Abstract
Streptogramin antibiotics are divided into types A and B, which in combination can act synergistically. We compared the molecular interactions of the streptogramin combinations Synercid (type A, dalfopristin; type B, quinupristin) and NXL 103 (type A, flopristin; type B, linopristin) with the Escherichia coli 70S ribosome by X-ray crystallography. We further analyzed the activity of the streptogramin components individually and in combination. The streptogramin A and B components in Synercid and NXL 103 exhibit synergistic antimicrobial activity against certain pathogenic bacteria. However, in transcription-coupled translation assays, only combinations that include dalfopristin, the streptogramin A component of Synercid, show synergy. Notably, the diethylaminoethylsulfonyl group in dalfopristin reduces its activity but is the basis for synergy in transcription-coupled translation assays before its rapid hydrolysis from the depsipeptide core. Replacement of the diethylaminoethylsulfonyl group in dalfopristin by a nonhydrolyzable group may therefore be beneficial for synergy. The absence of general streptogramin synergy in transcription-coupled translation assays suggests that the synergistic antimicrobial activity of streptogramins can occur independently of the effects of streptogramin on translation.
INTRODUCTION
Bacterial infections caused by antibiotic-resistant clinical isolates are an emerging medical threat. Based on conservative assumptions made by the Centers for Disease Control and Prevention (CDC), at least two million people acquire life-threatening infections caused by antibiotic-resistant bacterial strains in the United States every year, resulting in 23,000 deaths. Despite the constant need for new antibiotics, the number of new antibiotics approved by the FDA has significantly decreased over the last decade (1, 2). Antibiotics that target the bacterial ribosome specifically interfere with key processes of protein synthesis, such as mRNA decoding and peptide bond formation (3). The streptogramin antibiotics produced by some Streptomyces strains inhibit protein synthesis by interfering with peptide bond formation and by blocking the peptide exit tunnel in the large (50S) ribosomal subunit, which prevents the extension of the polypeptide chain (Fig. 1A). Streptogramin antibiotics are depsipeptides consisting of two chemically distinct types, a smaller type A and a larger type B. Streptogramin antibiotics have been used as growth promoters in food-producing animals for >50 years (4) but only began to be used to treat human infections with the approval of dalfopristin-quinupristin (Synercid), an injectable pair of streptogramin antibiotics (5), in 1999.
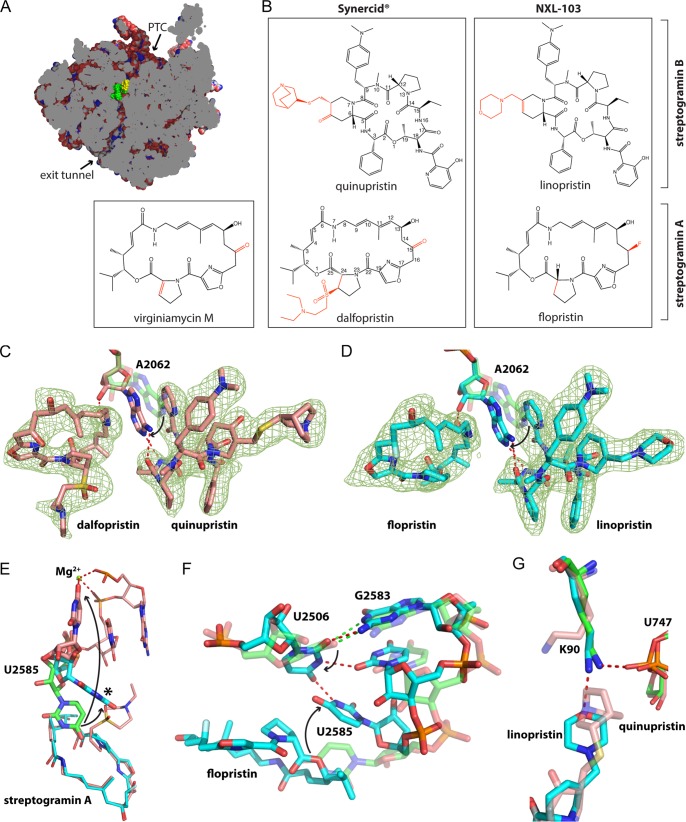
FIG 1.
