Abstract
The influence of UV irradiation on pigmentation is well established, but the molecular and cellular mechanisms controlling dendrite formation remain incompletely understood. MicroRNAs (miRNAs) are a class of small RNAs that participate in various cellular processes by suppressing the expression of target mRNAs. In this study, we investigated the expression of miRNAs in response to UVB irradiation using a microarray screen and then identified potential mRNA targets for differentially expressed miRNAs among the genes governing dendrite formation. We subsequently determined the ability of miRNA 340 (miR-340) to suppress the expression of RhoA, which is a predicted miR-340 target gene that regulates dendrite formation. The overexpression of miR-340 promoted dendrite formation and melanosome transport, and the downregulation of miR-340 inhibited UVB-induced dendrite formation and melanosome transport. Moreover, a luciferase reporter assay demonstrated direct targeting of RhoA by miR-340 in the immortalized human melanocyte cell line Pig1. In conclusion, this study has established an miRNA associated with UVB irradiation. The significant downregulation of RhoA protein and mRNA expression after UVB irradiation and the modulation of miR-340 expression suggest a key role for miR-340 in regulating UVB-induced dendrite formation and melanosome transport.
INTRODUCTION
Melanocytes originate from neural crest-derived melanoblasts, which migrate and differentiate in the basal layer of the epidermis (1). A hallmark of melanocytes is their ability to form dendrites, which are specialized cell structures that transport melanosomes to their tips for transfer to the surrounding keratinocytes in response to growth factors and UV irradiation. Following skin penetration by UV rays and subsequent DNA damage, thymidine dinucleotide fragments induce melanogenesis and cause the melanocyte to produce melanosomes (2).These melanosomes are then transferred to neighboring keratinocytes through the intricate network of melanocyte dendrites, contributing to skin darkening and thereby providing protection from UV radiation (3, 4). Melanocyte dendrites can vary markedly in length and number in response to different growth factors and, in a manner analogous to the way in which neural cells seek out target neurons, these dendrites form growth cone-like structures that attach to keratinocytes. Melanocyte-keratinocyte adhesion is a prerequisite for the transfer of melanosomes to keratinocytes; therefore, the formation of melanocyte dendrites, particularly of the appropriate length and number, is essential for efficient melanosome transfer. Due to the importance of dendricity for melanocyte activity, cutaneous pigmentation, and photoprotection, it is critical to determine the precise mechanisms involved in the regulation of melanocyte dendrite formation.
It is well known that melanocytes are sensitive to UV irradiation, with substantial evidence suggesting that it plays a pivotal role in regulating melanocyte dendricity. Studies have shown that UV irradiation induces the production of melanogenic stimulators such as nitric oxide, endothelin 1,α-melanocyte-stimulating hormone, adrenocorticotropic hormone, and prostaglandin E2 (5, 6, 7). In vivo reflectance confocal imaging of murine skin following exposure to UVB light highlighted its effect on the dendricity of melanocytes (8). Furthermore, UV light exposure along with genetic factors was found to be a strong predictor for the development of melasma (9, 10), a condition in which epidermal melanocytes within lesions exhibit higher levels of melanin and more dendrites than melanocytes in adjacent normal skin (11).
An additional effect of UV irradiation on human cells is the alteration of microRNA (miRNA) expression profiles. miRNAs are evolutionarily conserved, small noncoding RNAs that regulate gene expression through sequence-specific base pairing with the 3′ untranslated region (3′UTR) of target mRNAs. Diverse biological functions are regulated by miRNAs, including the coordination of cell differentiation, proliferation, apoptosis, tumorigenesis, and the immune response (12–17). Recent studies have shown that miRNAs are also important for regulating tissue morphogenesis, for example, the rescue of brain morphogenesis in maternal-zygotic dicer mutant zebrafish by miRNA 340 (miR-340) (18). Indeed, miR-34a has been shown to regulate cross talk between the GTPases RhoA and Rac1 and negatively modulate the reorganization of the actin cytoskeleton, which is essential for establishing chondrocyte-specific morphology (19). Other studies have shown that miR-134, miR-132, and miR-138 are involved in regulating dendritic spine morphogenesis during neuronal development (20–22). Although these varied roles for miRNAs have recently been elucidated, little is known about the function of miRNAs in the regulation of melanocyte dendrite formation.
The miRNA expression profiles of UV-irradiated cells have been investigated using miRNA microarrays, and in one such study, differential miRNA expression profiles were found in UVB-irradiated NIH 3T3 cells compared with nonirradiated cells (23). The miRNA expression changes were most pronounced within the first hours following UVB irradiation, suggesting that miRNA-mediated gene regulation operates earlier than most transcriptional responses (24).
In this study, to further elucidate the effect of UVB irradiation on miRNA expression in melanocytes, we utilized the immortalized human melanocyte cell line Pig1 (25). Specifically, we compared the miRNA expression profiles of UVB-treated Pig1 cells with those of untreated Pig1 cells using miRNA arrays. To predict miRNA targets, TargetScan and gene ontology analyses were conducted, which revealed that miR-340 was upregulated 4.8-fold in Pig1 cells treated with UVB irradiation. Overexpression of miR-340 significantly repressed RhoA protein expression, increased the number and total length of dendrites per cell, and promoted melanosome aggregation in dendritic tips of melanocytes. Conversely, transfection of cells with miR-340 inhibitors blocked UVB-induced miR-340 expression and upregulated RhoA mRNA and protein expression. This reduced the number and total length of dendrites per cell and caused melanosomes to aggregate around the cell nucleus. We concluded that miR-340 plays a key role in regulating UVB-induced dendrite formation and melanosome transport.
MATERIALS AND METHODS
Cell line culture and treatment.
The human epidermal melanocyte cell line Pig1 (kindly provided by Caroline Le Poole, Skin Cancer Laboratories, Loyola University Medical Center, Maywood, IL) and normal human epidermal melanocytes (NHEMs; isolated from human foreskin specimens obtained during circumcision surgery) were maintained in medium 254 supplemented with human melanocyte growth supplement (Life Technologies) and 5% fetal bovine serum (FBS; Gibco). The human HaCaT keratinocyte cell line was grown in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum. Cocultures were established by seeding Pig1 melanocytes and HaCaT keratinocytes in a ratio of 1:20 in medium containing 50% medium 254 supplemented with human melanocyte growth supplement and 50% DMEM supplemented with 10% FBS. Cells were grown to 80% confluence in six-well culture plates and used for subsequent experiments. All cell lines were cultured in a 37°C humidified atmosphere containing 5% CO2.
Pig1 cells and cocultured cells were evenly divided into a control group and a UVB-irradiated group. UVB irradiation was carried out using a UV light therapy apparatus (Sigma). Before the cells were irradiated, the culture medium was removed and cells were washed twice with phosphate-buffered saline (PBS), and then 1 ml PBS was added for the irradiation process. Cells were exposed to 100 mJ · cm−2 UVB, and then PBS was replaced with the appropriate cell culture medium.
miRNA microarray analysis.
Total RNA was extracted from cells in the control and UVB-irradiated groups using RNAiso Plus (TaKaRa Bio) according to the manufacturer's instructions. The concentration and quality of total RNA were measured by UV absorbance at 260 nm and 280 nm (A260/280) and confirmed by gel electrophoresis. The extracted RNA was Cy3 labeled and hybridized to miRNA microarrays. The hybridized microarray images were quantified with GenePix Software (Molecular Devices). All data normalization and selection of differentially regulated genes were performed using GeneSpring GX 10.0 (Agilent Technologies). The averages of normalized ratios were calculated by dividing the average normalized signal channel intensity by the average normalized control channel intensity. Gene expression by the control group was used as the control. The signal intensity of experimental groups was compared with that of the control group, and the ratio of signal intensity was calculated. The ratios of signal intensity for each miRNA were compared, and a differential expression of >3-fold was regarded as significant. All experiments were conducted in triplicate.
Validation of microarray expression by qRT-PCR.
Total RNA was isolated from the Pig1 cell line and primary melanocytes from the UV-treated and control groups. Real-time reverse transcription PCR (qRT-PCR) analysis was performed using a Chrom 4 or iQ5 real-time PCR detection system (Bio-Rad Laboratories) and the All-in-One miRNA qRT-PCR detection kit (GeneCopoeia). Amplified product levels were detected by real-time monitoring of SYBR green II dye fluorescence (TaKaRa Bio) under the following conditions: 95°C for 2 min, followed by 45 cycles of denaturation at 95°C for 5 s, annealing at 60°C for 10 s, and extension at 72°C for 15 s. All reactions were run in triplicate in a minimum of three independent experiments. Relative quantification was performed according to the comparative threshold cycle (CT) method, in which the mRNA fold change was generated using the equation 2−ΔΔCT. The expression of primary miR-340 was performed using TaqMan Pri-miRNA assays according to the manufacturer's protocol (Life Technologies).
