Abstract
Survival of fungal species depends on the ability of these organisms to respond to environmental stresses. Osmotic stress or high levels of reactive oxygen species (ROS) can cause stress in fungi resulting in growth inhibition. Both eukaryotic and prokaryotic cells have developed numerous mechanisms to counteract and survive the stress in the presence of ROS. In many fungi, the HOG signaling pathway is crucial for the oxidative stress response as well as for osmotic stress response. This study revealed that while the osmotic stress response is only slightly affected by the master regulator veA, this gene, also known to control morphological development and secondary metabolism in numerous fungal species, has a profound effect on the oxidative stress response in the aflatoxin-producing fungus Aspergillus flavus. We found that the expression of A. flavus homolog genes involved in the HOG signaling pathway is regulated by veA. Deletion of veA resulted in a reduction in transcription levels of oxidative stress response genes after exposure to hydrogen peroxide. Furthermore, analyses of the effect of VeA on the promoters of cat1 and trxB indicate that the presence of VeA alters DNA-protein complex formation. This is particularly notable in the cat1 promoter, where the absence of VeA results in abnormally stronger complex formation with reduced cat1 expression and more sensitivity to ROS in a veA deletion mutant, suggesting that VeA might prevent binding of negative transcription regulators to the cat1 promoter. Our study also revealed that veA positively influences the expression of the transcription factor gene atfB and that normal formation of DNA-protein complexes in the cat1 promoter is dependent on AtfB.
INTRODUCTION
Fungi are capable of colonizing a large variety of ecological niches. These organisms efficiently disseminate by producing airborne asexual spores called conidiospores or conidia that propagate long distances to different environments, where they able to adapt and proliferate. Some fungi are also able to produce resistant structures, such as sclerotia, that allow them to survive adverse environmental conditions (1–3). The formation of these developmental structures is particularly important when considering dissemination and survival of fungi (including Aspergillus flavus) that have a detrimental impact on agriculture and health. A. flavus is a saprophyte and an opportunistic pathogen (4) that can infect a variety of oil seed crops, such as corn, peanuts, tree nuts, cotton, and sorghum. In previous work, we showed that A. flavus pathogenicity is influenced by the global regulator VeA (5). This cellular regulator also controls fungal morphogenesis and secondary metabolism (6–11). While VeA is necessary for sclerotial production, it is a repressor of conidiation (8). Furthermore, this regulator is required for the synthesis of numerous deleterious natural products, including the A. flavus mycotoxin aflatrem, cyclopiazonic acid, and aflatoxins (AF) (8). The latter, in particular aflatoxin B1 (AFB1), is considered among the most carcinogenic natural products (12). Ingestion of foods contaminated with AF has been associated with acute toxicoses, while chronic low levels of exposure to AF can cause immune suppression and liver cancer (13, 14). The impact of AF contamination in crops is especially serious in developing countries, where a large proportion of the crop, because of the lack of legislation to control AF contamination in the grain, is directed to human consumption (15). Currently implemented control strategies fail to effectively control the A. flavus dissemination and survival that lead to its detrimental effects.
The development of novel control strategies to prevent or reduce the negative impact caused by A. flavus requires a better understanding of the effects of different environmental factors on this fungus and its adaptive response. Fungi respond to environmental changes, including biotic and abiotic stresses. Changes in nutrient type and abundance, temperature, pH, and oxidative and osmotic stresses activate signal transduction pathways that lead to adaptive responses. Most of the studies involving the molecular regulation of cellular responses to oxidative and osmotic stresses have been performed using yeast as a model organism, in particular Saccharomyces cerevisiae (i.e., reviewed by Hohmann et al. [16]), and the filamentous model fungus Aspergillus nidulans (17–22). The current availability of genome sequence databases has provided valuable tools for the identification of genetic elements putatively involved in the HOG pathway in several fungal species (17); the HOG pathway is a key signaling pathway that mediates the responses to both osmotic and oxidative stresses. In A. nidulans, the Hog1 homolog, SakA, has been experimentally characterized (18–21). Homolog genes of those involved in the HOG signaling pathway have also been identified in A. flavus (17).
In our present study, we investigated the A. flavus response to oxidative and osmotic stresses and the role of the master regulator veA and its gene product, VeA, in mediating such a response, which leads to physiological changes allowing the organism to survive these environmental stresses. We show that VeA plays an important role in the oxidative stress response in A. flavus. We also demonstrate that hogA is dispensable for survival in oxidative and osmotic environments, suggesting an alternative route to activate the genes necessary to counteract the stresses imposed. Importantly, VeA is a positive regulator in the expression of these genes, also affecting atfB expression and AtfB-dependent protein-DNA interactions in their promoter regions, particularly the promoter of the catalase gene cat1.
MATERIALS AND METHODS
Aspergillus flavus strains and culture conditions.
Aspergillus flavus 70S (wild type), psl82 (the transformation control), and the ΔveA mutant were used in this study. The generation of these strains has been previously described (8). Aspergillus flavus hogA deletion mutants (ΔhogA::pyrG) were generated in this study as described below. Strains were cultured on potato dextrose agar (PDA) or YGT medium (0.5% yeast extract, 2% dextrose, and 1 ml of trace elements prepared as described by Käfer [23]) as indicated in each case. For osmotic and oxidative stress-induced cultures, fungal strains were grown on PDA or YGT medium supplemented with NaCl, KCl, or sorbitol for osmotic stress and H2O2 or menadione for oxidative stress, respectively, as described below. Appropriate supplements for the corresponding auxotrophic markers (23) were added as needed. Solid medium was prepared by adding 10 g/liter agar. Strains were stored as 30% glycerol stocks at −80°C.
Osmotic stress bioassay.
The Aspergillus flavus 70S, psl82 control, and ΔveA mutant strains were point inoculated or top agar inoculated (approximately 106 spores per plate) on PDA medium supplemented with 0.6 M NaCl, 0.7 M KCl, or 1.0 M sorbitol and compared to PDA cultures without supplementation. Cultures were incubated at 30°C.
Oxidative stress bioassay.
In order to elucidate whether the A. flavus response to oxidative stress is influenced by veA, we performed experiments in which hydrogen peroxide was added to the culture medium. We also performed a similar experiment in which menadione, a compound commonly used to generate reactive oxygen species (ROS) (24), was added.
(i) Treatment with hydrogen peroxide.
A. flavus strains were point inoculated on YGT medium supplemented with hydrogen peroxide at the following concentrations: 0 (negative control), 5, 10, 15, 20, 25, and 30 mM. Hydrogen peroxide was added to the medium after autoclaving. The plates were incubated at 30°C in the dark. The experiment was repeated two times with three replicates each, obtaining similar results.
(ii) Treatment with menadione.
A. flavus strains were point inoculated on YGT medium supplemented with menadione at the following concentrations: 0, 0.5, 0.6, 0.7, and 0.8 mM. As in the case of hydrogen peroxide, menadione was added to YGT medium after autoclaving. The plates were incubated at 30°C in the dark.
Aflatoxin analysis.
