Abstract
Teichoic acid (TA), together with peptidoglycan (PG), represents a highly complex glycopolymer that ensures cell wall integrity and has several crucial physiological activities. Through an insertion-deletion mutation strategy, we show that ΔrafX mutants are impaired in cell wall covalently attached TA (WTA)-PG biosynthesis, as evidenced by their abnormal banding patterns and reduced amounts of WTA in comparison with wild-type strains. Site-directed mutagenesis revealed an essential role for external loop 4 and some highly conserved amino acid residues in the function of RafX protein. The rafX gene was highly conserved in closely related streptococcal species, suggesting an important physiological function in the lifestyle of streptococci. Moreover, a strain D39 ΔrafX mutant was impaired in bacterial growth, autolysis, bacterial division, and morphology. We observed that a strain R6 ΔrafX mutant was reduced in adhesion relative to the wild-type R6 strain, which was supported by an inhibition assay and a reduced amount of CbpA protein on the ΔrafX mutant bacterial cell surface, as shown by flow cytometric analysis. Finally, ΔrafX mutants were significantly attenuated in virulence in a murine sepsis model. Together, these findings suggest that RafX contributes to the biosynthesis of WTA, which is essential for full pneumococcal virulence.
INTRODUCTION
Streptococcus pneumoniae is a major human pathogen that affects mainly children and the elderly and can cause life-threatening diseases (1, 2). Asymptomatic colonization of the upper respiratory tract with pneumococci is a major risk factor in disease development and horizontal transmission within populations (3), highlighting the importance of bacterial adherence to epithelial cells in the pathogenesis of pneumococcal disease. Prevention with the pneumococcal polysaccharide vaccine (PPV23) and pneumococcal conjugate vaccine (PCV7) has greatly decreased the disease burden caused by pneumococci, but the appearance of non-vaccine-covered serotypes and/or a lack of immune memory call for new strategies to combat this pathogen (4). Besides, the emergence of vancomycin-resistant pneumococcal strains and multidrug-resistant strains raises great concern for human health and demands the development of novel antibiotics (5, 6).
As a fundamental component of the cell wall of Gram-positive bacteria, teichoic acids (TAs) can be divided, according to their anchors, into cell wall covalently attached TA (WTA) and membrane-anchored lipoteichoic acid (LTA) (7). The role of TAs has been well described in a previous review but could display species-specific variation (7). In Bacillus subtilis, WTA is implicated in bacterial growth and cell division and morphogenesis (7, 8). In Staphylococcus aureus, WTA is an alternative nonprotein adhesin involved in nasal colonization (9, 10). In contrast to the previously reported Toll-like receptor 2 agonist activity of LTA (11, 12), a recent report indicated that this effect is actually caused by a contaminating lipoprotein(s) (13). As an essential element in the biology of S. pneumoniae, the phosphorylcholine (P-Cho) moiety is unique to pneumococcal TAs (14), providing anchors for choline-binding proteins (CBPs) (15). The CBP family consists of several members, including LytA, -B, and -C (three autolysins); CbpA (an adhesin); and PspA (a protective antigen formerly named CbpC) (15). Moreover, specific binding of P-Cho to the receptor of platelet-activating factor (rPAF) has been reported to contribute to pneumococcal adherence and invasion (16).
In contrast to structures of TAs from other bacterial species, the chemical composition of pneumococcal TA is more complex in its repeating units (RU), containing the amino sugar 2-acetamido-4-amino-2,4,6-trideoxy-d-galactose, glucose, ribitol phosphate, N-acetylgalactosamine, and P-Cho (17, 18). Despite inconsistency among several reports regarding the backbone structure (13, 17, 19), pneumococcal WTA and LTA were recognized to consist of a maximum of eight or nine identical pentasaccharide RU with a maximal molecular mass of 10 kDa, as determined by mass spectrometry (MS) analysis (13, 18, 20). However, the molecular mass of WTA can be extended to 12.5 to 22.5 kDa because of the attachment of 25 or more disaccharide units (approximately 12.5 kDa) of peptidoglycan (PG) following digestion with LytA amidase (17).
Pneumococcal TA biosynthesis is a complex process. Choline metabolism has been fully described (21, 22) while the biosynthesis of the TA backbone is largely unknown. A recent review presented bioinformatic evidence that as many as 16 known genes and other hypothetical genes, primarily clustered in lic loci, are involved in the biosynthesis of pneumococcal TAs (23). So far, it has been accepted that products of these genes responsible for the biosynthesis of TAs are localized predominantly in the cell membrane (23, 24). Briefly, TAs were synthesized inside the cytoplasm, polymerized by rafX (SPD_1198) inside the cytoplasm, translocated to the outside of the cell by a flippase (TacF protein), and eventually linked to the PG to form WTA or attached to the glycolipid anchor to form LTA by an unknown mechanism (23). Despite recent progress, several key steps remain obscure, preventing a clear understanding of the biosynthesis of pneumococcal TAs.
Here, we report that RafX, a hypothetical protein, is involved in the biosynthesis of pneumococcal WTA, by showing that ΔrafX mutants display an abnormal banding pattern of the WTA-PG complex and have a reduced amount of cell WTA. ΔrafX mutants were strongly impaired in some physiological features, including autolysin, bacterial division, and morphology. We also show that ΔrafX mutants exhibit strongly reduced adherence to epithelial and endothelial cells, as well as bacterial virulence. Together, these findings may contribute to a better understanding of the biosynthesis of TAs in streptococci.
MATERIALS AND METHODS
Ethics statement.
All of the animals used in this study were purchased from the Laboratory Animal Center of Chongqing Medical University [certificate no. SYXK (yu) 2007-0001]. The research described here was carried out in accordance with the Declaration of Helsinki and with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the experimental protocols were approved by the Ethics Committee of Chongqing Medical University (reference no. 2011-032).
Bacterial strains, plasmids, and growth conditions.
All of the bacterial strains, except the clinical isolates, used in this study are listed in Table S1 in the supplemental material. All of the clinical isolates are listed in Table S3. Escherichia coli was grown in Luria-Bertani (LB) broth with shaking or on LB agar plates at 37°C. S. pneumoniae and S. mitis W1 strains were grown in semisynthetic casein hydrolysate medium supplemented with 5% yeast extract (C+Y, pH 7.0) medium or blood agar plates at 37°C under 5% CO2. All bacterial growth environments were supplemented with appropriate antibiotics as described in Table S1.
Construction of in-frame nonpolar deletion mutations.
The sequences of the primers used are listed in Table S2 in the supplemental material. The rafX and lgt genes were deleted from the chromosome of S. pneumoniae by insertion-deletion mutagenesis, which was performed essentially according to an established protocol (25). For example, upstream and downstream fragments of the rafX gene were PCR amplified with primers rafX UP P1-2 and rafX DW P3-4, respectively. Overlap extension with primers rafX UP P1 and rafX DW P4 was performed to generate the Up-Erm-Dw fusion fragment, which was then used to transform S. pneumoniae strains D39, R6, TIGR4, and CMCC(B)31203. Recombinants were selected on blood agar plates containing erythromycin. Successful replacement with the erm cassette was confirmed by sequencing of chromosomal DNA in the region containing the deletion.
Construction of vectors for complementation experiments.
For pEVP3 plasmid-based complementation, a fragment including the 633 bp upstream of rafX and the full-length rafX gene was cloned into plasmid pEVP3 at the BglII and SmaI restriction sites. This plasmid was used to transform S. pneumoniae ΔrafX mutants to obtain full-length-RafX-complemented strains (Compl or WL).
Elimination of the enzymatic site (residues 255 to 311) and replacement of critical active-site residues with alanine were performed as described elsewhere (26). pYYB5 (pEVP3-rafX) was used as the template. All of the primers used are listed in Table S2 in the supplemental material. PCR products generated with Pfu polymerase were digested with 1 U of DpnI at 37°C overnight. The resulting products were transformed into E. coli DH5α competent cells. Viable mutants were selected on LB agar plates supplemented with appropriate antibiotics.
Transmission electron microscopy (TEM).