Oligonucleotide synthesis and transient transfection.
miR-340 mimics and negative-control miRNA were chemically synthesized by Life Technologies. miR-340 inhibitors (Life Technologies) were used to inhibit the function of miR-340, and a scrambled RNA sequence was used as an additional negative control. The oligonucleotides were transfected into Pig1 cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. The sequences of these RNA oligonucleotides were as follows: for miR-340 mimics, sense, 5′-UUAUAAAGCAAUGAGACUGAUU-3′, and antisense, 5′-UUAAUAUUUCGUUACUCUGACU-3′; for miRNA control, sense, 5′-UUGUACUACACAAAAGUACUG-3′; for miR-340 inhibitors, 5′-AAUCAGUCUCAUUGAUUUAUAA-3′; and for scrambled RNA, 5′-CAGUACUUUUGUGUAGUACAA-3′ . For transfection, 1 × 105 Pig1 cells were seeded into culture plates and incubated in medium 254 supplemented with human melanocyte growth supplement overnight.
Plasmid construct and luciferase reporter assay.
The putative targets of miR-340 were predicted using TargetScan. To determine whether miR-340 was able to directly bind to the RhoA 3′UTR and suppress RhoA protein expression, the wild-type 3′UTR of RhoA (NM_001664) and an miR-340 binding site mutant of the RhoA 3′UTR were chemically synthesized by Life Technologies (see Fig. 3D for these two sequences). Each sequence was subcloned into the psiCHECK-2 vector (Promega), and these plasmids were designated Psicheck-2-RhoA-wt and Psicheck-2-RhoA-mut after sequencing. For the luciferase assay, cells were seeded in 24-well plates 1 day before transfection and then transiently cotransfected with 0.3 μg of either Psicheck-2-RhoA-wt or Psicheck-2-RhoA-mut and 20 pmol of a miR-340 mimic or control miRNA. Cotransfections were performed using Lipofectamine 2000 (Life Technologies), according to the manufacturer's protocol. Twenty-four hours after transfection, the cell culture medium was removed and cells were lysed in passive lysis buffer (Promega). The firefly and Renilla luciferase activities were measured consecutively using the dual-luciferase assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized against Renilla luciferase activity, and the relative luciferase activity was calculated using the following formula: relative luciferase activity = firefly luciferase activity/Renilla luciferase activity. All experiments were performed twice, in triplicate.
FIG 3.
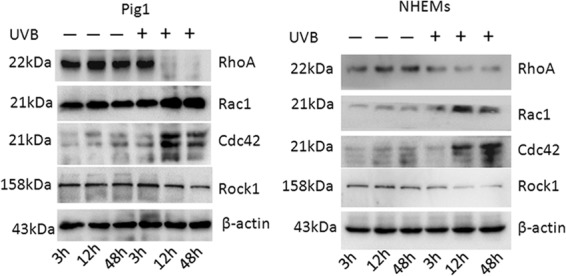
Effect of UVB irradiation on regulators of melanocyte dendricity. Rac1, RhoA, Cdc42, and Rock1 protein expression in UVB-irradiated and control cells, detected by Western blotting.
Protein extraction and Western blotting.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-HCl [pH7.4], 1 mM EDTA, and 0.1% NP-40) containing protease inhibitors (20 mg · ml−1 leupeptin, 10 mg · ml−1 pepstatin A, 10 mg · ml−1 chymostatin, 2 mg · ml−1 aprotinin, and 1 mM phenylmethylsulfonyl fluoride). After vigorous pipetting, extracts were centrifuged for 15 min at 10,000 × g at 4°C, and then total protein was measured using a bicinchoninic acid (BCA) protein assay kit (Bio-Rad Laboratories). Equal amounts of each sample (30 μg) were run on 12% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride (PVDF) membrane using the Bio-Rad semidry transfer system. After blocking of nonspecific binding using a solution of 5% (wt/vol) nonfat dry milk in Tris-buffered saline containing 0.05% Tween 20, membranes were incubated with the appropriate antibodies overnight at 4°C with gentle agitation. Blots were then incubated with peroxidase-conjugated secondary antibodies for 60 min at room temperature and visualized by enhanced chemiluminescence using the ChemiLucent Plus Western blot enhancing kit (Millipore).
Immunofluorescence and DOPA staining.
For immunofluorescence staining, cells on coverslips were fixed in cold acetone for 10 min at room temperature. The cells were permeabilized by incubation with 0.1% Triton X-100 in PBS for 10 min at room temperature. Nonspecific binding of antibodies was blocked by incubation with 5% bovine serum albumin for 1 h at room temperature. After washing twice with PBS, a primary antibody against gp100 (Thermo) was applied to cells, and they were incubated overnight for staining of melanosomes. Cells were examined for staining using a confocal microscope (Bio-Rad Laboratories). For 3,4-dihydroxyphenylalanine (DOPA) staining, cells on coverslips were fixed with 5% formalin in phosphate buffer (pH 7.0) at 4°C for 30 min, rinsed with distilled water, and then incubated with 0.1% L-DOPA (Sigma) in phosphate buffer at 37°C for 3 h. After one change of solution, the samples were fixed with 10% formalin in phosphate buffer at 25°C for 1 h, air dried, and examined by light microscopy (Niko).
Statistical analysis.
All data are expressed as the means ± standard deviations (SD). Data were analyzed using Student's t test or one-way analysis of variance (ANOVA) using Prism 5 software (GraphPad Software, San Diego, CA), with statistical significance defined as P values of <0.05. For each analysis, one representative experiment is shown from duplicate or triplicate analyses.
RESULTS
UVB irradiation promotes melanocyte dendricity.
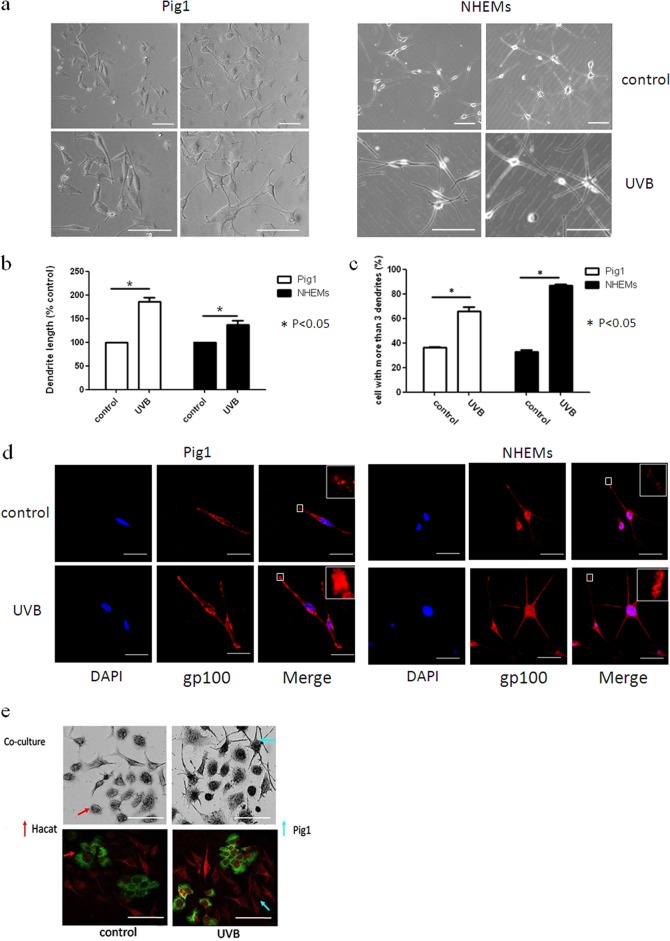
Although it is likely that UV light is a primary stimulus for melanocyte dendrite formation, the molecular mechanisms underlying this effect have not been fully elucidated. In this study, we first investigated the effect of UVB irradiation on dendrite formation using normal human epidermal melanocytes (NHEMs) and the Pig1 cell line. Cells were irradiated with 100 mJ · cm−2 UVB and then cultured for 48 h. The dendrites of NHEMs and Pig1 cells from each experiment were manually counted, and the dendrite lengths were measured by AxioVision Rel 4.8.2 software. Morphological changes induced by UVB irradiation were evident within Pig1 cells (Fig. 1a), with significant elongation of dendrite length (control, 42.37 ± 2.96 μm; UVB irradiated, 76.33 ± 4.06 μm) (Fig. 1b) and a marked increase in the number of dendrites (66% ± 3.46%) compared with untreated cells (36.27% ± 0.4%) (Fig. 1c). NHEMs showed statistically significant elongation of dendrite length (control, 101.32 ± 4.32 μm; UVB group, 137.48 ± 11.23 μm) (Fig. 1b) and a marked increase in the number of dendrites (86.77% ± 1.83%) compared with untreated cells (32.88% ± 2.68%) (Fig. 1c). Immunofluorescence staining analysis further verified dendrite elongation (Fig. 1d), and immunofluorescence and DOPA staining revealed that the number of dendrites was also increased by UVB irradiation when using a melanocyte-keratinocyte coculture model (Fig. 1e). In addition, increased transport of melanosomes to the dendritic tips of NHEMs and Pig1 cells (Fig. 1d) and transfer from Pig1 melanocytes to HaCat keratinocytes (Fig. 1e) were apparent after UVB treatment.
