Abstract
Fluorescence in situ hybridization (FISH) is a common technique for identifying cells in their natural environment and is often used to complement next-generation sequencing approaches as an integral part of the full-cycle rRNA approach. A major challenge in FISH is the design of oligonucleotide probes with high sensitivity and specificity to their target group. The rapidly expanding number of rRNA sequences has increased awareness of the number of potential nontargets for every FISH probe, making the design of new FISH probes challenging using traditional methods. In this study, we conducted a systematic analysis of published probes that revealed that many have insufficient coverage or specificity for their intended target group. Therefore, we developed an improved thermodynamic model of FISH that can be applied at any taxonomic level, used the model to systematically design probes for all recognized genera of bacteria and archaea, and identified potential cross-hybridizations for the selected probes. This analysis resulted in high-specificity probes for 35.6% of the genera when a single probe was used in the absence of competitor probes and for 60.9% when up to two competitor probes were used. Requiring the hybridization of two independent probes for positive identification further increased specificity. In this case, we could design highly specific probe sets for up to 68.5% of the genera without the use of competitor probes and 87.7% when up to two competitor probes were used. The probes designed in this study, as well as tools for designing new probes, are available online (http://DECIPHER.cee.wisc.edu).
INTRODUCTION
The use of the small subunit rRNA (SSU rRNA) as a phylogenetic marker for microbial classification, identification, and quantification was readily embraced after its discovery as a useful molecule to reconstruct microbial evolution (1). Fluorescence in situ hybridization (FISH), introduced in the late 1980s (2, 3), remains the technique of choice for cultivation-independent quantification of taxonomically relevant groups within microbial communities (4). Since its introduction, the wealth of knowledge surrounding microbial diversity has expanded tremendously, aided by rapid advances in DNA sequencing techniques. As a result, the SSU rRNA databases used for the early design of FISH probes were incomplete, and therefore, specificity and coverage may not be the same as originally thought for many probes that are still commonly used. For instance, Amann and Fuchs (4) reevaluated coverage and specificity of several group-specific probes more than 15 years after they were originally designed. They found that group coverage was generally smaller than the original expectation (e.g., 38 to 94%), and in most cases, phylogenetic groups outside the targeted group had the potential to cause false-positive hybridizations.
Problems with probe specificity are exacerbated when considering the hybridization of mismatched targets, for which the location and type of mismatch affect the strength of hybridization (5). Thermodynamic models that describe the hybridization of FISH probes to target sites with and without mismatches (6, 7) are helpful in identifying cross-hybridizations with nontargets, which may potentially be eliminated with competitor probes (8). However, the manual application of such models may be impractical in view of the large number of potential mismatches of concern when using modern databases, making it more difficult to select a sufficient set of competitor probes to use. These difficulties are amplified during the design of new probes when other factors such as probe length, nucleotide permutations, and multiple potential target sites are considered.
A catalog of FISH probes that have been designed and utilized throughout the years can be found in probeBase (9), and in the best scenario, a probe for the target group of interest may have already been designed and documented in the literature. In cases where de novo probe design is needed, a wide variety of design approaches are employed using software such as ARB (10), PRIMROSE (11), and mathFISH (6), as well as public databases such as SILVA (12) and RDP (Ribosomal Database Project) (13). These approaches often use a simplified subset of sequences to evaluate probe specificity, have different ways of predicting cross-hybridizations, and in many cases, require extensive experimental trial and error for probe optimization. Often potential target and nontarget organisms for a FISH probe are unculturable, which requires difficult optimization using a mixed community or Clone-FISH (14). High-throughput design approaches that minimize the amount of experimental optimization are needed to keep FISH at the forefront of microbial ecology as our awareness of microbial diversity continues to increase.
Group-specific FISH probes are typically hybridized under stringent conditions to minimize cross-hybridization with mismatched nontargets. The stringency of hybridization during FISH experiments is controlled by the concentration of formamide in the hybridization buffer, where more formamide will result in greater DNA/RNA denaturation. Increasing stringency during hybridization is a common tactic employed to minimize mismatched hybridization at the expense of reducing signal intensity from targeted organisms (i.e., reducing sensitivity). A practice that mitigates this sensitivity reduction is to block mismatched nontargets from hybridizing by using unlabeled competitor oligonucleotides, which dim or completely eliminate signal from nontargets (15). A third strategy is to require the hybridization of two different probes labeled with distinct fluorophores, as has been suggested previously (16–18). This strategy requires optimization of two different probes for simultaneous hybridization but may substantially reduce the number of nontargets. In this study, we explored these three strategies for the elimination of nontargets when using probes for genus-level identification.
Our first objective was to update the available mathematical models of FISH to improve the predictions of equilibrium formamide melting profiles. We characterized different models using cross-validation with several data sets of perfectly matched probes and an independent data set of mismatched probes. The best model was then used to systematically analyze the probe data set available in probeBase (9) and to update coverage and specificity for existing probes. Next, we developed a design tool that can optimize probe sensitivity, target group coverage, and probe specificity by two different approaches. The first approach depends on a single probe for identification, whereas the second approach uses two probes and the true identification is obtained from the hybridization overlap. The usefulness of this tool was demonstrated by massively designing genus-specific probes targeting every one of the 1,943 named genera in the RDP database. Our findings demonstrate that thermodynamics-based probe design can be automated to simultaneously optimize probe coverage, sensitivity, and specificity, while allowing the use of comprehensive rRNA databases to exhaustively evaluate perfect matches and potential mismatched cross-hybridizations.
MATERIALS AND METHODS
Microbial strains and growth conditions.
Xenorhabdus nematophila (ATCC 19061), Photorhabdus asymbiotica (ATCC 43949), Serratia marcescens (ATCC 13880), Aquabacterium parvum (ATCC BAA208), and Escherichia coli K-12 (ATCC MG1655) were used to form artificial communities in this study. The respective GenBank accession numbers for the 16S rRNA sequences of these five strains are D78009, Z76752, M59160, AF035052, and U00096 (gene rrsA). Strains grown aerobically in lysogeny broth (LB) (Sigma-Aldrich, St. Louis, MO) included X. nematophila at 30°C, P. asymbiotica at 28°C, S. marcescens at 25°C, and E. coli at 37°C. A. parvum was grown aerobically in R2A medium (Fisher Scientific, Waltham, MA) at 20°C. All cultures were harvested during mid-exponential growth phase (optical density at 600 nm of 0.3 to 0.4).
Flow cytometry FISH.
