Abstract
The Woronin body is a Pezizomycotina-specific organelle that is typically tethered to the septum, but upon hyphal wounding, it plugs the septal pore to prevent excessive cytoplasmic loss. Leashin (LAH) is a large Woronin body tethering protein that contains highly conserved N- and C-terminal regions and a long (∼2,500-amino-acid) nonconserved middle region. As the involvement of the nonconserved region in Woronin body function has not been investigated, here, we functionally characterized individual regions of the LAH protein of Aspergillus oryzae (AoLAH). In an Aolah disruptant, no Woronin bodies were tethered to the septum, and hyphae had a reduced ability to prevent excessive cytoplasmic loss upon hyphal wounding. Localization analysis revealed that the N-terminal region of AoLAH associated with Woronin bodies dependently on AoWSC, which is homologous to Neurospora crassa WSC (Woronin body sorting complex), and that the C-terminal region was localized to the septum. Elastic movement of Woronin bodies was observed when visualized with an AoLAH N-terminal-region–enhanced green fluorescent protein (EGFP) fusion protein. An N- and C-terminal fusion construct lacking the nonconserved middle region of AoLAH was sufficient for the tethering of Woronin bodies to the septum. However, Woronin bodies were located closer to the septum and exhibited impaired elastic movement. Moreover, expression of middle-region-deleted AoLAH in the Aolah disruptant did not restore the ability to prevent excessive cytoplasmic loss. These findings indicate that the nonconserved middle region of AoLAH has functional importance for regulating the position, movement, and function of Woronin bodies.
INTRODUCTION
Filamentous fungi grow via a polarized tip extension, which forms tubular filaments called hyphae that are further divided into distinct cells by the formation of a septum. Although filamentous fungi are classified as multicellular organisms, septa do not completely separate adjacent hyphal cells due to the presence of a septal pore, which allows the passage of cytoplasm and organelles between adjacent cells (1–3). Cytoplasmic continuity through the septal pore is associated with the catastrophic risk of cytoplasmic loss in the event of hyphal wounding. Therefore, Pezizomycotina species have evolved a septal pore-associated organelle known as the Woronin body (4), which plugs the septal pore upon hyphal damage to limit the loss of cytoplasm.
Jedd and Chua (5) first identified Hex1 as a major protein of the Woronin body in Neurospora crassa, and the encoding gene, hex1, is conserved in Pezizomycotina species (5–10). Hex1 self-assembles to form the core of the Woronin body and provides mechanical resistance against cytoplasmic streaming pressure (5, 11). Deletion of the hex1 gene leads to a defect of Woronin body formation and severe cytoplasmic bleeding upon hyphal wounding (5, 9, 12). Although in a limited number of species, including N. crassa, the hexagonal crystal structure of the Woronin body is clearly visible by light microscopy, the fusion of Hex1 with a fluorescent protein is necessary to visualize and track Woronin bodies in most Pezizomycotina species (9, 10). Such localization studies have revealed that Woronin bodies reversibly close the septal pore during normal growth and impede the cytoplasmic continuity between adjacent cells, thereby creating hyphal heterogeneity with regard to gene expression activity (13).
In most Pezizomycotina species, Woronin bodies are tethered to the septum at a distance of 100 to 200 nm (9, 14); however, in a small group of species, which includes members of the genera Neurospora and Sordaria, Woronin bodies exhibit a delocalized pattern of cortex association (15). In N. crassa, a leashin locus, consisting of the adjacent genes leashin-1 (lah-1) and leashin-2 (lah-2), is required for the cortex association of Woronin bodies (3). LAH-1 binds to Woronin bodies through its N-terminal region via the membrane protein Woronin sorting complex (WSC), while the C-terminal region of LAH-1 mediates the cell cortex association of Woronin bodies (3). In contrast, LAH-2 is localized to the hyphal tip and septum and is not functionally related to Woronin bodies.
Optical trapping experiments with Nectria haematococca have revealed that Woronin bodies are tethered to the septum by an elastic filament (16). Woronin bodies reversibly plug the septal pore during normal growth but quickly plug the septal pore upon hyphal wounding. These processes require the proper positioning of the Woronin body and sufficient flexibility of the tethering linker. For species with Woronin bodies tethered to the septum, it is speculated that the lah gene locus expresses a single polypeptide with the capacity to bind to both Woronin bodies and the septum (3, 15). Although Hex1 localization studies with Aspergillus fumigatus demonstrated that deletion of the C-terminal region of LAH impairs the tethering of Woronin bodies to the septum (17), it is not known whether septal tethering by the LAH protein alone is sufficient for Woronin body function.
For the filamentous fungus Aspergillus oryzae, a quantitative assay for evaluating Woronin body function upon hyphal wounding has been established (9). In this assay, hyphal tip bursting is induced by flooding A. oryzae colonies with water, and the ability of hyphal cells to prevent the excessive loss of cytoplasm is assessed. This ability is lost in hyphae defective for Woronin body formation (9), and excessive cytoplasmic loss also partially occurs as a result of a deficiency in Woronin body differentiation from peroxisomes (18). Using this quantitative assay, here, the roles of the A. oryzae LAH (AoLAH) protein in the tethering and septal plugging functions of Woronin bodies were investigated.
MATERIALS AND METHODS
Strains, DNA materials, and media.
A. oryzae wild-type strain RIB40 (19) was used as a DNA donor. Escherichia coli DH5α was used for DNA manipulation. A. oryzae NSRKu70-1-1 (18) was used as a host strain for gene disruption. Strains and primers used in this study are listed in Table 1 and Table S1 in the supplemental material, respectively. DPY medium (2% dextrin, 1% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, and 0.05% MgSO4 · 7H2O [pH 5.5]) was used for liquid cultivation and growth analyses of the A. oryzae strains. Czapek Dox (CD) medium (2% glucose, 0.3% NaNO3, 0.2% KCl, 0.1% KH2PO4, 0.05% MgSO4 · 7H2O, 0.002% FeSO4 · 7H2O [pH 5.5]), M medium [2% glucose, 0.2% NH4Cl, 0.1% (NH4)2SO4, 0.05% KCl, 0.05% NaCl, 0.1% KH2PO4, 0.05% MgSO4 · 7H2O, 0.002% FeSO4 · 7H2O (pH 5.5)], and their methionine-supplemented media were used for transformation and growth analyses of A. oryzae. Transformation of A. oryzae was carried out as described previously (20). Potato dextrose (PD) agar medium (Nissui, Tokyo, Japan) was used for the harvesting of conidia.
TABLE 1.
Strains used in this study
| Strain | Parental strain | Genotype |
|---|---|---|
| RIB40a | Wild type | |
| NSRKu70-1-1b | niaD− sC− adeA− ΔargB::adeA− Δku70::argB | |
| NSK-Δlah2 | NSRKu70-1-1 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA |
| NSK-Δwsc1 | NSRKu70-1-1 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAowsc::adeA |
| NSK-Δhex1-1 | NSRKu70-1-1 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAohex1::adeA |
| NSRKu70-1-1Ab | NSRKu70-1-1 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB pAdeA (adeA) |
| NSK-LAH[1–2039]G | NSRKu70-1-1A | niaD− sC− adeA− ΔargB::adeA− Δku70::argB pAdeA (adeA) pgAoLAH[1–2039]G (P-amyB::Aolah[1–2039]-egfp::T-amyB::AosC) |
| NSK-Δlah-LAH[1–2039]G | NSK-Δlah2 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA pgAoLAH[1–2039]G (P-amyB::Aolah[1–2039]-egfp::T-amyB::AosC) |
| NSK-Δwsc-LAH[1–2039]G | NSK-Δwsc1 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAowsc::adeA pgAoLAH[1–2039]G (P-amyB::Aolah[1–2039]-egfp::T-amyB::AosC) |
| NSK-LAH[4710–5727]G | NSRKu70-1-1A | niaD− sC− adeA− ΔargB::adeA− Δku70::argB pAdeA (adeA) pgAoLAH[4710–5727]G (P-amyB::Aolah[4710–5727]-egfp::T-amyB::niaD) |
| NSK-Δlah-LAH[4710–5727]G | NSK-Δlah2 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA pgAoLAH[4710–5727]G (P-amyB::Aolah[4710–5727]-egfp::T-amyB::niaD) |
| NSK-Δlah-LAH-3HA | NSK-Δlah2 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA pgAoLAHN (P-amyB::Aolah-3×HA::T-amyB:: niaD) |
| NSK-Δlah-LAH[1–2039]G-LAH-3HA | NSK-Δlah-LAH[1–2039]G | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA pgAoLAH[1–2039]G (P-amyB::Aolah[1–2039]-egfp::T-amyB::AosC) pgAoLAHN (P-amyB::Aolah-3×HA::T-amyB:: niaD) |
| NSK-LAH[(1–2039)+(4710–5727)]-3HA | NSK-Δlah2 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA pgAoLAH[(1–2039)+(4710–5727)]N (P-amyB::Aolah[(1–2039)+(4710–5727)]-3×HA::T-amyB::niaD) |
| NSK-LAH[1–2039]G-LAH[(1–2039)+(4710–5727)]-3HA | NSK-Δlah-LAH[1–2039]G | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA pgAoLAH[1–2039]G (P-amyB::Aolah[1–2039]-egfp::T-amyB::AosC) pgAoLAH[(1–2039)+(4710–5727)]N (P-amyB::Aolah[(1–2039)+(4710–5727)]-3×HA::T-amyB::niaD) |
| NSRKu70-1-1AN | NSRKu70-1-1A | niaD− sC− adeA− ΔargB::adeA− Δku70::argB pAdeA (adeA) pNR10 (niaD) |
| NSK-ΔlahN | NSK-Δlah2 | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA pNR10 (niaD) |
| NSK-LAH[1–2039]G-DPTS1 | NSK-Δlah-LAH[1–2039]G | niaD− sC− adeA− ΔargB::adeA− Δku70::argB ΔAolah::adeA pgAoLAH[1–2039]G (P-amyB::Aolah[1–2039]-egfp::T-amyB::AosC) pgDPTS1N (P-amyB::mdsred-pts1::T-amyB::niaD) |
Construction of an Aolah disruptant.