Structures of streptogramins bound to the E. coli 70S ribosome. (A) Cross-section through the bacterial 50S subunit with the peptidyl transferase center (PTC) and the exit tunnel labeled. Streptogramin A (yellow) and streptogramin B (green) are shown. (B) Chemical structure of streptogramin antibiotics, with differences indicated in red. (C and D) Unbiased Fobs − Fcalc difference density shown as mesh with chemical structure of Synercid and NXL 103, respectively. Residue A2062 in the vacant 70S ribosome is shown in transparent green. (E) U2585 assumes different positions in the vacant 70S ribosome (green) and the 70S ribosome in complex with either NXL 103 (cyan) or Synercid (salmon) and is shown by thick bonds. Adjacent nucleotides, the flopristin, and the dalfopristin component are shown by thin bonds. The asterisk indicates the position of U2585 when bound to either dalfopristin under hydrolytic conditions or NXL 103. (F) Conformational changes between the vacant ribosome (green) and the NXL 103 bound ribosome (cyan) are shown. Identical changes are observed in the structure of dalfopristin under hydrolyzing conditions. (G) Interaction of linopristin (cyan) and quinupristin (salmon) with K90 of ribosomal protein L22. (C to G) Hydrogen bonds are indicated by red dashed lines. Hydrogen bonds in the vacant ribosome in panel F are shown by green dashed lines. Arrows indicate conformational changes of rRNA nucleotides upon binding of the different streptogramin compounds.
To counteract the spread of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in the 1990s, Synercid was developed as a 70:30 (wt/wt) mixture of dalfopristin to quinupristin. Synercid was approved in 1999 for the treatment of life-threatening infections caused by vancomycin-resistant Enterococcus faecium (VREF) and complicated skin and skin structure infections (cSSSIs) caused by S. aureus or Streptococcus pyogenes and is currently the only clinically used streptogramin antibiotic. To overcome venous irritation caused by Synercid, and in order to reduce health care costs, a new and orally available streptogramin combination, flopristin-linopristin (NXL 103), a 70:30 (wt/wt) mixture of flopristin to linopristin, has been developed for use in the outpatient setting. NXL 103 has been shown to be more effective than Synercid in treating a large number of Gram-positive bacteria and their clinical isolates (6–9) and also has the potential to become an important drug in the treatment of community-acquired pneumonia and complex skin and soft tissue infections, including MRSA (10).
Both the streptogramin type A (dalfopristin and flopristin) and type B components (quinupristin and linopristin) differ between Synercid and NXL 103. The streptogramin A components dalfopristin of Synercid and flopristin of NXL 103 differ by a carbonyl oxygen at position 15 in dalfopristin, which is replaced by a fluorine in flopristin. Furthermore, dalfopristin is derivatized on its pyrrolidine group with a diethylaminoethylsulfonyl group (Fig. 1B). Rapid hydrolysis of the diethylaminoethylsulfonyl group at a physiological pH converts dalfopristin to virginiamycin M (11). In vitro studies have shown that the streptogramin A flopristin in NXL 103 has higher antimicrobial activity than that of its counterpart in Synercid, dalfopristin, suggesting that the increased activity of NXL 103 is mainly due to flopristin (6, 7). The streptogramin B component quinupristin of Synercid carries a quinuclidinylthiomethyl group compared to a methylmorpholine group in linopristin of NXL 103 (Fig. 1B). Both quinupristin and linopristin are derived from virginiamycin S, which is not functionalized at its oxopiperidinyl group.
Streptogramins A and B must be used in combination, due to the fact that the individual streptogramin components exert a bacteriostatic effect, whereas their combination is bactericidal (12). Streptogramin A and B antibiotics act synergistically in vitro and in animal models of infection (13–15). Synergy between two antibiotics may be advantageous because it reduces the likelihood of bacterially acquired resistance mutations at their binding sites. Furthermore, in order to achieve a given bactericidal effect, smaller doses of the individual compounds are required, reducing potential side effects caused by the toxicity of the components. As opposed to experiments in vivo and in vitro, synergy has not been studied extensively in transcription-coupled translation (TT) assays with natural mRNAs. In cell-free translation assays using poly(U)-directed polyphenylalanine synthesis, streptogramin A compounds showed activity, whereas streptogramin B compounds were inactive. However, streptogramin B antibiotics were active in inhibiting the translation of poly(A·C) synthetic messages (16–18). In cell-free translation studies using cell extracts of the Gram-positive bacterium Bacillus subtilis infected with phage 2C, translation inhibition by a combination of virginiamycin M plus virginiamycin S was higher than the sum of the inhibitory effects of the individual virginiamycin components (19). Furthermore, in different studies, the binding affinity of streptogramin B was shown to increase in the presence of streptogramin A compared to the binding affinity of streptogramin B alone (20, 21). However, these measurements were performed on purified ribosomes, which may not reflect the properties of actively translating ribosomes in intact cells or in transcription-coupled translation systems. The crystal structures of virginiamycin antibiotics that are bound to the large ribosomal subunit of the extreme halophilic archaeon Haloarcula marismortui and of Synercid to the extremophile Deinococcus radiodurans (22–24) reveal that nucleotide A2062 in 23S rRNA, which is present in the binding pocket of both streptogramins, changes conformation upon streptogramin A binding only, which may explain the increased affinity for streptogramin B in the presence of streptogramin A.