Fixation, hybridization, and washing steps of the flow cytometry protocol were carried out as described previously (19), with minor modifications. In brief, cell cultures were fixed with 3 volumes of 4% paraformaldehyde in phosphate-buffered saline (PBS) buffer (130 mM NaCl, 10 mM Na2HPO4) (pH 7.2) per volume cell culture. After 30 min, cells were centrifuged and resuspended in PBS to wash out fixative. Fixed cultures were then centrifuged again and stored in 50% ethanol and PBS solution at −20°C. Before hybridization, ethanol and PBS were removed, and cells were resuspended in 800 μl of hybridization buffer (1 M NaCl, 0.05 M EDTA, 20 mM Tris HCl [pH 8], 0.1% SDS, variable formamide concentrations) with 250 nM 5′-labeled probe. The following probes were used: Xeno-188 (Cy5 labeled, 5′-GCC ACC GTT TCC AGT GG) and Xeno-1279 (fluorescein labeled, 5′-AGG TCG CTT CTC TTT GTA TCY G). An unlabeled competitor oligonucleotide probe, with sequence (5′-AGG TCG CTT CAC TTT GTA TCY G) was used to block hybridization of Xeno-1279 to P. asymbiotica. All probes were synthesized by Integrated DNA Technologies (Coralville, IA). Samples were incubated overnight at 46°C in hybridization buffer. Excess probe was washed with hybridization buffer for 20 min at 46°C and then resuspended in PBS with 0.01% Tween 20 (Fisher Scientific) at 4°C.
Measurement of cell brightness was performed as previously described (20). In brief, three replicate hybridizations at each formamide concentration were measured using a FACSCalibur flow cytometer (Becton, Dickinson, San Jose, CA). A total of 10,000 events falling into the bacterial gate were collected, and probe brightness was determined from the channel corresponding to the mode of the smoothed fluorescence histogram (19). Negative controls were performed using the complement to the EUB probe (nonEUB; 5′-ACT CCT ACG GGA GGC AGC). Cy5- and fluorescein-labeled versions of the nonEUB probe were separately hybridized with the samples to determine the background for each dye. Net brightness was calculated by subtracting background fluorescence from each brightness measurement at the respective formamide concentration.
Microscopy FISH.
Microscopy FISH was performed following established protocols (21) on an artificial community composed of an equal mixture of the cultures listed above. In an additional set of experiments, FISH was performed with activated sludge collected from the Nine Springs Wastewater Treatment Plant (Madison, WI) on 3 April 2013. Microbial communities were filtered onto 0.22-μm Nuclepore filters (Millipore, Billerica, MA) and then hybridized in hybridization buffer (35% formamide, 0.9 M NaCl, 0.05 M EDTA, 20 mM Tris HCl [pH 8.0], 0.01% SDS) overnight at 46°C. After hybridization, excess probe was removed with wash buffer (80 mM NaCl, 0.05 M EDTA, 20 mM Tris HCl [pH 8.0], 0.01% SDS) for 20 min at 48°C. Samples were incubated in PBS for 15 min at room temperature and then mounted on a glass slide in Vectashield (Vector Laboratories, Burlingame, CA).
Samples were viewed using a Zeiss Axio Imager 2 (Zeiss, Oberkochen, Germany) with microscope settings (i.e., exposure time) held constant for each fluorescence channel across all samples. Representative images with the most even distribution and variety of cells were collected. Images were analyzed with ImageJ (22) by using the “Subtract Background” and “Color Balance” commands. Background was subtracted with a 500-pixel rolling ball radius using a sliding paraboloid. Color balance was used to normalize the three colors (Cy5, fluorescein, and 4′,6-diamidino-2-phenylindole [DAPI]), while keeping the same settings across all images.
RESULTS
Improved thermodynamic models for predicting formamide dissociation profiles.
The design of optimal probes requires reasonable predictions of probe sensitivity and specificity. To theoretically optimize probes, we have previously developed mechanistic models that predict probe affinity (20), formamide dissociation profiles for perfect-match targets (19), and offsets in these profiles due to mismatches (5). In this study, while we maintained the definition of probe affinity, we updated the parameters of the original formamide dissociation model, developed a new model for perfect matches, and used this new model to conservatively predict mismatch effects. These tools all feed into a probe design scheme that is depicted in Fig. 1A, summarized below, and described in detail in the supplemental material.
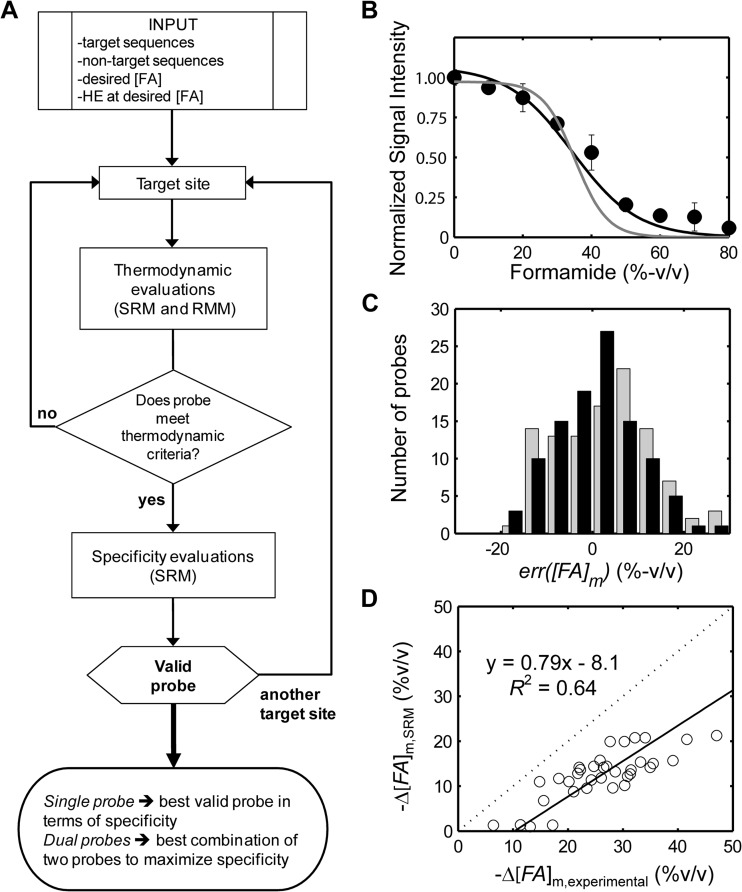
FIG 1.
Probe design algorithm and modeling. (A) Flow chart summary of design algorithm. The feedback loop on the right indicates the iterative procedure to find all possible probe candidates for a target, all of which are used in the output decision. See Fig. S6 in the supplemental material for a more detailed representation. [FA], concentration of formamide; HE, hybridization efficiency. (B) Example predictions (probe St796-813) based on leave-one-probe-out cross validation (LOPOCV) (supplemental material) for the single-reaction model (SRM) (black) and retrained mechanistic model (RMM) (gray). Experimental data are included as data points with standard deviations (error bars). See Fig. S1 for all plots. (C) Distribution of the melting prediction errors during LOPOCV for SRM (black bars) and RMM (light gray bars). The majority of probes are associated with predictions of less than 10% formamide error for both models. SRM errors are smaller by about 1.4% formamide on average, and they are more normally distributed due to improvements in the model's goodness of fit. (D) Correlation of experimental and theoretical offsets in melting points due to incorporation of a single mismatch. The equation indicates that SRM is a conservative predictor of mismatch stability, with the experimental offset being higher than predicted.