For disruption of the Aolah gene, plasmid pgdAoLAH was constructed by using the MultiSite Gateway cloning system (Invitrogen, Carlsbad, CA). The 1.5-kb upstream region of the Aolah gene was amplified by PCR using primers (attB4)Aolah-up-F and (attB1)Aolah-up-R, and the 1.5-kb downstream region was amplified by using primers (attB2)Aolah-down-F_2 and (attB3)Aolah-down-R_2. The upstream and downstream DNA fragments of the Aolah gene were cloned into the entry vectors pDONR-P4-P1R and pDONR-P2R-P3, respectively, using the BP clonase reaction of the MultiSite Gateway system. The obtained 5′ and 3′ entry clones and the center entry clone pgEaA, containing the adeA marker gene (21), were combined for the LR clonase reaction of the MultiSite Gateway system with the destination vector pDEST R4-R3 to obtain plasmid pgdAoLAH. A deletion DNA fragment for the Aolah gene was amplified by PCR using plasmid pgdAoLAH as the template and primers (attB4)Aolah-up-F and (attB3)Aolah-down-R_2. The obtained PCR product was introduced into strain NSRKu70-1-1 (18), and M agar medium supplemented with 0.15% methionine was used for the selection of adeA+ transformants. All six Aolah disruptants obtained in this study showed nearly identical phenotypes, and strain NSK-Δlah2 was used as the representative Aolah disruptant. Strain NSRKu70-1-1A, which was constructed by introducing plasmid pAdeA, containing the adeA marker gene, into strain NSRKu70-1-1 (18), was used as a wild-type strain. An Aohex1 disruptant strain, NSK-Δhex1-1, was obtained by using strain NSRKu70-1-1, as previously described (13), and was used for comparative phenotype analysis in the same genetic background as that for the Aolah disruptant.
Construction of strains expressing AoLAH[1–2039]-EGFP.
A fusion protein consisting of the first 2,039 amino acids of AoLAH and enhanced green fluorescent protein (EGFP) (AoLAH[1–2039]-EGFP) was constructed as follows. Aolah[1–2039] was amplified by PCR using primers (attB1)Aolah_1-F and (attB2)Aolah_6243-R and cloned into the entry vector pDONR221 using the BP clonase reaction, generating the center entry clone pgAoLAH[1–2039]. The 5′ entry clone pg5′PaB (PamyB) (22), center entry clone pgAoLAH[1–2039], 3′ entry clone pg3′E (egfp) (22), and destination vector pgDSO, containing the A. oryzae sC gene as a selectable marker (18), were subjected to the LR clonase reaction. The generated plasmid, designated pgAoLAH[1–2039]G, was introduced into the wild-type strain (NSRKu70-1-1A) and the Aolah disruptant (NSK-Δlah2), which were then plated onto M agar medium for the selection of transformants. The resulting transformants were named NSK-LAH[1–2039]G and NSK-Δlah-LAH[1–2039]G, respectively.
Construction of strains for visualization of Woronin bodies and peroxisomes.
A DNA fragment for mDsRed (monomeric DsRed) fused with PTS1 (peroxisomal targeting signal 1) was amplified by PCR using primers (attB1)-DsRed-M-F and (attB2)-PTS1-R and cloned into pDONR221 by the BP clonase reaction, generating the center entry clone pgmDsRed-PTS1. The 5′ entry clone pg5′PaB, containing the amyB promoter; center entry clone pgmDsRed-PTS1; 3′ entry clone pg3′TaN, containing the amyB terminator and niaD marker; and destination vector pDEST R4-R3 were mixed for the LR clonase reaction. The generated plasmid was named pgDPTS1N and introduced into strain NSK-LAH[1–2039]G. One of the resulting transformants was named NSK-LAH[1–2039]G-DPTS1 and was used in experiments to allow the visualization of Woronin bodies and peroxisomes.
Construction of strains expressing AoLAH[4710–5727]-EGFP.
A fusion protein consisting of the C-terminal region of AoLAH (amino acids 4710 to 5727) and EGFP was constructed as follows. Aolah[4710–5727] was PCR amplified by using primers Aolah-Cter-F(14254) and Aolah-Cter-R(17400) and cloned into pDONR221 by using the BP clonase reaction, generating the center entry clone pgAoLAH[4710–5727]. The 5′ entry clone pg5′PaB (PamyB), center entry clone pgAoLAH[4710–5727], 3′ entry clone pg3′E (egfp), and destination vector pgDN (22), containing the A. oryzae niaD gene as a selectable marker, were subjected to the LR clonase reaction. The generated plasmid, designated pgAoLAH[4710–5727]G, was introduced into the wild-type strain (NSRKu70-1-1A) and the Aolah disruptant (NSK-Δlah2), which were then plated onto CD agar medium supplemented with 0.0015% methionine for the selection of transformants. The resulting transformants were named NSK-LAH[4710–5727]G and NSK-Δlah-LAH[4710–5727]G, respectively.
Complementation of the Aolah disruptant.
For complementation of the Aolah disruptant (NSK-Δlah2), a plasmid expressing full-length AoLAH was constructed. Briefly, the Aolah coding region (not including the stop codon) was PCR amplified by using primers (attB1)Aolah_1-F and Aolah-Cter-R(17400) and then cloned into the entry vector pDONR-P221 by the BP clonase reaction. The obtained center entry clone; 5′ entry clone pg5′PaB, containing the amyB promoter; and 3′ entry clone pg3′HA3, containing the 3×hemagglutinin (3×HA) gene, were subjected to the LR clonase reaction with the destination vector pgDN to obtain plasmid pAoLAHN, which contained niaD as a selectable marker. Plasmid pgAoLAHN was introduced into strains NSK-Δlah2 and NK-Δlah-LAH[1–2039]G, and positive transformants were selected on CD agar medium supplemented with 0.0015% methionine and unsupplemented CD agar medium, respectively. Plasmid pNR10, containing the niaD marker, was transformed into wild-type strain NSRKu70-1-1A and Aolah disruptant strain NSK-Δlah2 as controls.
Construction of a strain expressing Aolah with a truncated middle region.
DNA fragments for AoLAH[1–2039] and AoLAH[4710–5727] were amplified by using primers (attB1)Aolah_1-F and Aolah_N-R-fusion and primers Aolah_C-F-fusion and Aolah-Cter-R(17400), respectively. The two DNA fragments were fused by PCR using primers (attB1)Aolah_1-F and Aolah-Cter-R(17400), and the resulting DNA fragment for AoLAH[(1–2039)+(4710–5727)] was cloned by the BP clonase reaction into the entry vector pDONR-P221. The obtained center entry clone was mixed with the 5′ entry clone pg5′PaB, containing the amyB promoter, and 3′ entry clone pg3′HA3, containing the 3×HA gene, for the LR clonase reaction in the presence of the destination vector pgDN to obtain plasmid pAoLAH[(1–2039)+(4710–5727)]N, containing niaD as a selectable marker. Plasmid pAoLAH[(1–2039)+(4710–5727)]N was introduced into the Aolah disruptant strains NSK-Δlah2 and NSK-Δlah-LAH[1–2039]EGFP, and positive transformants were selected on CD agar medium supplemented with 0.0015% methionine and unsupplemented CD agar medium, respectively. The expression of truncated AoLAH was confirmed by Western blotting (see the supplemental material).