FIG 1.
Effects of UVB irradiation on melanocyte dendrite formation and melanosome transport. (a) The morphology of Pig1 cells and NHEMs 48 h after 100 mJ · cm−2 UVB irradiation was acquired by a bright-field microscope using a 20× or 40× objective lens and compared with that of untreated control cells. Scale bar, 100 μm. (b) The dendrite lengths of Pig1 cells that were UVB irradiated or untreated were measured by AxioVisionRel 4.8.2 software. Data are depicted relative to the control, and graphs depict the means ± SEM. Student's t test was used to compare dendrite lengths between groups. *, P < 0.05. (c) The dendrites in Pig1 cells from each experiment were manually counted, and sample groups were compared using the Student t test. Graphs depict the means ± SEM; *, P < 0.05. (d) Control and UVB-treated cells were stained with anti-gp100 antibody (red) and DAPI (4′,6-diamidino-2-phenylindole) (blue) and then viewed using a confocal microscope under a 60× objective lens to determine the localization of melanosomes within the cells. Scale bar, 50 μm. (e) Melanosome transfer was observed in the melanocyte-keratinocyte coculture model by light and immunofluorescence microscopy. Pig1 melanocytes or HaCaT keratinocytes were stained by L-DOPA or labeled with antibodies against gp100 (red) or keratin 10 (green), respectively, and then visualized by light or confocal microscopy using a 40× objective lens. Scale bar, 100 μm.
Identification of miR-340 as a key regulator of dendrite formation.
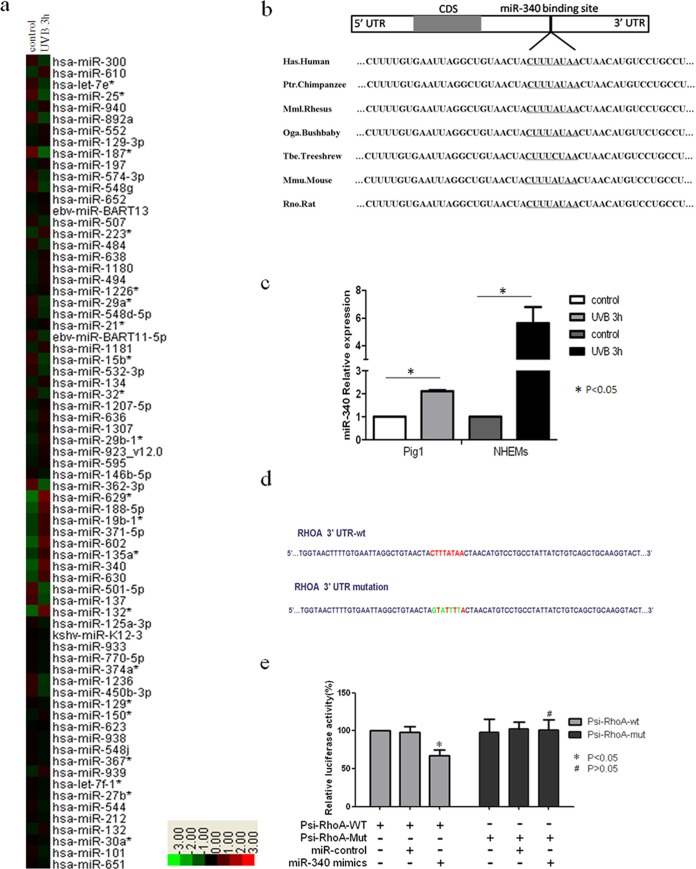
miRNAs are evolutionarily conserved small noncoding RNAs that regulate gene expression and play important roles in diverse biological functions, including cell differentiation, proliferation, apoptosis, tumorigenesis, and the immune response (12–17). Previous studies have shown that miRNA expression profiles change in response to UVB irradiation, and these changes were most pronounced within the first few hours following UVB exposure (23, 24). Thus, we wanted to determine whether miRNA is involved in UVB-induced melanocyte dendricity at an early stage. To investigate this, Pig1 cells were irradiated with 100 mJ · cm−2 UVB and total RNA was extracted after 3 h. An Agilent miRNA microarray was then used to screen differentially expressed miRNAs, and clustering analysis of the data was performed using Cluster 3.0 software (Fig. 2a). As expected, a number of miRNAs were differentially expressed (fold change, equal to or more than ±1.5) in treated cells compared with the control (Table 1). Specifically, the expression of 31 miRNAs was upregulated following UVB irradiation, while 19 miRNAs were downregulated.
FIG 2.
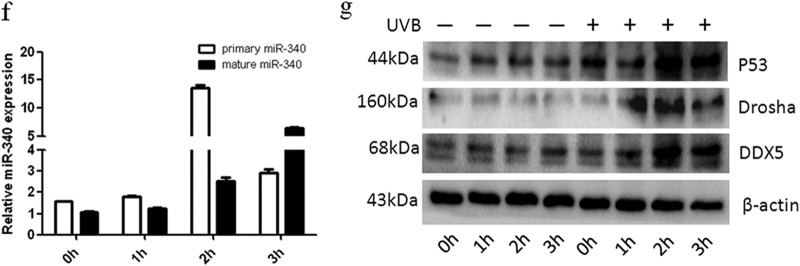
Identification of miRNAs involved in UVB-induced dendrite formation. (a) Hierarchical clustering analysis of miRNA expression in Pig1 cells without UVB irradiation or 3 h after UVB treatment. Red, +1.5-fold change in gene expression (high expression); green, −1.5-fold change in gene expression (low expression). (b) miR-340 target binding site in RhoA, which was predicted as a target of miR-340 using computational methods. The miR-340 seed binding region downstream of the coding sequence (CDS) is located in the 3′UTR of RhoA, and the sequence of this miR-340 seed binding region is identical in different species. (c) qRT-PCR analysis of miR-340 expression 3 h after UVB radiation in Pig1 cells and human epidermal melanocytes (NHEMs). The expression of miR-340 was normalized relative to the expression of U6 snRNA. Expression is presented relative to the controls, as the means ± SEM. *, P < 0.05, Student's t test. (d) Two luciferase reporter constructs that contained either the wild-type RhoA 3′UTR, including the putative binding site for miR-340, or the RhoA 3′UTR containing mutations within the putative targeting region were generated. The wild-type and mutated RhoA 3′UTR sequences are shown. (e) Wild-type and mutated RhoA 3′UTR sequences were inserted into the psiCHECK-2 vector to create Psi-RhoA-wt and Psi-RhoA-mut constructs, respectively. Each of these constructs was transiently cotransfected into Pig1 cells, along with either miR-340 mimics or control miRNA (miR-control), and luciferase activity was then measured 24 h after transfection. Luciferase activity is shown relative to that in Pig1 cells transfected with Psi-RhoA-wt alone, and data are presented as the means ± SEM. *, P < 0.05; #, P > 0.05 (Student's t test). (f) qRT-PCR analysis of primary and mature miR-340 expression UVB radiation in NHEMs. The expression of primary or mature miR-340 was normalized relative to the expression of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or U6 snRNA. Expression is presented relative to the controls, as the means ± SEM. *, P < 0.05, Student's t test. (g) P53, Drosha, and p68 (DDX5) protein expression in UVB-irradiated and control cells, detected by Western blotting.
TABLE 1.