Cells were grown in C+Y medium at 37°C to the mid-exponential phase. Bacteria were harvested by centrifugation at 4°C for 10 min at 10,000 × g. Samples were fixed in 2% glutaraldehyde in sodium cacodylate buffer (pH 7.4) for 12 h and then processed by the Electron Microscopy Research Service of Chongqing Medical University. For visualization, cells were sectioned and imaged with a Hitachi H-7500 transmission electron microscope.
Growth curves.
The wild-type (WT) D39, ΔrafX mutant, and complemented strains were grown in C+Y medium at 37°C to an optical density at 620 nm (OD620) of 0.4. Equal numbers of bacteria were cultured in 5 ml of fresh C+Y medium at 37°C. OD600 values were recorded with a cell density meter (Ultrospec 10; Amersham Biosciences) every 30 min.
Computer techniques.
Primers premier 5 and DNAMAN were used for DNA sequence analysis, oligonucleotide primer design, and sequence alignment. BLAST searches were performed with the tools provided by the National Center for Biotechnology Information (NCBI). Amino acid conservation was determined with hidden Markov model-based protein structure prediction server SAM-T02 by using the input peptide sequence from Arg-255 to Asp-311 (see Fig. 3C).
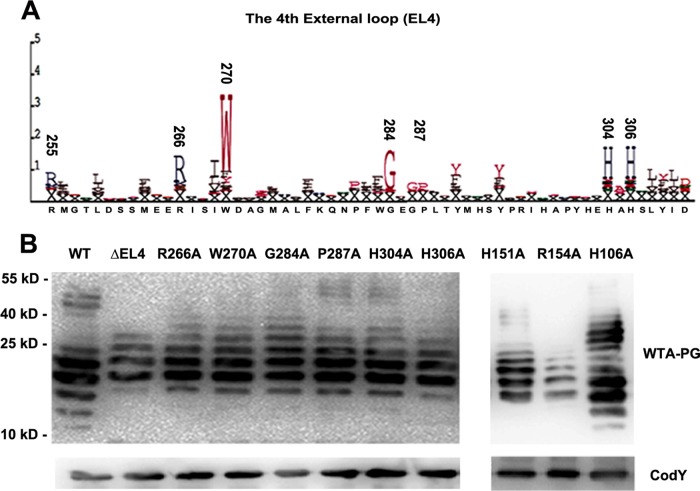
FIG 3.
Amino acid substitutions of the predicted conserved residues and deletion of EL4 impair the function of RafX. (A) Representation of conserved amino acids in EL4 generated by the SAM-T02 protein structure prediction server. The positions of the most-conserved residues are indicated above the letters. The size of each letter is proportional to the level of conservation of the amino acid. (B) Contributions of the predicted EL4 domain and conserved key amino acid residues to the WTA-PG banding pattern. Conserved residues R266, W270, G284A, P287A, H304, and H306, as well as H151 and R154 in the second external loop, were replaced with alanines to prepare ΔrafX mutant D39, and H106A (internal loop) served as a predicted negative control. Samples from the mutants were subjected to immunoblotting with antibodies specific for P-Cho and CodY. CodY was used as a loading control. The positions of size markers (molecular masses in kilodaltons are shown) are indicated.
Isolation and immunoblot assays of pneumococcal TAs.
LTA purification was performed as described previously but with some modifications (13). Pneumococcal cells were harvested from 200 ml of a culture with an OD600 of 0.5 by centrifugation at 6,000 × g for 30 min at 4°C. The cell pellet was resuspended in 4 ml of ice-cold 50 mM Tris-HCl buffer (pH 7.0). The cell suspension was introduced dropwise into a flask with 20 ml of boiling 5% sodium dodecyl sulfate (SDS) solution. The solution was boiled for another 30 min. The cell suspension was repeatedly pelleted by ultracentrifugation at 4°C for 45 min at 50,000 × g. The supernatant was collected and lyophilized. The resulting solid was repeatedly washed SDS free with 90% alcohol (10,000 × g, 15 min, 20°C). The resulting sediment was resuspended in citric buffer and extracted with an equal volume of n-butanol (Merck) for 30 min at room temperature under vigorous stirring. Phases were separated at 4,000 × g and 4°C for 10 min. The aqueous phase was removed, and the extraction procedure was repeated twice. The combined LTA-containing water phases were lyophilized to form crude LTA preparations. Lyophilized LTA was suspended in 50 mM Tris-HCl buffer for further analysis.
S. pneumoniae strains were grown to the mid-exponential phase. Bacterial cells were harvested at 9,000 × g for 5 min, and the cell pellets were washed twice with phosphate-buffered saline (PBS). Pneumococcal cells were then lysed by incubating cultures at 37°C for 30 min in the presence of a 0.4% sodium deoxycholate (DOC) solution containing proteinase inhibitors (Roche) (27) or digested with 300 U/ml mutanolysin (Sigma-Aldrich) and 10 mg/ml lysozyme (Sigma-Aldrich) in 50 mM Tris buffer (pH 7.5) containing 20 mM MgCl2 and 1 × proteinase inhibitors (Roche) at 37°C for the times indicated. The lysate was stored at −80°C immediately after digestion.
For detection of TA, samples were treated with 5 μl/ml proteinase K (10 mg/ml; Invitrogen) for 3 h at 51°C and protein loading buffer was added. For detection of CodY, bacterial lysate was mixed directly with protein loading buffer. After being boiled for 15 min, the samples were subjected to 16% Tricine SDS-polyacrylamide gel electrophoresis (PAGE) or 10% SDS-PAGE accordingly. After electrophoresis, the gels were transferred onto a polyvinylidene difluoride membrane (Millipore) with a wet-transfer apparatus (Bio-Rad) at a constant current of 210 mA for 70 min. After transfer, the membrane was fixed in 0.2% glutaraldehyde for 45 min and blocked with 5% bovine serum albumin in 0.1% Tween 20 in Tris-buffered saline, followed by sequential reaction with a primary mouse IgA monoclonal antibody specific for P-Cho (1:1,000; clone TEPC-15, catalog number M1421; Sigma-Aldrich) or polyclonal mouse anti-CodY serum (1:1,000). The secondary antibody was a goat anti-mouse IgA–horseradish peroxidase (HRP) conjugate (1:8,000; Santa Cruz Biotech) or a goat anti-mouse IgG–HRP conjugate (1:4,000; Millipore). After washes, the blots were developed with chemiluminescence reagents (Millipore).
Quantitation of TA by enzyme-linked immunosorbent assay (ELISA).
Bacteria were grown in C+Y medium to an OD600 of 0.5 (Ultrospec 10; Amersham Biosciences). Two-milliliter aliquots of bacteria were centrifuged at 13,000 × g for 10 min. The supernatants were transferred to a fresh tube (supernatant fraction). The remaining bacterial pellet was suspended in 2 ml of cell wall digestion buffer (1× protease inhibitor cocktail [Roche], 300 U/ml mutanolysin [Sigma-Aldrich], 10 mg/ml lysozyme [Sigma-Aldrich], 30% sucrose, 20 mM MgCl2, 50 mM Tris buffer [pH 7.5]) and incubated at 37°C for 2 h with shaking. The supernatant (cell wall fraction) was collected by centrifugation at 13,000 × g for 10 min. Supernatants were filtered through a 0.2-μm-pore-size syringe filter (Millipore) to remove any residual protoplasts or whole cells. The cell wall fraction was normalized for β-galactosidase activity (28). To test the total amount of TAs (cells plus supernatant), Triton X-100 was added directly to the cultures (OD600 = 0.5) to a final concentration of 0.02% (vol/vol) and the mixture was incubated for 30 min at 37°C.
The quantity of TA was determined by using a sandwich ELISA that was performed essentially as described previously (29). In this study, the polyclonal anti-cell wall polysaccharide (anti-CWPS) serum was used as the immobilized capture antibody and monoclonal antibody TEPC-15 was used as the secondary sandwich antibody. Samples were diluted appropriately to OD values in the detection ranges. After washings, antibodies were detected with HRP-conjugated goat anti-mouse IgA (Santa Cruz Biotechnology) and the absorbance at 450 nm was recorded. Experiments were performed in duplicate. TA concentrations are expressed in arbitrary units, and values were obtained by multiplying the ratio of the OD value of a test sample to the background value obtained with normal C+Y medium by the dilution factor.
CRP binding assays.