Plates containing 25 ml of solid PDA were inoculated with 5 ml of top agar containing approximately 106 conidia/ml of the A. flavus 70S, psl82, and ΔveA strains. Three cores (16 mm in diameter) from each replicate plate were extracted with CHCl3. The extracts were dried overnight and then resuspended in 200 μl of CHCl3. Samples were analyzed by thin-layer chromatography (TLC) using toluene-ethyl acetate-formic acid (5:4:1 [vol/vol]) as a solvent system. The plates were then sprayed with aluminum chloride (15% in ethanol) and baked for 10 min at 80°C. AF bands on TLC silica plates were viewed by exposing the plates under UV light (375 nm).
Gene expression analysis.
Conidia (106 spores/ml) from the A. flavus 70S, psl82, and ΔveA strains were inoculated into 400 ml of liquid YGT medium. The cultures were grown for 16 h at 30°C in a shaker incubator at 300 rpm. Mycelial samples were collected at different time points after 16 h of incubation. Approximately 2 g of wet mycelium was added to 500-ml Erlenmeyer flasks containing 250 ml of fresh liquid YGT medium with 15 mM hydrogen peroxide. Cultures were further grown for 6 and 24 h at 30°C in a shaker incubator at 300 rpm. For analysis of hogA expression in A. flavus 70S ΔhogA mutants and the pyrG-1 isogenic control, conidia (106 spores/ml) were inoculated into 250 ml potato dextrose broth (PDB) (BD Biosciences) and incubated for 36 h at 30°C with shaking at 250 rpm. Mycelia were collected by filtration through Miracloth, and 1.0 g was transferred to 125 ml PDB supplemented with 0.2 mM menadione. Cultures were incubated at 30°C with shaking for an additional 4 h. Mycelia were harvested, frozen in liquid nitrogen, and stored at −80°C for RNA extraction. Total RNA was isolated as previously described (25). Approximately 4 μg of total RNA was treated with DNase I RQI (Promega) and reverse transcribed with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega) to synthesize the cDNA. Finally quantitative reverse transcription-PCR (qRT-PCR) was performed using specific primers for different genes of interest listed in Table S1 in the supplemental material. The relative expression levels were calculated by the method described by Kenneth and Schmittgen (26), and all values were normalized to the expression of the A. flavus 18S rRNA gene.
Sequence alignment of HogA orthologs.
The protein sequence of A. flavus HogA (accession no. AFL2G_06243.2) was compared against those of HogA orthologs from other Aspergillus species. The sequences were obtained from the Broad Institute (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html). Multiple sequence alignment was performed with A. flavus HogA and ortholog sequences using the EMBOSS NEEDLE pairwise sequence alignment tool from EMBl-EBI (http://www.ebi.ac.uk/Tools/psa/emboss_needle/), followed by shading using Box Shade v3.2.1 (http://www.ch.embnet.org/software/BOX_form.html) for presentation purposes.
Generation of the A. flavus hogA deletion strain.
A method was used to construct the hogA gene knockout plasmid following essentially the same protocol described by Cary et al. (27) and as depicted in Fig. S4 in the supplemental material. Briefly, 5′ and 3′ regions of the A. flavus hogA gene were PCR amplified using oligonucleotide primers listed in Table S1 in the supplemental material. The locations of the primers within and flanking all of the A. flavus genes used in this study are based on the Broad Institute Aspergillus Comparative Database nucleotide sequence data for the A. flavus 3357 genome. Following amplification of A. flavus genomic DNA with Ex Taq HS polymerase (TaKaRa), PCR products of the expected size for both the 5′ (890 bp) and 3′ (922 bp) hogA gene regions were obtained. PCR products were subcloned into TOPO pCR2.1 (Invitrogen, Carlsbad, CA) and verified by DNA sequencing. In a two-step ligation procedure, the 5′ and 3′ PCR products were digested with the restriction enzymes engineered onto the ends of the PCR primers and subcloned into restriction enzyme-digested pPG2.8 such that the 5′ and 3′ gene regions flanked the Aspergillus parasiticus pyrG gene. This generated the deletion vector pHogA-pyrG.
EMSA.
Approximately 106 conidia/ml from the A. flavus 70S, psl82, and ΔveA strains were inoculated in 400 ml of liquid YGT medium in a 1-liter flask and grown at 30°C in the dark for 16 h in a shaker incubator at 300 rpm. Approximately 2 g of wet mycelium was then transferred to 250 ml of liquid YGT medium with or without 15 mM H2O2 in a 500-ml flask. The cultures were further incubated for 24 h. Preparation of cell extracts enriched in nuclear proteins from the A. flavus 70S, psl82, and ΔveA strains for electrophoretic mobility shift assays (EMSAs) was performed as previously described (28). Briefly, the mycelia cultured as detailed above were harvested by filtration through Miracloth, ground in liquid nitrogen with a mortar and pestle, and resuspended in lysis buffer as described previously (28). The proteins were precipitated with ammonium sulfate (10% and then 70%) and collected by centrifugation. After that, proteins were resuspended in dialysis buffer and dialyzed against the same buffer. The dialyzed protein solution was aliquoted and stored at −80°C.
EMSA and shift inhibition EMSA were performed as described previously (28). Double-stranded DNA fragments derived from the gene promoter regions were generated by PCR using the primers listed in Table S1 in the supplemental material. The DNA fragments were 5′-end labeled with [γ-32P]ATP using USB OptiKinase (Affymetrix, Cleveland, OH), purified using a Micro Bio-Spin P-30 chromatography column (Bio-Rad, Hercules, CA), and used as probes.
Electrophoresis on 5% nondenaturing polyacrylamide (80:1 acrylamide-bisacrylamide) gels was used to separate DNA-protein complexes. Twenty femtomoles of each 32P-labeled probe was incubated for 15 min at 30°C with 2 μg of poly(dI-dC) and 7.5 μg of bovine serum albumin with 5 μg of nuclear protein extract in a DNA binding buffer (15% glycerol, 15 mM HEPES [pH 7.9], 100 mM KCl, 1 mM EDTA, 2 mM dithiothreitol [DTT]) in 25 μl of final binding reaction mixture. After separation, the gels were dried and exposed to X-ray film.
Polyclonal anti-AtfB antibodies (YSR) were used to block formation of DNA-protein complexes (shift inhibition EMSA) as described previously (29). Five micrograms of nuclear protein extracts was incubated in a DNA binding buffer for 15 min at 30°C with 5 μl of anti-AtfB antibody (5.5 μg/μl), or preimmune serum obtained from the same rabbit as the antibody was generated. Then a 32P-labeled probe was added, and the mixture was incubated for additional 15 min at 30°C. Finally, DNA-protein complexes were resolved by electrophoresis. For shift inhibition EMSA with anti-VeA antibodies, VeA-specific polyclonal antibodies were used to block formation of DNA-protein complexes as described above.
RESULTS
Effect of osmotic stress.