Here, we report the crystal structures of Synercid and NXL 103, as well as their individual components bound to the intact Escherichia coli 70S ribosome, which is more closely related to the ribosomes of pathogenic bacteria. Furthermore, we comprehensively tested the activities of individual streptogramin components in biochemical and microbiological assays and the synergy between the streptogramin A and B components. Antimicrobial assays were used to determine the MICs of individual streptogramin antibiotics, and “checkerboard” assays (25) were used to quantify the synergistic effects between the streptogramin A and B components. We also measured the activities of the individual streptogramin components and their combinations on protein synthesis in transcription-coupled translation extracts. Finally, we measured the affinity of streptogramin B to purified intact E. coli ribosomes either alone or in the presence of streptogramin A in order to investigate the synergistic effect at the molecular level.
MATERIALS AND METHODS
Ribosome purification, ribosome crystallization, and antibiotic binding experiments.
Ribosomes were purified from E. coli strain MRE600 cells as described previously (26). Crystals were grown from purified ribosomes as described previously (27). For antibiotic soaking experiments, ribosome crystals were soaked overnight in cryoprotection buffer supplemented with either premixed Synercid (348 μM dalfopristin plus 101 μM quinupristin), NXL 103 (120 μM flopristin plus 70 μM linopristin), or with individual components (dalfopristin, quinupristin, flopristin, or linopristin) at a concentration of 100 μM. All components were dissolved in dimethyl sulfoxide (DMSO) to a concentration of ∼100 mM, except for premixed Synercid, which was dissolved in DMSO to a concentration of 348 mM dalfopristin and 101 mM quinupristin. The DMSO stocks were diluted 1,000-fold in cryoprotection buffer for the soaking experiments, and the crystals were flash-frozen in liquid nitrogen for the diffraction experiments. The premixed Synercid (30% quinupristin and 70% dalfopristin [wt/wt]) was a generous gift from Pfizer, flopristin and linopristin were provided by AstraZeneca, and dalfopristin and quinupristin were purchased from International Laboratory USA. Virginiamycin M1 was purchased from Sigma-Aldrich.
X-ray diffraction experiments and model building.
The X-ray diffraction data were measured at beamlines 8.3.1 and 12.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory, and at beamline 11-1 at the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, with oscillation ranges of 0.2 to 0.3° at 100 K recorded on an ADSC Q315r detector or a Dectris Pilatus 6M detector. The diffraction data were reduced, scaled, and converted using the XDS Program Package (28). Structure factors for the difference electron density maps were calculated using the phenix.refine component of the PHENIX software suite (29). Antibiotics were modeled in the unbiased difference electron density maps obtained from each complex after molecular replacement. Changes to the rRNA structure were made using Coot (30), followed by individual atomic displacement parameter (ADP) refinement using phenix.refine. The restraint files for each antibiotic structure were generated using phenix.elbow of the PHENIX software suite.
Dalfopristin hydrolysis.
DMSO stocks of dalfopristin and virginiamycin M1 at 4 mM were dissolved in 0.01 M sodium phosphate buffer (pH 7.4) or in cation-adjusted Mueller-Hinton II (CAMH II) medium adjusted to pH 6.0 to yield a final concentration of 200 μM target compound. The samples were prepared and monitored continuously for up to 24 h at 37°C. The hydrolysis of the target compounds was monitored via liquid chromatography-mass spectrometry (LC-MS) selected-ion monitoring (SIM) in electrospray ionization (ESI)+ and ESI− mode. The natural log of the area responses for dalfopristin and virginiamycin M1 were plotted against time, and first-order kinetics were used to derive the half-life (t1/2) of each compound.