Construction of an Aowsc disruptant.
For disruption of the Aowsc gene, plasmid pgdAoWSC was constructed by using the MultiSite Gateway cloning system. Briefly, the 1.5-kb upstream and downstream regions of the Aowsc gene were amplified by PCR using primers (attB4)Aowsc-up-F and (attB1)Aowsc-up-R and primers (attB2)Aowsc-down-F and (attB3)Aowsc-down-R, respectively. The amplified upstream and downstream DNA fragments were cloned into the entry vectors pDONR-P4-P1R and pDONR-P2R-P3, respectively, by the BP clonase reaction. The obtained 5′ and 3′ entry clones were mixed with the center entry clone pgEaA, containing the adeA marker gene, for the LR clonase reaction in the presence of the destination vector pDEST R4-R3 to obtain plasmid pgdAoWSC. The deletion fragment for the Aowsc gene was amplified by PCR using plasmid pgdAoWSC as the template and primers (attB4)Aowsc-up-F and (attB3)Aowsc-down-R. The PCR product was introduced into strain NSRKu70-1-1 (18), and an adeA+ transformant, designated NSK-Δwsc1, was selected on M agar medium supplemented with 0.15% methionine.
Transmission electron microscopy (TEM).
Mycelia grown on DPY agar medium at 30°C for 4 days were fixed in 0.1 M phosphate buffer (pH 7.0) containing 4% glutaraldehyde at 4°C for 4 h and were then washed three times in 0.1 M phosphate buffer (pH 7.0) for 10 min. The washed samples were treated with a solution containing 1% osmium tetroxide, 1% potassium bichromate, and 0.85% sodium chloride (adjusted to pH 7.2 with potassium hydroxide) for 1 h. The fixed mycelia were washed with water at 4°C for 2 h and then incubated in 30%, 50%, 70%, 80%, 90%, and 100% ethanol for 10 min. After incubation overnight in a mixture of dehydrated acetone and epoxy resin (4:6), the samples were embedded in epoxy resin and heated at 40°C, 50°C, 60°C, and 70°C for 1 day. Ultrathin sections were prepared by using an ultramicrotome fitted with a glass knife and then stained with uranyl acetate and lead citrate. Samples were observed with a JEM-1010 transmission electron microscope (JEOL, Tokyo, Japan), and images were analyzed with ImageJ v. 1.44 software (National Institutes of Health, Bethesda, MD [http://rsb.info.nih.gov/ij]). The nearest distance of the Woronin body margin from the septum was measured, and 44 and 48 independent Woronin bodies were analyzed for the strains expressing full-length AoLAH and truncated AoLAH, respectively.
Hypotonic shock experiment.
To induce hyphal tip bursting, 1 ml water was added to A. oryzae colonies cultured on a thin layer of DPY agar medium at 30°C for 24 h. Fifty randomly selected hyphae showing hyphal tip bursting in each culture were observed by differential interference contrast (DIC) microscopy, as described previously (9). The ability to prevent the excessive loss of cytoplasm was analyzed by judging whether the cytoplasm in the cell adjacent to the burst tip cell was retained.
Fluorescence microscopy.
Conidia (1 × 103) of the strain expressing an EGFP fusion protein were preinoculated into 100 μl CD liquid medium supplemented with 0.0015% methionine in a glass-bottom dish. After incubation at 30°C for 18 h, the cultures were observed by confocal laser scanning microscopy using an IX71 inverted microscope (Olympus, Tokyo, Japan) equipped with a 100× Neofluor objective lens (1.40 numerical aperture), a 488-nm semiconductor laser (Furukawa Electric, Tokyo, Japan), GFP filters (Nippon Roper, Chiba, Japan), a CSU22 confocal scanning system (Yokogawa Electronics, Tokyo, Japan), and an Andor iXon cooled digital charge-coupled-device (CCD) camera (Andor Technology PLC, Belfast, United Kingdom). Images were analyzed with Andor iQ software (Andor Technology PLC) and Meta-morph software (Molecular Devices, Sunnyvale, CA).
RESULTS
Identification of the Aolah gene.
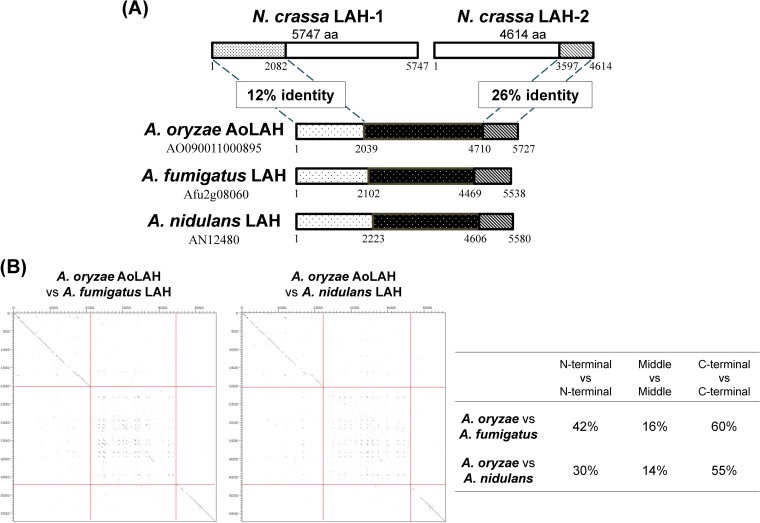
A gene (AO090011000895) encoding a protein with similarity to the N. crassa LAH-1 and LAH-2 proteins was identified in the A. oryzae genome database (DOGAN [Database of the Genomes Analyzed at NITE {National Institute of Technology and Evaluation, Japan}] [http://www.bio.nite.go.jp/dogan/Top]). The encoding region of this gene, designated Aolah, was again predicted to contain an additional 1,198 amino acids extending from the N terminus based on comparison with the gene sequences for other Aspergillus LAH proteins (see Fig. S1 in the supplemental material). In contrast to the N. crassa leashin (lah) locus, which consists of two unidirectionally aligned genes for LAH-1 and LAH-2 (3), the Aolah gene was predicted to encode a single polypeptide of 5,727 amino acids (Fig. 1A). The N-terminal region of AoLAH shared 12% identity with that of N. crassa LAH-1, whereas the C-terminal region displayed 26% identity with that of LAH-2, and few conserved sequences were found in the middle region of AoLAH. Dot plot analysis revealed that the N- and C-terminal regions of AoLAH shared homologies with those of A. fumigatus and Aspergillus nidulans (Fig. 1B). The AoLAH N-terminal region had 42% and 30% identities with those of the LAH proteins of A. fumigatus and A. nidulans, respectively, whereas the C-terminal region had 60% and 55% identities with the respective LAH proteins. However, the middle region of AoLAH had low identities with the LAH proteins of A. fumigatus and A. nidulans. Rectangular patterns between the middle regions in dot plots (Fig. 1B) indicate that several polylysine stretches are commonly present in these proteins (see Fig. S2 in the supplemental material).
FIG 1.
Comparison between AoLAH and LAH proteins of other fungal species (Neurospora crassa, Aspergillus fumigatus, and Aspergillus nidulans). (A) Identities between AoLAH and N. crassa LAH proteins. The ClustalW program (http://www.genome.jp/tools/clustalw/) was used for sequence identity analysis. (B) Dot plot analysis and identities between AoLAH (vertical axis) and LAH proteins of A. fumigatus and A. nidulans (horizontal axis). Amino acid sequence data for LAH proteins of A. fumigatus and A. nidulans were taken from the Aspergillus Genome Database (AspGD) (http://www.aspgd.org/). Sequence similarity comparisons were performed with Dotter software (33). When the residues of compared sequences match, a dot is plotted at the corresponding position. Note that diagonal lines are seen in N- and C-terminal regions, while in middle regions, rectangular patterns were found. The scale is in amino acid (aa) residues. Red lines indicate borders between the N-terminal, middle, and C-terminal regions.
Phenotypic analysis of an Aolah disruptant.
To investigate the function of AoLAH, the Aolah gene was disrupted by replacing the 17.4-kb open reading frame (ORF) with the adeA marker gene, and the successful disruption was confirmed by Southern blotting (see Fig. S3 in the supplemental material).
