skeletal muscle tissue of different gain-of-function (6) and loss-of-function (7, 8) mouse models in vivo. In skeletal muscle-specific PGC-1α knockout mice, the expression of several of these genes was reduced significantly (Fig. 6A to F). Surprisingly, however, some of the predicted transcripts were not altered in this loss-of-function model for PGC-1α, for example, Nr0b2 (Fig. 6E). To further clarify the role of PGC-1α in the regulation of these genes, relative transcript levels were next assessed in muscle-specific transgenic mice for PGC-1α (Fig. 6G to L). In most cases, the genes with a reduction in their transcription in the PGC-1α muscle-specific knockout animals were inversely elevated in the PGC-1α muscle-specific transgenic mice. Moreover, some of these genes were likewise induced by exercise (Fig. 6G to L), and at least in some cases, for example, Twf2 and Nr0b2 (Fig. 6J and K), PGC-1α overexpression and physical activity synergistically boosted gene expression; for Nr0b2, this occurred even in the absence of any effect of the muscle-specific PGC-1α transgene per se (Fig. 6K).
FIG 6.
PGC-1α controls the hypoxic gene program in muscle in vivo. (A to F) Quantitative real-time PCR analysis of hypoxic genes in sedentary control (ctrl) and muscle-specific knockout (MKO) mice. The control group is set as 1. Error bars represent SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (G to L) Quantitative real-time PCR analysis of hypoxic genes in treadmill-running mice. Control (ctrl) and muscle-specific transgenic (TG) mice were used under sedentary and exercise conditions. The control group under sedentary conditions is set as 1. Error bars represent SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (M) Schematic representation depicting the downstream effects of the functional interaction between PGC-1α and the AP-1 complex in the context of the hypoxia gene program. Direct targets of PGC-1α and AP-1 are indicated in bold.
DISCUSSION
Exercise-induced skeletal muscle cell plasticity is a highly complex biological program that involves the remodeling of a number of fundamental cellular properties. Since PGC-1α function has been strongly linked to the induction of an endurance-trained muscle phenotype, we here dissected the PGC-1α-controlled transcriptional network in muscle cells. First, our results reveal a broad recruitment of PGC-1α to many different sites in the mouse genome (7,512 peaks), the majority of which were either not located within 10 kb from a promoter or close to a gene that was not regulated by PGC-1α overexpression at the time of harvest of the cells, as has analogously been observed in many other ChIP-Seq experiments (for example, see reference 37). Apart from the fact that PGC-1α could mediate long-range enhancer effects that were excluded in our peak-gene assignment, it is conceivable that PGC-1α recruitment is transcriptionally silent in some binding peaks because it requires the recruitment of additional cofactors for activation, which are not present under the conditions or in the cell type in which our experiments were performed. In addition, it is possible that a large fraction of PGC-1α binding peaks are “neutral” in the sense of not having any direct role in regulating gene expression.
Second, while an almost equally strong effect of PGC-1α on gene induction and repression has been reported (31), our analysis now indicates that direct PGC-1α-mediated gene expression is restricted almost exclusively to positively regulated PGC-1α target genes, whereas the vast majority of gene repression is indirect, i.e., not associated with PGC-1α recruitment within a 10-kb distance to the genes' promoters. Thus, the fact that almost 95% of all repressed genes were not linked to PGC-1α recruitment strongly implies that this coregulator primarily acts as a coactivator, and not as a corepressor, as suggested by the data of some studies (38–40). Importantly, indirect repression of PGC-1α target genes was also supported by the MARA prediction. The strong indirect inhibition of genes, many of which are involved in inflammatory processes, is predicted by MARA to be mediated by TFs such as NF-κB and interferon regulatory factor (IRF). Such an indirect inhibition of NF-κB and proinflammatory genes by PGC-1α in muscle cells has been reported previously (33).