Prehydrolyzed dalfopristin for antimicrobial assays and transcription-coupled translation assays was prepared by dissolving dalfopristin to a concentration of 2.5 mM in 50 mM ammonium acetate (pH 7.5) containing 30% DMSO. The hydrolysis of dalfopristin was performed over 24 h at 4°C.
Antimicrobial susceptibility testing.
The MIC against each isolate was determined by the broth microdilution technique in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines found in document M07-A9 (31). The susceptibility breakpoint interpretations for reference compounds along with the quality control (QC) ranges for reference strains are described in CLSI document M100-S23 (32). Following incubation, MIC values were determined visually and reported as the lowest concentration of drug that completely inhibited the growth of the strain. A comparison of the streptogramins dalfopristin, quinupristin, flopristin, linopristin, hydrolyzed dalfopristin, and virginiamycin M was run versus five ATCC reference strains, E. coli ATCC 29417 (MRE 600), Enterococcus faecalis ATCC 29212, S. aureus ATCC 29213, Haemophilus influenzae ATCC 49247, and Streptococcus pneumoniae ATCC 49619.
“Checkerboard” assay.
A checkerboard assay described by Pillai et al. (33) was used to determine the microbiological interaction with pairs of agents in a two-dimensional array. The MICs of the streptogramin combinations dalfopristin-quinupristin and flopristin-linopristin, as well as of the individual components, were determined by the broth microdilution technique in accordance with Clinical and Laboratory Standards Institute guidelines in document M07-A9 (31) against three ATCC cultures, E. coli ATCC 29417 (MRE600), E. faecalis ATCC 29212, and S. aureus ATCC 29213. By comparing the MIC of an agent alone to the MIC combination, a series of fractional inhibitory concentrations (FICs) can be calculated, followed by FIC indices (34). The MIC values were determined for each isolate against combinations of streptogramins. The agent combinations were measured as fold reductions in the MIC based on the original MIC of the agent alone. The FICs were calculated by dividing the MIC of the agent alone by the MIC of the agent when tested in combination. The FIC index was obtained by adding the FICs. The FIC indices were interpreted as synergistic when values were ≤0.5, additive/indifferent when values were >0.5 to 4.0, and antagonistic when values were >4.0. The following equation was used to calculate the FIC indices: FICA + FICB = (MIC of A in combination/MIC of A alone) + (MIC of B in combination/MIC of B alone). A mean FIC index for synergic values was used to determine the possibility of drug interactions and to interpret the results according to accepted criteria (35).
Transcription-coupled translation assay.
Plasmid pKK3535 containing the E. coli rrnB operon encoding rRNA and a chloramphenicol resistance cassette was used for mutagenesis. U1782 and U2586 were mutagenized using the QuikChange kit from Agilent, and the sequence of the entire operon was verified by sequencing. E. coli SCB 53 cells, in which all endogenous rRNA genes were deleted and replaced by a pKK3535 plasmid with an ampicillin resistance cassette (36), were transformed with the mutagenized pKK3535 plasmid. The cells were first grown in liquid culture supplemented with chloramphenicol and then plated on LB agar plates containing chloramphenicol. Successful plasmid shuffling was confirmed by the absence of growth on an LB agar plate containing ampicillin for single colonies picked from an LB agar plate supplemented with chloramphenicol.
Transcription-coupled translation assays were performed essentially according to Buurman et al. (37). E. coli MRE600 cells were used to prepare wild-type S30 cell extract, as described previously (37). Mutant E. coli S30 cell extract was prepared as wild-type extract but using an E. coli strain bearing a U1782C U2586C double mutant. For the transcription-coupled translation assay, the following reagents were used: reagent 1 consisted of 0.5 mM (each) ATP, CTP, UTP, and GTP (Chem-Impex International, Wood Dale, IL), 20 mM phosphoenolpyruvate (Chem-Impex International), 100 μg/ml E. coli tRNA (Roche Diagnostics Corp., Indianapolis, IN), 20 μg/ml folinic acid, 1 mM cyclic AMP (cAMP), 0.8 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 0.2 mM dithiothreitol (DTT), 30 mg/ml polyethylene glycol 8000, 0.5 mM (each) all 20 of the translated amino acids, 2 U/ml pyruvate kinase, and 40 μg/ml pLH1824 reporter plasmid DNA; reagent 2 contained S30 extract prepared either from the E. coli MRE600 strain (Paragon Bioservices, Baltimore, MD) or from a mutant E. coli strain, as described above. The S30 extract was diluted to 2 mg/ml in S30 buffer consisting of 10 mM Tris-acetate (pH 7.4), 60 mM potassium acetate, and 14 mM magnesium acetate. Both reagents were allowed to preincubate at room temperature for 1 h.