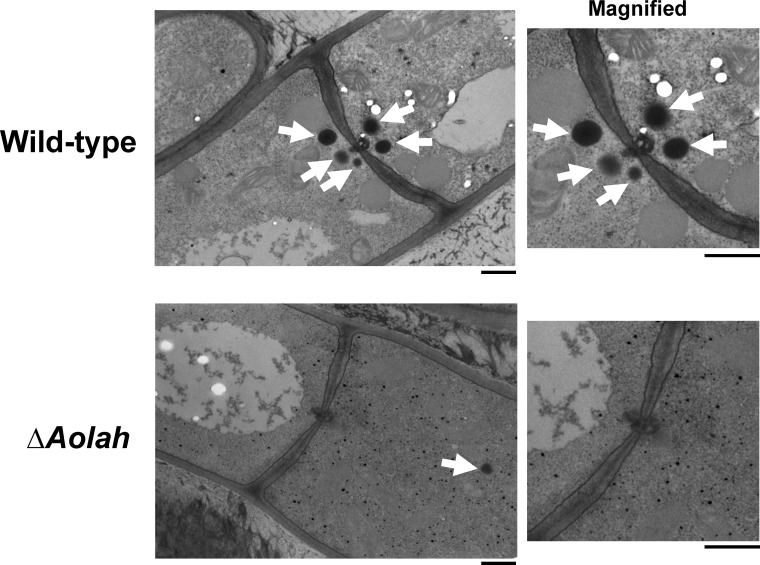
The localization of Woronin bodies in the Aolah disruptant (NSK-Δlah2) was analyzed by transmission electron microscopy (TEM) (Fig. 2). In the wild-type strain, Woronin bodies were observed as electron-dense spherical structures located near the septum. In contrast, in the Aolah disruptant, only untethered Woronin bodies located away from the septum were found (Fig. 2). Based on these imaging findings, it was concluded that AoLAH is required for the tethering of Woronin bodies to the septum.
FIG 2.
Transmission electron microscopic images of the wild-type A. oryzae and Aolah disruptant strains. Images around the septal pore are magnified. Arrows indicate Woronin bodies. Bars, 500 nm.
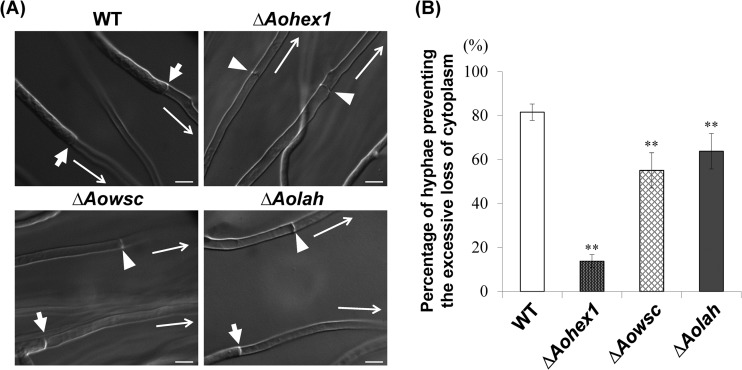
We next investigated whether the absence of AoLAH led to the excessive loss of cytoplasm upon hyphal wounding. Hyphal tip bursting was induced by the hypotonic shock of colonies grown on agar medium (9, 18), and cytoplasmic loss in the cells adjacent to the burst tip cell was examined by DIC microscopic observation. In this assay, it was considered that the septal pore was effectively plugged by the Woronin bodies if the cytoplasm was retained by the cells, and conversely, the excessive loss of cytoplasm was attributed to Woronin body dysfunction (Fig. 3A). Although 81% of wild-type cells retained their cytoplasmic constituents upon hypotonic shock, only 63% of cells of the Aolah disruptant retained their cytoplasmic constituents (Fig. 3B). As expected, the Aohex1 disruptant, which lacks Woronin bodies, showed a significantly impaired ability (14%) to prevent the excessive loss of cytoplasm (Fig. 3B).
FIG 3.
Ability of Aolah disruptant cells to prevent excessive loss of cytoplasm upon hyphal wounding. (A) Differential interference contrast images of wounded hyphae capable or incapable of preventing the excessive loss of cytoplasm from the second cell upon hyphal tip bursting. A colony grown on agar medium was subjected to hypotonic shock by flooding with water to induce hyphal tip bursting, and the first septum adjacent to the burst tip cell was observed by differential interference contrast microscopy. Arrows indicate the first septa in the hyphae preventing the excessive loss of cytoplasm, and arrowheads show the septa in the hyphae not preventing excessive cytoplasmic loss. Long arrows indicate the directions toward the burst hyphal tips. Bars, 10 μm. (B) Assay of the ability of cells to prevent excessive loss of cytoplasm upon hyphal wounding. The wild-type (WT) strain and Aohex1, Aowsc, and Aolah gene disruptant strains were subjected to hypotonic shock, and hyphae with cells adjacent to the burst tip cell that retained the cytoplasm were counted by differential interference contrast microscopy. Average percentages of hyphae preventing the excessive loss of cytoplasm are shown in the graph. Fifty randomly selected hyphae with burst hyphal tips were observed for each culture. The error bars indicate standard deviations. **, P < 0.01 (by one-way analysis of variance with Dunnett's post hoc test for comparison with the wild-type strain; n = 9).
The Aowsc gene (AO090102000111), which encodes a protein with identity to WSC proteins from N. crassa (50%), A. fumigatus (80%), and A. nidulans (71%), was also disrupted in A. oryzae (see Fig. S4 in the supplemental material). Disruption of the Aowsc gene was confirmed by Southern blotting (see Fig. S5 in the supplemental material). The Aowsc disruptant showed a reduced ability (55%) to prevent the excessive loss of cytoplasm compared to wild-type cells (Fig. 3B), suggesting that AoWSC is involved in Woronin body function. Taken together, these results indicate that AoLAH-mediated tethering of Woronin bodies to the septum is involved in their ability to prevent the excessive loss of cytoplasm upon hyphal wounding.
Localization analysis of AoLAH N- and C-terminal regions.
AoLAH was predicted to be a large protein consisting of 5,727 amino acid residues. To analyze the contribution of individual regions of AoLAH to its localization, the AoLAH protein was divided into N-terminal (amino acids 1 to 2039), middle (amino acids 2040 to 4709), and C-terminal (amino acids 4710 to 5727) regions based on the results of dot plot analysis (Fig. 1B).
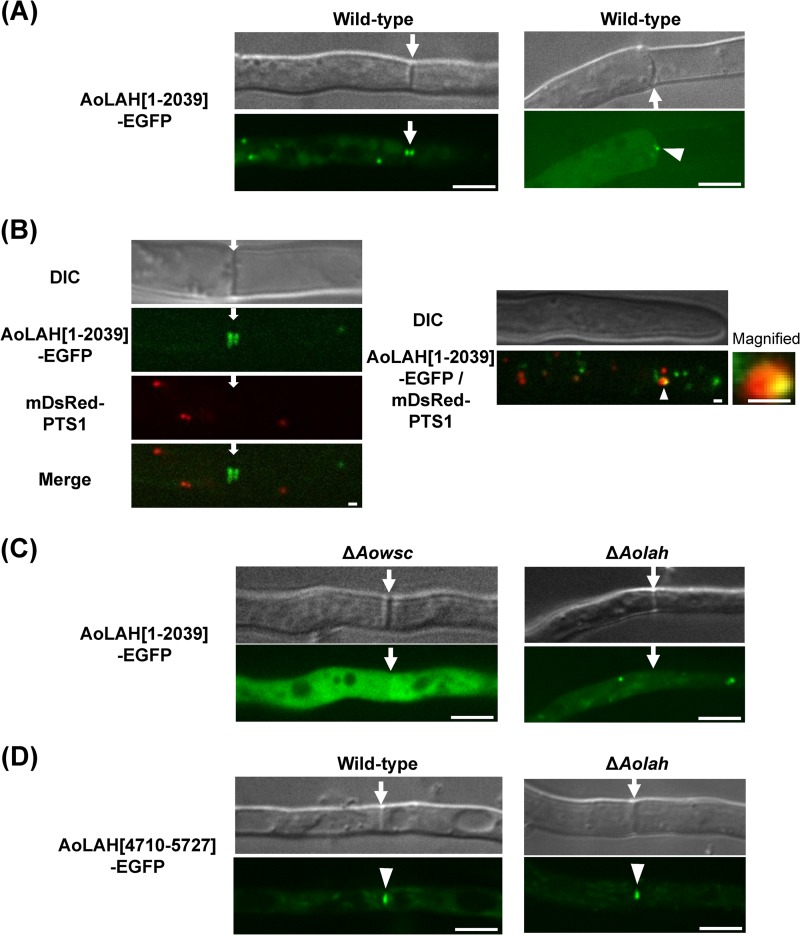
In N. crassa, LAH-1 binds and tethers Woronin bodies to the cell cortex (3). As the N-terminal region of AoLAH (AoLAH[1–2039]) showed homology to LAH-1, the localization of an AoLAH[1–2039]-EGFP fusion protein was examined in the wild-type strain. The fluorescence of AoLAH[1–2039]-EGFP was intensely localized at both sides of the septum and was also observed as punctate structures in the cytoplasm (Fig. 4A, left), a pattern that is reminiscent of the localization of Woronin bodies shown by TEM (Fig. 2). To determine whether the N-terminal region of AoLAH was associated with Woronin bodies, we induced hyphal tip bursting by hypotonic shock to observe whether AoLAH[1–2039]-EGFP moves together with Woronin bodies to the septal pore upon hyphal wounding. Upon hyphal tip bursting, the fluorescence of AoLAH[1–2039]-EGFP was detected at the septal pore (Fig. 4A, right). This localization behavior is similar to that observed during septal plugging by Woronin bodies, as previously demonstrated by the expression of AoHex1 fused with a fluorescent protein (9).