Liquichek Elevated CRP Control (level 2; Bio-Rad Laboratories) was used as the source of C-reactive protein (CRP). Bacteria were grown in C+Y medium to an OD600 of 0.5. Equal numbers of bacteria were inactivated in a water bath at 56°C for 1 h and then incubated with 300 μl of CRP Control at 37°C for 4 h. The bacteria were sedimented by centrifugation at 13,000 × g for 10 min, and the supernatants were collected to determine the concentration of CRP by an automatic biochemical analyzer (cobas system; Roche Diagnostics).
Fluorescence-activated cell sorting (FACS).
S. pneumoniae strains were grown to mid-exponential phase. A total of 5 × 106 bacterial cells in PBS supplemented with 5% fetal calf serum (HyClone, Logan, UT) were treated with mouse antiserum against CbpA recombinant protein (anti-CbpA antibody; dilution, 1:100) in blocking buffer (5% fatal bovine serum, 1× PBS) for 60 min at 37°C. The bacterial pellets were washed three times with PBS and incubated with a goat anti-mouse IgG phycoerythrin (PE)-labeled secondary antibody at 1:200 (Santa Cruz Biotech) for 60 min at 4°C. After three additional washes, bacteria were resuspended in PBS containing 1% bovine serum albumin and 1% paraformaldehyde before detection by flow cytometry (FACSVantage; Becton, Dickinson Biosciences). Serum from naive animals was used as a negative control.
Triton X-100-induced lysis.
S. pneumoniae strains were grown to an OD600 of 0.9. Triton X-100 was added directly to the growing cultures in growth medium to a final concentration of 0.01 or 0.02%, respectively, and then the cultures were incubated at 37°C. Cellular lysis was measured turbidimetrically at 600 nm after the addition of Triton X-100.
Adherence assays.
Bacterial adherence assays were carried out essentially as described previously (30). Briefly, MLE12 cells and human umbilical vein endothelial cells (HUVEC) (1 × 105; American Type Culture Collection) were incubated with 107 CFU of mid-exponential-phase bacteria, respectively, at 37°C for 1 h. After washes and lysis, the cell suspensions were properly diluted and plated onto blood agar plates to obtain colonies with easily recognizable sizes and the numbers of bacteria were determined. To test inhibition by antiserum against CWPS, bacteria were suspended at a concentration of 1 × 107 CFU in 1 ml of Dulbecco's modified Eagle's medium containing 1:100 anti-CWPS serum (Statens Serum Institut, Copenhagen, Denmark). Cells in the control group were pretreated with normal rabbit serum.
Animal studies.
D39, TIGR4, and the respective mutants were grown in C+Y medium to an OD620 of 0.4 at 37°C under 5% CO2. Bacteria were adjusted to obtain the desired concentration. Groups of nine 6-week-old female BALB/c mice were intraperitoneally (i.p.) challenged with 2 × 108 CFU of TIGR4 and mutant forms thereof or 1 × 107 CFU of D39 and mutant forms thereof. Survival was recorded every 6 h for the first 3 days and then daily until the end of the observation period.
Statistical analysis.
Differences between groups were statistically compared with Student's t test or the nonparametric Mann-Whitney or Wilcoxon test with GraphPad Prism 5 (GraphPad Software, San Diego, CA). Statistical significance was defined as a P value of <0.05.
RESULTS
Evidence by Western blotting for the TAs in S. pneumoniae.
The isolation strategy used for pneumococcal LTA was based on the fact that WTA could be precipitated with heavily cross-linked PG following treatment with a boiling 5% SDS solution and ultracentrifugation (12, 29). Recent MS results indicated that pneumococcal LTA has a maximum molecular mass of 10 kDa (13, 18, 19). Therefore, LTA samples of strain R6 were separated by 16% Tricine SDS-PAGE to allow the resolution of low-molecular-mass TAs and immunoblotted with a mouse IgA(κ) monoclonal antibody specific for P-Cho [IgA(κ) was purified from murine myeloma clone TEPC-15] (31, 32) or a polyclonal antiserum raised in rabbits against CWPS (structurally, a PG-attached TA complex of pneumococci [27]). We observed a ladder-like banding pattern for the LTA ranging from 10 to 25 kDa (Fig. 1A), demonstrating the polymeric nature of the TAs. Interestingly, no TEPC-15- or CWPS-reactive bands lower than 10 kDa could be detected in Western blot assays (Fig. 1A). All experiments yielded reproducible banding pattern of TAs, similar to those previously recognized as LTA in S. pneumoniae (12, 21, 29, 33).
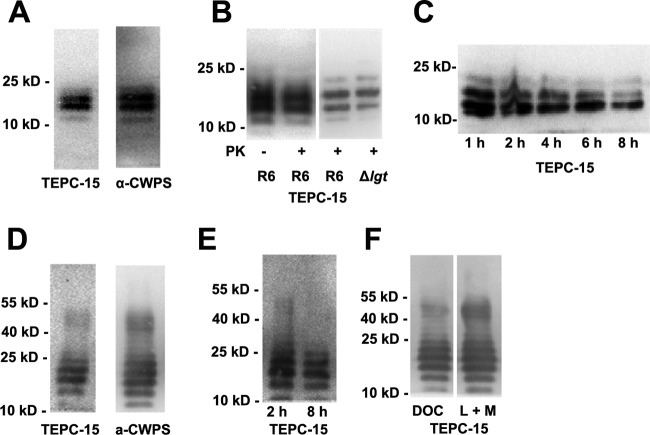
FIG 1.
Western blot analysis of S. pneumoniae TAs. (A) SDS-soluble TA was separated by 16% Tricine SDS-PAGE and probed with the antibodies indicated. TEPC-15 is a mouse IgA monoclonal antibody specific for P-Cho; α-CWPS is a rabbit CWPS (structurally, a PG-TA complex of pneumococci) polyclonal antiserum. (B) The banding pattern of SDS-soluble TAs was not influenced by proteinase K digestion and lgt mutation in strain R6, showing that neither proteinase K nor lgt mutation impaired the TA banding pattern. (C) Banding pattern of SDS-soluble TAs from strain R6 following digestion with mutanolysin plus lysozyme for the times indicated, showing reduced TA banding intensity with increasing digestion times. (D) Banding pattern of TAs from whole R6 bacteria digested with mutanolysin plus lysozyme for 2 h and probed with the antibodies indicated. (E) Banding pattern of TAs from whole R6 bacteria digested with mutanolysin plus lysozyme for the times indicated. (F) Banding pattern of TAs from whole R6 bacteria treated with DOC or lysozyme plus mutanolysin (L + M). The positions of size markers (molecular masses in kilodaltons are shown) are indicated.
We noticed the discrepancy between Western blotting results and MS analysis data regarding the molecular mass of LTA. A possible explanation is that some protein(s), probably the lipoprotein, is attached to the LTA, hence contributing to the high-molecular-mass glycomers (10 to 25 kDa). To test this hypothesis, LTA samples were digested with proteinase K to remove proteins. Moreover, we inactivated the lgt gene in the pneumococcal strain R6 background to block the formation of lipoproteins. Samples were separated by 10% SDS-PAGE and immunoblotted with TEPC-15. We found that the ladder-like banding pattern or the amount of TAs was not influenced by proteinase K digestion and lgt gene deletion (Fig. 1B), indicating that the increased mass of LTA is not caused by the attachment of a protein component. We also examined whether the increased molecular mass of TA was due to the attachment of PG. It is well known that mutanolysin and lysozyme specifically cleave the PG between the N-acetylmuramic acid and N-acetylglucosamine residues of the glycan chains. LTA samples were digested with the two enzymes for the times indicated. We show that TA bands decreased in intensity with increasing incubation times (Fig. 1C), demonstrating that TA bands, at least in part (10 to 25 kDa), are the WTA-PG complex.