To determine the 50% inhibitory concentrations (IC50s) of the streptogramins, the compounds were dissolved in DMSO to 2.5 mM and serially diluted 2-fold in DMSO. Hydrolyzed dalfopristin was dissolved in hydrolyzing buffer (see “Dalfopristin hydrolysis” above) and serially diluted in DMSO. The diluted compound (0.3 μl) was added to 384-well white polystyrene assay plates (Corning, Inc., Lowell, MA) using the Bravo automated liquid handling platform (Agilent Technologies, Santa Clara, CA). To determine additive/synergistic effects between class A and B streptogramins, two experiments were carried out. In the first experiment, the effect of streptogramin B on the activity of streptogramin A was determined. Class A streptogramins were serially diluted and added to the assay plate using the Bravo platform, as described above. Subsequently, class B streptogramins dissolved in DMSO were added to the assay plate using the Echo liquid handler (Labcyte, Inc., Sunnyvale, CA) to yield a final concentration in the reaction mixture equal to their respective IC50 values. In a second experiment, the effect of streptogramin A on the activity of streptogramin B was determined. Class A compounds were added to the assay plates using the Bravo platform to yield a concentration of their respective IC50 values in the final reaction mixture. In a second step, serially diluted class B compounds (0.3 μl) were added to the assay plate using the Echo liquid handler. Therefore, in the first experiment, the Bravo platform was used to deliver a serial dilution of the streptogramin A compound, whereas in the second experiment, the same platform was used to deliver the streptogramin A compound fixed at its IC50. For the percent inhibition calculations, luminescence intensity from the samples receiving the maximum dose of compounds were defined as 100% inhibition, and the samples with minimum dose of compounds were defined as 0% inhibition. The mean IC50 values from a representative experiment performed in triplicate are shown with standard deviation (Fig. 2B to H; see Tables S2 to S4 in the supplemental material).
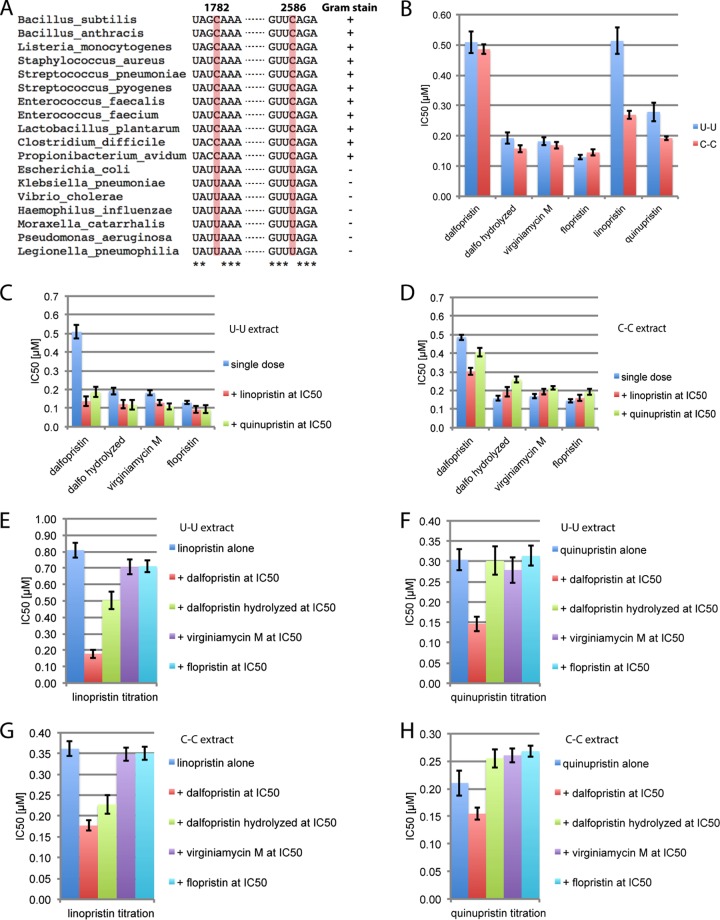
FIG 2.