FIG 4.
Localization of AoLAH N- and C-terminal regions fused with EGFP. (A) Localization of AoLAH[1–2039]-EGFP in the wild-type background around the septum (left) and upon hyphal wounding (right). Note that the fluorescent signal was located at a distance from the septum during normal growth (see left). The right image shows the localization of AoLAH[1–2039]-EGFP to the septal pore upon hyphal wounding. Hyphal tip bursting was induced by hypotonic shock, and the septal pore adjacent to the burst tip cell was observed by fluorescence microscopy. Arrows indicate the septum, and arrowheads show the localization of AoLAH[1–2039]-EGFP to the septal pore. Bars, 5 μm. (B) Subcellular localization analysis of AoLAH[1–2039]-EGFP and peroxisomes. mDsRed fused with PTS1 was expressed in an A. oryzae strain expressing AoLAH[1–2039]-EGFP. Fluorescence microscopic analysis revealed an independent distribution of the two fluorescent fusion proteins around the septum (left) and in the apical cell (right). The septum is shown by arrows. A peripheral association of AoLAH[1–2039]-EGFP with the peroxisome was also observed in the apical cell, as shown by the arrowhead. Bars, 1 μm. (C) Localization of AoLAH[1–2039]-EGFP in the Aowsc (left) and Aolah (right) disruptants. Arrows indicate the septum. Bars, 5 μm. (D) Localization of AoLAH[4710–5727]-EGFP in the wild-type strain (left) and Aolah disruptant (right). Arrows indicate the septum, and arrowheads show the close association of AoLAH[4710–5727]-EGFP with the septal pore. Bars, 5 μm.
Woronin bodies differentiate from the peroxisome via a budding process (2, 18, 23). In previous reports on A. oryzae and A. fumigatus, the Hex1 protein fused with a fluorescent protein was found to localize near the septum, but the fusion protein often colocalized or associated with peroxisomes (10, 18), demonstrating that its localization does not always correspond to Woronin bodies. To examine whether the localization of AoLAH[1–2039]-EGFP was independent of peroxisomes, mDsRed fused with peroxisomal targeting signal 1 (PTS1) was coexpressed with AoLAH[1–2039]-EGFP in A. oryzae. Fluorescence microscopic analysis revealed that AoLAH[1–2039]-EGFP was localized near the septum and in the cytoplasm independently of peroxisomes (Fig. 4B, left). It was previously reported that in N. crassa, hyphal apical cells contain undifferentiated Woronin bodies that associate with peroxisomes (2). Here, the peripheral association of AoLAH[1–2039]-EGFP with peroxisomes was also observed, in addition to numerous fluorescent punctate structures in the cytoplasm (Fig. 4B, right). These findings demonstrated that AoLAH[1–2039]-EGFP was localized independently of peroxisomes, with the exception of the observed association in apical cells, which likely represented actively budding Woronin bodies.
As N. crassa LAH-1 binds to Woronin bodies via WSC (3), we also investigated whether the localization patterns of the AoLAH N-terminal region were dependent on AoWSC. In the Aowsc disruptant, no intense localization of AoLAH[1–2039]-EGFP was detected in the vicinity of the septum; rather, the fusion protein was completely dispersed in the cytoplasm, and no punctate structures were observed (Fig. 4C, left). This result suggested that the recruitment of the AoLAH N-terminal region to Woronin bodies is dependent on AoWSC, a finding that is in agreement with data for N. crassa reported previously by Ng et al. (3). If AoLAH[1–2039]-EGFP localization represents the septal tethering of Woronin bodies, such localization would not occur in the Aolah disruptant background (Fig. 2). As expected, no fluorescence was observed near the septum when AoLAH[1–2039]-EGFP was expressed in the Aolah disruptant, although punctate structures were found in the cytoplasm (Fig. 4C, right). Time-lapse tracking revealed that the punctate structures found in the cytoplasm randomly moved in the wild-type and Aolah disruptant strains (see Videos S1 and S2, respectively, in the supplemental material), indicating that they were not attached to the septum. This finding demonstrated that the presence of endogenous AoLAH protein is needed for the localization of AoLAH[1–2039]-EGFP near the septum. As the AoLAH N-terminal region exhibited localization patterns typical of those of Woronin bodies, the AoLAH[1–2039]-EGFP fusion protein was used as a marker of Woronin bodies in subsequent experiments.
The C-terminal conserved region of AoLAH shares similarity with N. crassa LAH-2, which is closely associated with the septal pore (3). To examine its localization, the C-terminal region of AoLAH tagged with EGFP (AoLAH[4710–5727]-EGFP) was expressed in the wild-type and Aolah disruptant strains. As shown in Fig. 4D, AoLAH[4710–5727]-EGFP was localized to the septal pore in both strains, indicating that the C-terminal region of AoLAH associates with the septal pore.
Woronin body tethering mediated by AoLAH lacking the middle region.
According to the results of the localization analysis described above, it can be assumed that the AoLAH N- and C-terminal fusion construct without the nonconserved middle region may be functionally sufficient to tether Woronin bodies to the septum. To test this speculation, a middle-region-deleted AoLAH construct {AoLAH[(1–2039)+(4710–5727)]} and full-length AoLAH were expressed in the Aolah disruptant. The two constructs were expressed with a 3×HA tag, which was verified by Western blotting (see Fig. S6 in the supplemental material). Full-length AoLAH-3×HA was predicted to be a polypeptide of ∼620 kDa, and a band was detected at a mass much higher than the maximum molecular mass (460 kDa) of the protein marker. Middle-region-deleted AoLAH-3×HA was predicted to be ∼330 kDa but was detected at around 460 kDa.
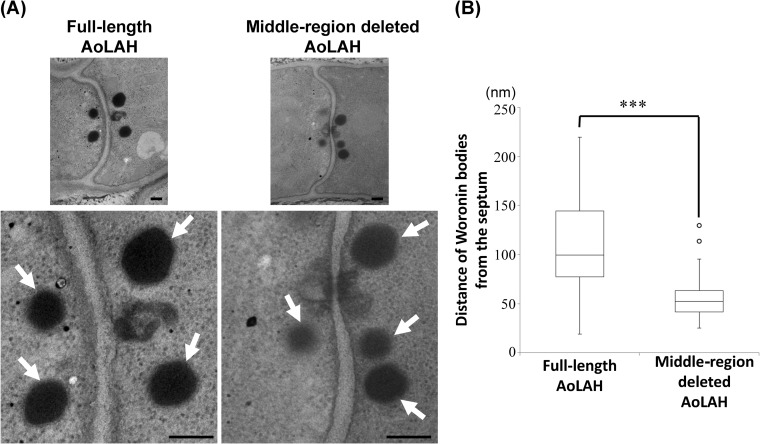
TEM revealed that Woronin bodies were located near the septal pore of the Aolah disruptant strain expressing full-length AoLAH (Fig. 5A). Similarly, in the strain expressing AoLAH[(1–2039)+(4710–5727)], the Woronin bodies were also observed near the septal pore (Fig. 5A), indicating that the N- and C-terminal fusion construct without the middle region is sufficient to tether Woronin bodies to the septum.
FIG 5.
Transmission electron microscopy of Woronin bodies in the Aolah disruptant strain expressing full-length or middle-region-deleted AoLAHs. (A) Transmission electron microscopy of Woronin bodies located around the septum in the strain expressing full-length or middle-region-deleted AoLAH. Arrows indicate Woronin bodies. Bars, 200 nm. (B) Distance of Woronin bodies from the septum in the strain expressing full-length or middle-region-deleted AoLAH. The data are presented in the box plot chart. The top, bottom, and middle lines correspond to the 75th percentile, 25th percentile, and median, respectively; bars represent the limits of the upper (top) and lower (bottom) quartiles. The whiskers show the highest and lowest readings within a 1.5× interquartile range. The outliers are indicated by ○ (n = 44 and 48 for strains expressing full-length AoLAH and middle-region-deleted AoLAH, respectively; ***, P < 0.001).