To further investigate the nature of the TA polymers, R6 pneumococcal cells harvested at the mid-exponential phase were digested with mutanolysin plus lysozyme in an osmotic stabilizing buffer for 2 h. Aliquots of the digestion mixture were digested with proteinase K to remove protein before separated by 10% SDS-PAGE and immunoblotting with either TEPC-15 or anti-CWPS serum. We showed that immunoblotting with either TEPC-15 or anti-CWPS serum gave rise to identical TA bands that were located predominantly in two ranges, 10 to 25 and 40 to 55 kDa (Fig. 1D). In another set of experiments, the cells were sedimented after 2 h of enzyme digestion and aliquots of the supernatants were incubated at 37°C for another 2 or 8 h. We found that the high-molecular-mass TA bands (40 to 55 kDa) disappeared and TA bands (10 to 25 kDa) also showed a less intense banding pattern after prolonged digestion (Fig. 1E). Together, these results suggest that Western blotting with anti-CWPS serum or TEPC-15 actually shows ladder-like polymers of the WTA-PG complex in S. pneumoniae.
Having demonstrated the banding patterns of WTA-PG, we decided to use Western blot analysis to examine the banding pattern of the TAs after treatment of R6 pneumococci with DOC. For this purpose, bacteria were treated with a 0.4% (vol/vol) DOC solution for 30 min. It is known that DOC-mediated pneumococcal lysis is accomplished by inducing the release of the intracellular LytA amidase, which hydrolyzes the bonds between the glycan chain and the stem peptides (34–37). Rapid and complete lysis of strain R6 occurred within 15 min after the addition of DOC. We found that the banding pattern of TAs from DOC-treated R6 pneumococci was similar to that of the TAs prepared by digestion with mutanolysin plus lysozyme (Fig. 1F).
The rafX gene contributes to TA biosynthesis.
rafX (accession no. AE007480) was identified as an in vivo-inducible gene in a model of murine pneumonia and sepsis in our laboratory (38). rafX is part of the raf operon with five upstream genes that have been shown to be involved in raffinose metabolism; nevertheless, the role of the RafX protein has not been elucidated (39). By searching the NCBI conserved-domain server, we found that RafX harbors O-antigen ligase activity belonging to pfam13425. Since there is no O antigen for pneumococci, we focused our investigation on the potential contribution of rafX to the formation of pneumococcal TAs.
rafX (SPD_1672) could be successfully deleted from S. pneumoniae strains by erm cassette replacement, which was a nonpolar event verified by reverse transcription (RT)-PCR (data not shown). By using Western blotting, we found that the ΔrafX mutants exhibited a moderate decrease in the amount of WTA-PG and an abnormal banding pattern relative to that of the WT strains (Fig. 2A and B, lanes 2). CodY was used as an internal standard (40). This WTA biosynthesis defect and the abnormal banding pattern of ΔrafX mutant could be reversed by complementation with the integrated rafX-pEVP3 plasmid encoding the full-length RafX protein (also described as Compl or WL) (Fig. 2A and B, lanes 3).
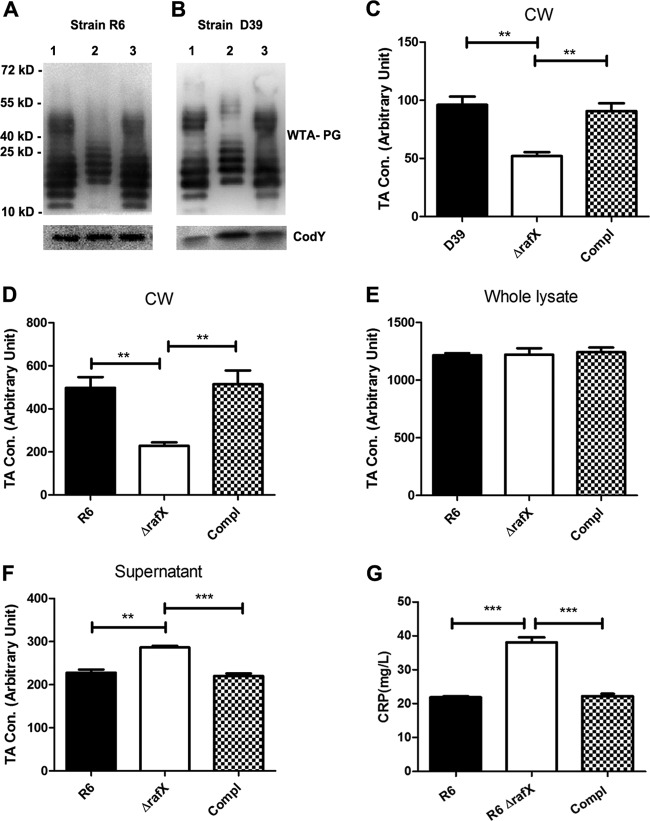
FIG 2.
The rafX gene contributes to WTA biosynthesis in S. pneumoniae. (A and B) Representative analyses of TAs of S. pneumoniae strains by Western blotting and probing with a P-Cho-specific monoclonal antibody (TEPC-15). Bacteria were treated with a 0.5% sodium DOC solution to release TAs. Lanes 1 to 3 contained TAs from the WT, ΔrafX mutant, and complemented strains. CodY was used as a loading control. The positions of size markers (molecular masses in kilodaltons are shown) are indicated. (C) The amounts of TA in the cell wall (CW) fraction from the WT, ΔrafX mutant, and complemented (Compl) D39 strains were determined by sandwich ELISA. The amounts of TAs in the cell wall fraction (D), whole lysate (cells plus supernatant, E), supernatant (F) obtained from the WT, ΔrafX mutant, and complemented (Compl) R6 strains were determined by sandwich ELISA. TA concentrations (Con.) are expressed in arbitrary units, and values were obtained by multiplying the ratio of the OD value of a test sample to the background value obtained with normal C+Y medium by the dilution factor. (G) CRP binding assays. The inactivated WT, ΔrafX mutant, and complemented (Compl) R6 strains were used to absorb CRP, and the free CRP was determined after 4 h of absorption. Results are from three independent experiments (n = 3). Error bars indicate standard deviations. **, P < 0.01; ***, P < 0.001.
To confirm the reduced amount of TAs in ΔrafX mutants observed in Western blot assays, the amount of WTA was determined by ELISA. Bacteria were digested with mutanolysin plus lysozyme to remove WTA, which was subsequently determined by ELISA. β-Galactosidase activity in cell wall preparations was quantitatively determined as the internal control. We showed that the amounts of WTA in ΔrafX mutants were about half of the levels in the parental strains (Fig. 2C and D). The total amounts of TAs (cells plus supernatant) in the WT, ΔrafX mutant, and complemented R6 strains were identical (Fig. 2E). There was a slightly greater amount of TAs in the cultural supernatant of ΔrafX mutant R6 than in the cultural supernatants of WT R6 and the complemented strain (Fig. 2F). The amounts of WTA were restored in the complemented strains. To further confirm these results, CRP (with P-Cho binding specificity [41]) adsorption assays were used to determine the amounts of surface-exposed TAs in the WT, ΔrafX mutant, and complemented R6 strains. Bacterial cells were incubated with CRP, and the amounts of free CRP in the supernatants were determined by ELISA. The greater amount of free CRP found after incubation with ΔrafX mutant strain R6 than after incubation with an equal number of parental strain R6 bacteria indicated that ΔrafX mutant R6 bound less CRP because of the smaller amount of surface-exposed TAs (Fig. 2G).
To examine whether the effect of rafX on WTA-PG biosynthesis was influenced by the host strain, deletion mutants lacking the rafX gene were constructed in the TIGR4, CMCC(B)31203, and S. mitis W1 (smi0513, annotation in S. mitis B6) backgrounds. The ΔrafX mutants also showed an abnormal WTA-PG banding pattern and a reduced amount of TAs relative to the WT strains (see Fig. S1 in the supplemental material).
Site-directed mutagenesis of predicted external loop 4 and conserved active-site residues confirms their importance for RafX function.
