Abstract
The peptidyl nucleoside arginomycin is active against Gram-positive bacteria and fungi but displays much lower toxicity to mice than its analog blasticidin S. It features a rare amino acid, β-methylarginine, which is attached to the deoxyhexose moiety via a 4′-aminoacyl bond. We here report cloning of the complete biosynthetic gene cluster for arginomycin from Streptomyces arginensis NRRL 15941. Among the 14 putative essential open reading frames, argM, encoding an aspartate aminotransferase (AAT), and adjacent argN, encoding an S-adenosyl methionine (SAM)-dependent methyltransferase, are coupled to catalyze arginine and yield β-methylarginine in Escherichia coli. Purified ArgM can transfer the α-amino group of l-arginine to α-ketoglutaric acid to give glutamate and thereby converts l-arginine to 5-guanidino-2-oxopentanoic acid, which is methylated at the C-3 position by ArgN to form 5-guanidino-3-methyl-2-oxopentanoic acid. Iteratively, ArgM specifically catalyzes transamination from the donor l-aspartate to the resulting 5-guanidino-3-methyl-2-oxopentanoic acid, generating β-methylarginine. The complete and concise biosynthetic pathway for the rare and bioactive amino acid revealed by this study may pave the way for the production of β-methylarginine either by enzymatic conversion or by engineered living cells.
INTRODUCTION
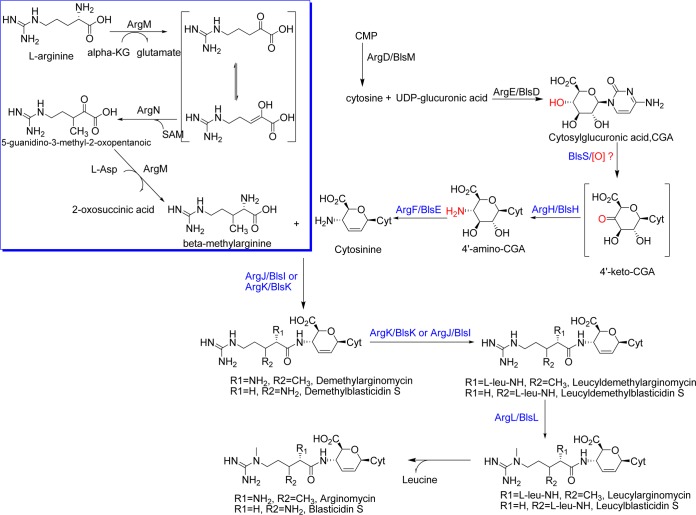
Arginomycin (Fig. 1) is a peptidyl nucleoside antibiotic isolated from Streptomyces arginensis NRRL 15941 that shows bioactivity against Gram-positive bacteria and fungi, such as Micrococcus luteus and Penicillium oxalicum (1). Although the overall skeleton of arginomycin is highly similar to that of blasticidin S (BS) (Fig. 1), a representative peptidyl nucleoside antibiotic exhibiting strong inhibitory activity against rice blast caused by Pyricularia oryzae Cavara (2), arginomycin showed much lower toxicity to mice (1). They differ from each other only in the modification of the l-arginine-derived guanidino side chain (Fig. 1). A feeding experiment indicated that β-arginine in BS is derived from an intramolecular amino migration of α-arginine (3). BlsG, an arginine 2,3-aminomutase homologous to lysine 2,3-aminomutase (4), was assumed to govern this migration in the BS biosynthesis pathway. In contrast, δ-N-methylation was demonstrated to be the ultrabiosynthetic step, as the conversion of demethylblasticidin S to BS was observed in a cell extract of the BS producer Streptomyces griseochromogenes supplied with radiolabeled S-adenosyl-l-methionine (SAM) (5). Likewise, δ-N-methylation of the guanidyl group of arginomycin should also occur after attachment of the guanidino side chain to the nucleoside core moiety. In addition to this, arginomycin bears at the relatively unreactive β position of arginine the β-methylarginine moiety for which β-methylation is a commitment step and is of considerable interest to this study in uncovering new enzymology. There are two possibilities for the timing of the β-methylation of arginine: (i) l-arginine is first converted to free β-methylarginine and is coupled with the nucleoside and (ii) β-methylation occurs after incorporation of l-arginine into the nucleoside. β-Methylarginine, a rare amino acid, was revealed as a constituent of the peptide lavendomycin from Streptomyces lavendulae subsp. brasilicus (6). Recently, it was isolated as a free biocontrol toxin from Pseudomonas syringae pv. Syringae 22d/93, which can strongly inhibit the growth of plant pathogen P. syringae pv. Glycinea, a relative of the producer (7). A biosynthesis study demonstrated that three clustered genes, mrsA, mrsB, and mrsC, can produce β-methylarginine in Escherichia coli. A catalytic assay of MrsA, a methyltransferase, can convert 5-guanidino-2-oxo-pentanoic acid into 5-guanidino-3-methyl-2-oxo-pentanoic acid in vitro (8). The resulting product was proposed as a promising substrate for MrsB, an aminotransferase, to give the final product β-methylarginine, which was discharged from the cell by the exporter MrsC.
FIG 1.
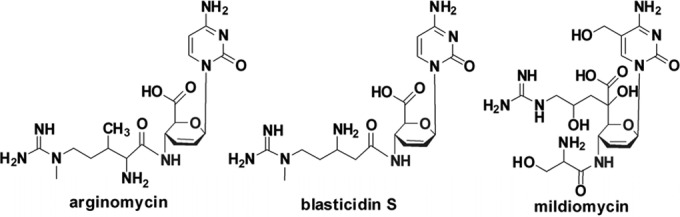
Chemical structures of arginomycin, blasticidin S, and mildiomycin. They all share the cytosylglucuronic acid moiety, which was derived from cytosine and UDP-glucuronic acid.