Our initial model of formamide dissociation calculated the hybridization efficiency of FISH probes based on the effect of formamide on three free energy values representing the reactions for probe-target duplex formation, probe folding, and target folding (19). This original model was calibrated and validated using 27 probes, all targeting E. coli. In the current study, we expanded the model training set, with probes from another study (23), yielding a total of 106 probes targeting five different organisms (see Table S1 in the supplemental material). This extended data set was used to retrain the original model and update the parameters that describe the linear relationship between free energy changes and formamide concentration (19). The new parameters defined the retrained mechanistic model (RMM) and were consistent with the confidence intervals provided in the original model (Table S2), thereby validating the approach. However, predicted curves were still steeper than experimental profiles (Fig. 1B; see Fig. S1 in the supplemental material), although formamide melting point ([FA]m) predictions were within 10% of experimental observations in most cases (Fig. 1C).
To improve predictions of formamide denaturation in FISH, we adopted a modeling approach recently used for microarray hybridizations (24). In this approach, hybridization is represented by a single reaction. The duplex sequence determines the free energy change of this reaction based on specific nearest neighbor rules that represent average values for all nucleic acid interactions taking place, and these rules are to be determined as modeling parameters. Although this approach provided better fits (see Table S2 in the supplemental material), the addition of 17 different DNA/RNA nearest neighbor parameters (Table S3) overparameterized the model. We therefore developed a single-reaction model (SRM) that converted the free energy change predicted from regular DNA/RNA nearest neighbor rules to a FISH-specific free energy using a linear relationship with only two parameters (Table S2). This model captured the slope of experimental profiles better than RMM (Fig. 1B and Fig. S1) and also had smaller errors in the prediction of melting points (Fig. 1C). However, since this model excluded probe and target folding free energies, it cannot predict probe affinity, which is the overall free energy change of hybridization obtained using probe-specific free energy values. Technically, SRM can be applied only to probes that have been designed a priori to achieve a reasonable level of probe affinity, as was the case with all of the probes used in model training (Table S1). Thus, our probe design scheme (Fig. 1A) uses both SRM and RMM, with the former predicting hybridization efficiency of the probe with the target and the latter providing a checkpoint based on all three free energy changes related to FISH (19).
Next, we sought a model to predict the effect of mismatches on melting point. Due to the large number of different mismatch conformations (24) compared to available data sets, it was not possible to develop models that can predict dissociation profiles with specific mismatch parameters for FISH. Instead, conservative predictors of the offset in melting point upon mismatch insertion (Δ[FA]m) are preferred (5). Using a previously established data set of melting points for mismatched probes (5), we checked the predictive ability of SRM against other predictors (see supplemental material). While the predictive power of SRM was not significantly better than any other model, it offered the advantage of not requiring the calculation of the free energy of target folding, which made the computation of Δ[FA]m orders of magnitude faster than the equivalent computation with RMM. In addition, SRM is a conservative predictor, since it systematically underestimates the experimental offsets (Fig. 1D). Accordingly, we defined qualitative thresholds based on Δ[FA]m calculated by SRM (Δ[FA]m,SRM) as follows. Nontargets can be considered at very high risk of hybridization if Δ[FA]m,SRM is more than −5%, high risk if Δ[FA]m,SRM is between −10% and −5%, moderate risk if Δ[FA]m,SRM is between −15% and −10%, low risk if Δ[FA]m,SRM is between −20% and −15%, and no risk if Δ[FA]m,SRM is below −20%.
Overall, the mathematical modeling background of the new probe design tool was established with improved and efficient predictors that reflect the best of our knowledge. Since SRM was selected as the predictor for perfect matched and mismatched probes, we integrated it into a program for computing free energy, formamide melting point, hybridization efficiency, and Δ[FA]m,SRM. This tool, named ProbeMelt, has been made accessible online on the DECIPHER website (http://DECIPHER.cee.wisc.edu/ProbeMelt.html). Given a set of probe and target sequences, ProbeMelt will return their hybridization efficiencies at different levels of stringency, with mismatched target sites color-coded by their risk of cross-hybridization. This output can then be downloaded and easily used for plotting formamide denaturation curves. As we show in the next section, ProbeMelt can be used in combination with comprehensive database searches to efficiently identify potential targets and nontargets.
In silico analysis of FISH probes in probeBase.
Having established an updated model for perfect match hybridizations and risk levels for cross-hybridization of mismatched hybrids based on Δ[FA]m,SRM, we next performed an in silico evaluation of published FISH probes. For this, we downloaded (13 August 2013) the set of all 816 probes that had been “tested for FISH” available from probeBase (9). For a subset of probes that had no nucleotide permutations and included the formamide concentration used experimentally (649 probes), we computed the differences between the predicted melting point (using ProbeMelt with perfectly matched targets) and the experimental formamide concentration. The difference was approximately normally distributed with a mean of 6.5% and a standard deviation of 14%. This indicated that probes are generally hybridized about 7% formamide below the melting point, where stringency is reasonably high and the target's brightness is between the maximum (100%) and half-maximum (50%). The large standard deviation was expected, given the considerable variability in experimental procedures and objectives, as well as prediction error.
Next we asked whether probes available in literature had reasonably high coverage (arbitrarily defined here as >75%) of their intended target group. We identified 138 probes in probeBase that were designed specifically for targeting named taxonomic groups present in the RDP database (version 10.28). For each of these probes and their target groups, we calculated the fraction of sequences containing the target site for which Δ[FA]m,SRM was −10% to 0%. That is, we estimated coverage taking into account perfect matches as well as stable mismatches within the target group. Using this method, we identified 15 probes with low coverage (<50%) and 15 probes with moderate coverage (50 to 75%) of their stated target group (see Table S4 in the supplemental material). Thus, the in silico analysis showed that 22% of probes in probeBase targeting the SSU rRNA of common taxonomic groups have less than desirable coverage. Many of these probes were designed more than 10 years ago when rRNA databases were substantially smaller and before many taxonomic redefinitions took place. Other probes in this group have been designed more recently, and therefore, the lack of sufficient coverage may represent either an inaccurate annotation or a systematic bias resulting from specific probe design approaches.