To further substantiate the role of the RafX protein in pneumococcal TA biosynthesis, we constructed pEVP3-rafX1-765:934-1194, which encodes a ΔEL4 deletion derivative of RafX that lacks the predicted EL4 domain. Moreover, to explore the key residues in EL4, alanine substitution mutagenesis was used to replace predicted conserved residues R266, W270, G284, P287, H304, and H306 (the numbers denote the positions of these residues in S. pneumoniae RafX) as predicted by SAM-T02 (Fig. 3A). In addition, H151 and R154 in the second external loop and H106 (internal loop) in an internal domain were mutated to alanine. The H106A mutation was considered a negative control. The expression of rafX variants was confirmed by RT-PCR and sequencing (data not shown). The complementation with RafX ΔEL4 showed an abnormal TA banding pattern, indicating that EL4 is essential for its function. In mutants complemented with the RafX H151A, R154A, R266A, W270A, G284A, P287A, H304A, and H306A variants, the TAs shifted to a higher molecular mass than those in the parental strain (Fig. 3B). TAs could be successfully restored in rafX mutants by complementation with the RafX H106A variant (Fig. 3B). We concluded from these data that the external domains and these active-site residues are essential for RafX function.
rafX-mediated TA biosynthesis is highly conserved in closely related streptococci.
rafX is >99.9% conserved among all 28 sequenced S. pneumoniae strains and other closely related species, including S. mitis B6 and S. pseudopneumoniae VT162, available for analysis in the NCBI DNA database. Moreover, sequencing data suggested that the rafX gene was 99.93% conserved among 20 different sequence types of pneumococcal clinical isolates included in this study (Fig. 4, top; see Fig. S2 in the supplemental material). We next examined the TA banding patterns of all of the clinical isolates. TAs from all of the pneumococcal strains displayed characteristic TA ladders on Western blot assays, and the TA of TIGR4 showed a lower molecular mass (Fig. 4, bottom). Together, these data suggest that the rafX gene is required for TA biosynthesis and is conserved in streptococci.
FIG 4.
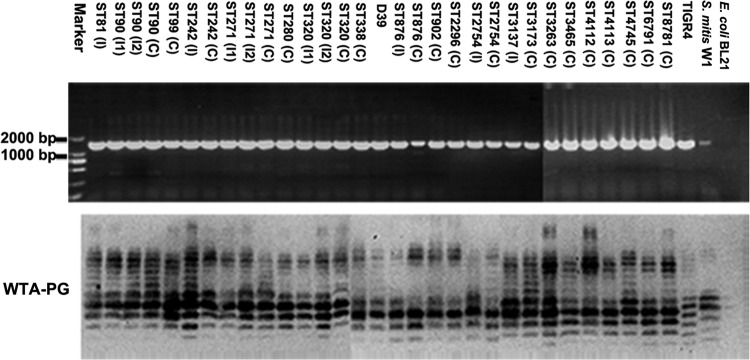
RafX-mediated WTA biosynthesis is highly conserved in closely related streptococci. rafX is conserved in pneumococcal strains and S. mitis W1. A 1,433-bp rafX fragment composed of the full-length open reading frame of rafX and the upstream sequence was PCR amplified from pneumococcal strains. E. coli BL21 served as the negative control. The positions of size markers are indicated on the left of the top panel. The bottom panel shows the WTA-PG banding patterns of clinical strains obtained by Western blotting with TEPC-15. The letter I in parentheses denotes an invasive strain, and the following number indicates the assigned strain number. The letter C in parentheses denotes a carriage strain. ST, sequence type.
The D39 ΔrafX mutant is impaired in bacterial growth, autolysis, and cell shape.
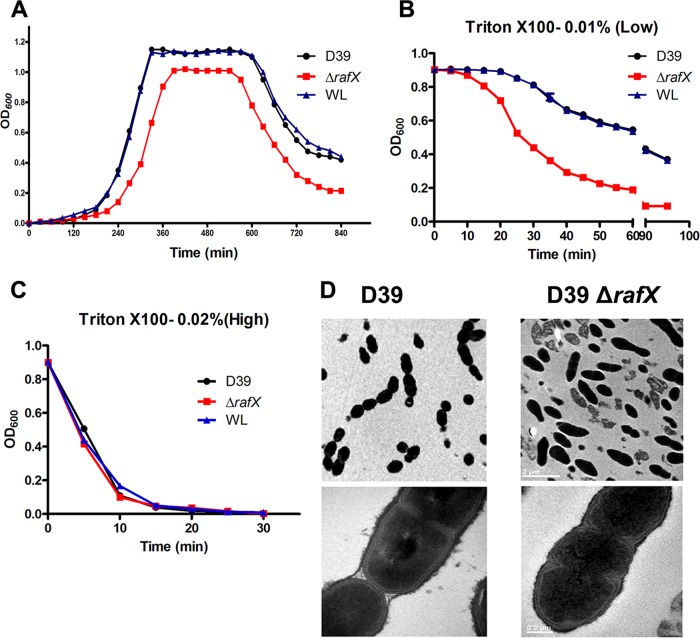
The growth curve of pneumococcal strain D39 (NCTC 7466) was typical for S. pneumoniae, while the ΔrafX mutant grew at a much lower rate and showed a shortened stationary phase and premature autolysis compared to WT strain D39 (Fig. 5A). Besides, colonies of the ΔrafX mutant on blood agar plates were consistently smaller than those of WT strains (data not shown). Introduction of a recombinant rafX-pEVP3 plasmid (the full-length rafX gene) into the D39 ΔrafX mutant fully restored the growth rate to the level of WT strain D39 (Fig. 5A). Moreover, lysis induced by a high concentration of 0.02% Triton X-100 was identical among the D39 WT, ΔrafX mutant, and complemented strains (Fig. 5B), whereas lysis induced by 0.01% Triton X-100 was quicker in the ΔrafX mutant (Fig. 5C). Examination of the bacterial morphology by TEM demonstrated that the D39 ΔrafX mutant displayed a rod-like shape (Fig. 5D). The rod-like cells showed striking cell wall defects, including multiple septa and asymmetrical divisions, compared to WT cells. Together, these results suggest that RafX-mediated TA biosynthesis is important for bacterial growth, autolysis, and proper cell shape and division.
FIG 5.
rafX gene is required for bacterial growth, autolysis, and cell shape and division in S. pneumoniae. (A) Representative bacterial growth rate experiment. Bacteria were grown in a chemically defined medium. OD600 was measured at the times indicated. WL represents the D39 ΔrafX mutant complemented with the full-length RafX protein. Bacteria were grown to an OD600 of 0.9, and autolysis was induced by 0.01% Triton X-100 (B), resulting in slightly quicker autolysis of the D39 ΔrafX mutant than the WT or complemented strain, or by 0.02% Triton X-100 (C), resulting in indistinguishable autolysis rates. Results are from three independent experiments (n = 3). Error bars indicate standard deviations. (D) Ultrastructures of S. pneumoniae WT and ΔrafX mutant D39 strains. Bacteria were harvested in the mid-exponential phase of growth and conventionally embedded in thin sections for examination by TEM. Bar, 2 (top) or 0.2 (bottom) μm.
ΔrafX mutant R6 is defective in adherence to epithelial and endothelial cells.
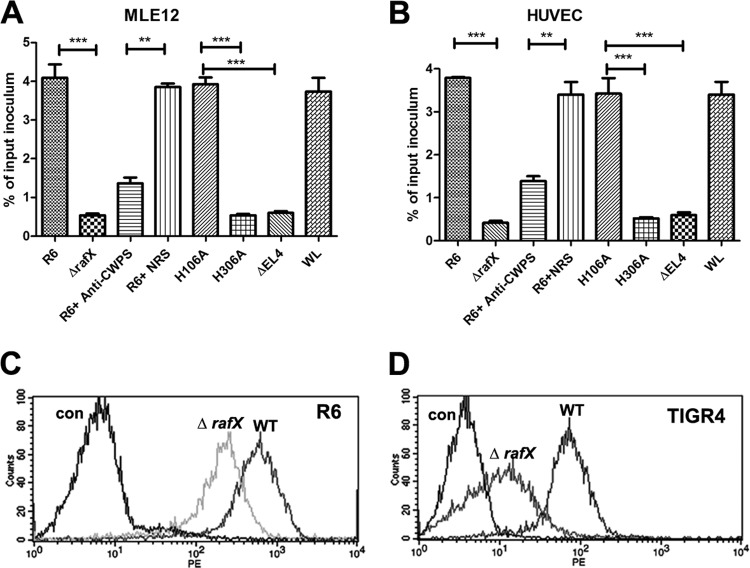
TAs are abundant in pneumococci, and a previous study found that the interaction of P-Cho with the rPAF was responsible for the adhesion of S. pneumoniae to cytokine-activated cells (16). The majority of the TAs in D39 were covered by the thick capsule cultured in vitro and showed a different phenotype during nasopharyngeal colonization (42), where surface-exposed TAs are much more abundant. Upon learning of this, we used the avirulent unencapsulated R6 strain to study adherence to the mouse lung epithelial cell line MLE12 and HUVEC. The adherence ability of ΔrafX mutant R6 was only 10 to 15% of the adherence of R6 to both cell types (Fig. 6A and B). Additional evidence that pretreatment of S. pneumoniae with polyclonal rabbit anti-CWPS serum resulted in significant inhibition of the capacity of R6 to adhere to MLE12 cells and HUVEC (Fig. 6A and B), supporting the requirement of TAs in adhesion. To explore whether the presence of RafX protein is required for bacterial adhesion, we compared the adherence of the mutants harboring the ΔEL4, H306A, and H106A variants. The H106A mutant was as adhesive as WT strain R6. The ΔEL4 and H306A mutants were significantly impaired in adherence, like the ΔrafX mutant. This observation further suggests that TA itself is important for pneumococcal adhesion. Moreover, FACS analysis demonstrated that the amount of CbpA protein on the surface of R6 and TIGR4 ΔrafX mutant cells was significantly reduced (Fig. 6C and D), suggesting the contribution of CbpA to the reduced adherence of ΔrafX mutants.