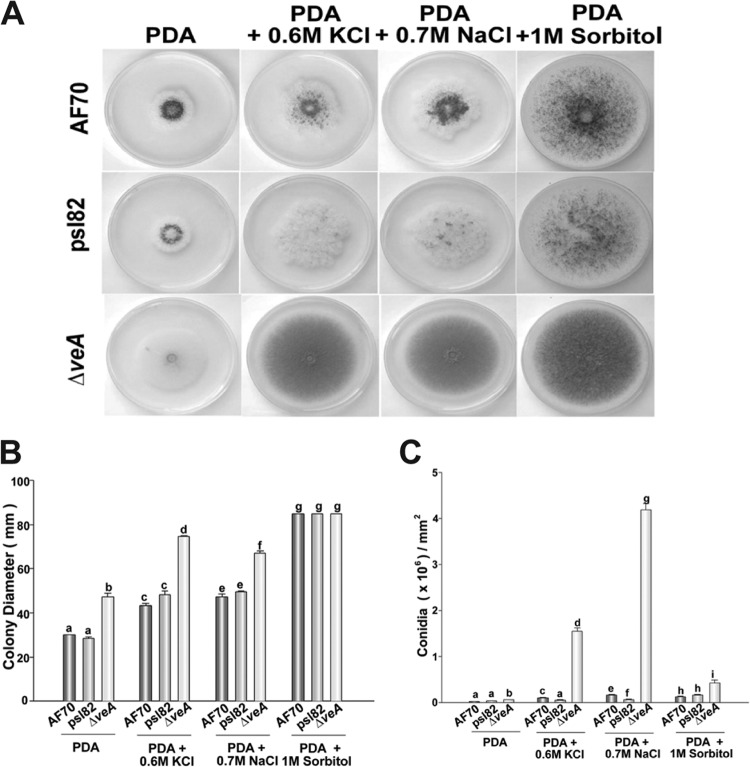
Our study showed that in A. flavus, the osmotic stress response is linked to colony growth and development (Fig. 1A and B). Osmotic stress did not inhibit growth; on the contrary, the colony diameter increased in all strains tested when the cultures were supplemented with 0.6 M NaCl, 0.7 M KCl, or 1.0 M sorbitol. Conidiation was also promoted under osmotic stress (Fig. 1A and C) in all the cultures, particularly in the ΔveA colonies. Formation of sclerotia and the sclerotial maturation rate decreased in a hypertonic environment (Fig. 2) in the control strains. It is known that veA is necessary for the production of sclerotia (7, 8). In this study, addition of 0.6 M NaCl, 0.7 M KCl, or 1.0 M sorbitol did not remediate this phenotype, and ΔveA colonies did not produce sclerotia on PDA with or without osmotic stress.
FIG 1.
Effect of osmotic stress on A. flavus colony growth and conidiation. (A) Photographs of A. flavus 70S, psl82, and ΔveA point-inoculated cultures incubated for 5 days. The medium was supplemented with 0.6 M NaCl, 0.7 M KCl, or 1.0 M sorbitol to generate osmotic stress. (B) Colony growth measured as colony diameter. (C) Quantification of conidial production. Cores (16 mm in diameter) were taken 1 cm from the center of the plates and homogenized in water. Spores were counted with a hemocytometer. Error bars represent standard errors. Experiments were carried out with three replicates. Different letters indicate samples that are significantly different (P ≤ 0.05).
FIG 2.
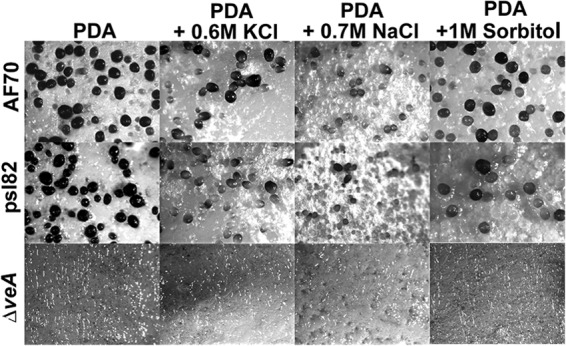
Effect of osmotic stress on sclerotial production. Microscopic examination of sclerotia. Sclerotial production on top agar-inoculated A. flavus 70S, psl82, andΔveA cultures was observed after 5 days of incubation at 30°C. The medium was supplemented with 0.6 M NaCl, 0.7 M KCl, or 1.0 M sorbitol to generate osmotic stress. Images (×32) were captured using an upright Leica MZ75 stereomicroscope.
We previously demonstrated that veA is also required for the biosynthesis of several secondary metabolites, including AFB1 in A. flavus (5, 8). The biosynthesis of this carcinogenic mycotoxin was not recovered in the ΔveA mutant under the impact of a hypertonic environment (see Fig. S1 in the supplemental material). Both control strains produced AF under the same experimental conditions. Biosynthesis of this mycotoxin in the controls was not altered under osmotic stress. Interestingly, two unknown veA-dependent metabolites were also detected, and the synthesis of one of them was suppressed in a hypertonic environment (see Fig. S1).
Effect of oxidative stress.
In order to elucidate whether veA is involved in the oxidative stress response in A. flavus, we exposed the veA deletion mutant strain and the two isogenic control strains, 70S and pls82, to increasing concentrations of hydrogen peroxide (Fig. 3). Our results revealed that addition of this compound lead to a dramatic reduction in colony growth in the ΔveA mutant cultures at a concentration of 25 mM. Furthermore, no growth was observed in ΔveA mutant cultures in the presence of 30 mM H2O2, while the wild-type 70S and psl82 controls were able to form colonies.
FIG 3.
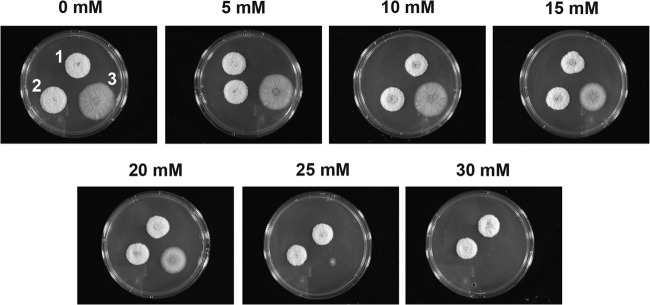
veA is necessary for a normal response to oxidative stress in A. flavus. Photographs of point-inoculated A. flavus 70S, psl82, and ΔveA strains growing on YGT supplemented with increasing amounts of hydrogen peroxide. Culture numbers: 1, 70S; 2, psl82; and 3, ΔveA mutant. Cultures were incubated at 30°C for 3 days.
We performed a similar experiment using menadione (see Fig. S2 in the supplemental material). Addition of this compound inhibited the growth of the veA deletion mutant at a concentration of 0.8 mM, a condition that still allowed the colonies of the two controls to form.
hogA is dispensable for osmotic and oxidative stress responses in A. flavus.