One of the main limitations in the design of FISH probes is the impracticality of experimentally testing all possible nontargets that may have the potential to cross-hybridize. We propose that ProbeMelt can be used to programmatically test potential false-positive results in silico, thus helping in the probe design process as a tool for screening and detecting cross-hybridizations. To illustrate the benefit of the model in this regard, we created color-coded phylogenetic trees to graphically represent the in silico predictions of probe hybridization to different genera (see Fig. S9 in the supplemental material). In this analysis, we used the set of 677 SSU rRNA-targeting probes from probeBase having only a single permutation (i.e., no degeneracy). For each probe, we scored the predicted level of hybridization to members of each genus by first identifying target sequences in the RDP database that had a chance of forming either perfectly matched or mismatched hybrids. We then multiplied the fraction of the genus represented by these target sequences with the calculated chance of hybridization, which was defined as a linear value from 0 (Δ[FA]m,SRM of less than or equal to −15%) to 1 (Δ[FA]m,SRM = 0% [perfect match]). The sum of these products for all the potential targets within a genus became a weighted score for the chance that the probe would hybridize with members of the genus, a metric that was useful to graphically represent the potential hybridization of probes to each genus in the phylogenetic tree (Fig. S9).
Out of the 677 probes analyzed, 178 probes had no in silico prediction of hybridization with more than 1% coverage to any named genus in the RDP database, which reflects the high number of probes designed to target very specific subgroups within a genus or groups that are unclassified in the RDP database. For other probes, an assessment of whether the probe is adequate for the specific identification of the targeted group can be obtained by comparing the graphical representations in Fig. S9 in the supplemental material with the intended targets. For instance, probe Nso1225, originally designed to target ammonia-oxidizing bacteria in the Nitrosomonas and Nitrosospira genera within the Betaproteobacteria (25), is predicted to have a low to moderate chance of hybridization with many other genera within and outside the proteobacteria, and a high chance of hybridization with members of the Bacteroidetes phylum (Fig. S9). Examples of other probes for which the in silico predictions indicated a significant discrepancy between the targeted genera and the predicted hybridizations are found in the supplemental material (Table S5).
Integrated automatic probe design.
To facilitate the design of new probes consistent with modern databases, we integrated SRM into a program that can optimize probes' coverage, specificity, and sensitivity, as well as evaluate a large number of nontarget groups for potential cross-hybridizations (Fig. 1A). This program for the design of FISH probes, described in detail in the supplemental material, has been made available as part (DesignProbes function) of the DECIPHER package (26) for R (27) and also online as the Design Probes tool (http://DECIPHER.cee.wisc.edu/DesignProbes.html). The objectives of the probe design program are to (i) use the thermodynamic principles formulated in SRM and RMM to design probes with high affinity to the target sequences, (ii) use multiple permutations per probe to maximize coverage of the targeted group, (iii) use the thermodynamic principles of formamide denaturation embedded in SRM to comprehensively detect nontarget sequences with the potential for cross-hybridization, and (iv) evaluate the use of two probes targeting the same group as a way to further maximize specificity.
The program accepts user-defined target and nontarget groups, and it can also search for potential nontarget cross-hybridizations in a comprehensive rRNA database (available online at http://DECIPHER.cee.wisc.edu/Download.html). Both 16S and 23S rRNA comprehensive databases are available for use in finding additional nontargets during probe design. In cases where a single probe is insufficient to achieve the desired level of specificity, the program is able to search the space of all combinations of dual probes to find the probe set with minimal cross-hybridization overlap. In a dual-probe experiment, the two probes would be labeled with different fluorophores and the overlap in fluorescence signal would be considered a positive identification.
For an example, we applied the program to design genus-specific probes for all 1,943 named genera in the RDP database (version 10.30) encompassing 1,696,150 SSU rRNA sequences (13). We designed probes specifically targeting each genus, with all 1,942 other genera declared as nontarget sequences. For input parameters, we chose to design probes (with up to 4 permutations allowed) that represent at least 90% of sequences classified as belonging to each genus. The lengths of the probes were adjusted to achieve at least 50% hybridization efficiency at standard FISH conditions: 46°C and 35% formamide. Initially, to estimate an upper bound on the number of genera for which it may be possible to completely prevent cross-hybridization, we performed a hypothetical simulation that assumed any mismatch completely blocked hybridization. By using a single probe, potential false-positive results could be prevented for 55.6% of genera, and with dual probes, the number increased to 76.3% of genera. This hypothetical demonstration epitomized the difficulty of designing genus-specific probes due to strong conservation of the 16S rRNA gene between closely related genera and the presence of polyphyletic genera in the database.
We next asked whether the actual probe design would result in probes near the hypothetical maximum and whether it was generally possible to design adequate probes for genus-level identification. Using a Δ[FA]m,SRM of greater than or equal to −20% as the threshold for potential cross-hybridizations, with a single probe it was possible to find probes with no false-positive results for only 13.4% of genera, and 35.4% of genera if up to 5 false-positive genera were permitted. With the dual-probe approach, the fraction rose substantially, with the ideal probe set having no cross-hybridizations for 35.5% of genera, and 64.1% of genera when 5 false-positive genera were allowed. Since a Δ[FA]m,SRM of greater than or equal to −20% is a conservative estimate of specificity, we repeated this analysis using Δ[FA]m,SRM of greater than or equal to −10% (only high-risk nontargets) as qualification for a potential false-positive result. With the relaxed definition of specificity, 25.6% of single probes had no predicted false-positive results and 58.5% were usable if 5 cross-hybridizations are allowed. For dual probes, these numbers increased to 50.1% and 81.5%, respectively.
Next, we incorporated the possibility of using two competitor oligonucleotides to block hybridization of mismatched nontargets with a high risk of cross-hybridization (defined as mismatches having Δ[FA]m,SRM of greater than or equal to −10%). Using the competitors, it was possible to design probes with no predicted false-positive genera for 35.6% of genera with a single probe and 60.9% of genera with dual probes. If up to 5 cross-hybridizations were allowed, adequate single probes could be identified for 68.5% of genera with one probe and 87.7% of genera with dual probes. The fraction of genera with adequate dual probes was comparable to that obtained with recently published in silico designs of genus-specific 16S primers for PCR (28). These results demonstrated the challenges associated with designing probes at the genus level, the advantage of using two probes over a single probe for identification, and the considerable benefit obtained from the use of competitor oligonucleotides. All of these predesigned genus-specific probes and the recommended competitor probes to achieve maximum specificity are available in the DECIPHER 16S Oligos database online (http://DECIPHER.cee.wisc.edu/16SOligos.html).