Expression of miRNA in UVB-treated cells compared with control untreated cells
| Direction of change in expression | miRNA name | Fold change |
|---|---|---|
| Upregulation | hsa-miR-629* | 7.46 |
| hsa-miR-132* | 5.64 | |
| hsa-miR-602 | 5.06 | |
| hsa-miR-340 | 4.87 | |
| hsa-miR-188-5p | 4.50 | |
| hsa-miR-630 | 3.56 | |
| hsa-miR-19b-1* | 3.08 | |
| hsa-miR-371-5p | 3.08 | |
| hsa-miR-135a* | 2.82 | |
| hsa-miR-610 | 2.64 | |
| hsa-miR-223* | 2.60 | |
| hsa-miR-29b-1* | 2.20 | |
| hsa-miR-636 | 2.14 | |
| hsa-miR-1181 | 2.09 | |
| hsa-miR-939 | 2.07 | |
| hsa-miR-940 | 1.97 | |
| hsa-miR-1180 | 1.94 | |
| hsa-miR-134 | 1.91 | |
| hsa-miR-494 | 1.90 | |
| hsa-miR-132 | 1.81 | |
| hsa-miR-923_v12.0 | 1.80 | |
| hsa-miR-638 | 1.78 | |
| hsa-miR-552 | 1.72 | |
| hsa-miR-1307 | 1.71 | |
| ebv-miR-BART13 | 1.67 | |
| hsa-miR-197 | 1.67 | |
| hsa-miR-1207-5p | 1.65 | |
| hsa-miR-595 | 1.61 | |
| hsa-miR-150* | 1.61 | |
| hsa-miR-1226* | 1.59 | |
| hsa-miR-129-3p | 1.58 | |
| Downregulation | hsa-miR-187* | −3.82 |
| hsa-miR-362-3p | −3.30 | |
| hsa-miR-501-5p | −2.79 | |
| hsa-miR-25* | −2.62 | |
| hsa-miR-137 | −2.36 | |
| hsa-miR-892a | −2.29 | |
| hsa-miR-15b* | −1.98 | |
| hsa-miR-548g | −1.97 | |
| hsa-miR-300 | −1.88 | |
| hsa-miR-1236 | −1.79 | |
| hsa-miR-29a* | −1.78 | |
| hsa-let-7e* | −1.77 | |
| ebv-miR-BART11-5p | −1.74 | |
| hsa-miR-450b-3p | −1.73 | |
| hsa-miR-484 | −1.64 | |
| hsa-miR-32* | −1.61 | |
| hsa-miR-574-3p | −1.59 | |
| hsa-miR-548d-5p | −1.57 |
The prediction of miRNA-regulated gene targets is a necessary step in understanding the functions of miRNA; therefore, we predicted the targets of all miRNAs that were differentially expressed in response to UVB irradiation using a public database (TargetScan). Fourteen genes that are known to be associated with dendrite formation and contain recognition sequences for miR-340 were identified (Table 2). As RhoA is a key regulator of melanocyte dendricity, it was selected from the identified miR-340 target genes for further analysis. This analysis identified a conserved miR-340 binding site in the 3′UTR of RhoA from a number of different species (Fig. 2b). Real-time PCR was used to measure the expression of miR-340 in order to confirm the array data. This analysis revealed that miR-340 expression was significantly upregulated within 3 h of UVB irradiation in both Pig1 melanocytes and normal human epidermal melanocytes compared with untreated controls (Fig. 2c).
TABLE 2.
Potential targets of miR-340 associated with dendrite formation
| Target gene | Name | Function in dendrite formation |
|---|---|---|
| TIAM-1 | T-cell lymphoma invasion and metastasis 1 | Modulates the activity of RHO-like proteins and connects extracellular signals to cytoskeletal activities; activates RAC1, CDC42, and to a lesser extent RHOA. |
| ARHGEF6 | Rac/Cdc42 guanine nucleotide exchange factor 6 | Acts as a RAC1 guanine nucleotide exchange factor. |
| RHOA | Ras homolog gene family, member A | Regulates a signal transduction pathway linking plasma membrane receptors to the assembly of focal adhesions and actin stress fibers; represses dendrite formation in melanocyte cells. |
| CDC42SE2 | CDC42 small effector 2 | Likely involved in the organization of the actin cytoskeleton by acting downstream of CDC42, inducing actin filament assembly; alters CDC42-induced cell shape changes. |
| RAB27B | Member RAS oncogene family | Involved in vesicular fusion and trafficking and melanosome transport |
| MYO10 | Myosin X | Functions as an actin-based molecular motor and plays a role in the integration of F-actin and microtubule cytoskeletons during meiosis. |
| MACF1 | Microtubule-actin cross-linking factor 1 | May function in microtubule dynamics to facilitate actin-microtubule interactions at the cell periphery and couple the microtubule network to cellular junctions. |
| ARHGEF26 | Rho guanine nucleotide exchange factor 26 | Activates RhoG GTPase by promoting the exchange of GDP by GTP; required for the formation of membrane ruffles during macropinocytosis. |
| ROCK1 | Rho-associated, coiled-coil-containing protein kinase 1 | Regulates the actin cytoskeleton and cell polarity. |
| ARHGAP24 | Rho GTPase activating protein 24 | Involved in cell polarity, cell morphology and cytoskeletal organization |
| MYRIP | Myosin VIIA and Rab interacting protein | Involved in melanosome transport, serving as link between melanosome-bound RAB27A and the motor proteins MYO5A and MYO7A. |
| ARHGAP5 | Rho GTPase activating protein 5 | Mediates cytoskeleton changes by stimulating the hydrolysis of bound GTP. |
| ARHGAP29 | Rho GTPase activating protein 29 | Acts as a GTPase activator for the Rho-type GTPases by converting them to an inactive GDP-bound state; has strong activity toward RHOA and weaker activity toward RAC1 and CDC42. |
| CDC42BPA | CDC42 binding protein kinase alpha (DMPK-like) | Plays a role in the regulation of cytoskeleton reorganization and cell migration. |
Next, we performed a luciferase reporter assay to verify the sequence region of the RhoA 3′UTR that directly interacts with miR-340. Two luciferase reporter constructs were made, containing either the wild-type RhoA 3′UTR, including the putative binding site for miR-340, or a RhoA 3′UTR that contained mutations within the putative targeting region that were predicted to abolish miR-340 binding. These sequences (Fig. 2d) were inserted downstream of the open reading frame of the Renilla luciferase gene in the psiCHECK-2vector (Promega), and the constructs were designated Psi-RhoA-wt and Psi-RhoA-mut, respectively. Transient cotransfection of Pig1 cells with Psi-RhoA-wt and miR-340 mimics resulted in a significant decrease in luciferase activity, compared with cotransfection with Psi-RhoA-wt and control miRNA. Moreover, the mutation of the miR-340 putative binding site within the RhoA 3′UTR prevented this inhibition of luciferase expression (Fig. 2e). These results indicate that miR-340 can directly regulate RhoA expression through the predicted sequence sites.
To determine whether the expression changes of miR-340 were occurring only in mature miR-340 or also in primary miRNA, real-time PCR was used to measure the expression of both primary miR-340 and mature miR-340. Primary and mature miR-340 expression was significantly upregulated within 2 h after UVB irradiation, compared with untreated controls (Fig. 2f). P53 is not only a major response protein in UVB-induced DNA damage but also a modulator of microRNA processing through interaction with the Drosha processing complex member p68 (26); thus, we evaluated the expression of p53, Drosha, and p68 (DDX5). The data showed that primary and mature miR-340s were upregulated within 2 h following p53, Drosha, and p68 (DDX5) upregulation (Fig. 2g). These results reveal that p53, Drosha, and p68 (DDX5) may be involved in UVB-induced miR-340 processing.
UVB irradiation modifies the expression of melanocyte dendricity regulators.