One of the main functions of PGC-1α in all cells and organs is to boost mitochondrial gene transcription and oxidative metabolism. Accordingly, we observed that Gene Ontology terms related to these pathways were highly enriched when analyzing positively regulated PGC-1α target genes in muscle cells. Based on previous studies, the regulation of this core function could have been assigned to the direct interaction of PGC-1α and ERRα binding to regulatory elements of these genes (31, 32). Surprisingly, our data indicate that many of the genes that are involved in oxidative metabolic pathways are indirectly controlled by PGC-1α and, hence, do not require PGC-1α recruitment to enhancer and promoter elements. Likewise unexpectedly, the MARA implies ERRα action on direct and indirect PGC-1α-induced target genes, i.e., in the presence or absence of PGC-1α coactivation. Thus, while these observations might obviously reflect a temporally distinct control of different PGC-1α target genes that is not represented in our simultaneous analysis of DNA binding and gene expression at one time point, it is conceivable that PGC-1α acts primarily as an upstream regulator of other factors that are subsequently controlling more downstream PGC-1α target genes without direct involvement of PGC-1α itself.
In skeletal muscle, PGC-1α has been reported to interact with ERRs, peroxisome proliferator-activated receptors (PPARs), and other nuclear receptors, as well as myocyte enhancer and nuclear respiratory factors, to mediate transcriptional regulation (3). Accordingly, ERRα and other nuclear receptor binding motifs were among the most highly significant binding elements in our present report. Importantly, however, we also predict a number of so-far unknown TFs to functionally interact with PGC-1α and thereby contribute to PGC-1α-controlled gene expression in skeletal muscle. Since a complete functional validation of all new putative TF partners is beyond the scope of this paper, we combined the high-throughput results with several computational analyses (Table 1) to select and test some of the potentially most important factors together with predicted target genes. Notably, in siRNA-based knockdown experiments, we could show that depletion of FOS and its putative AP-1 multimerization partners JUN and ATF3 as well as NFE2L2, ZFP143, and GTF2I in muscle cells reduced the ability of PGC-1α to positively regulate target genes. Second, we could provide evidence of a corecruitment of FOS and PGC-1α to the same regulatory sites in the vicinity of AP-1/PGC-1α target genes, confirming a functional interaction between these TFs and PGC-1α. Thus, our results indicate that the coactivation repertoire of PGC-1α in muscle exceeds the prediction of previous studies by far. For example, even in our list of the top 15 motifs, several predicted TFs have not yet been investigated in the context of PGC-1α-controlled gene expression, including BPTF, FOSL2, REST, or RREB1. Future studies will aim at a more detailed dissection of the global functional consequences of PGC-1α coactivation of these TFs in muscle cells.
Curiously, almost all of our analyses, and in particular the principal component analysis, highlighted the relevance of FOS-JUN-like motifs. In fact, the largest amount of variation in TFBS occurrence within PGC-1α binding peaks results from either ESRRA-like or FOS-JUN-like motifs. The FOS-JUN-like motif, in particular, embodies the main binding elements of the AP-1 complex, which consist of different configurations of FOS, JUN, ATF, and MAF proteins (35, 41). Our data comparing gene expression in cells with reduced FOS, JUN, and ATF3 levels indicate that PGC-1α functionally interacts with the AP-1 complex in different configurations in the regulation of specific genes. The differential requirement observed for distinct AP-1 components might provide an additional layer of control for specific PGC-1α target gene regulation.
AP-1 function itself is regulated by a variety of stimuli, including cytokines, growth factors, and stress, and subsequently controls a number of cellular processes, including apoptosis, cell proliferation and differentiation, stress response, and hypoxia (41, 42). Mechanistically, we classified PGC-1α-induced/AP-1-knockdown targets in either direct or indirect genes. Most interestingly, functional analysis of these two groups of genes revealed that when AP-1 and PGC-1α act disjointedly, they are involved in the regulation of mitochondrial and other metabolic genes, while when coactivated by PGC-1α, AP-1 distinctly alters the expression of genes that are enriched in the ontology terms “response to hypoxia” and “mTOR signaling” (Fig. 5F). Intriguingly, a closer analysis of all 47 direct AP-1/PGC-1α target genes revealed 24 genes that are induced by