In this study, the arginomycin biosynthetic gene cluster was cloned and heterologously expressed in Streptomyces coelicolor M145. Comparison of its biosynthetic gene cluster to blasticidin S unveiled that two arginomycin-specific genes, namely, argM and argN, are involved in the biosynthesis of β-methylarginine. Particularly, we demonstrated that ArgM, a pyridoxal phosphate (PLP)-dependent aspartate aminotransferase, along with ArgN, a SAM-dependent methyltransferase, can produce β-methylarginine in E. coli. The complete biosynthetic pathway of β-methylarginine was demonstrated by in vitro catalysis in which ArgM catalyzes two rounds of transaminations in an iterative way by using l-arginine and l-aspartate as the donors of the amino group and by using α-ketoglutaric acid and 5-guanidino-3-methyl-2-oxopentanoic acid as the receptors. The ArgN can transfer one molecule of the methyl group from SAM to 5-guanidino-2-oxo-pentanoic acid to give 5-guanidino-3-methyl-2-oxopentanoic acid.
MATERIALS AND METHODS
Strains and culture.
Strains used in this study are listed in Table S3 in the supplemental material. S. arginensis NRRL 15941 was obtained from the Agricultural Research Service, U.S. Department of Agriculture (http://www.ars.usda.gov/). S. arginensis NRRL 15941 was grown at 30°C on mannitol soy flour medium (SFM; 2% mannitol, 2% soy flour, 2% agar) agar plates for harvest of spores or production of arginomycin in 10.3% TSBY (TSB liquid medium supplemented with sucrose [10.3%, wt/vol] and yeast extract [1%, wt/vol]). All E. coli strains were grown in liquid Luria-Bertani (LB) medium or LB agar medium at 37°C. Conjugation was performed, and exconjugants were cultivated in SFM agar plates containing an appropriate antibiotic (apramycin, 50 mg ml−1). For arginomycin fermentation and analysis from S. arginensis NRRL 15941, the seed culture was prepared by inoculating stock culture into a 250-ml baffled flask containing seed medium (blackstrap molasses [5.8 g], Difco peptone [10 g], Difco yeast extract [4 g], dextrin [4 g], l-asparagine [0.2 g], CoCl2·6H2O [1 mg], and tap water [1 liter]; the mixture was adjusted to pH 7.2 before being autoclaved). The flask was incubated in a rotary shaker for 48 h at 30°C (220 rpm), and the seed culture of 5 ml was transferred into a 500-ml flask incubated with fermentation medium (glucose monohydrate [20 g], soybean meal [20 g], brewer's yeast [2 g], tap water [1 liter]; the mixture was adjusted to pH 7.2 before being autoclaved). The culture was incubated under the same conditions for another 7 days. The authentic arginomycin was obtained by large-scale preparation of fermentation and confirmed by nuclear magnetic resonance (NMR) and high-resolution electrospray ionization mass spectrometry (HR-ESI-MS).
General molecular biology methods.
PCR amplifications were performed on a Veriti thermal cycler (Applied Biosystems, Carlsbad, CA) using Taq DNA polymerase purchased from TaKaRa Co. Ltd. (Dalian, China) or Q5 high-fidelity DNA polymerase purchased from New England BioLabs. Restriction enzymes and DNA ligase were purchased from Fermentas (Thermo Fisher Scientific Inc.) or NEB. A gel extraction kit was purchased from Omega Company (USA). Unless otherwise stated, other biochemicals and chemicals were purchased from standard commercial sources. All DNA manipulations in E. coli and Streptomyces were performed according to standard procedures. Cosmid DNA sequencing was accomplished at the Majorbio Biotech Co. Ltd. (Shanghai, China). All primers used in this work were synthesized by Sangong Biotech Co. Ltd. (Shanghai, China) and are listed in Table S1 in the supplemental material.
LC-MS analysis.
The fermentation broth was extracted with ethyl acetate twice, and the water phase was concentrated in vacuo. The residue was dissolved in 0.5 ml methanol, filtered, and centrifuged. The supernatant was then subjected to high-performance liquid chromatography MS (HPLC-MS) analysis. The mobile phase used for HPLC-MS was comprised of solvent A (H2O containing 20 mM ammonium acetate) and solvent B (CH3CN) at a ratio of 90:10 (vol/vol). Analytical HPLC was performed on an Agilent series 1100 HPLC using a Zorbax 300SB C18 column (250 by 4.6 mm, 5-μm particle size; Agilent), and the flow rate was 0.3 ml min−1 at 25°C. UV detection at 272 nm and MS analysis (ESI) were carried out in the positive mode.
Genomic library construction, screening, and sequence analysis.
A genomic library of S. arginensis NRRL 15941 was constructed according to a standard protocol with a CopyControl fosmid library production kit. About 600 clones were picked up and transferred into 96-well microtiter plates containing 120 μl LB medium supplemented with chloramphenicol (12.5 μg ml−1) and incubated overnight at 37°C; 120 μl 40% glycerol was added to store the samples for 24 h at −80°C. The clones in 96-well microtiter plates were screened by using primers Argscr-For and -Rev (see Table S1 in the supplemental material), which were designed according to the conserved regions of CGA genes. Assignments of open reading frames (ORFs) and their functional predictions were accomplished with the ORF finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), FramePlot 4.0 beta online program (http://watson.nih.go.jp/∼jun/cgi-bin/frameplot-3.0b.pl), and BLAST program (http://blast.ncbi.nlm.nih.gov/). Multiple nucleotide sequence alignments and analyses were performed using the BioEdit sequence alignment editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) or Vector NTI (Invitrogen). The deduced biosynthetic gene cluster is shown in Table S3.