The HOG pathway has been best characterized in the model yeast S. cerevisiae (reviewed by Hohmann et al. [16]), where the Hog1 kinase plays an important role in the activation of effector genes involved in osmotic and oxidative stress response. The deduced amino acid sequence of A. flavus HogA is highly conserved with respect to other HogA homologs in Aspergillus spp. or Hog1 in S. cerevisiae (Table 1; see Fig. S3 in the supplemental material). We analyzed whether hogA is important for A. flavus to survive when exposed to osmotic and oxidative stresses. With this goal, we generated ΔhogA deletion mutants. Verification of the hogA deletion in the fungal transformants was carried out by PCR analysis of genomic DNA from three Af70 ΔhogA mutants using primers (hog1 Up and Down) that flanked the hogA coding sequence (see Fig. S4A and B and Table S1 in the supplemental material). Successful disruption of the hogA coding region by the pyrG gene was confirmed by the presence of a PCR product of about 4.7 kb, while the Af70 pyrG-1 control gave a product of about 3.3 kb, as expected for the wild-type hogA gene. Real-time quantitative PCR of the ΔhogA mutants and isogenic control demonstrated that hogA expression was inactivated in the mutants (see Fig. S4C). Two ΔhogA mutants were selected to test their tolerance to both osmotic and oxidative stresses. Aspergillus flavus ΔhogA mutants and their isogenic control strain were incubated on medium containing 0.6 M NaCl, 0.7 M KCl, or 1.0 M sorbitol. The ΔhogA colonies grew as much as the wild-type control (see Fig. S5 in the supplemental material). Furthermore, when these strains were cultured on medium containing a range of menadione concentrations (0, 0.2, 0.4, and 0.6 mM), the ΔhogA mutants had similar tolerance to oxidative stress to that of the control strain (see Fig. S6 in the supplemental material). These results indicate that in A. flavus, the hogA homolog is not essential for growth under the oxidative or osmotic stress conditions tested in this study.
TABLE 1.
Comparison of the putative amino acid sequence of A. flavus HogA to those of other Aspergillus HogA homologs and S. cerevisiae Hog1
| Protein and species | Accession no.a | Identity (%) | Similarity (%) |
|---|---|---|---|
| HogA | |||
| A. oryzae | AO090701000642 | 84.7 | 85.0 |
| A. niger | fge1_pg_C_5000454 | 54.2 | 69.4 |
| A. terreus | ATEG_00489.1 | 43.7 | 56.0 |
| A. fischerianus | NFIA_012610 | 54.5 | 68.8 |
| A. fumigatus | Afu1g12940 | 54.4 | 68.8 |
| A. clavatus | ACLA_022520 | 51.1 | 64.1 |
| A. nidulans | ANID_04668 | 51.1 | 63.9 |
| Hog1 | |||
| S. cerevisiae | SCRG_05095.1 | 45.5 | 59.5 |
Broad Institute Aspergillus Comparative Database, Broad Institute, Cambridge, MA.
Expression of oxidative stress response genes is veA dependent.
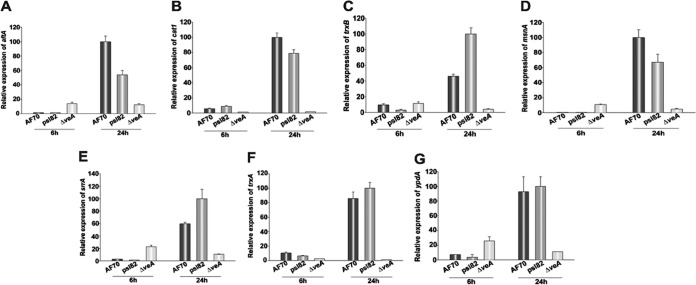
Our study showed that veA is necessary for oxidative stress tolerance in A. flavus. In order to gain further insight into the veA-dependent mechanism involved in the oxidative stress response, we analyzed the expression of genes known to play a role in oxidative stress tolerance in the ΔveA mutants and compared that with expression levels in the control strains. Specifically using qRT-PCR, we examined the expression of A. flavus srrA, msnA, atfA, ypdA, trxA, trxB, and cat1. Note that with regard to catB, the deduced amino acid sequence of A. flavus mycelial cat1 is highly conserved with those of the catB genes from other fungal species, such as A. nidulans, Histoplasma, and Penicillium spp. (30–32) (Fig. 4). Our results indicated that after addition of hydrogen peroxide to the culture, the expression of all of these genes increased in the 70S and psl82 control strains; however, this was not the case in the ΔveA strain cultures, where the expression levels did not show an increase over time. These results were also verified by semiquantitative PCR (data not shown). Analysis of the effect of veA on the expression of atfB, encoding a transcription factor known to bind to the promoter regions of oxidative stress response genes (29, 33), was done separately. Our results indicated that veA is also necessary for the increase in the expression of atfB in the presence of hydrogen peroxide over time (see Fig. S7 in the supplemental material).
FIG 4.
veA is required for wild-type expression levels of A. flavus oxidative stress response genes. Relative expression levels of A. flavus genes involved in the oxidative stress response 6 and 24 h after addition of 15 mM hydrogen peroxide. The relative expression levels were calculated by the method described by Kenneth and Schmittgen (26), and all values were normalized to the expression of the A. flavus 18S rRNA gene and to the greatest expression, considered 100.
Detection of DNA-protein complexes at the promoters of oxidative stress response genes.
In our present studies, we screened promoter fragments of the atfA, srrA, cat1, ypdA, trxA, trxB, and msnA genes for complex formation with protein extracts from the A. flavus wild-type strain by using electrophoretic mobility shift assays (EMSAs). DNA-protein complex formation was assessed by the mobility and intensity of the shifted band compared with those of the free DNA probe lanes, which contained only the DNA fragment, with no protein extract. We detected formation of prominent DNA-protein complexes with all promoters, except for the atfA and srrA promoters (data not shown). EMSA detected a strong DNA-protein complex formation between nuclear protein extracts from A. flavus 70S grown in YGT medium for 24 h and DNA fragments derived from the promoter regions of the cat1 gene (518 bp upstream from ATG) and the trxB gene (521 bp upstream from ATG). In order to further characterize these DNA-protein interactions, the large DNA fragments were divided into smaller pieces (Fig. 5A; see Table S2 in the supplemental material). The cat1 promoter region was divided into cat1-1 (259 bp) and cat1-2 (259 bp). The trxB promoter region was divided into trxB-1 (263 bp) and trxB-2 (258 bp). The cat1-2 and trxB-2 fragments demonstrated prominent DNA-protein complexes in EMSAs using nuclear protein extracts from A. flavus 70S and psl82. For this reason, these fragments were selected to perform the following EMSA experiments described below.
FIG 5.