Activities of different streptogramin combinations for wild-type and mutant 70S ribosomes. (A) Sequence alignment of 23S rRNA from various Gram-positive and Gram-negative pathogens. Nucleotides 1782 and 2586 (shaded in red) form a base pair (see Fig. S5 and S6 in the supplemental material). Nucleotides that are conserved in all shown pathogens are indicated by an asterisk. (B to H) IC50 values in μM for different streptogramin antibiotics determined in transcription-coupled translation assays from E. coli bearing either its intrinsic U-U base pair (B, C, E, and F) at position 1782 to 2586 or a C-C base pair (B, D, G, and H). (C and D) Comparison of IC50 values of streptogramin A components in transcription-coupled translation assays either alone or in the presence of streptogramin B at its IC50. (E to H) Comparison of IC50 values of streptogramin B components in transcription-coupled translation assays either alone or in the presence of various streptogramin A components at their IC50 value. The standard deviation of the IC50 values, measured in triplicate, is indicated by the error bars. Dalfo, dalfopristin.
After streptogramin compounds were added to the assay plate, 15 μl of reagent 1 was added, followed by 15 μl of reagent 2. The plate was then briefly shaken in an Eppendorf MixMate plate shaker. The plates were sealed with foil and incubated at room temperature for 30 min, avoiding temperature fluctuation. Subsequently, 15 μl of luciferin developer consisting of 0.4 μM lithium salt of coenzyme A (Sigma), 0.7 μM d-luciferin (Gold Biotechnology, St. Louis, MO), 0.8 μM ATP, 20 mM Tricine (pH 7.8), 1 mM magnesium carbonate, 0.1 mM EDTA, 2.3 mM magnesium sulfate, and 33 mM DTT was added, and light production was measured immediately using a PHERAstar plate reader (BMG Labtech, Ortenberg, Germany).
Isothermal titration calorimetry.
Isothermal titration calorimetry was performed in a MicroCal Auto-iTC200 system (GE Healthcare). Purified E. coli 70S ribosomes were dialyzed against 20 mM Tris HCl (pH 7.5), 60 mM NH4Cl, 6 mM MgCl2, 0.5 mM EDTA, and 2 mM Tris(2-carboxyethyl)phosphine (TCEP) at 4°C either without or in the presence of 1.5 equivalent dalfopristin or flopristin in the dialysis buffer. Quinupristin or linopristin (39 μM) dissolved in dialysis buffer was titrated into the cell containing 2.2 to 3.9 μM 70S ribosome either alone or in the presence of dalfopristin or flopristin, with a first injection of 0.5 μl volume, followed by 12 injections of 3.1 μl volume at 4°C.
PDB accession codes.
The coordinates and structure factors for 70S ribosomes bound to streptogramin antibiotics were deposited in the Protein Data Bank (PDB). There are four entries for each complex. The electron density maps of the first ribosomes are better than those of the second ribosome in the asymmetric unit. Therefore, entries for the subunits of the first ribosome are of better quality than those of the second ribosome. The codes are 4TP8, 4TP9, 4TPA, and 4TPB (dalfopristin and quinupristin [Synercid]), 4TPC, 4TPD, 4TPE, and 4TPF (flopristin and linopristin [NXL 103]), 4TOU, 4TOV, 4TOW, and 4TOX (flopristin), 4TOL, 4TOM, 4TON, and 4TOO (linopristin), 4TP0, 4TP1, 4TP2, and 4TP3 (dalfopristin), 4PE9, 4PEB, 4PEA, and 4PEC (quinupristin), and 4TP4, 4TP5, 4TP6, and 4TP7 (dalfopristin under hydrolyzing conditions, which is chemically equivalent to virginiamycin M).
RESULTS
NXL 103 has been shown to be more potent than Synercid against a variety of Gram-positive pathogens and H. influenzae (8, 38). In order to shed light on the increased activity of NXL 103 versus that of Synercid, we solved crystal structures of the 70S E. coli ribosome in complex with either Synercid or NXL 103, or their individual components, at 2.8- to 3.0-Å resolution (see Table S1 in the supplemental material). Positive difference electron density was observed in an unbiased Fobs − Fcalc map for both the streptogramin A and B components. The chemical structures of streptogramin A and B were modeled into the electron density, thereby unambiguously identifying their locations, orientations, and conformations (Fig. 1C and D; see also Fig. S1 in the supplemental material).
Binding mode of Synercid.