To more precisely investigate the effect of the deletion of the middle region of AoLAH on the tethering of Woronin bodies, the average distance of Woronin bodies from the septum was measured by TEM (Fig. 5B). In the Aolah disruptant strain expressing full-length AoLAH, Woronin bodies were located approximately 99 nm from the septum, whereas in the strain expressing middle-region-deleted AoLAH, the position of Woronin bodies had shifted approximately 50 nm closer to the septum (Fig. 5B). These results suggest that the middle region of AoLAH has a role in regulating the distance of Woronin bodies from the septum.
Elastic movement of Woronin bodies visualized with an AoLAH N-terminal-region–EGFP fusion protein.
The tethering of Woronin bodies to the septum in N. haematococca was previously reported to exhibit elasticity, as demonstrated by the return of the Woronin bodies to the septum after their physical separation by laser trapping (16). As shown in Fig. 5B, the distance of Woronin bodies from the septum was more variable in the Aolah disruptant strain expressing full-length AoLAH than in the strain expressing the middle-region-truncated form of AoLAH. Based on this finding, it was hypothesized that the middle region of AoLAH might confer the observed elastic characteristics of the Woronin body tether.
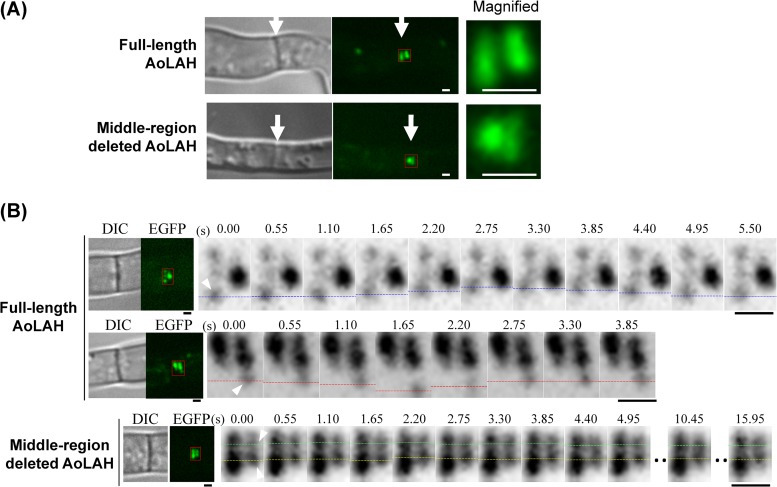
To answer this question, the movement of Woronin bodies in living cells expressing AoLAH[1–2039]-EGFP was examined by fluorescence microscopy (Fig. 6). In the strain expressing middle-region-deleted AoLAH, Woronin bodies were located near the septum, and fluorescent dots were closer together across the septum than in the strain expressing full-length AoLAH (Fig. 6A). Time-lapse analysis revealed that Woronin bodies located near the septum exhibited positional changes in cells expressing full-length AoLAH (Fig. 6B). Microscopic observation of the first septum of hyphae expressing full-length AoLAH showed that at least one of the tethered Woronin bodies moved rapidly back and forth between two positions in approximately 5 s (see Video S3 in the supplemental material). Two distinct movement patterns of Woronin bodies were observed: movement toward the septal pore and then back to the starting position (Fig. 6B, top) and movement away from the septal pore and then back to the starting position (Fig. 6B, middle). These observational analyses demonstrated that tethered Woronin bodies are capable of elastic movement. In the strain expressing middle-region-deleted AoLAH, however, such movement was not observed for Woronin bodies tethered to the septum (Fig. 6B, bottom; see also Video S4 in the supplemental material). This finding indicates that the middle region of AoLAH is required for the elastic movement of Woronin bodies.
FIG 6.
Fluorescence microscopy of Woronin bodies visualized with the AoLAH N-terminal-region–EGFP fusion protein in the Aolah disruptant strain expressing full-length or middle-region-deleted AoLAH. (A) Woronin bodies visualized by expression of AoLAH[1–2039]-EGFP near the septum in the strain expressing full-length or middle-region-deleted AoLAH. Arrows indicate the septum. Bars, 1 μm. (B) Back-and-forth movement of the Woronin bodies visualized by using AoLAH[1–2039]-EGFP. Woronin body movement at the first hyphal septum was analyzed by time-lapse analysis with a time interval of 550 ms. Tracked Woronin bodies are indicated by arrowheads in the first frame of time-lapse analysis. Blue, red, green, and yellow dotted lines indicate the positions of Woronin bodies in individual frames. Note that the back-and-forth movement was not clearly detected in the strain expressing middle-region-deleted AoLAH. Bars, 1 μm.
Involvement of the AoLAH middle region in Woronin body function.
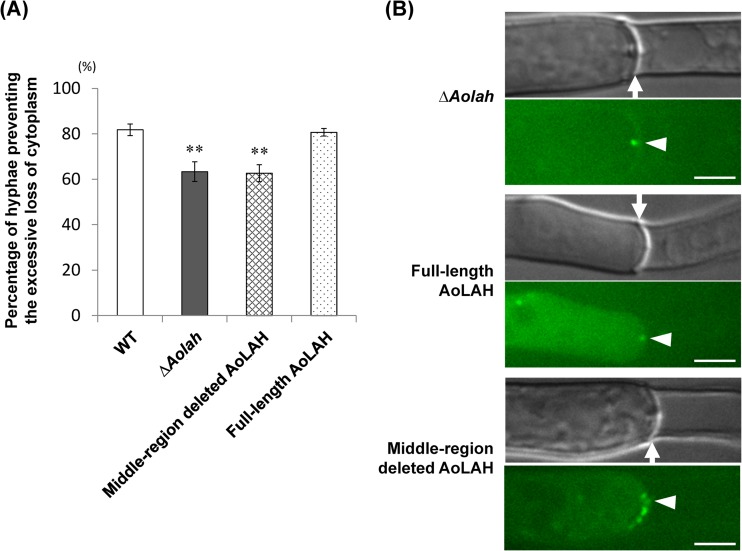
As it was demonstrated that the middle region of AoLAH influences the positional variation and elastic motility of tethered Woronin bodies (Fig. 5B and 6B), we next investigated if the function of Woronin bodies was affected by deleting the middle region of AoLAH. Complementation analysis of the Aolah disruptant was performed with various AoLAH constructs, and marker-introduced strains in the wild-type and Aolah disruptant backgrounds were used as positive and negative controls, respectively. The ability of cells to prevent the excessive loss of cytoplasm was analyzed by inducing hyphal tip bursting in each strain through hypotonic shock. Although 82% of wild-type cells prevented the excessive loss of cytoplasm in cells adjacent to the hyphal tip upon lysis, cells of the Aolah disruptant showed a reduced ability (63%). Cells of the Aolah disruptant expressing full-length AoLAH displayed a restored ability (81%) (Fig. 7A) that was comparable to that of the wild-type strain, indicating the functional complementation of the disruption. However, the expression of middle-region-deleted AoLAH in the Aolah disruptant did not restore the resistance of cells to cytoplasmic loss through the septum (62%) (Fig. 7A). These results indicate that the middle region of AoLAH is needed for proper Woronin body position to prevent the excessive loss of cytoplasm upon hyphal wounding.
FIG 7.
Functional analysis of Woronin bodies in the Aolah disruptant strain expressing middle-region-deleted AoLAH. (A) Colonies formed on agar medium were subjected to hypotonic shock, and the ability of hyphae to prevent the excessive loss of cytoplasm was analyzed as described in the legend of Fig. 3B. The error bars indicate standard deviations. **, P < 0.01 (by one-way analysis of variance with Dunnett's post hoc test for comparison with the wild-type strain; n = 9). (B) Woronin body localization around the septum upon hyphal wounding in the Aolah disruptant strain and Aolah disruptant strains expressing full-length AoLAH or middle-region-deleted AoLAH. Woronin bodies were visualized by using AoLAH[1–2039]-EGFP. Arrows indicate the septum, and arrowheads show the septal pore. Bars, 5 μm.
As the Woronin bodies visualized with the EGFP fusion of the AoLAH N-terminal region plugged the septal pore upon hyphal wounding (Fig. 4A, right), we also examined whether Woronin bodies plug the septal pore in strains expressing full-length or middle-region-deleted AoLAH. In the Aolah disruptant, although Woronin bodies did not localize near the septum (Fig. 4C, right), a fluorescent dot was observed in the septal pore upon hyphal wounding (Fig. 7B), suggesting that untethered Woronin bodies were pushed into the septal pore by cytoplasmic flow. In the strain expressing full-length AoLAH, a fluorescent dot was also observed in the septal pore (Fig. 7B), a finding that is consistent with the results presented in Fig. 4A. In the Aolah disruptant and the strain complemented with full-length AoLAH, almost no fluorescence was detected near the septum, with the exception of the dot plugging the septal pore. In contrast, in the strain expressing middle-region-deleted AoLAH, Woronin bodies tethered to the septum were frequently observed after hyphal wounding, in addition to a fluorescent dot within the septal pore (Fig. 7B). Taken together, these results indicated that the nonconserved middle region of AoLAH is involved in the proper spacing of tethered Woronin bodies around the septal pore, and this spacing is important for efficient plugging of the septal pore upon hyphal wounding.