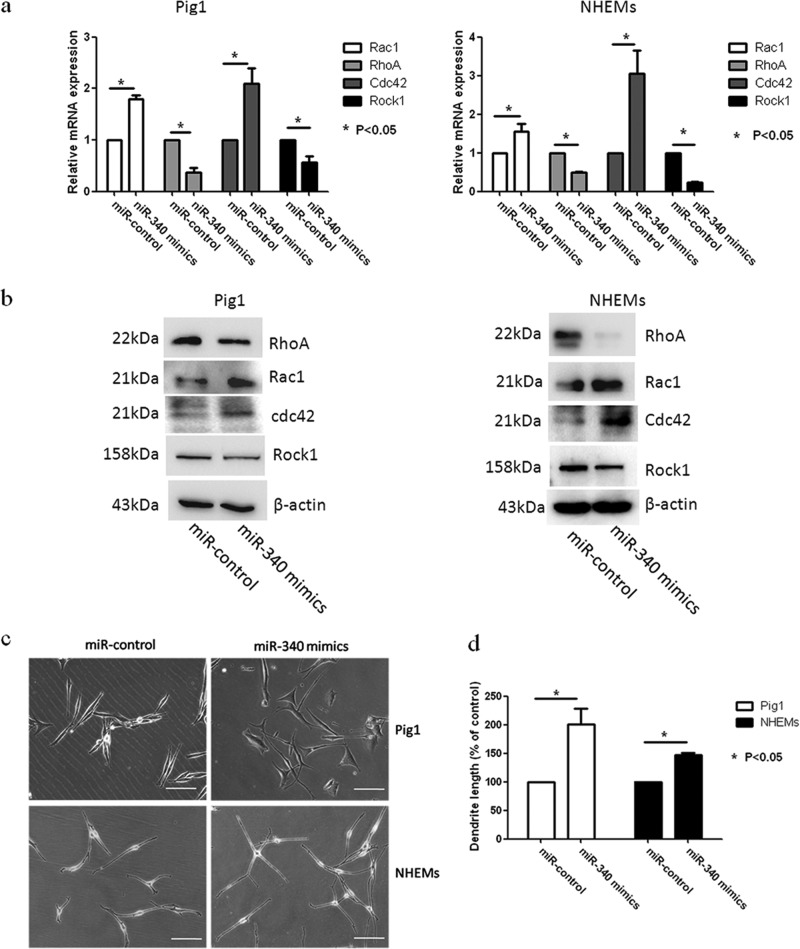
Rac1, RhoA, and Cdc42 are small GTPases that play a key role in the regulation of cell morphology (27). Several studies have shown that these GTPases act in opposition to control dendrite formation and elongation in human melanocytes and melanoma cells, with Rac1 and Cdc42 promoting dendrite formation and RhoA inhibiting this process (28–30). Thus, we aimed to further clarify the role of Rac1, RhoA, and Cdc42 in dendricity regulation by monitoring their presence in NHEMs and Pig1 cells following UVB irradiation. As shown in Fig. 3, we found that expression of Rac1 and Cdc42 protein was upregulated 12 h and 48 h after 100 mJ · cm−2 UVB exposure, whereas RhoA protein expression was downregulated. We also investigated the changes in the expression of Rock1, which constitute the main downstream pathway of RhoA signaling, in response to UVB irradiation. The results showed that the protein expression of Rock1 was downregulated following exposure to UVB. These results suggest that UVB irradiation may promote the expression of Rac1and Cdc42 and repress RhoA signaling, leading to dendrite elongation and an increase in the number of dendrites in melanocytes.
miR-340 promotes dendrite extension and melanosome transport by modulating the expression of RhoA.
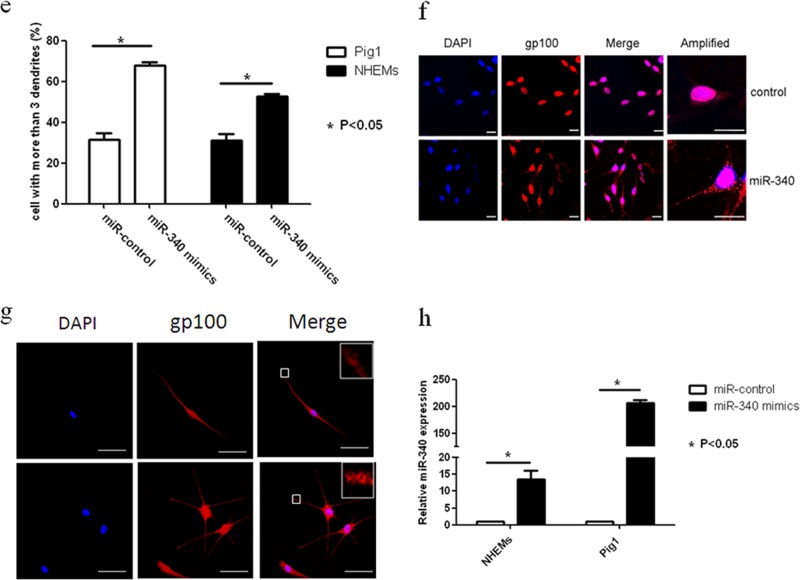
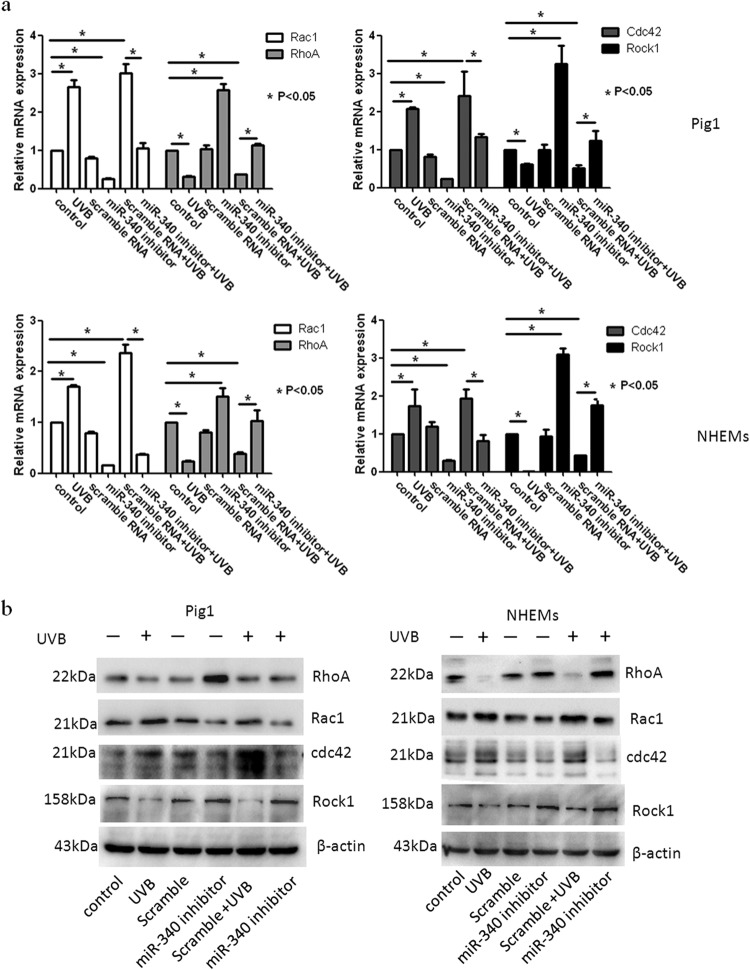
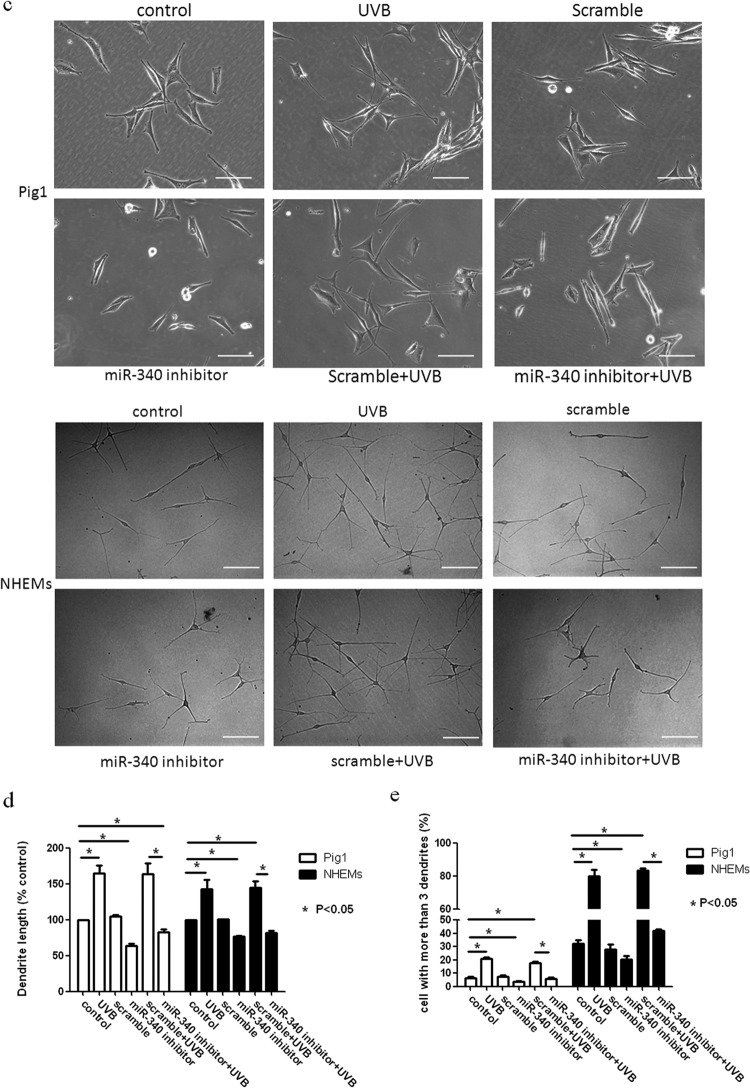
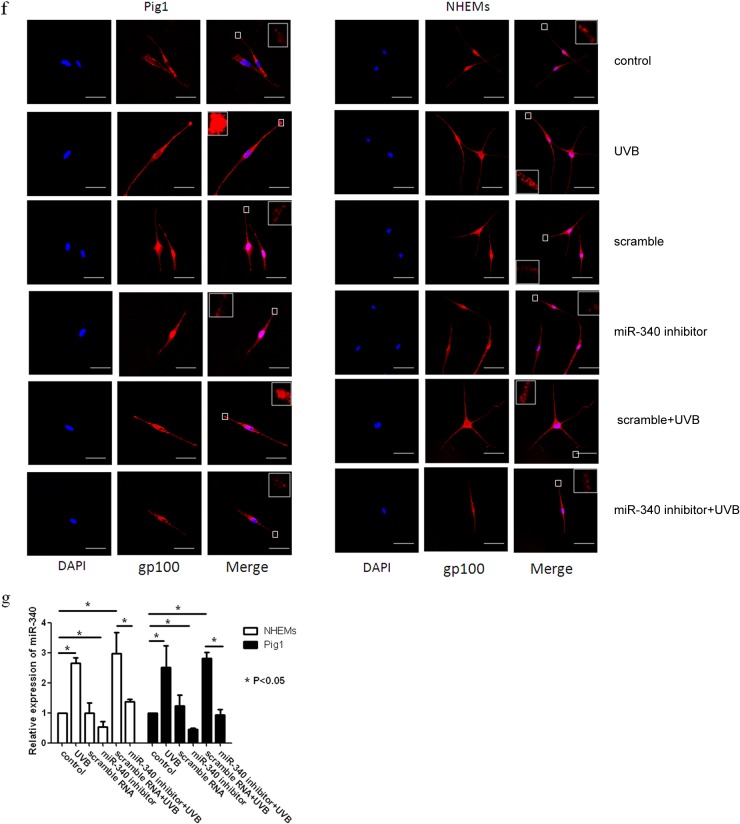
To determine whether miR-340 can affect the dendricity of melanocytes, miR-340 mimics were transfected into Pig1 cells and NHEMs (Fig. 4h). Morphological changes were seen in Pig1 cells transfected with miR-340 mimics compared with control miRNA (Fig. 4c), with significant elongation of dendrites (miR-340 mimics, 104.4 ± 10.76 μm; control, 49 ± 3.03 μm) (Fig. 4d) and a marked increase in the number of dendrites (miR-340 mimics, 68 ± 1.53; control, 31.33 ± 3.28) (Fig. 4e). NHEMs showed a statistically significant elongation of dendrite length (miR-340 mimics, 147.17 ± 4.36 μm; control, 110.9 ± 4.48 μm) (Fig. 4d) and a marked increase in the number of dendrites (52.63% ± 1.92%) compared with untreated cells (31.11% ± 4.55%) (Fig. 4e). Then, two molecules involved in RhoA signaling, Rac1 and Cdc42, were detected. The results showed that miR-340 can negatively regulate the expression of RhoA and Rock1 while upregulating Rac1 and Cdc42 expression at both the mRNA and protein levels (Fig. 4a and b). Immunofluorescence analysis further demonstrated the elongation of dendrites in cells transfected with miR-340 mimics and revealed that the dendrites of melanocytes contain more melanosomes and the dendritic tips aggregated more melanosome (Fig. 4f and g). These data provided evidence that miR-340 promotes dendrite extension and melanosome transport by modulating the expression of RhoA signaling.
FIG 4.
miR-340 alters dendrite formation and melanosome transport in melanocytes. Pig1 cells and NHEMs were transfected with control miRNA and miR-340 mimics, and then the cells were analyzed after 48 h. (a and b) The cells from each group were harvested and lysed, and then Rac1, RhoA, Cdc42, and Rock1 mRNA expression was analyzed by qRT-PCR (a) and the protein expression was analyzed by Western blotting (b). (c) Morphology of melanocytes after transfection, as assessed by bright-field microscopy using a 10× or 20× objective lens; scale bar, 100 μm. (d) Sum of the lengths of cell dendrites, as