Dalfopristin binds to the large ribosomal subunit at the entrance of the exit tunnel to a hydrophobic pocket (Fig. 1A). The macrolactone ring stacks under the bases of G2061 and A2451, thereby forming favorable hydrophobic interactions between its aliphatic stretch (C-9 to C-12) and the base of G2061, as well as between its oxazole group and the base of A2451. Upon binding of the streptogramin pair, the base of A2062 moves toward the streptogramin A molecule to form stacking interactions with both streptogramin A and B (Fig. 1C). As a result, the amide bond of dalfopristin stacks on the base of A2062. The streptogramin-induced movement of A2062 positions the 2′-OH group of A2062 to form a hydrogen bond with the amide oxygen of dalfopristin (Fig. 1C). Furthermore, the carbonyl oxygen of C-6 and C-26 each form a hydrogen bond with the exocyclic amino group of G2061, and the only hydroxyl group of dalfopristin forms a hydrogen bond with the nonbridging phosphate oxygen of G2505. The structure reveals that acetylation of the C-13 hydroxyl group, a known resistance mechanism (39), would interfere with dalfopristin binding by steric hindrance.
Quinupristin, the streptogramin B component of Synercid, binds to the large ribosomal subunit at the entrance of the exit tunnel adjacent to dalfopristin (Fig. 1A and C). The hydroxypicoline group of quinupristin stacks on the opposite face of the base of A2062 compared to dalfopristin. This positions the base of A2062 to form two hydrogen bonds with quinupristin, between the N-1 and N-6 of A2062 and the N-16 amide nitrogen and the C-14 carbonyl oxygen of quinupristin, respectively (Fig. 1C). These contacts and favorable stacking interactions are likely disrupted by an A2062C mutation found in S. pneumoniae, which confers resistance to streptogramins and macrolides (40). Quinupristin stacks with its aliphatic stretch comprising atoms C-17 to C-19, including its C-19 methyl group on the base of U2586, and also with its phenyl group under the ribose of U2609. The dimethylaminophenyl group of quinupristin stacks on the edge of A2058. This hydrophobic interaction is likely crucial for streptogramin B binding, since A2058 N6 methylation by erm methyltransferases and mutation of A2058 to either G, U, or C leads to streptogramin B-resistant phenotypes in Helicobacter pylori (41) and E. coli (42).
The binding of Synercid to the large ribosomal subunit causes dramatic structural rearrangements in the peptidyl transferase center (PTC). In the vacant E. coli ribosome, the base of U2585 reaches into the entrance of the exit tunnel to a position that is occupied by the lactone group of dalfopristin when Synercid is bound. The binding of Synercid causes nucleotide U2585 to flip roughly 160° away from the peptidyl transferase center. In this alternate conformation, the O-4 of U2585 coordinates a magnesium ion, which in turn coordinates one nonbridging phosphate oxygen of each of the two proceeding residues, U2584 and G2583 (Fig. 1E). This magnesium ion, which is not present in the vacant ribosomal structure, and the base flip of U2585 force the G2583·U2506 wobble base pair to break and nucleotide U2506 to move out of the plane of the G·U base pair. Although the vacant archaeal H. marismortui 50S ribosomal subunit and vacant E. coli 70S ribosomal structures differ in the positioning of A2503 in the exit tunnel when either virginiamycin M or both virginiamycin M and virginiamycin S are bound, A2503 and A2062 assume identical positions in the H. marismortui 50S subunit (22, 24), compared to their positions observed in the E. coli ribosome bound to Synercid.
Binding mode of NXL 103.
Flopristin binds to the same site as dalfopristin and adopts similar hydrophobic interactions and hydrogen bonding patterns with the ribosome. Flopristin differs from dalfopristin by a fluorine atom bonded to an sp3 hybridized C-15 carbon, which is a carbonyl group in dalfopristin. Neither the carbonyl of dalfopristin nor the fluorine of flopristin are within hydrogen bonding distance of the ribosomal components. Despite the highly similar binding modes of dalfopristin and flopristin, the absence of the diethylaminoethylsulfonyl group in flopristin (Fig. 1B) has a dramatic effect on the conformation of the peptidyl transferase center (PTC). Unlike the binding of Synercid, NXL 103 causes nucleotide U2585 to flip only by about 45° and its base to rotate by ∼90° compared to the vacant 70S ribosomal structure (Fig. 1E). This conformational change causes the G2583·U2506 wobble base pair to break. U2506 flips out of the plane of the G·U wobble base pair and clamps together U2583 to G2585 by an intricate network of hydrogen bonds, thereby stabilizing this alternate conformation of the PTC (Fig. 1F). Interestingly, if the complex of dalfopristin bound to the E. coli 70S is crystallized under conditions that favor the hydrolysis of the diethylaminoethylsulfonyl group, U2585, U2584, and G2583 together with their base pairing partners assume a conformation identical to that seen in the complex with flopristin (Fig. 1E).