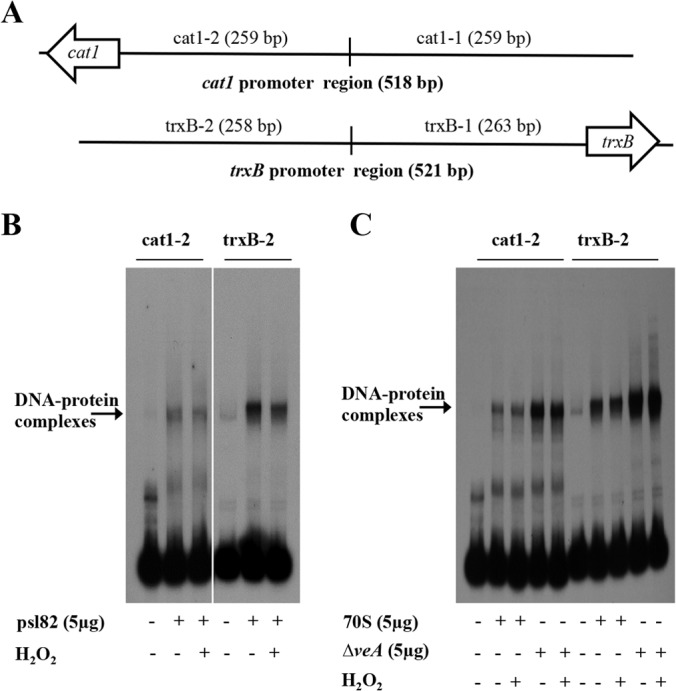
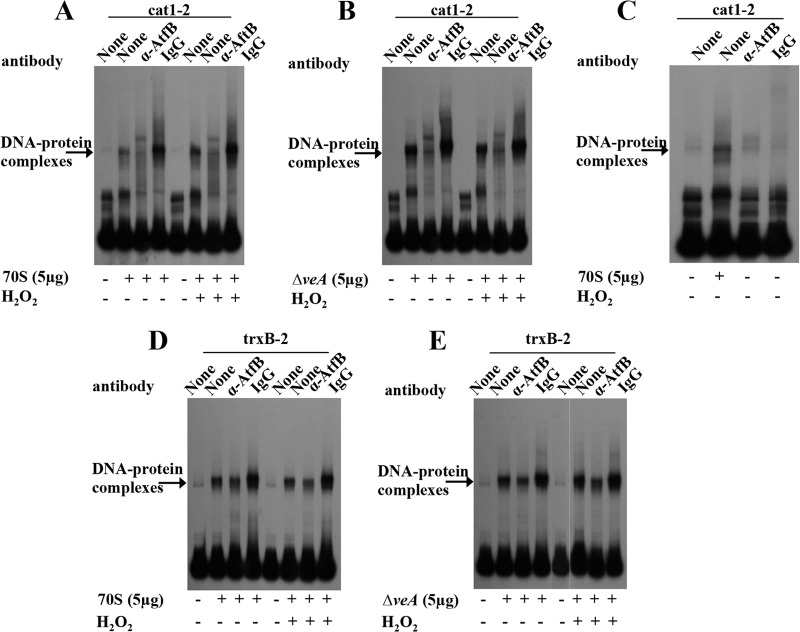
The formation of protein-DNA complexes at the cat1 and trxB promoters is influenced by VeA. (A) Schematic representation of promoter regions of the cat1 and trxB genes used in EMSA. The cat1 promoter region was divided into two fragments designated cat1-1 and cat1-2. The trxB promoter region was also divided into two fragments, designated trxB-1 and trxB-2. (B and C) DNA-protein complex formation on cat1-2 and trxB-2 promoters by EMSA using 70S, psl82, or ΔveA strain protein extracts with or without H2O2 treatment. Cell extracts enriched in nuclear proteins were prepared as described in Materials and Methods. Five micrograms of enriched nuclear protein extracts was added to a 32P-labeled promoter probe for each gene.
Protein extracts from the ΔveA strain show strong DNA-protein complex formation on cat1-2 and trxB-2 promoter fragments.
We examined cat1-2 and trxB-2 fragments for DNA-protein complex formation using protein extracts obtained from controls and ΔveA mycelia grown with or without H2O2 treatment. Hydrogen peroxide treatment of 70S, psl82, and ΔveA mutant cultures did not affect DNA-protein complex formation with either cat1-2 or trxB-2 fragments compared to the same cultures without H2O2 treatment (Fig. 5B and C). However, ΔveA protein extracts showed DNA-protein complex formation with significantly higher intensity on cat1-2 and trxB-2 fragments than that for the corresponding 70S protein extracts regardless of H2O2 treatment (Fig. 5C). Based on the fact that these and previous studies including the 70S and psl82 controls yielded similar results, the 70S wild-type strain was selected for further promoter analyses.
AtfB binds more intensely to cat1-2 than to the trxB-2 promoter fragment.
Previously we showed that AtfB, a basic leucine zipper (bZIP) transcription factor, binds not only to aflatoxin gene promoters but also to promoters of oxidative stress response genes, including mycelium-specific cat1 and the mitochondrion-specific Mn superoxide dismutase (SOD) gene (Mn sod) in A. parasiticus (33). In addition, we also demonstrated that VeA is necessary for histone H4 acetylation and AtfB binding to occur at normal levels in the aflatoxin gene cluster (29, 34). To examine the differences between DNA-protein complexes formed on cat1-2 and trxB-2 fragments by 70S and ΔveA protein extracts, AtfB- or VeA-specific polyclonal antibodies were used to block DNA-protein complex formation (by shift inhibition EMSA). Formation of a specific DNA-protein complex was significantly inhibited by pretreatment of the enriched 70S or ΔveA protein extracts with anti-AtfB antibodies before addition of the cat1-2 fragment but not by preimmune serum obtained from the same rabbit from which the antibody was generated (Fig. 6A to C). Addition of anti-AtfB antibody alone or preimmune serum alone to the cat1-2 fragment did not produce any shifted complexes. However, DNA-protein complex formation on the trxB-2 fragment using the enriched protein extracts prepared from 70S or the ΔveA mutant was significantly less inhibited by pretreatment with anti-AtfB antibodies compared with shift inhibition on the cat1-2 fragment (Fig. 6D and E). Shift inhibition EMSA with anti-VeA antibodies demonstrated no apparent inhibition of the DNA-protein complex formation on either the cat1-2 or trxB-2 fragment using 70S and ΔveA protein extracts (Fig. 7), suggesting that VeA does not directly bind to cat1-2 or trxB-2 promoter fragments.
FIG 6.
Analysis of AtfB binding to the cat1 and trxB promoters and its relationship with VeA. Shown are results from EMSA of AtfB binding in the cat1-2 (A to C) and trxB-2 (D to E) promoter fragments using 70S or ΔveA mutant protein extracts with or without H2O2 treatment. Enriched nuclear protein extracts were prepared as described in Materials and Methods. Five micrograms of enriched nuclear protein extracts was added to a 32P-labeled promoter probe for each gene. Anti-AtfB antibodies (YSR) or preimmune serum was added to determine whether these could block protein-DNA interaction (shift inhibition). (A) cat1-2 probe and 70S protein extracts. (B) cat1-2 probe and ΔveA protein extracts. (C) cat1-2 probe and anti-AtfB antibodies alone (without protein extracts). (D) trxB-2 probe and 70S protein extracts. (E) trxB-2 probe and ΔveA protein extracts.
FIG 7.