FIG 6.
Decreased adherence of ΔrafX mutant R6 to MLE12 cells and HUVEC. Adherence of bacterial strains to MLE12 cells (A) or HUVEC (B). ΔEL4, H106A, H306A, and WL denote the ΔrafX mutant complemented with the RafX ΔEL4, RafX H106A, and RafX H306A variants and full-length RafX, respectively. For blocking experiments with rabbit anti-CWPS serum, bacteria were treated with a 1:100 dilution of anti-CWPS serum or normal rabbit serum (NRS) for 30 min before infection. Data are from four independent experiments. Error bars indicate standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Flow cytometry histograms for pneumococcal surface protein CbpA in strains R6 (C) and TIGR4 (D) are shown. The control (con) was WT bacteria incubated with normal mouse serum and probed with a PE-labeled secondary antibody.
Virulence of S. pneumoniae TA-deficient mutants.
The virulence of TA-deficient mutant bacterial strains with two genetic backgrounds, D39 (1 × 107 CFU) and TIGR4 (2 × 108 CFU), was tested. BALB/c mice were challenged i.p. with the indicated numbers of TIGR4 or D39 background strains, and deaths were recorded at 6-h intervals over a period of 7 days. Animals infected with the WT, RafX H106A, and RafX WL complemented strains eventually succumbed to the infections, whereas mice challenged with the ΔrafX and RafX H306A-complemented strains survived much longer than those infected with the WT strains (Fig. 7A and B). All of the WTA-deficient mutants were less virulent than the WT strains, suggesting that pneumococcal WTA is important for full bacterial virulence.
FIG 7.
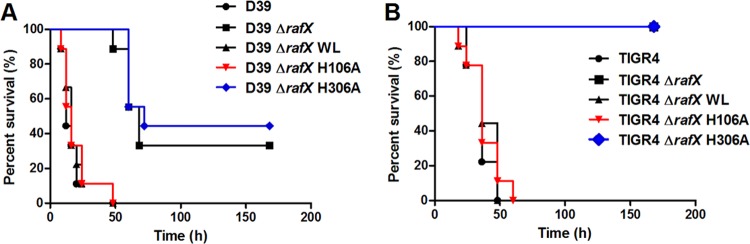
Virulence of WTA-deficient pneumococcal mutants. (A) Survival of mice after i.p. infection with 1 × 107 CFU of the WT, ΔrafX mutant, or RafX variant D39 strain. The D39 ΔrafX and ΔrafX H306 mutants were significantly less virulent than the D39 WT strain (P < 0.001). The D39 ΔrafX H106 mutant was as virulent as the D39 WT strain. (B) Survival of mice after i.p infection with 2 × 108 CFU of either strain TIGR4 or the TIGR4 ΔrafX mutant. The TIGR4 ΔrafX and TIGR4 ΔrafX H306 mutants were significantly less virulent than the WT strain (P < 0.001). ΔrafX WL is a ΔrafX mutant complemented with full-length RafX. ΔrafX H106A and ΔrafX H306A are ΔrafX mutants complemented with the RafX H106A and H306A variants, respectively.
DISCUSSION
S. pneumoniae produces an unusual TA, and MS analyses of pneumococcal TA have shown that both WTA and LTA have a maximal molecular mass of 10 kDa (13, 18). This observation disagrees with the SDS-PAGE and Western blot results showing that the LTA bands were located in the 10- to 40-kDa range (12, 29). In the present study, we examined the nature of the TAs in Western blot assays. Neither proteinase K nor lgt gene deletion impaired the banding pattern of TAs. However, the band intensity of SDS-soluble TAs of strain R6 was reduced following digestion with lysozyme plus mutanolysin. This difference was more apparent in ΔrafX mutant R6 (see Fig. S3 in the supplemental material). These results suggest that Western blotting with TEPC-15 or anti-CWPS serum actually detects the polymers of WTA-PG ranging from 10 to 25 kDa, which was further supported by previous data that PG with LytA amidase or lysozyme produced glycan chains with at least 25 disaccharide units, which corresponds to a molecular mass of approximately 12.5 kDa (18). On the basis of these findings, we speculated that the SDS-soluble TAs displayed in Western blot assays are newly synthesized WTA-PG polymers that are thought to be soluble before being attached to the mature cell wall.
Having demonstrated the banding patterns of WTA-PG in Western blot assays, this work also presents evidence that the rafX gene contributes to the biosynthesis of pneumococcal WTA. We show that ΔrafX mutants have impaired WTA-PG banding patterns and produce reduced amounts of WTA. To determine the role of RafX in WTA biosynthesis, we compared the total amounts and cell wall fractions of TAs of the WT, ΔrafX mutant, and complemented R6 strains, as well as those in the cultural supernatants. The results presented in Fig. 2E show that the total amount of TAs (cells plus supernatant) was identical in the ΔrafX mutant and WT R6 strains. We observed an increase in the amount of TAs in the cultural supernatant of ΔrafX mutant R6 (Fig. 2F); however, this slight increase is not equivalent to the reduction in the cell wall fraction of ΔrafX mutant R6 (Fig. 2D). Together these data suggest that RafX contributes to the attachment of WTA to the cell wall.
The sequence length of RafX is 397 amino acids, which is comparable to that of Pseudomonas aeruginosa PAO1 WaaL, which is 401 amino acids. Although the amino acid sequence of RafX shows 18.6% identity to P. aeruginosa PAO1 WaaL, secondary-structure prediction with the TMHMM Server v.2.0 program revealed that, similar to P. aeruginosa PAO1 WaaL, RafX contains multiple transmembrane domains and a large external loop (EL4). In addition, alignment of the amino acid sequences of RafX and P. aeruginosa WaaL revealed that amino acid residues R266 to V325 of RafX, belonging to the predicted EL4 domain, showed 24% identity and 46% similarity to the region of WaaL (R264 to V322) belonging to the catalytic periplasmic loop 5 domain (see Fig. S4 in the supplemental material). This analysis also predicted the existence of positively charged conserved amino acids, including R266 and H306 (see Fig. S4). Previous studies indicated the essential role of the positively charged His residue of WaaL (H303) in the ligation of O antigen to the lipid A core (43, 44). In this work, we show that EL4 domain deletion and H306A mutagenesis rendered RafX nonfunctional by showing that the ΔEL4 and H306A mutants had impaired WTA-PG banding patterns. Combined with the reduced amount of WTA (Fig. 2C and D), these data may suggest a ligase function for RafX in the biosynthesis of TAs but further enzymatic evidence is required.
In this work, immunoblotting was used to identify TAs. On the basis of currently available data, this method actually detects the WTA-PG complex. We initially believed that LTA could be prepared by using SDS plus n-butanol (12) and its amount could be determined by ELISA (29); however, as presented, SDS-soluble LTA could be contaminated with soluble WTA-PG (Fig. 1). In addition, the quantitation of LTA could be complicated by the presence of the undecaprenyl pyrophosphoryl-linked TA precursors. Thus, whether RafX is also involved in the biosynthesis of LTA remains an open question. Other analysis strategies are required to address this question.