measured by AxioVisionRel 4.8.2 software. Total dendrite length is presented relative to the control, as the means ± SEM. *, P < 0.05, Student's t test. (e) The dendrites in Pig1 cells from each experiment were manually counted. The percentage of cells with >3 dendrites is shown for each group, as the mean ± SEM. *, P < 0.05, Student's t test. (f and g) miR-control (top) and miR-340-mimic (bottom)-transfected Pig1 cells (f) or NHEMs (g) were stained with an anti-gp100 antibody (red) and DAPI (blue) and then viewed under a confocal microscope using a 20× objective lens to determine melanosome localization within cells. Scale bar, 20 μm. (h) miR-340 expression was analyzed by qRT-PCR after transfection with control miRNA or miR-340 mimics in both Pig1 cells and NHEMs.
Inhibition of miR-340 blocks UVB-induced dendrite formation and melanosome transport via RhoA.
In order to identify the role of miR-340 in UVB-induced dendrite formation and melanosome transport, Pig1 cells and NHEMs were transfected with miR-340 inhibitors or scrambled-sequence miRNA and then treated with UVB. Compared with untreated control cells, miR-340 was upregulated following UVB irradiation and transfection with scrambled-sequence miRNA. However, miR-340 was downregulated following transfection with miR-340 inhibitors, and UVB-induced miR-340 upregulation was blocked by transfection with the miR-340 inhibitors (Fig. 5g). The expression of RhoA and Rock1 was downregulated by treatment with UVB (with or without scrambled-sequence miRNA transfection), whereas the expression of Rac1 and Cdc42 was upregulated. Transfection with miR-340 inhibitors notably upregulated expression of RhoA and Rock1 and downregulated the expression of Rac1 and Cdc42 (Fig. 5a and b). We also found that transfection of miR-340 inhibitors reversed the effect of UVB on the morphology of melanocytes (Fig. 5c). As shown in Fig. 5d and e, UVB treatment significantly increased the total length of dendrites per cell (UVB Pig1, 104.5 ± 3.48 μm; control, 49.78 ± 1.41 μm; UVB NHEMs, 159.67 ± 7.47 μm; control, 103.8 ± 5.43 μm) and markedly increased the number of dendrites compared with those in untreated cells (UVB Pig1, 22.8% ± 1.5%; control Pig1, 5.0% ± 0.8%; UVB NHEMs, 80% ± 6.67%; control NHEMs, 32.3% ± 4.5%). Transfection with scrambled-sequence miRNA did not significantly enhance the effect of UVB irradiation, but transfection with miR-340 inhibitors evidently decreased the total length of dendrites (Pig1, 25.28 ± 4.48 μm; NHEMs, 79.15 ± 5.62 μm) and the number of dendrites (Pig1, 22.88% ± 0.56%; NHEMs, 20.56% ± 4.19%). In contrast, UVB treatment in conjunction with miR-340 inhibitor transfection decreased the total length of the dendrites (Pig1, 51.36 ± 1.49 μm; NHEMs, 84.96 ± 6.92 μm) and the number of dendrites (Pig1, 4.9% ± 1.1%; NHEMs, 41.9% ± 1.65%), compared with UVB treatment alone. Immunofluorescence analysis confirmed these findings and also revealed aggregation of melanosomes in the dendritic tips of Pig1 cells or NHEMs after UVB irradiation (with or without scrambled-miRNA transfection) (Fig. 5f). However, melanosomes were found to aggregate around the nucleus when miR-340 inhibitors were transfected into Pig1 cells or NHEMs, and a similar phenomenon was found in transfection by the miR-340 inhibitor and in UVB irradiation (Fig. 5f). These data suggest that downregulation of miR-340 inhibits UVB-induced dendrite formation and melanosome transport via RhoA signaling.
FIG 5.