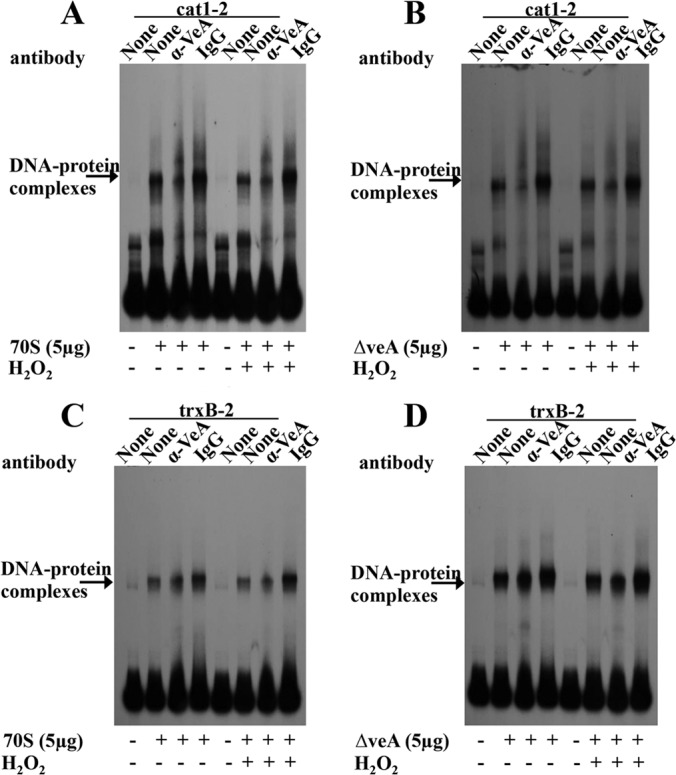
The presence of a VeA antibody did not prevent protein-DNA complex formation at the cat1 and trxB promoters in shift inhibition EMSA. Shown are results from EMSA of VeA binding in the cat1-2 and trxB-2 promoter fragments using 70S or ΔveA mutant protein extracts with or without H2O2 treatment. Enriched nuclear protein extracts were prepared as described in Materials and Methods. Five micrograms of enriched nuclear protein extracts was added to a 32P-labeled promoter probe for each gene. Anti-VeA antibodies or preimmune serum was added to determine whether these could block protein-DNA interaction (shift inhibition). (A) cat1-2 probe and 70S protein extracts. (B) cat1-2 probe and ΔveA protein extracts. (C) trxB-2 probe and 70S protein extracts. (D) trxB-2 probe and ΔveA protein extracts.
DISCUSSION
Fungi are versatile eukaryotic organisms capable of colonizing diverse environments and rapidly responding to changes, optimizing their survival. The negative effects of diverse types of abiotic stresses could be overcome in these organisms by the activation of signaling pathways that lead to a cellular adaptive response. Progress has been made in understanding the relationship between osmotic and oxidative stresses and the cellular response triggered by these stimuli, which is mediated by the HOG signaling pathway. The HOG pathway has been characterized in yeast in great detail (16, 35). Studies in filamentous fungi, mostly in aspergilli (20, 36, 37), suggest that the HOG pathway is semiconserved compared to that described in yeast (17). Interestingly, the regulatory output of this signaling pathway in filamentous fungi also seems to vary from that described in yeast (20). For instance, while HOG1 is necessary for the response and survival in S. cerevisiae, deletion of the sakA homolog in A. nidulans does not show sensitivity to high osmolarity (20). Previous studies indicate that mutations in genes involved in the HOG pathway in filamentous fungi result in changes in morphological differentiation, suggesting a link between the osmotic stress response and fungal development. For example, the sakA deletion mutant showed premature sexual development. Furthermore, cleistothecial formation was prevented when the A. nidulans steC gene, a homolog of the yeast gene STE11, was deleted (38). Some osmotic stress response mutants also show an effect on asexual development, such as reduction in conidium viability in the sakA mutant (20) or abnormality in conidiophore formation in the steC mutant (38). Furthermore, a decrease in conidiation was also observed in the A nidulans srrA and sskA mutants as well as in the A. fumigatus MA21 mutant (SHO S. cerevisiae homolog) (39), further illustrating the interaction between the osmotic stress response signaling pathway and fungal morphogenesis. The effect of osmotic stress on conidiation directly influences fungal dispersal. This is especially relevant when considering fungal species with an agricultural impact, such as the aflatoxin producer Aspergillus flavus. This fungus is an opportunistic plant pathogen of important oil seed crops (4) that disseminates mainly by producing airborne conidia, causing large infestations in the field.
As part of the present study, we investigated first whether suboptimal conditions caused by osmotic stress affect growth and morphogenesis in A. flavus and whether this could be influenced by veA. Our results revealed that osmotic stress positively affects vegetative growth and leads to an increase in conidiation in A. flavus. This is in agreement with a report from Han et al. (40) in which the authors showed an increase in conidiation in A. nidulans cultures with high levels of sorbitol. In contrast, Mert and Ekmekci (41) reported a reduction of conidia in A. flavus inoculated on NaCl medium. It is possible that this variability in conidiation could result from the use different A. flavus strains or medium conditions in these studies. In our studies, we observed that osmotic stress further enhances hyperconidiation in the A. flavus veA mutant, suggesting that veA is involved in modulating osmotic stress-induced conidiation.
In addition to the ability to efficiently disseminate by forming conidia, A. flavus is able to produce sclerotia. In other fungi, such as Sclerotinia sclerotiorum, Rhizoctonia solani, and Sclerotium rolfsii, salinity stress inhibits the formation of sclerotia (42). In Botrytis cinerea, sclerotial production was decreased as osmotic stress increased (43). However, in Aspergillus ochraceus, osmotic stress had little effect on sclerotial production (44). The results from our present study indicate that in A. flavus, hyperosmotic medium caused a delay in sclerotial production and maturation. Sclerotia are postulated to be vestiges of cleistothecia (45, 46). For this reason, it could be expected that the effects of hypertonic media on sclerotial formation could be similar to the effect on cleistothecial formation. Indeed, in A. nidulans cleistothecial production also decreased in the presence of 1 M sorbitol (40). As expected, based on our previous studies (7, 8), ΔveA cultures did not produce sclerotia or the carcinogenic mycotoxin AFB1, and the formation of these resistant structures and the synthesis of AFB1 were not rescued by exposure to a hypertonic environment.
Although the oxidative stress response has been extensively studied in prokaryotes and eukaryotes, it is possible that additional genetic elements unique to filamentous fungi, such as veA, could modulate this response in these organisms. Importantly, our study indicated that tolerance to oxidative stress decreased in the ΔveA strain compared to that in the control strains. Supplementation of medium with hydrogen peroxide or menadione resulted in complete growth inhibition of the veA mutant, whereas the 70S and psl82 strains were able to form colonies. These results indicate that veA has an important role in protecting A. flavus from oxidative stress. To gain insight into the mechanism by which veA influences the oxidative stress response in A. flavus, the expression patterns of several genes encoding proteins known to play a role in the HogA-related signaling pathway were analyzed in the ΔveA strain and compared to those of the control strains under oxidative stress. Our experiments revealed that veA is necessary for normal expression of critical genes in this adaptive genetic response, including atfA, srrA, msnA, and ypdA, which encode transcription factors, and structural genes, such as trxA, trxB, and cat1 (22).
To further elucidate the role of VeA in the transcription of the oxidative response genes, we performed electrophoretic mobility shift assays utilizing their promoters and protein extracts from A. flavus ΔveA mutant and control strains. The formation of DNA-protein complexes in cat1 and trxB promoters was particularly interesting; more prominent DNA-protein complex formation on cat1-2 and trxB-2 promoter fragments (Fig. 6) was detected when ΔveA protein extracts were used compared to the level of these complexes formed with wild-type protein extracts. When taken together with the lack of transcriptional induction of the oxidative stress response genes under oxidative stress in the ΔveA strain, these results suggest that the observed VeA-dependent DNA-protein complex formation on cat1-2 and trxB-2 promoter fragments could include repressor proteins negatively influencing the transcriptional activation of cat1 and trxB. Future proteomics studies in our laboratory will provide additional information on the VeA-dependent protein complexes associated with these promoters.