It is expected that the absence of some residue(s) would lead to a decreased molecular mass of TA. For example, Zhang et al. showed that deletion of the licD2 gene gave rise to lower-molecular-mass bands and an abnormal TA banding pattern because of the loss of P-Cho moieties (21). In this study, we observed that TAs from the TIGR4 strain migrated faster than those from the D39 strain and other clinical isolates, which is supported by the evidence that one or both P-Cho residues were missing from the terminus of the TA chain of the TIGR4 strain (13). Interestingly, ΔrafX mutants yielded ladder-like WTA-PG bands that shifted to a molecular mass higher than those of WT strains in Western blot assays. This observation may be explained by the assumption that TA was shared by more PG disaccharide units in the ΔrafX mutant but needs to be investigated further.
In agreement with previous results of cell wall-related glycopolymers in bacterial growth (8, 45), the D39 ΔrafX mutant grew more slowly at 37°C in chemically defined medium than the parental D39 strain (Fig. 5A). Additional Mg2+ partially restored the growth of the D39 ΔrafX mutant in C+Y medium but not to the WT level (see Fig. S5 in the supplemental material). Besides, the D39 ΔrafX mutant displays premature autolysis. Since a difference in the amount of LytA between the WT and ΔrafX mutant strains was not observed by Western blotting and FACS (see Fig. S6), a difference in LytA expression did not appear to account for the premature autolysis of the D39 ΔrafX mutant. A previous study reported that 1% choline delays the onset of autolysis, demonstrating the attachment of the excess LytA to the P-Cho moiety (46). One possible explanation is that abnormal or reduced nascent WTA-PG biosynthesis (less WTA in nascent PG) in the late stationary phase is quantitatively not sufficient to anchor the extracellular LytA, and excessive LytA binds to the surrounding mature PG, leading to the earlier onset of autolysis in the ΔrafX mutant.
It has previously been shown that the sensitivity of pneumococcal cell wall to amidase LytA is influenced by the amount of WTA (47). The wall of transparent cells (with a greater amount of TAs) is more resistant to hydrolysis by LytA than that of opaque cells (47). In this work, we show that autolysis induced by a low concentration of Triton X-100 (0.01%) is quicker in the ΔrafX mutant than in the WT strain, further suggesting differences between the amounts of WTA and probably the PG structures of the two strains.
Colonization of the upper respiratory tract represents the first step of infection with S. pneumoniae; however, the mechanisms involved are still not fully understood. Previous observations suggested that TA expression may be essential for successful colonization of the nasopharynx (47, 48). In agreement with these reports, ΔrafX mutant R6 showed significantly reduced adherence to cells. This observation was supported by FACS analysis demonstrating significantly reduced attachment of CbpA on the rafX mutant bacterial surface (49, 50). Furthermore, it was important to test whether WTA is important for invasive pneumococcal infection. We show that WTA-deficient mutants were significantly less virulent than the WT strain, indicating that WTA is an important determinant implicated in pneumococcal disease.
To conclude, RafX protein contributes to the biosynthesis of pneumococcal WTA. RafX protein is highly conserved among pneumococcal serotypes and has a primary amino acid sequence identical to those of homologs in other streptococci, such as S. pseudopneumoniae and S. mitis. Because of a WTA defect, S. pneumoniae displays septum and autolysis abnormalities. The results of earlier work, along with our results, suggest that WTA is essential for bacterial virulence. Hence, RafX inhibition is proposed as an attractive strategy to fight streptococcal infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jan-Willem Veening for providing plasmid pJWV25, Jing-Ren Zhang for providing genomic DNA of S. mitis NCTC 12261, and Donald A. Morrison for providing pEVP3 and S. pneumoniae CPM8. We thank Nicolas Gisch and Ulrich Zähringer for valuable discussion and Gregory Storch for editing the manuscript. We thank Jianyong Wu for FACS analysis and Hong Wang, Jingchuan Fan, and Xiaogang Liao for electron microscopy work.
This work was supported by National Natural Science Foundation of China grants 30970110 and 81171532 and the special fund of the Chongqing Key Laboratory (CSTC). This work was also supported by Natural Science Foundation Project of CQ CSTC 2011jjA10063.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01696-14.
REFERENCES
- 1.Montagnani F, Fanetti A, Stolzuoli L, Croci L, Arena F, Zanchi A, Cellesi C. 2008. Pneumococcal disease in a paediatric population in a hospital of central Italy: a clinical and microbiological case series from 1992 to 2006. J. Infect. 56:179–184. 10.1016/j.jinf.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Wang H, Chen M, Sun Z, Zhao R, Zhang L, Zhang H, Wang L, Chu Y, Ni Y. 2008. Serotype distribution and antimicrobial resistance patterns of Streptococcus pneumoniae isolated from children in China younger than 5 years. Diagn. Microbiol. Infect. Dis. 61:256–263. 10.1016/j.diagmicrobio.2008.02.004 [DOI] [PubMed] [Google Scholar]
- 3.Bogaert D, De Groot R, Hermans PW. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4:144–154. 10.1016/S1473-3099(04)00938-7 [DOI] [PubMed] [Google Scholar]
- 4.Hsu HE, Shutt KA, Moore MR, Beall BW, Bennett NM, Craig AS, Farley MM, Jorgensen JH, Lexau CA, Petit S, Reingold A, Schaffner W, Thomas A, Whitney CG, Harrison LH. 2009. Effect of pneumococcal conjugate vaccine on pneumococcal meningitis. N. Engl. J. Med. 360:144–156. 10.1056/NEJMoa0800836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590–593. 10.1038/21202 [DOI] [PubMed] [Google Scholar]
- 6.Whitney CG, Farley MM, Hadler J, Harrison LH, Lexau C, Reingold A, Lefkowitz L, Cieslak PR, Cetron M, Zell ER, Jorgensen JH, Schuchat A. 2000. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N. Engl. J. Med. 343:1917–1924. 10.1056/NEJM200012283432603 [DOI] [PubMed] [Google Scholar]
- 7.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6:276–287. 10.1038/nrmicro1861 [DOI] [PubMed] [Google Scholar]
- 8.Gründling A, Schneewind O. 2007. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104:8478–8483. 10.1073/pnas.0701821104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243–245. 10.1038/nm991 [DOI] [PubMed] [Google Scholar]
- 10.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. 2011. A widespread family of bacterial cell wall assembly proteins. EMBO J. 30:4931–4941. 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draing C, Pfitzenmaier M, Zummo S, Mancuso G, Geyer A, Hartung T, von Aulock S. 2006. Comparison of lipoteichoic acid from different serotypes of Streptococcus pneumoniae. J. Biol. Chem. 281:33849–33859. 10.1074/jbc.M602676200 [DOI] [PubMed] [Google Scholar]
- 12.Schröder NW, Morath S, Alexander C, Hamann L, Hartung T, Zahringer U, Gobel UB, Weber JR, Schumann RR. 2003. Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via Toll-like receptor (TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14, whereas TLR-4 and MD-2 are not involved. J. Biol. Chem. 278:15587–15594. 10.1074/jbc.M212829200 [DOI] [PubMed] [Google Scholar]
- 13.Gisch N, Kohler T, Ulmer AJ, Muthing J, Pribyl T, Fischer K, Lindner B, Hammerschmidt S, Zahringer U. 2013. Structural reevaluation of Streptococcus pneumoniae lipoteichoic acid and new insights into its immunostimulatory potency. J. Biol. Chem. 288:15654–15667. 10.1074/jbc.M112.446963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomasz A. 1967. Choline in the cell wall of a bacterium: novel type of polymer-linked choline in pneumococcus. Science 157:694–697. 10.1126/science.157.3789.694 [DOI] [PubMed] [Google Scholar]