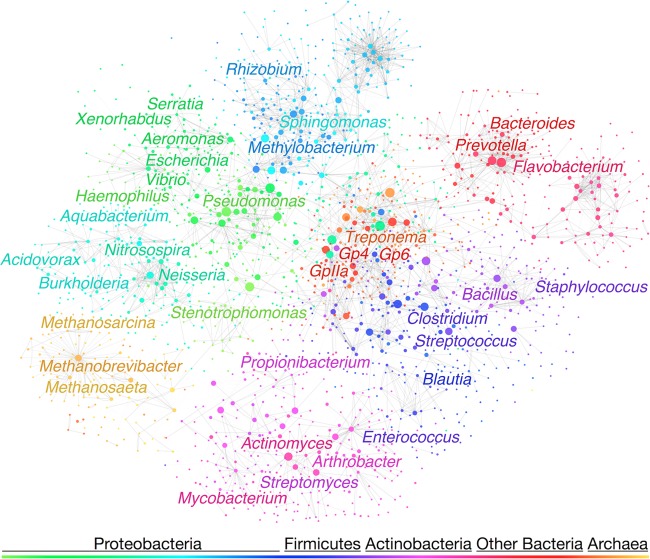
As a final analysis, we constructed networks representing the cross-hybridization of probes targeting each genus with other nontarget genera. Figure 2 shows the dual-probe network, where each node represents probes targeting a single genus, and the edges represent the cross-hybridization between the 1,943 genera. The network was visualized by using the magnitude of Δ[FA]m along each edge to guide a force-directed layout. Although the network's layout is based solely on its connectivity, the network structure is clearly organized by phylogenetic relationship as shown by like colors grouping together. To measure the network density, we calculated the average number of neighbors per node, which is the sum of all outgoing and incoming cross-hybridizations with other genera. The dual-probe network was substantially sparser (P < 1e−15 by the Mann-Whitney U test) than the network representing single-probe designs (not shown), having an average number of neighbors equal to 44 versus 101 for the single-probe network. This analysis illustrated the increased probability of cross-hybridization between related organisms and the large increase in specificity obtained by using two probes.
FIG 2.
Dual-probe cross-hybridization network visualized with Cytoscape (v2.8.3). Nodes of the network represent the probes designed to target each genus in the RDP database. Nodes are colored by their phylogenetic group and sized according to their number of potential cross-hybridizations with other genera. Edges represent cross-hybridizations with a high risk of causing a false-positive identification (Δ[FA]m,SRM of more than −10%). Labeled genera have more than 5,000 sequences in the RDP database (version 10.28).
Dual probes designed by the algorithm successfully distinguish targets from potential nontargets.
We experimentally evaluated with flow cytometry and microscopy one set of probes designed by the algorithm. We chose the proteobacterium X. nematophila as the target, because it is a model organism and had easily cultured nontargets for each of the dual probes. The dual-probe design output (see Fig. S7 in the supplemental material) for the genus Xenorhabdus was the Xeno-188 (5′-GCC ACC GTT TCC AGT GG) and Xeno-1279 (5′-AGG TCG CTT CTC TTT GTA TCY G) probes, which were labeled with Cy5 and fluorescein, respectively. This dual-probe set was predicted to have 10 potential overlapping cross-hybridizations, in contrast to the most specific single probe available for Xenorhabdus, which was predicted to have 30 false-positive genera (Fig. S8).
We constructed an artificial community of nontargets by mixing four different proteobacteria: A. parvum, E. coli, P. asymbiotica, and S. marcescens. Table 1 shows that both E. coli and S. marcescens had two mismatches to the Xeno-188 probe (Δ[FA]m,SRM of −23% for both; no risk of cross-hybridization predicted) and only one mismatch to the Xeno-1279 probe (Δ[FA]m,SRM of −10% and −11%, respectively; moderate cross-hybridization risk). In contrast, P. asymbiotica had no mismatches to Xeno-188 and only a single mismatch to Xeno-1279 (Δ[FA]m,SRM of −6%; high risk), which made it a candidate false-positive result even when both probes were used together. The distantly related A. parvum was added to the community as a negative control with 7 mismatches to Xeno-188 and 9 mismatches to Xeno-1279, resulting in very large Δ[FA]m,SRM magnitudes. The observed formamide melting curves were in agreement with the model's predictions (Fig. 3). Furthermore, use of an unlabeled competitor oligonucleotide probe almost completely blocked hybridization of Xeno-1279 with P. asymbiotica (Fig. 3B).
TABLE 1.
Target sites for probes targeting the artificial community
| Species | Sequence of the Xeno-188 target site (5′ to 3′)a,c | Xeno-188 probe |
Sequence of the Xeno-1279 target site (5′ to 3′)b,c | Xeno-1279 probe |
||
|---|---|---|---|---|---|---|
| Δ[FA]m,SRM | RFU (%) (35% FA)d | Δ[FA]m,SRM | RFU (%) (35% FA)d | |||
| X. nematophila | CCACTGGAAACGGTGGC | 0 | 79 | CRGATACAAAGAGAAGCGACCT | 0 | 71 |
| A. parvum | A...GTC.T..AA.... | −66 | N/A | .C.G....G..G.CT..C.A.C | −60 | N/A |
| E. coli | .T............A.. | −23 | 0.4 | ..C................... | −10 | 1.2 |
| P. asymbiotica | ................. | 0 | 37 | ...........T.......... | −6 | 15 |
| S. marcescens | .T............A.. | −23 | 0.3 | ..T................... | −11 | 4.9 |
Probe labeled with 5′-Cy5.
Probe labeled with 5′-fluorescein. R stands for A or G.
The nucleotides that are different from those in the X. nematophila sequence are shown; nucleotides that are identical to those in the X. nematophila sequence are indicated by dots.
Relative fluorescence units (RFU) at 35% formamide (FA) relative to the maximum X. nematophila fluorescence on the entire formamide curve (Fig. 3). N/A, not available.
FIG 3.
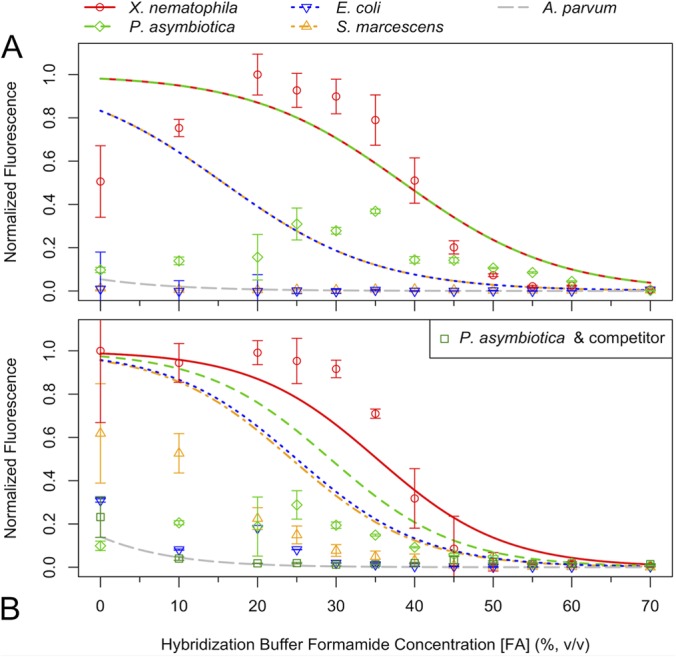
Denaturation curves illustrating the relationship between model predictions and experimental data. The Cy5-labeled Xeno-188 probe (A) and the fluorescein-labeled Xeno-1279 probe (B) were hybridized separately with four different organisms. Points show the median cell fluorescence values determined with a flow cytometer and then normalized to the maximum value observed in each channel. Error bars show the standard deviations from three replicate hybridizations. The model predictions (lines) adequately describe the melting points but do not reflect the plateau of each curve. Model predictions for X. nematophila and P. asymbiotica are superimposed in panel A, because Δ[FA]m,SRM is zero. S. marcescens and E. coli are superimposed in panel A, because they have the same Δ[FA]m,SRM. The addition of a competitor oligonucleotide probe effectively blocked hybridization of P. asymbiotica with Xeno-1279 in panel B.