hypoxia, are effectors of hypoxia, or attenuate the detrimental consequences of hypoxia (Fig. 6M). For example, several inhibitors of the mTOR signaling pathways are included in this group of genes, and hypoxia has been described as a suppressor of mTORC1 activity (43). Another group of genes contributes to the reduction of cellular stress, detrimental metabolites, and reactive oxygen species and an increase in cellular survival to reduce potential harmful consequences of prolonged hypoxia (44). Furthermore, several genes promote endothelial regeneration, vascular remodeling, and vascularization (45). In this context, PGC-1α has previously been shown to promote VEGF-induced angiogenesis in skeletal muscle in a hypoxia-inducible factor 1α (HIF-1α)-independent, ERRα-dependent manner (36). Similarly, PGC-1α regulates the hypoxic response of brown fat (46) and neuronal and endothelial cells (47), even though the mechanisms of cellular protection exerted by PGC-1α in these experimental contexts have not been elucidated. Our findings now indicate that, to ensure adequate oxygen and nutrient supplies for oxidative metabolism in skeletal muscle cells, PGC-1α might coordinate metabolic needs through ERRα-induced Vegf expression with a broad, stress-induced AP-1-dependent hypoxia program. Such a functional convergence was found for a subset of the direct AP-1/PGC-1α target genes that likewise seem to be under the control of ERRα together with PGC-1α (Fig. 5H and I to K). Inversely, for the complementary subset of these genes, the functional interaction between AP-1 and PGC-1α seems distinct from the ERRα-dependent PGC-1α target gene regulation. Finally, in vivo evidence supports our muscle cell culture-based prediction, considering that many of the AP-1/PGC-1α hypoxia-related target genes exhibit reduced and elevated transcript levels in PGC-1α muscle-specific knockout and transgenic animals, respectively. As previously demonstrated for VEGF and skeletal muscle vascularization (36), many aspects of the phenotypic consequences of exercise-induced muscle hypoxia occur in the muscle-specific transgenic mice even in the absence of physical activity. In an extension of these studies, we now, however, found additional genes involved in this process that show an additional, or in the case of Nr0b2, even an exclusive synergistic activation by exercise in the PGC-1α transgenic animals. Thus, combined with previous descriptions of muscle plasticity in these mice postexercise in regard to insulin sensitivity (29), our present findings reiterate the importance of bona fide exercise even in a genetic model for endurance training such as the PGC-1α muscle-specific transgenic animals.
In summary, our data provide a first insight into the transcriptional network controlled by PGC-1α in muscle cells. While one other study of global DNA recruitment of PGC-1α has been performed in the human hepatoma cell line HepG2 (48), our results highlight the importance of combining ChIP-Seq experiments and transcriptional data together with a comprehensive computational modeling approach and experimental validation of predicted key regulators, in order to be able to discover mechanistic as well as functional outcomes of such a network. Combined with the knowledge of transcriptional regulation, posttranslational modifications, alternative splicing, and recruitment of different chromatin remodeling protein complexes, a scenario can thus be conceived in which PGC-1α is able to control and integrate different signaling pathways using a multitude of different transcription factor binding partners (10, 11). A better understanding of such regulatory networks will eventually allow the targeting of whole biological programs or specific submodules in pathological states of dysregulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anastasia Kralli, Svenia Schnyder, Gesa Santos, Kristoffer Svensson, and Markus Beer for reagents, help, and input in the preparation of the manuscript. We are grateful to the [BC]2 Basel Computational Biology Center for providing computational resources.
This project was funded by ERC Consolidator grant 616830-MUSCLE_NET to C.H., the Swiss National Science Foundation (31003A_135397 to E.V.N. and 310030_132900 to C.H.), SystemsX.ch (CellPlasticity, StoNets, and BrainstemX research projects to E.V.N.), the Swiss Society for Research on Muscle Diseases (SSEM), the Neuromuscular Research Association Basel (NeRAB), the Gebert-Rüf Foundation “Rare Diseases” Program, the University of Basel, and the Biozentrum. S.S. was supported by an IPhD fellowship of the SystemsX.ch Swiss Initiative in Systems Biology.
We have no conflict of interest in regard to this paper.
Footnotes
Published ahead of print 9 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01710-13.