Expression and purification of ArgM and ArgN.
The argM and argN genes were amplified from the total DNA of S. arginensis NRRL 15941 with the primers listed in Table S1 in the supplemental material. The PCR product was cloned into the SK+ vector for sequencing. argM and argN were then excised by NdeI and EcoRI digestion and ligated with the corresponding sites of pET-28a for protein expression. The resulting plasmid was transformed into the Escherichia coli BL21(DE3)/pLysS expression strain supplied with kanamycin (50 μg ml−1) and chloramphenicol (20 μg ml−1). Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration at 0.2 mM when the cell optical density at 600 nm reached 0.6 to 0.8. The culture was then induced at 16°C for another 20 to 24 h. Cells were harvested by centrifugation at 6,000 rpm for 10 min, and the pellets were kept at −80°C until use.
For in vivo tandem protein vector construction, argM and argN were digested with NdeI and EcoRI and ligated into the corresponding sites of the PCT vector, which was derived from pET-28a, in which the XbaI site was mutated and its isocaudomer, a SpeI site with an end compatible to XbaI, was added. First, the 1.2-kb argM fragment was digested with NdeI and EcoRI and then ligated with the same site of PCT to yield the PCT-ArgM fusion protein (pFJ5). The PCT-ArgN (pFJ6) fusion protein was constructed using the same method as that used for pFJ5. The ArgM-PCT was digested with XbaI and SpeI, ligated into the XbaI site of ArgN-PCT to yield the tandem expression fusion protein PCT-ArgM-ArgN (pFJ7), and then transferred into E. coli BL21(DE3)/pLysS.
For in vitro assay and ArgM, ArgN, and TyrB purification, cells were resuspended in lysis buffer (50 mM HEPES, pH 8.0) and lysed using a Microfluidizer (model JY92-II DN; Ningbo Scientz Biotechnology Co., Ltd.); the cell debris were removed by centrifugation at 12,000 rpm for 60 min, and the supernatant was incubated with 5 ml of Ni-nitrilotriacetic acid (NTA) resin (Invitrogen) preequilibrated with lysis buffer. The resin was loaded onto a polypropylene column and washed with lysis buffer until there was no UV absorption at 280 nm. The column was washed successively with wash buffer (50 mM HEPES, 300 mM NaCl, 50 mM imidazole, 10% glycerol, pH 8.0) before we obtained the target His6-tagged protein with elution buffer (50 mM HEPES, 300 mM NaCl, 500 mM imidazole, 10% glycerol, pH 8.0). The resulting protein was concentrated with an ultracentrifugation filter onto a YM-10 membrane (Millipore), the protein purity was analyzed by 12% SDS-PAGE, and the concentration was determined with a NanoDrop 2000 spectrophotometer (Thermo Scientific).
In vivo assay of the transamination and methylation activities of the E. coli cell extract.
To confirm whether ArgM and coupled ArgM and ArgN were capable of converting arginine into 5-guanidino-2-oxopentanoic acid and 5-guanidino-3-methly-2-oxopentanoic acid into β-methylarginine, respectively, we constructed the expression vectors pFJ5 (argM) and pFJ7 (argM and argN). l-Arginine was added into the fermentation medium for E. coli cells containing pFJ5 and pFJ7 after 48 h. The fermentation lasted for another 6 h (37°C, 220 rpm), and the mixture was spun down at 12,000 rpm for 5 min. The supernatant was filtered with 0.22-μm filters (Millipore) and transferred and derivatized with dansyl chloride (DNS-Cl) to allow UV detection.
In vitro enzymatic assay.
For the in vitro ArgM assay, the 100-μl assay mixture containing 1 mM arginine, 0.5 mM pyridoxal phosphate (PLP), 1 mM α-ketoglutaric acid (α-KG), and 15 μM ArgM in 50 mM HEPES buffer (pH 8.0) was incubated at 25°C for 3 h. The reaction mixture with the boiled ArgM was used as a negative control. The reactions were quenched by 5% trifluoroacetic acid (TFA). The protein-free reaction was derivatized with DNS-Cl under dark conditions for 1 h and quenched by ethylamine (10%, vol/vol); the mixture was spun down, and the supernatant was subjected to HPLC analysis.
For coupled assays (ArgM and ArgN, and ArgM and TyrB plus ArgN), the mixture contained 1 mM arginine, 0.5 mM PLP, and 1 mM α-KG in the presence of 15 μM enzyme. For detection of the second amino donor, four amino group donors (l-glutamic acid, l-glutamine, l-aspartic acid, l-asparagine) were added to the reaction mixture at a final concentration of 1 mM. The reaction mixture was incubated at 25°C for 3 h and terminated by TFA. The protein-free reaction was derivatized with DNS-Cl for 1 h and detected by LC-MS. Three inorganic donors for the amino group were added to the mixture at a concentration of 1 mM.
Structural elucidation of the product.