Figure 4A and B show images of cells hybridized with both probes and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). X. nematophila (target) cells are easily identifiable by the white color resulting from superimposition of strong signals from DAPI and the two probes. Cells hybridized only with the Xeno-188 probe and DAPI appear purple (P. asymbiotica), whereas cells with signals from the Xeno-1279 probe and DAPI appear green (E. coli and S. marcescens), and cells not hybridizing to either probe appear blue (A. parvum). These results were expected based on the model predictions (Table 1) and formamide curves (Fig. 3), except for P. asymbiotica, which did not require an unlabeled competitor oligonucleotide to block hybridization of Xeno-1279. Here P. asymbiotica could hybridize to both probes, but the fluorescence signal from the perfect match Xeno-188 outweighed the signal from the mismatched Xeno-1279, resulting in purple cells. For this reason, the competitor probe tested with flow cytometry was not required for microscopy FISH.
FIG 4.
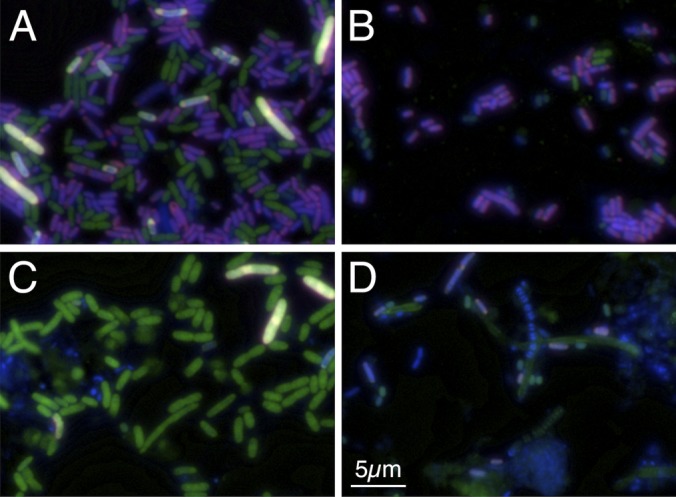
Dual probes successfully distinguish the target X. nematophila from predicted nontargets and wastewater sludge. Separate probes labeled with Cy5 (red channel) and fluorescein (green channel) were hybridized using an artificial community with four nontargets with (A) or without (B) the target organism (X. nematophila). The target appears white, while nontargets appear purple if they hybridized to only the Xeno-188 probe, green if they hybridize to only the Xeno-1279 probe, and blue (DAPI) if they hybridize to neither. To confirm the specificity of the probes with a wide array of organisms, the experiment was repeated using activated sludge from a wastewater treatment plant with (C) and without (D) adding in the target organism.
Additional tests with and without spiking X. nematophila into an activated sludge sample were also conducted. Activated sludge is an ideal negative control for the dual probes, because it contains a wide range of organisms, including many members of the family Enterobacteriaceae, which is the family of the target genus, yet likely has a negligible abundance of Xenorhabdus cells, which are obligate symbionts of the nematode Steinernema (29). In the absence of X. nematophila (Fig. 4D), we detected cells that separately hybridized to each of the two probes and cells that were not hybridized, but there were no cells that simultaneously hybridized with both probes, indicating that Xenorhabdus cells were not present in the activated sludge sample. In the spiked samples (Fig. 4C), the X. nematophila cells were clearly seen as hybridized with both probes, further demonstrating the advantage of using dual probes for the specific detection of organisms at the genus level.
DISCUSSION
While the use of single probes to detect specific microorganisms with FISH is commonplace, our analysis of genus-level probes demonstrates that significant increases in specificity can be obtained by using dual probes. This concept is not novel, as the use of multiple probes without a nested hierarchy has previously been proposed and used as a strategy to improve confidence levels of target detection (16–18). However, to our knowledge, this is the first study that systematically compares specificity when using single or dual probes. Furthermore, we present here the first computational tool for automated design of FISH probes that effortlessly incorporates target identification using dual probes.
Also, in this study we improved the prediction of formamide curves for perfectly matched hybrids in comparison to earlier models (19) by using new thermodynamic calculations and data sets with multiple organisms. We also used the computationally efficient model (SRM) to evaluate and classify cross-hybridizations according to thermodynamically based calculations of mismatch effects. This model offers a departure from other probe design programs that estimate potential cross-hybridizations based on the number of mismatches (11) or qualitative weighted scores (10). Thus, our model allows, for the first time, the systematic application of thermodynamic principles to evaluate the specificity of existing probes or to optimize the design of new probes. Our analysis predicted a much larger space of potential false-positive hybridizations for existing probes, an observation that was expected given the rapid expansion of the rRNA databases (30) and has been confirmed in specific situations (7), but to our knowledge has never been systematically evaluated.
As specificity can be improved with the application of dual probes, a desired condition is for both probes to have similar [FA]m values so that the benefits of increased specificity can be achieved with a single hybridization. If the probes have [FA]m values that are too different from each other, then successive hybridizations would be required (31). Thus, one of the advantages of mathematical modeling is to maximize the probability that a single hybridization will work, as the dual probes are designed to have similar melting points. More importantly, perfect match predictions should be accurate enough to identify true positive results with high confidence. To evaluate the confidence in modeling predictions, we performed statistical analyses (see supplemental material), which revealed that dual probes designed with SRM are expected to provide reasonable confidence in the identification of target organisms in more than two-thirds of all cases. Nonetheless, in silico predictions cannot completely substitute experimental evaluations of formamide curves. Thus, in agreement with common FISH practice (4, 32), experimental formamide curves should be obtained for both probes to determine the difference in experimental [FA]m values and establish the [FA]m errors of each probe. The final advantage of mathematical modeling is the determination of potential false-positive results based on mismatch thermodynamics. This not only minimizes the chance of cross-hybridizations but also provides a list of organisms (i.e., likely false-positive results) to use competitors against or to choose from during experimental optimization with pure cultures.