Another important finding in our studies includes the requirement of the transcription factor AtfB for the formation of the DNA-protein complex at the cat1 and trxB promoters. This was particularly notable in the case of the cat1 promoter. This bZIP transcription factor, which our current study also revealed to be transcriptionally regulated by veA, was previously studied in A. parasiticus (29). AtfB is known to contribute to conidium resistance to oxidative stress and activation the transcription of catA (47). Furthermore, AtfB has been demonstrated to bind to the promoter regions of oxidative stress response genes, such as the cat1 and Mn superoxide dismutase (SOD) genes, as well as genes within the aflatoxin gene cluster (29, 33), where AtfB may aid AflR, an endogenous transcription factor of this cluster, to activate aflatoxin biosynthetic genes (28, 48).
In conclusion, the present study revealed that while the global regulatory veA gene and its product have a mild effect on the response to osmotic stress, they have a critical role in survival under oxidative stress in A. flavus, an agriculturally important fungus with a robust oxidative stress response in which, interestingly, HogA is dispensable for growth under this stress, suggesting an alternative route for activation of the oxidative stress response genes from that described in yeast. Mechanistically, we found that VeA is a positive regulator in the expression of oxidative stress response genes. Furthermore, VeA modulates the formation of protein-DNA complexes in their promoter region of oxidative stress response genes, such as cat1, and perhaps contributes to coordinate production of toxin with the oxidative stress response. Also, we found that DNA-protein complex formation on the promoter is mediated by the veA-dependent transcription factor AtfB, known to bind to the promoters of aflatoxin genes. Due to the conserved nature of the master regulator VeA, it is likely that the role of VeA in the oxidative stress response could also be conserved in other fungal species of agriculture or medical importance. These studies could contribute to set the basis for the design of future control strategies that could decrease the negative impact of the aflatoxin producer A. flavus and other detrimental fungi.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by USDA-SCA 58-6435-9-386 and the Department of Biological Sciences at NIU.
We thank Timothy Satterlee and Barbara Ball for technical support.
Footnotes
Published ahead of print 20 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00099-14.
REFERENCES
- 1.Coley-Smith JR, Cooke RC. 1971. Survival and germination of fungal sclerotia. Annu. Rev. Phytopathol. 9:65–92. 10.1146/annurev.py.09.090171.000433 [DOI] [Google Scholar]
- 2.Malloch DM, Cain RF. 1972. The Trichocomataceae: ascomycetes with Aspergillus, Paecilomyces, and Penicillium imperfect states. Can. J. Bot. 50:2613–2628. 10.1139/b72-335 [DOI] [Google Scholar]
- 3.Wicklow DT. 1987. Survival of Aspergillus flavus sclerotia in soil. Trans. Br. Mycol. Soc. 89:131–134. 10.1016/S0007-1536(87)80073-6 [DOI] [Google Scholar]
- 4.Mellon JE, Cotty PJ, Dowd MK. 2007. Aspergillus flavus hydrolases: their role in pathogenesis and substrate utilization. Appl. Microbiol. Biotechnol. 77:497–503. 10.1007/s00253-007-1201-8 [DOI] [PubMed] [Google Scholar]
- 5.Duran RM, Cary JW, Calvo AM. 2009. The role of veA on Aspergillus flavus infection of peanuts, corn and cotton. Open Mycol. J. 3:27–36. 10.2174/1874437000903010027 [DOI] [Google Scholar]
- 6.Kato N, Brooks W, Calvo AM. 2003. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2:1178–1186. 10.1128/EC.2.6.1178-1186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo AM, Bok J, Brooks W, Keller NP. 2004. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 70:4733–4739. 10.1128/AEM.70.8.4733-4739.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duran RM, Cary JW, Calvo AM. 2007. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73:1158–1168. 10.1007/s00253-006-0581-5 [DOI] [PubMed] [Google Scholar]
- 9.Cary JW, O'Brian GR, Nielsen DM, Nierman W, Harris-Coward O, Yu J, Bhatnagar D, Cleveland TE, Payne GA, Calvo AM. 2007. Elucidation of veA-dependent genes associated with aflatoxin and sclerotial production in Aspergillus flavus by functional genomics. Appl. Microbiol. Biotechnol. 76:1107–1118. 10.1007/s00253-007-1081-y [DOI] [PubMed] [Google Scholar]
- 10.Calvo AM. 2008. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 45:1053–1061. 10.1016/j.fgb.2008.03.014 [DOI] [PubMed] [Google Scholar]
- 11.Dhingra S, Andes D, Calvo AM. 2012. VeA regulates conidiation, gliotoxin production, and protease activity in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell 11:1531–1543. 10.1128/EC.00222-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squire RA. 1981. Rating animal carcinogens: a proposed regulatory approach. Science 214:877–880. 10.1126/science.7302565 [DOI] [PubMed] [Google Scholar]
- 13.Probst C, Schulthess F, Cotty PJ. 2010. Impact of Aspergillus section Flavi community structure on the development of lethal levels of aflatoxins in Kenyan maize (Zea mays). J. Appl. Microbiol. 108:600–610. 10.1111/j.1365-2672.2009.04458.x [DOI] [PubMed] [Google Scholar]
- 14.Turner PC, Moore SE, Hall AJ, Prentice AM, Wild CP. 2003. Modification of immune function through exposure to dietary aflatoxin in Gambian children. Environ. Health Perspect. 111:217–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Widstrom ND. 1996. The aflatoxin problem with corn grain. Adv. Agron. 56:219–280. 10.1016/S0065-2113(08)60183-2 [DOI] [Google Scholar]
- 16.Hohmann S, Krantz M, Nordlander B. 2007. Yeast osmoregulation. Methods Enzymol. 428:29–45. 10.1016/S0076-6879(07)28002-4 [DOI] [PubMed] [Google Scholar]
- 17.Miskei MZ, Karanyi Z, Pocsi I. 2009. Annotation of stress-response proteins in the aspergilli. Fungal Genet. Biol. 46:105–120 [DOI] [PubMed] [Google Scholar]
- 18.Han KH, Prade RD. 2002. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol. Microbiol. 43:1065–1078. 10.1046/j.1365-2958.2002.02774.x [DOI] [PubMed] [Google Scholar]
- 19.Furukawa K, Hoshi Y, Maeda T, Nakajima T, Abe K. 2005. Aspergillus nidulans HOG pathway is activated only by two-component signaling pathway in response to osmotic stress. Mol. Microbiol. 56:1246–1261. 10.1111/j.1365-2958.2005.04605.x [DOI] [PubMed] [Google Scholar]