Inhibition of miR-340 blocks UVB-induced dendrite formation. Pig1 cells and NHEMs were exposed to 100 mJ · cm−2 UVB, with or without transfection with miR-340 inhibitors or control (scrambled-sequence) miRNA. (a) Rac1, RhoA, Cdc42, and Rock1 mRNA expression was assessed by qRT-PCR. Expression of mRNA is presented relative to the untreated control, and data are presented as the means ± SEM. *, P < 0.05, Student's t test. (b) Western blotting was used to detect the expression of Rac1, RhoA, Cdc42, and Rock1 proteins. (c) Inhibition of miR-340 blocks UVB-induced morphology change. The morphology of melanocytes was acquired after exposure to 100 mJ · cm−2 UVB, with or without transfection with miR-340 inhibitors or control (scrambled-sequence) miRNA by bright-field microscopy using a 40× objective lens; scale bar, 100 μm. (d) The lengths of cell dendrites were measured by AxioVisionRel 4.8.2 software. Dendrite length is shown relative to that of untreated control cells, and graphs depict the means ± SEM. Student's t test was used to compare dendrite lengths between groups. *, P < 0.05. (e) The dendrites in Pig1 cells from each experiment were manually counted, and sample groups were compared using the Student t test. The percentage of cells with >3 dendrites is shown for each group as the means ± SEM. *, P < 0.05. (f) Inhibition of miR-340 blocks UVB-induced melanosome transport. Pig1 cells and NHEMs were stained with anti-gp100 antibody (red) and DAPI (blue) and then viewed under a confocal microscope using a 60× objective lens to determine melanosome localization within cells. Scale bar, 50 μm. (g) miR-340 expression was assessed by qRT-PCR after exposure to 100 mJ · cm−2 UVB, with or without transfection with miR-340 inhibitors or control (scrambled-sequence) miRNA.
DISCUSSION
Melanocytes are derived from neural crest cells and morphologically resemble neuronal cells, especially as they possess dendrites. Melanocyte dendrites are the conduit for melanosome transfer to the surrounding keratinocytes in the epidermis and are intimately associated with the melanogenic potential of melanocytes (28). Although it has been recognized that UV irradiation can induce the production of melanogenic stimulators (e.g., nitric oxide, endothelin 1, α-melanocyte-stimulating hormone, adrenocorticotropic hormone, and prostaglandin E2), which can induce melanogenesis concomitantly with dendrite formation through downstream signaling (5–7), the precise mechanism remains to be determined. In this study, miR-340 was identified as a novel miRNA associated with UVB irradiation that was involved in melanocyte dendrite formation and melanosome transport and showed marked upregulation in melanocytes treated with UVB. Overexpression of miR-340 promoted dendrite formation and melanosome transport, whereas blocking miR-340 by transfection of miR-340 inhibitors prevented dendrite formation and melanosome transport and attenuated UVB-induced dendrite formation and melanosome transport. Collectively, these data highlight the importance of miR-340 in UVB-induced dendrite formation and melanosome transport.
miRNAs play diverse regulatory roles in cells, including development (31, 32), cell proliferation and differentiation (31–35), apoptosis (36, 37), tumorigenesis (38, 39), and many other physiological or pathological processes (40, 41). Notably, the first identified target for miR-340 was microphthalmia-associated transcription factor (MITF), which is a master regulator of melanocyte development and melanogenesis in melanoma cells. It was found that miR-340 interacts with two target sites on the 3′UTR of MITF mRNA, causing mRNA degradation and decreased expression and activity of MITF protein (42). Following this discovery, additional functions of miR-340 were identified. In breast cancer research, miR-340 was found to inhibit breast cancer cell migration and invasion through the targeting of the oncoprotein c-MET (43). miR-340 has also been reported to regulate colorectal cancer growth via inhibition of the Warburg effect, in conjunction with miR-124 and miR-137 (44), and to suppress osteosarcoma tumor growth and metastasis by directly targeting ROCK1 (45). In the current study, prediction data from the TargetScan database showed that miR-340 could target RhoA through only one potential binding site, located at the proximal region of the RhoA 3′UTR. We confirmed the direct binding of miR-340 to the RhoA 3′UTR using luciferase reporter assays and found through real-time PCR and Western blot assays that miR-340 could repress the mRNA and protein expression of RhoA, respectively.
The tumor suppressor p53 orchestrates multiple cellular pathways as a central node of antioncogenic programs in response to DNA damage, oncogene activation, and several stresses (46). p53 is an important response protein in UVB-induced DNA damage of melanocytes. Suzuki et al. reported that p53 promoted the processing of several primary miRNA transcripts through association with Drosha, a central RNase III in miRNA biogenesis, under DNA damage-inducing conditions (26). We determined the expression of p53, Drosha, and p68 (DDX5) when we analyzed the primary and mature miR-340 expression after UVB irradiation. The data showed that primary and mature miR-340s were upregulated within 2 h following p53, Drosha, and p68 (DDX5) upregulation. These results reveal that p53, Drosha, and p68 (DDX5) may be involved in UVB-induced miR-340 processing.
Rho family GTPases play a pivotal role in dendrite formation. It was previously established that RhoA is involved in dendrite retraction and stress fiber formation, while Rac1 promotes dendrite and lamellipodium formation, and Cdc42 mediates filopodium and neurite formation (8). Other studies have shown that UVB induced up- or downregulation of Rho family GTPases, resulting in morphological changes in B16 melanoma cells (47–50). Fan et al. reported that RhoA RNA interference (RNAi) could significantly reduce F-actin expression and induce neurite outgrowth of PC12 cells (51). In this study, we found that UVB irradiation promoted melanosome transport to the tips of melanocytes and subsequent melanocyte transfer to keratinocytes, induced upregulation of Rac1 andCdc42 and downregulation of RhoA and its downstream molecules (Rock1) within 12 h and 48 h of treatment, and upregulated primary miR-340 and mature miR-340 within 2 h or 3 h of treatment, respectively. Moreover, upregulation of miR-340 promoted dendrite formation and melanosome transport. However, blocking of miR-340 by transfection of miR-340 inhibitors prevented dendrite formation and melanosome transport and attenuated UVB-induced dendrite formation and melanosome transport. Considering the established role of miR-340 in RhoA regulation, it is plausible that miR-340 may have a direct effect on dendrite formation and melanosome transport via RhoA.
In conclusion, the results of the present study provide novel information on the expression of miRNAs in melanocytes in response to UVB irradiation and demonstrate a functional role for miR-340 in the regulation of dendrite formation and melanosome transport in melanocytes. These results establish a novel UVB-miRNA-mRNA regulatory network that could be associated with the control of skin pigmentation.
ACKNOWLEDGMENTS
We gratefully acknowledge the valuable suggestions and technical assistance provided by laboratory members within the Department of Dermatology at Xijing Hospital.
This work was funded by the National Natural Sciences Foundation of China (grants 30572099 and 30872264).
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 30 June 2014
REFERENCES
- 1.Dorsky RI, Moon RT, Raible DW. 2000. Environmental signals and cell fate specification in premigratory neural crest. Bioessays 22:708–716. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi Y, Brenner M, Hearing VJ. 2007. The regulation of skin pigmentation. J. Biol. Chem. 282:27557–27561. 10.1074/jbc.R700026200 [DOI] [PubMed] [Google Scholar]
- 3.Eller MS, Maeda T, Magnoni C, Atwal D, Gilchrest BA. 1997. Enhancement of DNA repair in human skin cells by thymidine dinucleotides: evidence for a p53-mediated mammalian SOS response. Proc. Natl. Acad. Sci. U. S. A. 94:12627–12632. 10.1073/pnas.94.23.12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sulaimon SS, Kitchell BE. 2003. The biology of melanocytes. Vet. Dermatol. 14:57–65. 10.1046/j.1365-3164.2003.00327.x [DOI] [PubMed] [Google Scholar]
- 5.Iyengar B. 1992. Neural differentiation as an expression of UV sensitivity of melanocytes. Acta Anat. (Basel) 143:236–240. 10.1159/000147254 [DOI] [PubMed] [Google Scholar]
- 6.Iyengar B. 1994. UV guided dendritic growth patterns and the networking of melanocytes. Experientia 50:669–672. 10.1007/BF01952870 [DOI] [PubMed] [Google Scholar]
- 7.Duval C, Régnier M, Schmidt R. 2001. Distinct melanogenic response of human melanocytes in mono-culture, in co-culture with keratinocytes and in reconstructed epidermis, to UV exposure. Pigment Cell Res. 14:348–355. 10.1034/j.1600-0749.2001.140506.x [DOI] [PubMed] [Google Scholar]