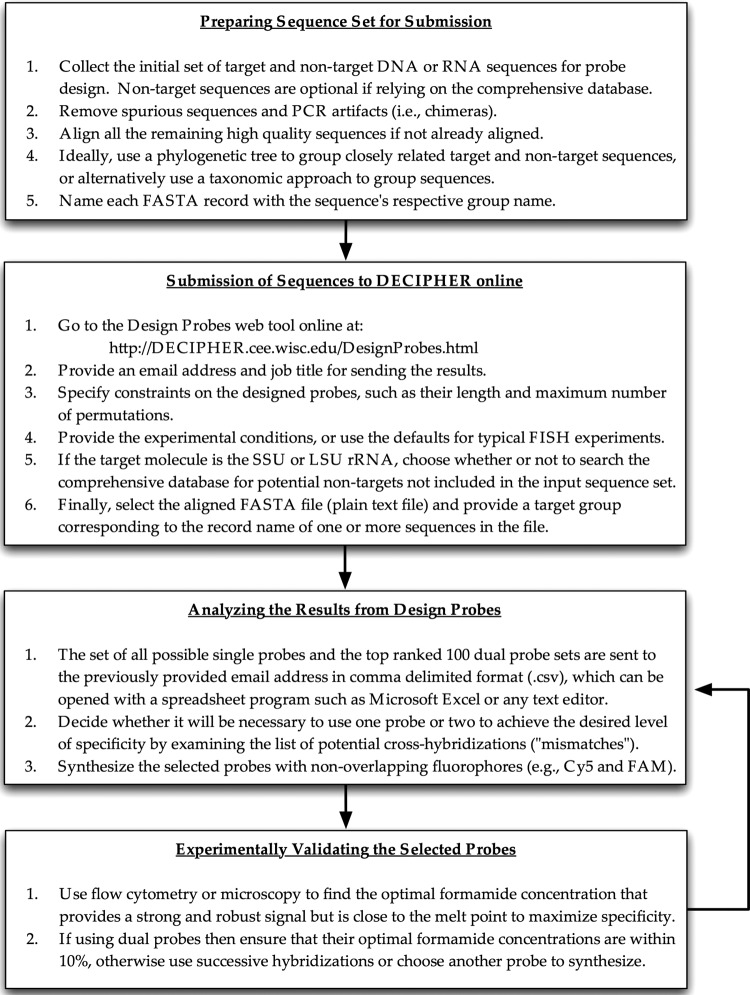
The overall probe design strategy that results from our model development and subsequent analyses is summarized in Fig. 5, which depicts the design and verification of new probes from the user's perspective. The user must first select target and nontarget sequences to consider during design. Here the user may take a phylogeny-based or a taxonomy-based approach to define the target and nontarget groups. Optionally, the user may consider letting the program find the nontarget groups of concern in a comprehensive reference database, which will employ a taxonomy-based grouping of nontargets, as we used here in the design of the genus-specific 16S Oligos. The user also specifies the desired formamide concentration at which the hybridizations will be carried out, as well as the minimum hybridization efficiency desired at the selected formamide concentration. This is important, since users may choose to hybridize with nonstringent buffers in order to maximize probe signal when the targeted organisms are expected to have low ribosomal content. After submission, the program will return lists of single and dual probes ranked by specificity. The user should carefully consider the outputs to decide whether single or dual probes will be necessary to achieve the desired level of specificity.
FIG 5.
Flow chart depicting how to design and validate probes step by step with the help of the Design Probes tool online. LSU, large subunit; FAM, 6-carboxyfluorescein.
After the selected probes are synthesized, it is necessary to generate experimental formamide curves for perfectly matched targets with the samples of interest. In some cases, pure cultures would be available, but in other cases, formamide curves need to be obtained using the mixed culture that contains the organisms of interest. Formamide curves are useful in determining the melting point, comparing model predictions with experimental observations, and deciding on an appropriate hybridization stringency for targets that may have low maximal brightness. Target organisms with low brightness should be hybridized at a formamide concentration closer to the point of maximum brightness, whereas very bright nontargets can be hybridized close to their melting point. Low-risk nontargets (Δ[FA]m,SRM between −15% and −20%) should be more carefully considered when choosing to hybridize significantly below the melting point. In the case of dual probes, if the experimental [FA]m values of the two probes are within 10% of each other, then the probe set is adequate for hybridization near the lower [FA]m. In other cases, the user has the option to go back to the design step, select a different set from already designed probes, or use two different formamide concentrations in successive hybridizations (31).
Running formamide curves for all potential cross-hybridizations is usually prohibitive, either because representatives are not available in pure culture, there are too many potential cross-hybridizations, or it is nearly impossible to obtain adequate mismatched formamide curves for nontargets in mixed cultures (except if there are substantial differences in morphology). Here the design outputs can be helpful in two ways. First, cross-hybridizations are classified according to risk, while taking into account the effects of mismatches, insertions, and deletions. Second, the design outputs include a dual-probe option that drastically reduces the list of potential cross-hybridizations so that the user can have a reduced set of cross-hybridizations to focus their attention on and potentially design competitor probes against. One aspect that is not included in the model but that works to the benefit of the user is that in many mismatched hybrids there is a reduction in the maximum signal obtained, and therefore, some predicted cross-hybridizations will have low maximum fluorescence in practice (Fig. 3).
Our high-throughput application in this study focused on designing probes at the genus level, but the same approach could be applied at lower or higher taxonomic levels. Inherent challenges at higher taxonomic levels may include needing a larger number of permutations to achieve reasonable coverage of the target group and maintaining high sensitivity across a wide variety of organisms, since probe signal at the same target site can vary significantly among organisms (33). To demonstrate probe design at higher taxonomic levels, we designed probes for all phyla in the RDP database while maintaining the same constraints used in genus-level probe design. The program was able to design probes for all 39 phyla except Euryarchaeota, OD1, and OP11, which contained too much sequence diversity to achieve 90% coverage with a maximum of four probe permutations. However, specificity was considerably lower than for genus-level probes, with many nontarget genera detected as potential cross-hybridizations in all cases. Reassuringly, four of the probes designed by the algorithm closely corresponded to the small number of phylum-specific probes already available in probeBase (see Table S6 in the supplemental material).
In conclusion, using the approach provided by this new model and the advantages of dual probes, FISH protocols can now be systematically designed with high sensitivity to the targeted group and higher specificity than previously obtainable. To this end, we have provided three online tools: (i) ProbeMelt for generating denaturation curves and quickly identifying potential nontargets of previously designed probes, (ii) 16S Oligos for FISH, which is a preprocessed database of genus-specific single and dual probes, and (iii) Design Probes for designing new probes targeting a user-defined set of sequences while minimizing cross-hybridization with other user-provided sequences and/or a comprehensive sequence database. For those users that prefer to use the stand-alone DECIPHER program for R, the functions CalculateEfficiencyFISH and DesignProbes are described in the downloadable documentation, including an extensive example of probe design in the vignette “Designing Group-Specific FISH Probes” available online. The main difference between the website and stand-alone program for probe design is that the latter assumes some experience with R, allows design of probes for multiple target groups at one time, and allows greater flexibility in the number and definition of nontarget groups in a comprehensive sequence database. It is our hope that these tools will enable new research in environmental microbiology by simplifying the accurate design of highly sensitive and specific probes.