REFERENCES
- 1.Handschin C, Spiegelman BM. 2008. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 454:463–469. 10.1038/nature07206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen BK, Febbraio MA. 2012. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 8:457–465. 10.1038/nrendo.2012.49 [DOI] [PubMed] [Google Scholar]
- 3.Handschin C. 2010. Regulation of skeletal muscle cell plasticity by the peroxisome proliferator-activated receptor gamma coactivator 1alpha. J. Recept. Signal Transduct. Res. 30:376–384. 10.3109/10799891003641074 [DOI] [PubMed] [Google Scholar]
- 4.Handschin C, Spiegelman BM. 2006. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 27:728–735. 10.1210/er.2006-0037 [DOI] [PubMed] [Google Scholar]
- 5.Finck BN, Kelly DP. 2006. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 116:615–622. 10.1172/JCI27794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. 2002. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418:797–801. 10.1038/nature00904 [DOI] [PubMed] [Google Scholar]
- 7.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. 2007. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J. Biol. Chem. 282:30014–30021. 10.1074/jbc.M704817200 [DOI] [PubMed] [Google Scholar]
- 8.Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, Kulkarni RN, Shulman GI, Spiegelman BM. 2007. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J. Clin. Invest. 117:3463–3474. 10.1172/JCI31785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lonard DM, O'Malley BW. 2006. The expanding cosmos of nuclear receptor coactivators. Cell 125:411–414. 10.1016/j.cell.2006.04.021 [DOI] [PubMed] [Google Scholar]
- 10.Lonard DM, O'Malley BW. 2007. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell 27:691–700. 10.1016/j.molcel.2007.08.012 [DOI] [PubMed] [Google Scholar]
- 11.Spiegelman BM, Heinrich R. 2004. Biological control through regulated transcriptional coactivators. Cell 119:157–167. 10.1016/j.cell.2004.09.037 [DOI] [PubMed] [Google Scholar]
- 12.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. 1999. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286:1368–1371. 10.1126/science.286.5443.1368 [DOI] [PubMed] [Google Scholar]
- 13.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. 2003. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 12:1137–1149. 10.1016/S1097-2765(03)00391-5 [DOI] [PubMed] [Google Scholar]
- 14.Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD. 2008. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 8:105–117. 10.1016/j.cmet.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin J, Handschin C, Spiegelman BM. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1:361–370. 10.1016/j.cmet.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 16.Handschin C. 2009. The biology of PGC-1alpha and its therapeutic potential. Trends Pharmacol. Sci. 30:322–329. 10.1016/j.tips.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- 19.Perez-Schindler J, Summermatter S, Santos G, Zorzato F, Handschin C. 2013. The transcriptional coactivator PGC-1alpha is dispensable for chronic overload-induced skeletal muscle hypertrophy and metabolic remodeling. Proc. Natl. Acad. Sci. U. S. A. 110:20314–20319. 10.1073/pnas.1312039110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnold P, Erb I, Pachkov M, Molina N, van Nimwegen E. 2012. MotEvo: integrated Bayesian probabilistic methods for inferring regulatory sites and motifs on multiple alignments of DNA sequences. Bioinformatics 28:487–494. 10.1093/bioinformatics/btr695 [DOI] [PubMed] [Google Scholar]
- 21.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Development Core Team. 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 23.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. 2004. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20:578–580. 10.1093/bioinformatics/btg455 [DOI] [PubMed] [Google Scholar]
- 24.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, Katayama S, Schroder K, Carninci P, Tomaru Y, Kanamori-Katayama M, Kubosaki A, Akalin A, Ando Y, Arner E, Asada M, Asahara H, Bailey T, Bajic VB, Bauer D, Beckhouse AG, Bertin N, Bjorkegren J, Brombacher F, Bulger E, Chalk AM, Chiba J, Cloonan N, Dawe A, Dostie J, Engstrom PG, Essack M, Faulkner GJ, Fink JL, Fredman D, Fujimori K, Furuno M, Gojobori T, Gough J, Grimmond SM, Gustafsson M, Hashimoto M, Hashimoto T, Hatakeyama M, Heinzel S, Hide W, Hofmann O, Hornquist M, Huminiecki L, et al. 2009. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 41:553–562. 10.1038/ng.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddharthan R, Siggia ED, van Nimwegen E. 2005. PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput. Biol. 1:e67. 10.1371/journal.pcbi.0010067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahony S, Benos PV. 2007. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35:W253–W258. 10.1093/nar/gkm272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summermatter S, Santos G, Perez-Schindler J, Handschin C. 2013. Skeletal muscle PGC-1alpha controls whole-body lactate homeostasis through estrogen-related receptor alpha-dependent activation of LDH B and repression of LDH A. Proc. Natl. Acad. Sci. U. S. A. 110:8738–8743. 10.1073/pnas.1212976110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summermatter S, Shui G, Maag D, Santos G, Wenk MR, Handschin C. 2013. PGC-1alpha improves glucose homeostasis in skeletal muscle in an activity-dependent manner. Diabetes 62:85–95. 10.2337/db12-0291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. 2004. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 24:9079–9091. 10.1128/MCB.24.20.9079-9091.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mootha VK, Handschin C, Arlow D, Xie X, St. Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM. 2004. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U. S. A. 101:6570–6575. 10.1073/pnas.0401401101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. 2004. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. U. S. A. 101:6472–6477. 10.1073/pnas.0308686101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisele PS, Salatino S, Sobek J, Hottiger MO, Handschin C. 2013. The peroxisome proliferator-activated receptor gamma coactivator 1alpha/beta (PGC-1) coactivators repress the transcriptional activity of NF-kappaB in skeletal muscle cells. J. Biol. Chem. 288:2246–2260. 10.1074/jbc.M112.375253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangelsdorf DJ, Evans RM. 1995. The RXR heterodimers and orphan receptors. Cell 83:841–850. 10.1016/0092-8674(95)90200-7 [DOI] [PubMed] [Google Scholar]
- 35.Hai T, Curran T. 1991. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. U. S. A. 88:3720–3724. 10.1073/pnas.88.9.3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, Baek KH, Rosenzweig A, Spiegelman BM. 2008. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature 451:1008–1012. 10.1038/nature06613 [DOI] [PubMed] [Google Scholar]
- 37.Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. 2011. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol. 18:120–127. 10.1038/nsmb.2000 [DOI] [PubMed] [Google Scholar]
- 38.Qian J, Chen S, Huang Y, Shi X, Liu C. 2013. PGC-1alpha regulates hepatic hepcidin expression and iron homeostasis in response to inflammation. Mol. Endocrinol. 27:683–692. 10.1210/me.2012-1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang WG, Kim EJ, Park KG, Park YB, Choi HS, Kim HJ, Kim YD, Kim KS, Lee KU, Lee IK. 2007. Glucocorticoid receptor mediated repression of human insulin gene expression is regulated by PGC-1alpha. Biochem. Biophys. Res. Commun. 352:716–721. 10.1016/j.bbrc.2006.11.074 [DOI] [PubMed] [Google Scholar]
- 40.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. 2006. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. U. S. A. 103:16260–16265. 10.1073/pnas.0607795103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaulian E, Karin M. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131–E136. 10.1038/ncb0502-e131 [DOI] [PubMed] [Google Scholar]
- 42.Curran T, Franza BR., Jr 1988. Fos and Jun: the AP-1 connection. Cell 55:395–397. 10.1016/0092-8674(88)90024-4 [DOI] [PubMed] [Google Scholar]
- 43.Cam H, Easton JB, High A, Houghton PJ. 2010. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Mol. Cell 40:509–520. 10.1016/j.molcel.2010.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majmundar AJ, Wong WJ, Simon MC. 2010. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40:294–309. 10.1016/j.molcel.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner PD. 2001. Skeletal muscle angiogenesis. A possible role for hypoxia. Adv. Exp. Med. Biol. 502:21–38. 10.1007/978-1-4757-3401-0_4 [DOI] [PubMed] [Google Scholar]
- 46.Pino E, Wang H, McDonald ME, Qiang L, Farmer SR. 2012. Roles for peroxisome proliferator-activated receptor gamma (PPARgamma) and PPARgamma coactivators 1alpha and 1beta in regulating response of white and brown adipocytes to hypoxia. J. Biol. Chem. 287:18351–18358. 10.1074/jbc.M112.350918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao J, Li L, Pei Z, Li C, Wei H, Zhang B, Peng Y, Wang Y, Tao Y, Huang R. 2012. Peroxisome proliferator activated receptor (PPAR)-gamma co-activator 1-alpha and hypoxia induced factor-1alpha mediate neuro- and vascular protection by hypoxic preconditioning in vitro. Brain Res. 1447:1–8. 10.1016/j.brainres.2012.01.059 [DOI] [PubMed] [Google Scholar]
- 48.Charos AE, Reed BD, Raha D, Szekely AM, Weissman SM, Snyder M. 2012. A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells. Genome Res. 22:1668–1679. 10.1101/gr.127761.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.