- 15.Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68:5690–5695. 10.1128/IAI.68.10.5690-5695.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cundell DR, Gerard NP, Gerard C, Idanpaan-Heikkila I, Tuomanen EI. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435–438. 10.1038/377435a0 [DOI] [PubMed] [Google Scholar]
- 17.Behr T, Fischer W, Peter-Katalinić J, Egge H. 1992. The structure of pneumococcal lipoteichoic acid. Eur. J. Biochem. 207:1063–1075. 10.1111/j.1432-1033.1992.tb17143.x [DOI] [PubMed] [Google Scholar]
- 18.Bui NK, Eberhardt A, Vollmer D, Kern T, Bougault C, Tomasz A, Simorre JP, Vollmer W. 2012. Isolation and analysis of cell wall components from Streptococcus pneumoniae. Anal. Biochem. 421:657–666. 10.1016/j.ab.2011.11.026 [DOI] [PubMed] [Google Scholar]
- 19.Seo HS, Cartee RT, Pritchard DG, Nahm MH. 2008. A new model of pneumococcal lipoteichoic acid structure resolves biochemical, biosynthetic, and serologic inconsistencies of the current model. J. Bacteriol. 190:2379–2387. 10.1128/JB.01795-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer W, Behr T, Hartmann R, Peter-Katalinić J, Egge H. 1993. Teichoic acid and lipoteichoic acid of Streptococcus pneumoniae possess identical chain structures: a reinvestigation of teichoic acid (C polysaccharide). Eur. J. Biochem. 215:851–857. 10.1111/j.1432-1033.1993.tb18102.x [DOI] [PubMed] [Google Scholar]
- 21.Zhang J-R, Idanpaan-Heikkila I, Fischer W, Tuomanen E. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31:1477–1481. 10.1046/j.1365-2958.1999.01291.x [DOI] [PubMed] [Google Scholar]
- 22.Kharat AS, Denapaite D, Gehre F, Bruckner R, Vollmer W, Hakenbeck R, Tomasz A. 2008. Different pathways of choline metabolism in two choline-independent strains of Streptococcus pneumoniae and their impact on virulence. J. Bacteriol. 190:5907–5914. 10.1128/JB.00628-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denapaite D, Bruckner R, Hakenbeck R, Vollmer W. 2012. Biosynthesis of teichoic acids in Streptococcus pneumoniae and closely related species: lessons from genomes. Microb. Drug Resist. 18:344–358. 10.1089/mdr.2012.0026 [DOI] [PubMed] [Google Scholar]
- 24.Eberhardt A, Wu LJ, Errington J, Vollmer W, Veening JW. 2009. Cellular localization of choline-utilization proteins in Streptococcus pneumoniae using novel fluorescent reporter systems. Mol. Microbiol. 74:395–408. 10.1111/j.1365-2958.2009.06872.x [DOI] [PubMed] [Google Scholar]
- 25.Wu K, Zhang X, Shi J, Li N, Li D, Luo M, Cao J, Yin N, Wang H, Xu W, He Y, Yin Y. 2010. Immunization with a combination of three pneumococcal proteins confers additive and broad protection against Streptococcus pneumoniae infections in mice. Infect. Immun. 78:1276–1283. 10.1128/IAI.00473-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez JM, McGarry MA, Marolda CL, Valvano MA. 2008. Functional analysis of the large periplasmic loop of the Escherichia coli K-12 WaaL O-antigen ligase. Mol. Microbiol. 70:1424–1440. 10.1111/j.1365-2958.2008.06490.x [DOI] [PubMed] [Google Scholar]
- 27.Skovsted IC, Kerrn MB, Sonne-Hansen J, Sauer LE, Nielsen AK, Konradsen HB, Petersen BO, Nyberg NT, Duus JO. 2007. Purification and structure characterization of the active component in the pneumococcal 22F polysaccharide capsule used for adsorption in pneumococcal enzyme-linked immunosorbent assays. Vaccine 25:6490–6500. 10.1016/j.vaccine.2007.06.034 [DOI] [PubMed] [Google Scholar]
- 28.Zähner D, Hakenbeck R. 2000. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 182:5919–5921. 10.1128/JB.182.20.5919-5921.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368–377 [DOI] [PubMed] [Google Scholar]
- 30.Yan W, Wang H, Xu W, Wu K, Yao R, Xu X, Dong J, Zhang Y, Zhong W, Zhang X. 2012. SP0454, a putative threonine dehydratase, is required for pneumococcal virulence in mice. J. Microbiol. 50:511–517. 10.1007/s12275-012-2014-8 [DOI] [PubMed] [Google Scholar]
- 31.Stuertz K, Merx I, Eiffert H, Schmutzhard E, Mader M, Nau R. 1998. Enzyme immunoassay detecting teichoic and lipoteichoic acids versus cerebrospinal fluid culture and latex agglutination for diagnosis of Streptococcus pneumoniae meningitis. J. Clin. Microbiol. 36:2346–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolberg J, Hoiby EA, Jantzen E. 1997. Detection of the phosphorylcholine epitope in streptococci, haemophilus and pathogenic neisseriae by immunoblotting. Microb. Pathog. 22:321–329. 10.1006/mpat.1996.0114 [DOI] [PubMed] [Google Scholar]
- 33.Sanchez CJ, Kumar N, Lizcano A, Shivshankar P, Dunning Hotopp JC, Jorgensen JH, Tettelin H, Orihuela CJ. 2011. Streptococcus pneumoniae in biofilms are unable to cause invasive disease due to altered virulence determinant production. PLoS One 6(12):e28738. 10.1371/journal.pone.0028738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CJ, Liu TY. 1977. The autolytic enzyme activity upon pneumococcal cell wall. Int. J. Biochem. 8:573–580. 10.1016/0020-711X(77)90078-7 [DOI] [Google Scholar]
- 35.Howard LV, Gooder H. 1974. Specificity of the autolysin of Streptococcus (Diplococcus) pneumoniae. J. Bacteriol. 117:796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goebel WF, Avery OT. 1929. A study of pneumococcus autolysis. J. Exp. Med. 49:267–286. 10.1084/jem.49.2.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avery OT, Cullen GE. 1923. Studies on the enzymes of pneumococcus. IV. Bacteriolytic enzyme. J. Exp. Med. 38:199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng JP, Yin YB, Zhang XM, Huang YS, Lan K, Cui F, Xu SX. 2008. Identification of Streptococcus pneumoniae genes specifically induced in mouse lung tissues. Can. J. Microbiol. 54:58–65. 10.1139/W07-117 [DOI] [PubMed] [Google Scholar]
- 39.Rosenow C, Maniar M, Trias J. 1999. Regulation of the alpha-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 9:1189–1197. 10.1101/gr.9.12.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price KE, Camilli A. 2009. Pneumolysin localizes to the cell wall of Streptococcus pneumoniae. J. Bacteriol. 191:2163–2168. 10.1128/JB.01489-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volanakis JE, Kaplan MH. 1971. Specificity of C-reactive protein for choline phosphate residues of pneumococcal C-polysaccharide. Proc. Soc. Exp. Biol. Med. 136:612–614. 10.3181/00379727-136-35323 [DOI] [PubMed] [Google Scholar]
- 42.Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Muller E, Rohde M. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73:4653–4667. 10.1128/IAI.73.8.4653-4667.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abeyrathne PD, Lam JS. 2007. WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core. Mol. Microbiol. 65:1345–1359. 10.1111/j.1365-2958.2007.05875.x [DOI] [PubMed] [Google Scholar]
- 44.Islam ST, Taylor VL, Qi M, Lam JS. 2010. Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. mBio 1(3):e00189–00110. 10.1128/mBio.00189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Elia MA, Millar KE, Beveridge TJ, Brown ED. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J. Bacteriol. 188:8313–8316. 10.1128/JB.01336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mellroth P, Daniels R, Eberhardt A, Rönnlund D, Blom H, Widengren J, Normark S, Henriques-Normark B. 2012. LytA, major autolysin of Streptococcus pneumoniae, requires access to nascent peptidoglycan. J. Biol. Chem. 287:11018–11029. 10.1074/jbc.M111.318584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiser JN, Markiewicz Z, Tuomanen E, Wani JH. 1996. Relationship between phase variation in colony morphology, intrastrain variation in cell wall physiology, and nasopharyngeal colonization by Streptococcus pneumoniae. Infect. Immun. 64:2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenow C, Ryan P, Weiser JN, Johnson S, Fontan P, Ortqvist A, Masure HR. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819–829. 10.1111/j.1365-2958.1997.mmi494.x [DOI] [PubMed] [Google Scholar]
- 50.Zhang JR, Mostov KE, Lamm ME, Nanno M, Shimida S, Ohwaki M, Tuomanen E. 2000. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102:827–837. 10.1016/S0092-8674(00)00071-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.