DISCUSSION
In this study, we investigated the role of the AoLAH protein in the tethering of and septal plugging by Woronin bodies. TEM analysis clearly demonstrated that AoLAH is required for the tethering of Woronin bodies to the septum (Fig. 2). Moreover, the excessive loss of cytoplasm was observed in an Aolah disruptant upon hyphal wounding induced by the hypotonic shock of A. oryzae colonies grown on agar media (9). Aohex1 gene disruption causes the loss of Woronin bodies and leads to a significant defect in the ability of cells to prevent the loss of cytoplasm (Fig. 3B) (9). Here, disruption of the Aolah gene also reduced this protective ability but to a lesser extent than that observed for the Aohex1 gene disruptant (Fig. 3B). This finding indicates that the tethering of Woronin bodies to the septum by AoLAH is important for their efficiency in plugging the septal pore upon hyphal wounding and thereby preventing the excessive loss of cytoplasm.
To functionally characterize the extremely large AoLAH protein, which is >5,000 amino acids, it was divided into N-terminal, middle, and C-terminal regions according to amino acid sequence homology with the LAH proteins of other Aspergillus species (Fig. 1B). The N-terminal conserved region of AoLAH (amino acids 1 to 2039) fused with EGFP was detected at a distance from both sides of the septal pore, and the C-terminal conserved region of AoLAH (amino acids 4710 to 5727) fused with EGFP was closely associated with the septal pore (Fig. 4), localizations that are in agreement with a recent report on A. fumigatus (17). The fluorescence of AoLAH[1–2039]-EGFP was observed at the septal pore upon hyphal wounding (Fig. 4A, right), which also induces the DsRed2-AoHex1 fusion protein to plug the septal pore (9). These data suggest that the N-terminal region of AoLAH exhibits a behavior typical of Woronin bodies. AoHex1 has been used as a Woronin body marker (9); however, fluorescent protein-tagged AoHex1 only partially complements an Aohex1 disruptant and often colocalizes to peroxisomes (18). Here, nearly all AoLAH[1–2039]-EGFP fluorescent dots observed in A. oryzae cells were independent of peroxisomes (Fig. 4B), and expression of the AoLAH[1–2039]-EGFP fusion protein did not affect the ability of cells to prevent the excessive loss of cytoplasm (data not shown). N. crassa LAH-1 is associated with Woronin bodies via binding to the WSC protein (3). Similarly, AoLAH[1–2039]-EGFP was dispersed in the cytoplasm of the Aowsc disruptant and did not exhibit dot-like localization patterns (Fig. 4C, left), confirming the AoWSC-dependent recruitment of the AoLAH N-terminal region to Woronin bodies. A model was recently proposed where WSC might assemble into a pore-like structure and cytosolically expose the N-terminal end of Hex1 for the recruitment of LAH to Woronin bodies in A. fumigatus (17), which could be in the same line of our genetic data. However, it remains to be determined whether Hex1 or WSC directly binds to LAH. Taken together, the findings from this study demonstrate that AoLAH[1–2039]-EGFP is a more reliable marker for the observation of Woronin bodies in A. oryzae.
In addition to the characterization of the conserved N- and C-terminal regions of AoLAH, the functional roles of the nonconserved middle region of AoLAH were also examined. When the AoLAH-EGFP fusion construct lacking the middle region (∼2,700 amino acids) of AoLAH was expressed in the Aolah disruptant, Woronin bodies were retethered to the septum (Fig. 5A), revealing that middle-region-deleted AoLAH is sufficient for the tethering of Woronin bodies to the septum. However, the Woronin bodies were located approximately 50 nm closer to the septum than those in the strain expressing full-length AoLAH (Fig. 5B). Based on the fact that the giant protein titin, which is 4 MDa, is 1 μm in length (24), the length of the deleted 2,670 amino acids of AoLAH is theoretically estimated to be 70 nm, which is consistent with the reduced distance of Woronin bodies from the septum that was observed for the Aolah disruptant expressing middle-region-deleted AoLAH.
In the Aolah disruptant, although no Woronin bodies were tethered to the septum, untethered Woronin bodies were found to plug the septal pore after hyphal wounding (Fig. 7B) and were likely pushed into position by cytoplasmic flow. However, untethered Woronin bodies were not able to plug the septal pore as quickly as the tethered Woronin bodies, which may explain why the loss of cytoplasm was greater in the Aolah disruptant than in the wild-type strain but was less than that in the Aohex1 disruptant (Fig. 3B and 7A). This is similar to the fact that an A. fumigatus lah mutant defective in the septal tethering of Woronin bodies exhibits no increased sensitivity to calcofluor white, whereas the hex1 disruptant is sensitive (17). Unexpectedly, the ability to prevent the excessive loss of cytoplasm was not restored by expressing the middle-region-deleted AoLAH construct in the Aolah disruptant (Fig. 7A), even though Woronin bodies were tethered to the septum in this strain (Fig. 5A and 6A). We specifically examined the septal plugging state by visualizing Woronin bodies bound to the AoLAH N-terminal-region–EGFP fusion protein. Upon hyphal wounding, nearly all Woronin bodies tethered to the septum contributed to septal plugging in the wild-type and full-length AoLAH-expressing strains (Fig. 4A, right, and 7B). However, the Woronin bodies remained tethered at a distance from the septal pore upon hyphal wounding in the strain expressing middle-region-deleted AoLAH (Fig. 7B), indicating that they failed to plug the septal pore. It was likely that the untethered Woronin bodies plugged the septal pore, as was observed for the Aolah disruptant. This result may explain why the cells expressing middle-region-deleted AoLAH had a reduced ability to prevent the excessive loss of cytoplasm (Fig. 7A), although it should be noted that the middle region is not essential for the overall ability of Woronin bodies to plug the septal pore.
According to the TEM observations, the distance between Woronin bodies and the septum displayed greater variability in the presence of full-length AoLAH (Fig. 5B), suggesting that Woronin bodies have positional flexibility. This speculation is consistent with the reported elasticity of the Woronin body tether in N. haematococca, as demonstrated by laser capturing experiments (16). It was recently reported that limited lateral mobility of Woronin bodies tethered at the septum was found for A. fumigatus (17). In this study, by visualizing Woronin bodies with an AoLAH N-terminal-region–EGFP fusion protein, we observed that some tethered Woronin bodies displayed dynamic movement that may reflect the presence of an elastic tether, a characteristic that is not seen in the strain expressing middle-region-deleted AoLAH (Fig. 6B). This finding suggests that the middle region of AoLAH confers elastic movement activity to Woronin bodies. Moreover, the fixed tethering of Woronin bodies to the septum (Fig. 7B) is attributable to the excessive loss of cytoplasm upon hyphal wounding and is due to decreased movement activity by deletion of the middle region of AoLAH. Collectively, the results presented here suggest that efficient septal plugging requires not only that Woronin bodies be tethered to the septum but also that they must have the movement activity, which is conferred by the middle region of AoLAH.
A number of intrinsically disordered proteins are localized to the septum, where they are often involved in intercellular connectivity (25). AoLAH is also predicted to be highly disordered, particularly in the N-terminal and middle regions (see Fig. S7 in the supplemental material). Detection of the full-length and middle-region-deleted AoLAHs with a higher molecular mass than that predicted by Western blotting (see Fig. S6 in the supplemental material) may be caused by the presence of intrinsically disordered characteristics in the N-terminal and middle regions, which is in agreement with previous reports (26, 27). The middle region of AoLAH does not share high similarity with other fungal LAH proteins (Fig. 1B); however, several polylysine stretches are commonly present in these proteins, although their interval length is not conserved (see Fig. S2 in the supplemental material). Glutamate was the most frequently found amino acid in the middle region of AoLAH (see Fig. S8 in the supplemental material), a feature that is also seen in other Aspergillus species. The giant protein titin exhibits molecular spring-like elasticity via its intrinsically disordered region and contributes to the contraction-relaxation cycle of skeletal muscle (28). Calcium reduces the length of titin, and this calcium-dependent process requires the presence of a glutamate-rich motif in the disordered region (29). The high content of glutamate in the middle region of AoLAH raises the possibility that this protein may also be calcium sensitive, given the fact that the calcium concentration would be increased upon hyphal wounding, as has been reported for other organisms (30). We speculate that glutamates frequently found in the middle region of AoLAH may be involved in the extension and contraction of the Woronin body tether. An increasing number of intrinsically disordered proteins have recently been reported to have many biological roles (31). However, with the exception of a few studies (25, 32), little knowledge about the function of disordered proteins in filamentous fungi has been accumulated. Here, we unexpectedly discovered the physiological importance of a nonconserved region in AoLAH that is predicted to be disordered and potentially functions as a molecular spring, although further analyses of this protein are needed to confirm these findings.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant-in-aid for young scientists from the Japan Society for the Promotion of Science (JSPS) and by funding from the Noda Institute for Scientific Research.