For 5-guanidino-2-oxopentanoic acid and 5-guanidino-3-methyl-2-oxopentanoic acid detection, HR-ESI-MS was used. To elucidate the structure of the final product, β-methylarginine, the peaks corresponding to putative DNS-βmArg were collected with an Agilent semipreparative column (Zorbax octadecylsilyl [ODS], 5-μm particle size, 9.4 by 250 mm). The column had its detector set at 275 nm, and products eluted for 30 min in a linear gradient of 2% to 40% acetonitrile with 20 mM ammonium acetate at a flow rate of 1.5 ml min−1. The same fragments were combined, concentrated, and then analyzed for their HR-ESI-MS spectra by the 1H and 13C NMR methods. For NMR analysis, the frozen sample was transferred into a 5-mm NMR tube. The NMR data, including 1H, 13C, and 1H-1H COSY and heteronuclear single quantum coherence (HSQC) spectra were recorded using Agilent instruments (1H frequency was at 500 MHz, which corresponds to a 13C frequency of 125 MHz). J values are expressed in parts per million relative to in hertz.
Nucleotide sequence accession number.
The DNA sequences for fosmid 3H10 insertion were deposited in the NCBI database under the GenBank accession number KC181124.
RESULTS AND DISCUSSION
Cloning and heterologous expression of the arginomycin biosynthetic gene cluster from Streptomyces arginensis NRRL 15941.
Arginomycin, blasticidin S, and mildiomycin share the core moiety cytosylglucuronic acid (CGA), which is catalyzed by the CGA synthase (BlsD/MilC) via coupling of the UDP-glucuronic acid and free cytosine or its analog (see Fig. S1A in the supplemental material) (9, 10). Degenerate primers SCR-For and -Rev (Table S1) were designed based on the multiple DNA sequence alignments with milC, blsD, and other homologous genes from the database (Fig. S1B). A 550-bp PCR product was amplified from Streptomyces arginensis NRRL 15941 (Fig. S2), and the deduced polypeptide of this DNA fragment shares 38% identity and 50% homology with BlsD and 48% identity and 57% homology with MilC. Within this DNA fragment, one nested primer pair, Argscr-For and -Rev (Table S1), was used to screen the genomic library of S. arginensis NRRL 15941. Two overlapping fosmids, 3F10 and 5H10, were identified to contain a milC- and blsD-homologous gene.
Sequencing of fosmid 3F10 revealed 29 ORFs in a continuous 31.1-kb insertion (Fig. 2). The orientations and deduced functions of these ORFs are detailed in Table S2 in the supplemental material. To verify whether the identified fosmid harbors all essential genes for arginomycin biosynthesis, a 29-kb DNA fragment spanning from orf1 to approximately orf9 was excised from 3F10 by HpaI and XbaI treatment and subsequently cloned into the integrative plasmid pSET152 to give pFJ1, which was introduced into S. coelicolor M145 by conjugation (Table S3); fermentation broth of the resultant mutant, FJ1, gave a peak with a retention time of 14.2 min, which is the same as that of authentic arginomycin in the liquid chromatography (LC) profile (sample I in Fig. 3). MS analysis of this peak from the extract of FJ1 (sample IV in Fig. 3) gives a dominant ion at [M + H]+ 437.2, which corresponds to the mass of protonated arginomycin. Concordant tandem mass spectroscopy (MS/MS) analysis of this ion revealed two fragmented ions at [M + H]+ 326 and at [M + H]+ 185, which separately correspond to the masses of the moieties indicated in Fig. S3 in the supplemental material. Heterologous production of arginomycin in S. coelicolor demonstrated that all essential genes are included in the 29-kb DNA fragment.
FIG 2.
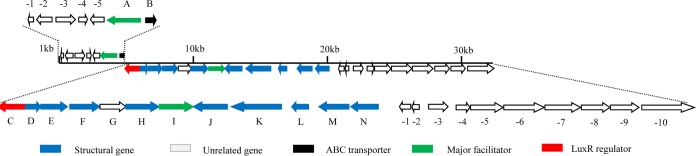
Arrangement of biosynthetic gene clusters for arginomycin by S. arginensis NRRL 15941. The colored arrows show the essential ORFs. The ABC transporter and major facilitator may encode self-resistance to arginomycin.
FIG 3.
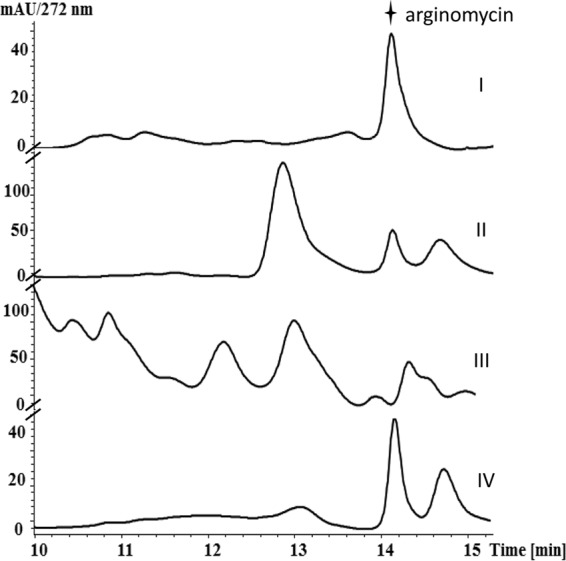
HPLC detection of heterologous production of arginomycin in S. coelicolor M145. I, arginomycin standard; II, fermentation broth of the arginomycin producer S. arginensis NRRL 15941; III, fermentation broth of S. coelicolor M145::pSET152 as a negative control; IV, fermentation broth of S. coelicolor M145::pFJ1, which encodes orf1 to approximately -9. Arginomycin is labeled with a star in the LC profile.
Comparative analysis of the biosynthetic pathways of arginomycin and blasticidin S. (i) Genes responsible for the generation of cytosinine.