Supplementary Material
ACKNOWLEDGMENTS
This research was partially supported by National Science Foundation grant CBET-0606894.
We thank Rowan Meara for conducting some of the preliminary experiments that were not shown herein, Heidi Goodrich-Blair for providing X. nematophila and P. asymbiotica isolates, Sri Ram for constructive feedback on the manuscript, and the Center for High Throughput Computing for providing computing resources.
Footnotes
Published ahead of print 13 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01685-14.
REFERENCES
- 1.Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 74:5088–5090. 10.1073/pnas.74.11.5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeLong EF, Wickham GS, Pace NR. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360–1363. 10.1126/science.2466341 [DOI] [PubMed] [Google Scholar]
- 3.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R, Fuchs BM. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6:339–348. 10.1038/nrmicro1888 [DOI] [PubMed] [Google Scholar]
- 5.Yilmaz LS, Bergsven LI, Noguera DR. 2008. Systematic evaluation of single mismatch stability predictors for fluorescence in situ hybridization. Environ. Microbiol. 10:2872–2885. 10.1111/j.1462-2920.2008.01719.x [DOI] [PubMed] [Google Scholar]
- 6.Yilmaz LS, Parnerkar S, Noguera DR. 2011. mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl. Environ. Microbiol. 77:1118–1122. 10.1128/AEM.01733-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McIlroy SJ, Tillett D, Petrovski S, Seviour RJ. 2011. Non-target sites with single nucleotide insertions or deletions are frequently found in 16S rRNA sequences and can lead to false positives in fluorescence in situ hybridization (FISH). Environ. Microbiol. 13:33–47. 10.1111/j.1462-2920.2010.02306.x [DOI] [PubMed] [Google Scholar]
- 8.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593–600. 10.1016/S0723-2020(11)80121-9 [DOI] [Google Scholar]
- 9.Loy A, Maixner F, Wagner M, Horn M. 2007. probeBase–an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 35:D800–D804. 10.1093/nar/gkl856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A, Lai T, Steppi S, Jobb G, Förster W. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371. 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashelford KE, Weightman AJ, Fry JC. 2002. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 30:3481–3489. 10.1093/nar/gkf450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schramm A, Fuchs BM, Nielsen JL, Tonolla M, Stahl DA. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713–720. 10.1046/j.1462-2920.2002.00364.x [DOI] [PubMed] [Google Scholar]
- 15.Kirschner AKT, Rameder A, Schrammel B, Indra A, Farnleitner AH, Sommer R. 2012. Development of a new CARD-FISH protocol for quantification of Legionella pneumophila and its application in two hospital cooling towers. J. Appl. Microbiol. 112:1244–1256. 10.1111/j.1365-2672.2012.05289.x [DOI] [PubMed] [Google Scholar]
- 16.Amann R, Ludwig W. 2000. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev. 24:555–565. 10.1111/j.1574-6976.2000.tb00557.x [DOI] [PubMed] [Google Scholar]
- 17.Fieseler L, Horn M, Wagner M, Hentschel U. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724–3732. 10.1128/AEM.70.6.3724-3732.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer KH. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yilmaz LS, Noguera DR. 2007. Development of thermodynamic models for simulating probe dissociation profiles in fluorescence in situ hybridization. Biotechnol. Bioeng. 96:349–363. 10.1002/bit.21114 [DOI] [PubMed] [Google Scholar]
- 20.Yilmaz LS, Noguera DR. 2004. Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Appl. Environ. Microbiol. 70:7126–7139. 10.1128/AEM.70.12.7126-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. 2003. An improved protocol for quantification of freshwater actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928–2935. 10.1128/AEM.69.5.2928-2935.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 23.Okten HE, Yilmaz LS, Noguera DR. 2012. Exploring the in situ accessibility of small subunit ribosomal RNA of members of the domains Bacteria and Eukarya to oligonucleotide probes. Syst. Appl. Microbiol. 35:485–495. 10.1016/j.syapm.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 24.Yilmaz LS, Loy A, Wright ES, Wagner M, Noguera DR. 2012. Modeling formamide denaturation of probe-target hybrids for improved microarray probe design in microbial diagnostics. PLoS One 7:e43862. 10.1371/journal.pone.0043862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mobarry BK, Wagner M, Urbain V, Rittmann BE, Stahl DA. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright E. 2013. DECIPHER: Database Enabled Code for Ideal Probe Hybridization Employing R. R package version 1.10.0. http://www.bioconductor.org/packages/release/bioc/html/DECIPHER.html
- 27.R Core Team. 2013. R: a language and environment for statistical computing, 3rd ed. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 28.Wright ES, Yilmaz LS, Ram S, Gasser JM, Harrington GW, Noguera DR. 2014. Exploiting extension bias in polymerase chain reaction to improve primer specificity in ensembles of nearly identical DNA templates. Environ. Microbiol. 16:1354–1365. 10.1111/1462-2920.12259 [DOI] [PubMed] [Google Scholar]
- 29.Goodrich-Blair H. 2007. They've got a ticket to ride: Xenorhabdus nematophila–Steinernema carpocapsae symbiosis. Curr. Opin. Microbiol. 10:225–230. 10.1016/j.mib.2007.05.006 [DOI] [PubMed] [Google Scholar]
- 30.Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. 2008. probeCheck - a central resource for evaluating oligonucleotide probe coverage and specificity. Environ. Microbiol. 10:2894–2898. 10.1111/j.1462-2920.2008.01706.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner M, Amann R, Kämpfer P, Assmus B, Hartmann A, Hutzler P, Springer N, Schleifer K-H. 1994. Identification and in situ detection of Gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405–417. 10.1016/S0723-2020(11)80058-5 [DOI] [Google Scholar]
- 32.Wagner M, Horn M, Daims H. 2003. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr. Opin. Microbiol. 6:302–309. 10.1016/S1369-5274(03)00054-7 [DOI] [PubMed] [Google Scholar]
- 33.Behrens S, Fuchs BM, Mueller F, Amann R. 2003. Is the in situ accessibility of the 16S rRNA of Escherichia coli for Cy3-labeled oligonucleotide probes predicted by a three-dimensional structure model of the 30S ribosomal subunit? Appl. Environ. Microbiol. 69:4935–4941. 10.1128/AEM.69.8.4935-4941.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.