We are grateful to Fumiko Ishizuna for her help with transmission electron microscopy.
Footnotes
Published ahead of print 9 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00060-14.
REFERENCES
- 1.Lew RR. 2005. Mass flow and pressure-driven hyphal extension in Neurospora crassa. Microbiology 151:2685–2692. 10.1099/mic.0.27947-0 [DOI] [PubMed] [Google Scholar]
- 2.Tey WK, North AJ, Reyes JL, Lu YF, Jedd G. 2005. Polarized gene expression determines Woronin body formation at the leading edge of the fungal colony. Mol. Biol. Cell 16:2651–2659. 10.1091/mbc.E04-10-0937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SK, Liu F, Lai J, Low W, Jedd G. 2009. A tether for Woronin body inheritance is associated with evolutionary variation in organelle positioning. PLoS Genet. 5: e1000521. 10.1371/journal.pgen.1000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markham P, Collinge AJ. 1987. Woronin bodies of filamentous fungi. FEMS Microbiol. Rev. 46:1–11. 10.1111/j.1574-6968.1987.tb02448.x [DOI] [Google Scholar]
- 5.Jedd G, Chua NH. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2:226–231. 10.1038/35008652 [DOI] [PubMed] [Google Scholar]
- 6.Asiegbu FO, Choi W, Jeong JS, Dean RA. 2004. Cloning, sequencing and functional analysis of Magnaporthe grisea MVP1 gene, a hex-1 homolog encoding a putative ‘woronin body' protein. FEMS Microbiol. Lett. 230:85–90. 10.1016/S0378-1097(03)00858-9 [DOI] [PubMed] [Google Scholar]
- 7.Curach NC, Te'o VS, Gibbs MD, Bergquist PL, Nevalainen KM. 2004. Isolation, characterization and expression of the hex1 gene from Trichoderma reesei. Gene 331:133–140. 10.1016/j.gene.2004.02.007 [DOI] [PubMed] [Google Scholar]
- 8.Soundararajan S, Jedd G, Li X, Ramos-Pamplona M, Chua NH, Naqvi NI. 2004. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell 16:1564–1574. 10.1105/tpc.020677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruyama J, Juvvadi PR, Ishi K, Kitamoto K. 2005. Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 331:1081–1088. 10.1016/j.bbrc.2005.03.233 [DOI] [PubMed] [Google Scholar]
- 10.Beck J, Ebel F. 2013. Characterization of the major Woronin body protein HexA of the human pathogenic mold Aspergillus fumigatus. Int. J. Med. Microbiol. 303:90–97. 10.1016/j.ijmm.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 11.Yuan P, Jedd G, Kumaran D, Swaminathan S, Shio H, Hewitt D, Chua NH, Swaminathan K. 2003. A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat. Struct. Biol. 10:264–270. 10.1038/nsb910 [DOI] [PubMed] [Google Scholar]
- 12.Tenney K, Hunt I, Sweigard J, Pounder JI, McClain C, Bowman EJ, Bowman BJ. 2000. Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31:205–217. 10.1006/fgbi.2000.1230 [DOI] [PubMed] [Google Scholar]
- 13.Bleichrodt RJ, van Veluw GJ, Recter B, Maruyama J, Kitamoto K, Wösten HA. 2012. Hyphal heterogeneity in Aspergillus oryzae is the result of dynamic closure of septa by Woronin bodies. Mol. Microbiol. 86:1334–1344. 10.1111/mmi.12077 [DOI] [PubMed] [Google Scholar]
- 14.Momany M, Richardson EA, Van Sickle C, Jedd G. 2002. Mapping Woronin body position in Aspergillus nidulans. Mycologia 94:260–266. 10.2307/3761802 [DOI] [PubMed] [Google Scholar]
- 15.Plamann M. 2009. Cytoplasmic streaming in Neurospora: disperse the plug to increase the flow? PLoS Genet. 5: e1000526. 10.1371/journal.pgen.1000526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berns MW, Aist JR, Wright WH, Liang H. 1992. Optical trapping in animal and fungal cells using a tunable, near-infrared titanium-sapphire laser. Exp. Cell Res. 198:375–378. 10.1016/0014-4827(92)90395-O [DOI] [PubMed] [Google Scholar]
- 17.Beck J, Echtenacher B, Ebel F. 2013. Woronin bodies, their impact on stress resistance and virulence of the pathogenic mould Aspergillus fumigatus and their anchoring at the septal pore of filamentous Ascomycota. Mol. Microbiol. 89:857–871. 10.1111/mmi.12316 [DOI] [PubMed] [Google Scholar]
- 18.Escaño CS, Juvvadi PR, Jin FJ, Takahashi T, Koyama Y, Yamashita S, Maruyama J, Kitamoto K. 2009. Disruption of the Aopex11-1 gene involved in peroxisome proliferation leads to impaired Woronin body formation in Aspergillus oryzae. Eukaryot. Cell 8:296–305. 10.1128/EC.00197-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. 10.1038/nature04300 [DOI] [PubMed] [Google Scholar]
- 20.Maruyama J, Kitamoto K. 2011. Targeted gene disruption in koji mold Aspergillus oryzae. Methods Mol. Biol. 765:447–456. 10.1007/978-1-61779-197-0_27 [DOI] [PubMed] [Google Scholar]
- 21.Jin FJ, Watanabe T, Juvvadi PR, Maruyama J, Arioka M, Kitamoto K. 2007. Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 76:1059–1068. 10.1007/s00253-007-1088-4 [DOI] [PubMed] [Google Scholar]
- 22.Mabashi Y, Kikuma T, Maruyama J, Arioka M, Kitamoto K. 2006. Development of a versatile expression plasmid construction system for Aspergillus oryzae and its application to visualization of mitochondria. Biosci. Biotechnol. Biochem. 70:1882–1889. 10.1271/bbb.60052 [DOI] [PubMed] [Google Scholar]
- 23.Liu F, Ng SK, Lu Y, Low W, Lai J, Jedd G. 2008. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J. Cell Biol. 180:325–339. 10.1083/jcb.200705049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nave R, Fürst DO, Weber K. 1989. Visualization of the polarity of isolated titin molecules: a single globular head on a long thin rod as the M band anchoring domain? J. Cell Biol. 109:2177–2187. 10.1083/jcb.109.5.2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai J, Koh CH, Tjota M, Pieuchot L, Raman V, Chandrababu KB, Yang D, Wong L, Jedd G. 2012. Intrinsically disordered proteins aggregate at fungal cell-to-cell channels and regulate intercellular connectivity. Proc. Natl. Acad. Sci. U. S. A. 109:15781–15786. 10.1073/pnas.1207467109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iakoucheva LM, Kimzey AL, Masselon CD, Bruce JE, Garner EC, Brown CJ, Dunker AK, Smith RD, Ackerman EJ. 2001. Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci. 10:560–571. 10.1110/ps.29401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uversky VN, Dunker AK. 2010. Understanding protein non-folding. Biochim. Biophys. Acta 1804:1231–1264. 10.1016/j.bbapap.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Oberhauser AF, Redick SD, Carrion-Vazquez M, Erickson HP, Fernandez JM. 2001. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. U. S. A. 98:10682–10686. 10.1073/pnas.191189098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labeit D, Watanabe K, Witt C, Fujita H, Wu Y, Lahmers S, Funck T, Labeit S, Granzier H. 2003. Calcium-dependent molecular spring elements in the giant protein titin. Proc. Natl. Acad. Sci. U. S. A. 100:13716–13721. 10.1073/pnas.2235652100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeil PL, Kirchhausen T. 2005. An emergency response team for membrane repair. Nat. Rev. Mol. Cell Biol. 6:499–505. 10.1038/nrm1665 [DOI] [PubMed] [Google Scholar]
- 31.Rauscher S, Pomès R. 2012. Structural disorder and protein elasticity. Adv. Exp. Med. Biol. 725:159–183. 10.1007/978-1-4614-0659-4_10 [DOI] [PubMed] [Google Scholar]
- 32.Hurley JM, Larrondo LF, Loros JJ, Dunlap JC. 2013. Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered Neurospora clock protein FRQ. Mol. Cell 52:832–843. 10.1016/j.molcel.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnhammer EL, Durbin R. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1–GC10 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.