Cytosinine was previously isolated as an intermediate during blasticidin S biosynthesis (11, 12) and postulated to be a common precursor for arginomycin and blasticidin S, given their structural similarity. Based on the BS biosynthetic pathway, four pairs of homologous counterparts, BlsM/ArgD, BlsD/ArgE, BlsH/ArgH, and BlsE/ArgF (13) (see Table S2 in the supplemental material), are assumed to be involved in the biosynthesis of this intermediate. Their putative roles in arginomycin and blasticidin S biosynthesis are shown in Fig. 4. Cytosinine biosynthesis includes the promising oxidation of 4′-hydroxyl CGA to 4′-keto CGA, which is the substrate for the subsequent transamination reaction. However, oxidoreductase or dehydrogenase counterparts are lacking from both of two gene clusters. A single oxidoreductase, BlsS, that is essential for BS biosynthesis (9) was postulated to govern this step; GouA in gougerotin is an arabinose dehydrogenase-like protein with little homology to BlsS that was proposed to be in charge of the oxidation of 4′-hydroxyl CGA (14). In contrast, no oxidoreductase/dehydrogenase gene(s) can be identified in either the arginomycin or the mildiomycin biosynthetic gene cluster. Two phosphatases, MilE and MilQ, that are essential for mildiomycin biosynthesis in vivo (10) can catalyze C-α–C-β dehydration on the guanidino side chain of mildiomycin in vitro (unpublished data). This dehydration seems unnecessary in mildiomycin biosynthesis and is thus regarded as a side reaction. A plausible functional assignment for MilE and/or MilQ might be their conversion of 4′-hydroxyl CGA to 4′-amino CGA, but the corresponding protein governing this step in arginomycin remains obscure. This observation reflects the possibility that the oxidation of 4′-hydroxyl CGA involves versatile biochemistry. In addition, BlsH, a degT dnrJ eryC1 strS aminotransferase homologous to ArgH, was proposed to be responsible for conversion of the 4′-keto CGA to 4′-amino CGA (15).
FIG 4.
Comparison of proposed biosynthesis pathways for arginomycin and blasticidin S. The counterpart genes are marked with black (known function) and blue (uncertain function) for arginomycin and blasticidin S.
(ii) Attachment and further modification of the β-methylarginine side chain.
One unique feature of arginomycin's structure is β-methylation at the guanidino side chain. It remains unclear whether this methylation occurs prior to or after attachment of arginine to the 4′-amino group of the cytosinine. This study provided solid evidence for the formation of free β-methylarginine (the following paragraphs will detail this result). With respect to the attachment of β-methylarginine to the 4′-amino group of the hexose via an atypical amide bond, ArgJ (a homolog of BlsI encoding a putative ligase) and ArgK (a homolog of BlsK bearing homology to lysyl-tRNA synthetase) might be related. Both blsI and blsK are essential to BS biosynthesis (9), which involves the formation of two amide bonds. As leucylblasticidin S (LBS) was proved to be the penultimate compound in the blasticidin S biosynthetic pathway (12), two enzymes were required to activate and attach leucine and β-arginine/or leucyl-β-arginine to BS. Given that ArgJ and ArgK are homologous to BlsI and BlsK, respectively, it is proposed that there also exists a leucylation of the α-amino group of the β-methylarginine in arginomycin biosynthesis (Fig. 4). Another pair of homologous counterparts for two antibiotics is BlsL/ArgL, both of which show clear similarity to guanidino methyltransferase from Solanum lycopersicum. This methylation is proposed to occur immediately following the leucylation of the β-amino group to yield LBS in the BS biosynthesis pathway (9). Accordingly, we propose that ArgL may catalyze the methylation of the guanidyl group in a similar way.
ArgM transfers the amino group from l-arginine to α-ketoglutaric acid.
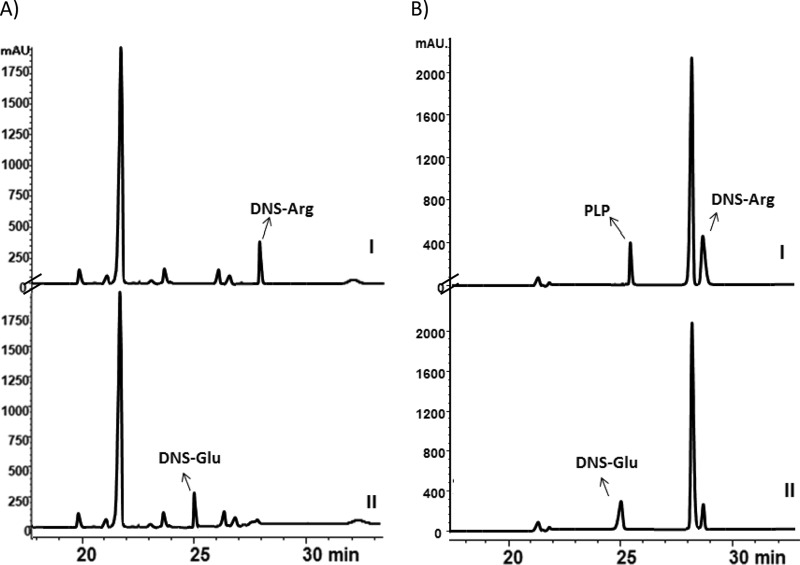