- 8.Scott G. 2002. Rac and rho: the story behind melanocyte dendrite formation. Pigment Cell Res. 15:322–330. 10.1034/j.1600-0749.2002.02056.x [DOI] [PubMed] [Google Scholar]
- 9.Jimbow M, Jimbow K. 1989. Pigmentary disorders in oriental skin. Clin. Dermatol. 7:11–27. 10.1016/0738-081X(89)90053-9 [DOI] [PubMed] [Google Scholar]
- 10.Pichardo R, Vallejos Q, Feldman SR, Schulz MR, Verma A, Quandt SA, Arcury TA. 2009. The prevalence of melasma and its association with quality of life in adult male Latino migrant workers. Int. J. Dermatol. 48:22–26. 10.1111/j.1365-4632.2009.03778.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang WH, Yoon KH, Lee ES, Kim J, Lee KB, Yim H, Sohn S, Im S. 2002. Melasma: histopathological characteristics in 56 Korean patients. Br. J. Dermatol. 146:228–237. 10.1046/j.0007-0963.2001.04556.x [DOI] [PubMed] [Google Scholar]
- 12.Miska EA. 2005. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 15:563–568. 10.1016/j.gde.2005.08.005 [DOI] [PubMed] [Google Scholar]
- 13.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. 2007. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 67:2456–2468. 10.1158/0008-5472.CAN-06-2698 [DOI] [PubMed] [Google Scholar]
- 14.Gottwein E, Cullen BR. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375–387. 10.1016/j.chom.2008.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonkoly E, Stahle M, Pivarcsi A. 2008. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 18:131–140. 10.1016/j.semcancer.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 16.Li T, Li D, Sha J, Sun P, Huang Y. 2009. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem. Biophys. Res. Commun. 383:280–285. 10.1016/j.bbrc.2009.03.077 [DOI] [PubMed] [Google Scholar]
- 17.Hicks JA, Trakooljul N, Liu HC. 2010. Discovery of chicken microRNAs associated with lipogenesis and cell proliferation. Physiol. Genomics 41:185–193. 10.1152/physiolgenomics.00156.2009 [DOI] [PubMed] [Google Scholar]
- 18.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308:833–838. 10.1126/science.1109020 [DOI] [PubMed] [Google Scholar]
- 19.Kim D, Song J, Kim S, Park HM, Chun CH, Sonn J, Jin EJ. 2012. MicroRNA-34a modulates cytoskeletal dynamics through regulating RhoA/Rac1 cross-talk in chondroblasts. J. Biol. Chem. 287:12501–12509. 10.1074/jbc.M111.264382 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. 2006. A brain-specific microRNA regulates dendritic spine development. Nature 439:283–289. 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- 21.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hübel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. 2009. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11:705–716. 10.1038/ncb1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Impey S, Davare M, Lesiak A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. 2010. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol. Cell. Neurosci. 43:146–156. 10.1016/j.mcn.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, Huang ZX, Chen XW, Deng QK, Yan W, Zhou MJ, Ou CS, Ding ZH. 2009. Differential expression profiles of microRNAs in NIH3T3 cells in response to UVB irradiation. Photochem. Photobiol. 85:765–773. 10.1111/j.1751-1097.2008.00482.x [DOI] [PubMed] [Google Scholar]
- 24.Pothof J, Verkaik NS, Hoeijmakers JH, van Gent DC. 2009. MicroRNA responses and stress granule formation modulate the DNA damage response. Cell Cycle 8:3462–3468. 10.4161/cc.8.21.9835 [DOI] [PubMed] [Google Scholar]
- 25.Le Poole van den Berg FM, van den Wijngaard RM, Galloway DA, van Amstel PJ, Buffing AA, Smits HL, Westerhof W, Das PK. 1997. Generation of a human melanocyte cell line by introduction of HPV16 E6 and E7 genes. In Vitro Cell. Dev. Biol. Anim. 33:42–49. 10.1007/s11626-997-0021-6 [DOI] [PubMed] [Google Scholar]
- 26.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. 2009. Modulation of microRNA processing by p53. Nature 460:529–533. 10.1038/nature08199 [DOI] [PubMed] [Google Scholar]
- 27.Burridge K, Wennerberg K. 2004. Rho and Rac take center stage. Cell 116:167–179. 10.1016/S0092-8674(04)00003-0 [DOI] [PubMed] [Google Scholar]
- 28.Kim MY, Choi TY, Kim JH, Lee JH, Kim JG, Sohn KC, Yoon KS, Kim CD, Lee JH, Yoon TJ. 2010. MKK6 increases the melanocyte dendricity through the regulation of Rho family GTPases. J. Dermatol. Sci. 60:114–119. 10.1016/j.jdermsci.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 29.Ito Y, Kanamaru A, Tada A. 2006. Centaureidin promotes dendrite retraction of melanocytes by activating Rho. Biochim. Biophys. Acta 1760:487–494. 10.1016/j.bbagen.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 30.Scott G, Leopardi S. 2003. The cAMP signaling pathway has opposing effects on Rac and Rho in B16F10 cells: implications for dendrite formation in melanocytic cells. Pigment Cell Res. 16:139–148. 10.1034/j.1600-0749.2003.00022.x [DOI] [PubMed] [Google Scholar]
- 31.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337–350. 10.1016/S1534-5807(03)00228-4 [DOI] [PubMed] [Google Scholar]
- 32.Fineberg SK, Kosik KS, Davidson BL. 2009. MicroRNAs potentiate neural development. Neuron 64:303–309. 10.1016/j.neuron.2009.10.020 [DOI] [PubMed] [Google Scholar]
- 33.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38:228–233. 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. 2004. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 279:52361–52365. 10.1074/jbc.C400438200 [DOI] [PubMed] [Google Scholar]
- 35.Nakahara K, Carthew RW. 2004. Expanding roles for miRNAs and siRNAs in cell regulation. Curr. Opin. Cell Biol. 16:127–133. 10.1016/j.ceb.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 36.Mott JL, Kobayashi S, Bronk SF, Gores GJ. 2007. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26:6133–6140. 10.1038/sj.onc.1210436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welch C, Chen Y, Stallings RL. 2007. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26:5017–5022. 10.1038/sj.onc.1210293 [DOI] [PubMed] [Google Scholar]
- 38.Hwang HW, Mendell JT. 2006. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 94:776–780. 10.1038/sj.bjc.6603023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skaftnesmo KO, Prestegarden L, Micklem DR, Lorens JB. 2007. MicroRNAs in tumorigenesis. Curr. Pharm. Biotechnol. 8:320–325. 10.2174/138920107783018390 [DOI] [PubMed] [Google Scholar]
- 40.Kasinath BS, Feliers D, Sataranatarajan K, Ghosh Choudhury G, Lee MJ, Mariappan MM. 2009. Regulation of mRNA translation in renal physiology and disease. Am. J. Physiol. Renal Physiol. 297:F1153–F1165. 10.1152/ajprenal.90748.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martino S, di Girolamo I, Orlacchio A, Datti A, Orlacchio A. 2009. MicroRNA implications across neurodevelopment and neuropathology. J. Biomed. Biotechnol. 2009:654346. 10.1155/2009/654346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goswami S, Tarapore RS, Teslaa JJ, Grinblat Y, Setaluri V, Spiegelman VS. 2010. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J. Biol. Chem. 285:20532–20540. 10.1074/jbc.M110.109298 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J, Zhao JJ, Mao SS, Zhang GH, Xu XC, Zhang N. 2011. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer 117:2842–2852. 10.1002/cncr.25860 [DOI] [PubMed] [Google Scholar]
- 44.Sun Y, Zhao X, Zhou Y, Hu Y. 2012. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol. Rep. 28:1346–1352 [DOI] [PubMed] [Google Scholar]
- 45.Zhou X, Wei M, Wang W. 2013. MicroRNA-340 suppresses osteosarcoma tumor growth and metastasis by directly targeting ROCK1. Biochem. Biophys. Res. Commun. 437:653–658. 10.1016/j.bbrc.2013.07.033 [DOI] [PubMed] [Google Scholar]
- 46.Suzuki HI, Miyazono K. 2013. p53 actions on microRNA expression and maturation pathway. Methods Mol. Biol. 962:165–181. 10.1007/978-1-62703-236-0_14 [DOI] [PubMed] [Google Scholar]
- 47.Buscà R, Bertolotto C, Abbe P, Englaro W, Ishizaki T, Narumiya S, Boquet P, Ortonne JP, Ballotti R. 1998. Inhibition of Rho is required for cAMP-induced melanoma cell differentiation. Mol. Biol. Cell 9:1367–1378. 10.1091/mbc.9.6.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh H, Aoki J, Ichikawa A, Negishi M. 1998. p160 RhoA-binding kinase ROKalpha induces neurite retraction. J. Biol. Chem. 273:2489–2492. 10.1074/jbc.273.5.2489 [DOI] [PubMed] [Google Scholar]
- 49.Scott GA, Cassidy L. 1998. Rac1 mediates dendrite formation in response to melanocyte stimulating hormone and ultraviolet light in a murine melanoma model. J. Investig. Dermatol. 111:243–250. 10.1046/j.1523-1747.1998.00276.x [DOI] [PubMed] [Google Scholar]
- 50.Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. 1999. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147:1009–1022. 10.1083/jcb.147.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan YM, Pang CP, Harvey AR, Cui Q. 2008. Marked effect of RhoA-specific shRNA-producing plasmids on neurite growth in PC12 cells. Neurosci. Lett. 440:170–175. 10.1016/j.neulet.2008.05.045 [DOI] [PubMed] [Google Scholar]