- 20.Kawasaki L, Sanchez O, Shiozaki K, Aguirre J. 2002. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45:1153–1163. 10.1046/j.1365-2958.2002.03087.x [DOI] [PubMed] [Google Scholar]
- 21.Hagiwara D, Asano Y, Marui J, Furukawa K, Kanamaru K, Kato M, Abe K, Yamashino T, Mizuno T, Kobayashi T. 2007. The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71:1003–1014. 10.1271/bbb.60665 [DOI] [PubMed] [Google Scholar]
- 22.Duran RM, Cary JW, Calvo AM. 2010. Role of the osmotic stress regulatory pathway in morphogenesis and secondary metabolism in filamentous fungi. Toxins 2:367–381. 10.3390/toxins2040367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kafer E. 1977. Anthranilate synthetase enzyme complex and trifunctional trpC gene of Aspergillus. Can. J. Genet. Cytol. 19:723–738 [DOI] [PubMed] [Google Scholar]
- 24.Criddle DN, Gillies SS, Baumgartner-Wilson HK, Jaffar M, Chinje EC, Passmore S, Chvanov M, Barrow S, Gerasimenko OV, Tepikin AV, Sutton R, Petersen OH. 2006. Menadione-induced reactive oxygen species generation via redox cycling promotes apoptosis of murine pancreatic acinar cells. J. Biol. Chem. 281:40485–40492. 10.1074/jbc.M607704200 [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26.Livak K J, Schmittgen TD. 2001. Analysis of real gene expression data using real time-quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 27.Cary JW, Ehrlich KC, Bland JM, Montalbano BG. 2006. The aflatoxin biosynthesis cluster gene, aflX, encodes an oxidoreductase involved in the conversion of versicolorin A to demethylsterigmatocystin. Appl. Environ. Microbiol. 72:1096–1101. 10.1128/AEM.72.2.1096-1101.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roze LV, Miller MJ, Rarick M, Mahanti N, Linz JE. 2004. A Novel cAMP-response element, CRE1, modulates expression of nor-1 in Aspergillus parasiticus. J. Biol. Chem. 279:27428–27439. 10.1074/jbc.M400075200 [DOI] [PubMed] [Google Scholar]
- 29.Roze LV, Chanda A, Wee J, Awad D, Linz JE. 2011. Stress related transcription factor AtfB integrates secondary metabolism with oxidative stress response in aspergilli. J. Biol. Chem. 286:35137–35148. 10.1074/jbc.M111.253468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawasaki L, Wysong D, Diamond R, Aguirre J. 1997. Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J. Bacteriol. 179:3284–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson CH, Klotz MG, York JL, Kruft V, McEwen JE. 2002. Redundancy, phylogeny and differential expression of Histoplasma capsulatum catalases. Microbiology 148:1129–1142 [DOI] [PubMed] [Google Scholar]
- 32.Vainshtein BK, Melik-Adamyan WR, Barynin VV, Vagin AA, Grebenko AI, Borisov VV, Bartels KS, Fita I, Rossmann MG. 1986. Three-dimensional structure of catalase from Penicillium vitale at 2.0 A resolution. J. Mol. Biol. 188:49–61. 10.1016/0022-2836(86)90479-1 [DOI] [PubMed] [Google Scholar]
- 33.Hong SY, Roze LV, Wee J, Linz JE. 2013. Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in aspergilli. MicrobiologyOpen 2:144–160. 10.1002/mbo3.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roze LV, Arthur AE, Hong SY, Chanda A, Linz JE. 2007. The initiation and pattern spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol. Microbiol. 66:713–726. 10.1111/j.1365-2958.2007.05952.x [DOI] [PubMed] [Google Scholar]
- 35.Hohmann S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300–372. 10.1128/MMBR.66.2.300-372.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du C, Sarfati J, Latge JP, Calderone R. 2006. The role of the sakA (Hog1) and tcsB (s1n1) genes in the oxidant adaptation of Aspergillus fumigatus. Med. Mycol. 44:211–218. 10.1080/13693780500338886 [DOI] [PubMed] [Google Scholar]
- 37.Angelova MB, Pashova SB, Spasova BK, Vassilev SV, Slokoska LS. 2005. Oxidative stress response of filamentous fungi induced by hydrogen peroxide and paraquat. Mycol. Res. 109:150–158. 10.1017/S0953756204001352 [DOI] [PubMed] [Google Scholar]
- 38.Wei H, Requena N, Fischer R. 2003. The MAPKK kinase SteC regulates conidiophore morphology and is essential for heterokaryon formation and sexual development in the homothallic fungus Aspergillus nidulans. Mol. Microbiol. 47:1577–1588. 10.1046/j.1365-2958.2003.03405.x [DOI] [PubMed] [Google Scholar]
- 39.Ma Y, Qiao JM, Liu W, Wan Z, Wang X, Calderone R, Li R. 2008. The sho1 sensor regulates growth, morphology, and oxidant adaptation in Aspergillus fumigatus but is not essential for development of invasive pulmonary aspergillosis. Infect. Immun. 76:1695–1701. 10.1128/IAI.01507-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han KH, Seo JA, Yu JH. 2003. A putative G protein-coupled receptor controls growth, germination and coordinated development in Aspergillus nidulans. Fungal Genet. Newsl. 50(Suppl):177 [Google Scholar]
- 41.Mert HH, Ekmekci S. 1987. The effect of salinity and osmotic pressure of the medium on the growth, sporulation and changes in the total organic acid content of Aspergillus flavus and Penicillium chrysogenum. Mycopathologia 100:85–89. 10.1007/BF00467099 [DOI] [PubMed] [Google Scholar]
- 42.El-Abyad MS, Hindorf H, Rizk MA. 1988. Impact of salinity stress on soil-borne fungi of sugarbeet. Plant Soil 110:33–37. 10.1007/BF02143536 [DOI] [Google Scholar]
- 43.Whipps JM, Magan N. 1986. Effects of nutrient status and water potential of media on fungal growth and antagonist-pathogen interactions. EPPO Bull. 17:581–591 [Google Scholar]
- 44.Ramos AJM, Magan N, Sanchis V. 1999. Osmotic and matric potential effects on growth, sclerotia and partitioning of polyols and sugars in colonies and spores of Aspergillus ochraceus. Mycol. Res. 103:141–147. 10.1017/S0953756298006819 [DOI] [Google Scholar]
- 45.Geiser DM, Timberlake WE, Arnold LM. 1996. Loss of meiosis in Aspergillus. Mol. Biol. Evol. 13:809–817. 10.1093/oxfordjournals.molbev.a025641 [DOI] [PubMed] [Google Scholar]
- 46.Horn BW, Moore GG, Carbone I. 2009. Sexual reproduction in Aspergillus flavus. Mycologia 101:275–280. 10.3852/08-205 [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto K, Arima TH, Iwashita K, Yamada O, Gomi K, Akita O. 2008. Aspergillus oryzae atfB encodes a transcription factor required for stress tolerance in conidia. Fungal Genet. Biol. 45:922–932. 10.1016/j.fgb.2008.03.009 [DOI] [PubMed] [Google Scholar]
- 48.Miller MJ, Roze LV, Trail F, Linz JE. 2005. Role of cis-acting sites NorL, a TATA box, and AflR1 in nor-1 transcriptional activation in Aspergillus parasiticus. Appl. Environ. Microbiol. 71:1539–1545. 10.1128/AEM.71.3.1539-1545.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.