Arginomycin contains a unique β-methylarginine moiety that is of considerable interest to us. Extensive work by Huang et al. revealed that TyrB, an S-adenosyl methionine (SAM)-dependent methyltransferase, and MppJ, an (S)-aromatic-amino acid aminotransferase, can convert phenylalanine to the (2S,3S)-β-methylphenylalanine moiety, which requires PLP and SAM, found in the mannopeptimycin biosynthetic pathway (16). Two genes in the arginomycin biosynthesis gene cluster encode ArgM with 30% identity and 43% similarity to TyrB, and ArgN with 25% identity and 37% similarity to MppJ. They have no homologous genes in the biosynthetic gene clusters of its structurally similar analog blasticidin S and therefore are proposed to be involved in the formation of β-methylarginine. ArgM and ArgN were individually expressed and purified from E. coli. We first tested their in vivo transamination activities with E. coli expressing ArgM, as the l-arginine has no UV absorption, so all the amino acids in the extract of E. coli cells were derivatized with dansyl chloride (DNS-Cl) and subjected to HPLC analysis. No DNS-Arg could be detected, but DNS-Glu was detected in the argM-expressing strain (Fig. 5A and S4). The failure of detection of l-arginine in the extract of E. coli cells may be attributed to the conversion of l-arginine to 5-guanidino-2-oxopentanoic acid, which could not be modified by DNS-Cl, but it was detected by high-resolution electrospray ionization MS (HR-ESI-MS) (Fig. S5). The presence of DNS-Glu implied that α-ketoglutaric acid may serve as the receptor for the amino group from l-arginine. To further verify this assumption, l-arginine and α-ketoglutaric acid were directly measured as the substrates for purified ArgM. DNS-Glu was again detected in the HPLC spectra (Fig. 5B), and MS data also support the existence of 5-guanidino-2-oxopentanoic acid in the reaction mixture (data not shown). These in vivo and in vitro results demonstrated that the amino group of the arginine was transferred to the α-ketoglutaric acid to form glutamate. Some arginine analogs, such as d-arginine, l-homoarginine, and N-methylarginine) together with α-ketoglutaric acid were individually assayed as the substrate for ArgM; conversion of any of these compounds was not observed, showing that ArgM is specific for natural l-arginine (Fig. S6) and also providing evidence that the final product may have a (2S)-amino configuration.
FIG 5.
Biochemical characterization of ArgM. (A) HPLC traces of in vivo ArgM. I represents the fermentation broth of BL21(DE3)/pLysS:pET-28a, II represents the fermentation broth of BL21(DE3)/pLysS:pFJ2. (B) HPLC traces of a reaction mixture of ArgM in an in vitro assay. I represents a boiled-ArgM-catalyzed reaction; II represents an ArgM-catalyzed reaction.
ArgM and ArgN in vivo can produce β-methylarginine.
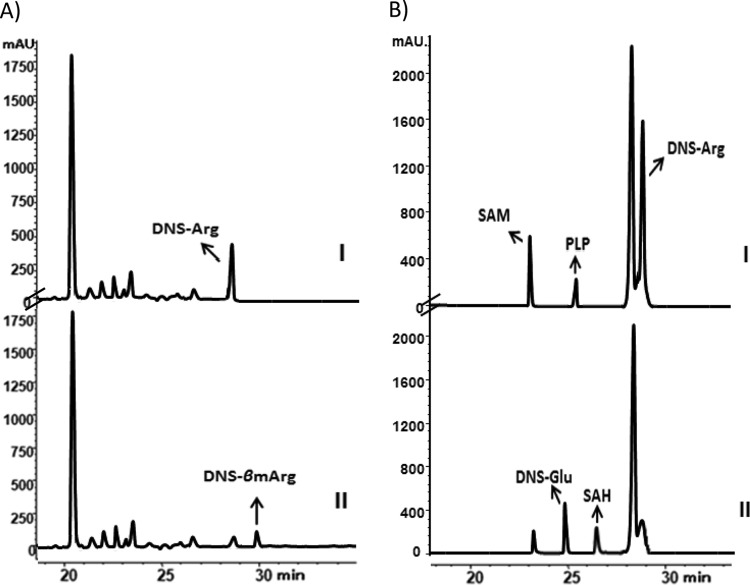
To confirm whether coupled ArgM and ArgN are enough to catalyze the formation of β-methylarginine, argM and argN genes were constructed and expressed in pET28-derived pFJ7 in a tandem manner (see Fig. S7A and B in the supplemental material). First, l-arginine was assayed as the substrate by using cell extract of BL21(DE3)/pFJ7. β-Methylarginine formation was detected in the HPLC spectrum (Fig. 6A) and further confirmed by HR-ESI-MS, in which the ions of fragments at 408.0 and 422.1 corresponded to DNS-Arg (incomplete conversion) and DNS-β-methylarginine (DNS-βmArg), respectively (Fig. S8). In addition, the proposed intermediate 5-guanidino-3-methyl-2-oxopentanoic acid was also detected by HR-MS assay (Fig. 4 and S9). This result supports the hypothesis that coupled ArgM and ArgN were capable of converting l-arginine to β-methylarginine in vivo.
FIG 6.
Biochemical characterization of coupled ArgM and ArgN reactions. (A) HPLC traces of in vivo ArgM and ArgN. I represents the fermentation broth of BL21(DE3)/pLysS:pET-28a, II represents the fermentation broth of BL21(DE3)/pLysS:pFJ7. (B) HPLC traces in vitro of coupled ArgM and ArgN. I represents the boiled-ArgM and -ArgN reaction. II represents the coupled ArgM and ArgN reaction.
ArgM transfers the amino group from l-aspartate to the 5-guanidino-3-methyl-2-oxopentanoic acid that is catalyzed by ArgN to form β-methylarginine.
When we used purified ArgM and ArgN, along with α-ketoglutaric acid, PLP, and SAM, to measure their activities toward l-arginine, surprisingly, no DNS-βmArg was detected in the HPLC spectra (Fig. 6B). However, the DNS-Glu and S-adenosylhomocysteine (SAH) from SAM were detected. The inconsistency between the in vivo catalysis and the in vitro catalysis by ArgM and ArgN raised two possibilities: (i) another aminotransferase in E. coli participated in the transamination from glutamate to 5-guanidino-3-methyl-2-oxopentanoic acid and (ii) the amino group donor is other than glutamate in the second transamination for ArgM. We first introduced a transaminase from E. coli BL21(DE3), named TyrB, which often converts tyrosine to 4-aminophenylpyruvate in the primary metabolism (17–19). tyrB from BL21(DE3) was amplified, cloned, and expressed, and the purified TyrB was coupled with ArgM and ArgN to measure their activities toward l-arginine. However, no β-methylarginine could be detected by either LC or MS (see Table S4 in the supplemental material). We then turned to the second possibility. Three inorganic amino group donors (ammonium sulfate, ammonium chloride, and ammonium acetate) and four amino acids (l-glutamate, l-glutamine, l-aspartate, and l-asparagine) were individually tested in the reaction mixture containing α-ketoglutaric acid, l-arginine, SAM, PLP, ArgM, and ArgN. HPLC spectral analysis showed that β-methylarginine was generated exclusively when l-aspartate was included in the reaction mixture (Fig. S10). This result coincides with the functional prediction of ArgM as an aspartate aminotransferase-like protein.
Stereoselectivity of the methyl group catalyzed by ArgN.
In order to confirm the structure of β-methylarginine generated by ArgM and ArgN in vitro and provide information for the stereoisomerism of the β-methyl group introduced by ArgN, correlation spectroscopy nuclear magnetic resonance (COSY NMR) was used to test the two sets of H · H correlations for the protons of the α-carbon and the β-methyl group in βmArg. The reaction was carried out in the presence of l-arginine, α-ketoglutaric acid, l-aspartate, SAM, PLP, ArgM, and ArgN at 25°C for 3 h. Reactions were derivatized with DNS-Cl and quenched by 10% ethylamine, and reactions mixtures were subjected to an Agilent semipreparative column (Zorbax ODS, 5-μm particle size, 9.4 by 250 mm). The same fragments of DNS-βmArg were combined, concentrated, and analyzed for their 1H NMR (500-MHz, methanol-d4) and 13C NMR (125-MHz, methanol-d4) spectra. The signals of DNS-βmArg are as follows: δH 8.78 (s, -NH2), 8.14 (s, 1H), 7.66 (s, 1H), 7.43 (s, 1H), 7.28 (s, 1H), 7.24 (s, 1H), 7.18 (s, 1H), 6.84 (s, 1H), 3.98 (m, 1H), 3.62 (d, 3H), 3.38 (d, 3H), 2.60 (m, 2H), 2.18 (m, 1H), 1.94 (m, 2H), 1.44 (d, 3H), and δc 178.66, 160.12, 157.45, 143.31, 132.15, 125.46, 124.32, 122.21, 118.55, 117.20, 115.61, 114.23, 64.33, 47.32, 46.11, 35.72, 28.51, 26.33, and 18.71 (see Fig. S11 to S12 in the supplemental material). Further COSY NMR clearly showed that the H · H protons of the α-carbon and β-methyl groups were correlated, thus proving the configuration of the (3R)-methyl group of DNS-βmArg (Fig. S13). Thereby, ArgN is considered to be an l-arginine (3R)-β-methyltransferase. Given all the experimental data, we have proposed the mechanism of (2S,3R)-β-methylarginine formation in arginomycin biosynthesis (inset in Fig. 4).
Conclusions.
The complete biosynthetic gene cluster for arginomycin was cloned and heterologously expressed in S. coelicolor M145. Comparative analysis to blasticidin S revealed two arginomycin-specific genes, argM and argN, which encode a PLP-dependent aminotransferase and SAM-dependent methyltransferase, respectively, involved in the formation of the rare amino acid β-methylarginine. ArgM can transfer the amino group from l-arginine to the α-ketoglutaric acid to give glutamate. Another product of this reaction derived from l-arginine is 5-guanidino-2-oxopentanoic acid, which can be immediately utilized as a substrate for ArgN to generate 5-guanidino-3-methyl-2-oxopentanoic acid. Surprisingly, following this methylation reaction, ArgM can catalyze another transamination from the amino group donor l-aspartate to 5-guanidino-3-methyl-2-oxopentanoic acid, generating the final product, β-methylarginine. This study presented all experimental proofs for the complete biosynthesis pathway for the nonproteinogenic β-methylarginine. It will provide a solid base for the engineering of microbial strains with high yields of β-methylarginine.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lin Shuangjun and Jiang Ming for their suggestive discussions on the experimental data.
This work was supported in part by a grant from the opening foundation of the State Key Laboratory of Microbial Metabolism to J.F. (MMLKF14-03) and by grants from the Ministry of Science and Technology of China (2012CB721004), the National Natural Science Foundation of China (31170083, 31130068, and 31121064), the Ministry of Education of China (20110073130011), and the Chen Xing Young Scholars Program of Shanghai Jiao Tong University, awarded to X.H.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 6 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01172-14.
REFERENCES
- 1.Argoudelis AD, Baczynskyj L, Kuo MT, Laborde AL, Sebek OK, Truesdell SE, Shilliday FB. 1987. Arginomycin: production, isolation, characterization and structure. J. Antibiot. (Tokyo) 40:750–760. 10.7164/antibiotics.40.750 [DOI] [PubMed] [Google Scholar]
- 2.Huang KT, Misato T, Asuyama H. 1964. Effect of blasticidin S on protein synthesis of Pyricularia oryzae. J. Antibiot. (Tokyo) 17:65–70 [PubMed] [Google Scholar]
- 3.Prabhakaran PC, Woo NT, Yorgey PS, Gould SJ. 1988. Biosynthesis of blasticidin S from l-α-arginine. Stereochemistry in the arginine 2,3-aminomutase reaction. J. Am. Chem. Soc. 110:5785–5791. 10.1021/ja00225a033 [DOI] [Google Scholar]
- 4.Seto H, Furihata K, Yonehara H. 1968. Studies on the biosynthesis of blasticidin S. Part I. Precursors of blasticidin S biosynthesis. J. Antibiot. (Tokyo) 32:1292–1298 [DOI] [PubMed] [Google Scholar]
- 5.Gould SJ, Guo J. 1991. Biosynthesis of blasticidin S. Cell-free demonstration of n-methylation as the last step. Bioorg. Med. Chem. Lett. 1:497–500 [Google Scholar]
- 6.Komori T, Ezaki M, Kino E, Kohsaka M, Aoki H, Imanaka H. 1985. Lavendomycin, a new antibiotic. I. Taxonomy, isolation and characterization. J. Antibiot. (Tokyo) 38:691–698 [DOI] [PubMed] [Google Scholar]
- 7.Braun SD, Volksch B, Nuske J, Spiteller D. 2008. 3-Methylarginine from Pseudomonas syringae pv. syringae 22d/93 suppresses the bacterial blight caused by its close relative Pseudomonas syringae pv. Glycinea. Chembiochem 9:1913–1920. 10.1002/cbic.200800080 [DOI] [PubMed] [Google Scholar]
- 8.Braun SD, Hofmann J, Wensing A, Ullrich MS, Weingart H, Volksch B, Spiteller D. 2010. Identification of the biosynthetic gene cluster for 3-methylarginine, a toxin produced by Pseudomonas syringae pv. syringae 22d/93. Appl. Environ. Microbiol. 76:2500–2508. 10.1128/AEM.00666-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cone MC, Yin X, Grochowski LL, Parker MR, Zabriskie TM. 2003. The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: sequence analysis, organization, and initial characterization. Chembiochem 4:821–828. 10.1002/cbic.200300583 [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Li L, Deng Z, Zabriskie TM, He X. 2012. Analysis of the mildiomycin biosynthesis gene cluster in Streptoverticillum remofaciens ZJU5119 and characterization of MilC, a hydroxymethyl cytosyl-glucuronic acid synthase. Chembiochem 13:1613–1621. 10.1002/cbic.201200173 [DOI] [PubMed] [Google Scholar]
- 11.Gould SJ, Zhang Q. 1995. Cytosinine: pyridoxal phosphate tautomerase, a new enzyme in the blasticidin S biosynthetic pathway. J. Antibiot. (Tokyo) 48:652–656. 10.7164/antibiotics.48.652 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Cone MC, Gould SJ, Zabriskie TM. 2000. Reevaluation of the final steps in the biosynthesis of blasticidin S by Streptomyces griseochromogenes and identification of a novel self-resistance mechanism. Tetrahedron 56:693–701. 10.1016/S0040-4020(99)01060-1 [DOI] [Google Scholar]
- 13.Feng J, Wu J, Dai N, Lin S, Xu HH, Deng Z, He X. 2013. Discovery and characterization of BlsE, a radical S-adenosyl-l-methionine decarboxylase involved in the blasticidin S biosynthetic pathway. PLoS One 8:e68545. 10.1371/journal.pone.0068545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu G, Li L, Wei J, Tan H. 2013. Cloning, heterologous expression, and characterization of the gene cluster required for gougerotin biosynthesis. Chem. Biol. 20:34–44. 10.1016/j.chembiol.2012.10.017 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Gould SJ, Zabriskie TM. 1998. A new cytosine glycoside from Streptomyces griseochromogenes produced by the use in vivo of enzyme inhibitors. J. Nat. Prod. 61:648–651. 10.1021/np970468o [DOI] [PubMed] [Google Scholar]
- 16.Huang YT, Lyu SY, Chuang PH, Hsu NS, Li YS, Chan HC, Huang CJ, Liu YC, Wu CJ, Yang WB, Li TL. 2009. In vitro characterization of enzymes involved in the synthesis of nonproteinogenic residue (2S,3S)-beta-methylphenylalanine in glycopeptide antibiotic mannopeptimycin. Chembiochem 10:2480–2487. 10.1002/cbic.200900351 [DOI] [PubMed] [Google Scholar]
- 17.Dietrich JB. 1992. Tyrosine aminotransferase: a transaminase among others? Cell. Mol. Biol. 38:95–114 [PubMed] [Google Scholar]
- 18.Szkop M, Bielawski W. 2013. tyrB-2 and phhC genes of Pseudomonas putida encode aromatic amino acid aminotransferase isozymes: evidence at the protein level. Amino Acids 45:351–358. 10.1007/s00726-013-1508-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Camakaris H, Pittard J. 2002. Molecular analysis of tyrosine-and phenylalanine-mediated repression of the tyrB promoter by the TyrR protein of Escherichia coli. Mol. Microbiol. 45:1407–1419. 10.1046/j.1365-2958.2002.03108.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.