Linopristin, the streptogramin B component of NXL 103, binds to the same site as its counterpart quinupristin of Synercid and maintains all of the hydrophobic interactions and hydrogen bonding interactions with A2062. Linopristin differs from quinupristin by the replacement of the quinuclidinylthiomethyl group in quinupristin with a methylmorpholine group in linopristin (Fig. 1B). The morpholine of linopristin does not penetrate the exit tunnel as far as the quinuclidinyl group of dalfopristin, and it forms a hydrogen bond to the ε-amino group of lysine 90 in ribosomal protein L22, which in turn forms an additional hydrogen bond with a nonbridging phosphate oxygen of U747. In the vacant ribosomal structure, K90 forms a salt bridge with a nonbridging phosphate oxygen of U747, and in the quinupristin structure, the quinuclidinyl group displaces the side chain of K90, thereby disrupting its interaction with U747 (Fig. 1G). Interestingly, when linopristin alone is bound to the ribosome, the morpholine group no longer forms a hydrogen bond to K90, and furthermore, A2058 and A2059 move apart by 1 Å. The dimethylaminophenyl group of linopristin moves over the newly formed gap and no longer stacks on the edge of A2058 (see Fig. S2 in the supplemental material). This suggests that the binding mode of linopristin shifts due to the presence of the streptogramin A component.
The structures of Synercid and NXL 103 and their individual components (except for linopristin alone) reported here superimpose within the limits of coordinate error with virginiamycin M + S bound to the H. marismortui 50S ribosomal subunit (22, 24), whereas the present structures differ from those reported for Synercid bound to the D. radiodurans 50S ribosomal subunit (23). For example, the macrolactone rings of both the streptogramin A and B components in the D. radiodurans 50S subunit are shifted to different extents with respect to their counterparts in the structures reported here and in the H. marismortui 50S subunit structures. In the D. radiodurans 50S subunit, both the pyrrolidine and oxazole groups of the streptogramin A component are displaced compared to their counterparts in the E. coli and H. marismortui structures. Further, A2062 assumes a different conformation in the D. radiodurans context and therefore fails to stack on the amide group of streptogramin A and the hydroxypicoline group of streptogramin B. Finally, the dalfopristin-specific diethylaminoethylsulfonyl group and U2585 in the D. radiodurans structure assume different conformations compared to the structure of Synercid bound to the E. coli 70S ribosome. The observed differences between the Synercid structures of E. coli and D. radiodurans might be due to the fact that the ribosomes are from divergent bacterial species or due to the low resolution of the D. radiodurans structure (3.4 Å) compared to the resolutions of the present structures (see Table S1 in the supplemental material). Given the similarity of the present E. coli structures with those of the H. marismortui 50S subunit, the binding mode of Synercid and the associated conformational changes in the ribosome observed here probably reflect the relevant structures for understanding streptogramin interactions with ribosomes in pathogenic bacteria.
Dalfopristin hydrolysis.
Dalfopristin has been reported to hydrolyze rapidly to virginiamycin M under physiological conditions (11). In order to estimate the effects of dalfopristin hydrolysis on its potency, we first determined the hydrolysis rate of dalfopristin at pH 7.4 and pH 6.0 using mass spectrometry (see Fig. S4 in the supplemental material). Dalfopristin hydrolyzes to virginiamycin M with a half-life (t1/2) of about 11 min at pH 7.4 and 77 min at pH 6.0. The hydrolysis product virginiamycin M is very stable at pH 7.4, with a half-life of about 45 h. The antimicrobial assays performed in this study ran over the course of 16 to 24 h; thus, the effect of dalfopristin in these experiments can be attributed to its hydrolysis product virginiamycin M. The transcription-coupled translation assays lasted 30 min at pH 7.5 and, therefore, substantial hydrolysis of dalfopristin has occurred by that point. However, due to the shorter time frame of the transcription-coupled translation assays, the activity of dalfopristin can in part be attributed to its unhydrolyzed form.
Antimicrobial activities of streptogramin antibiotics.
In order to determine the antimicrobial activities of streptogramin antibiotics on living cells, the MICs were determined by growing different Gram-positive and Gram-negative pathogens in the presence of different concentrations of streptogramin antibiotics. The MICs for streptogramin antibiotics were generally in the low μg/ml range, with the exception of streptogramin B in Gram-negative species and streptogramin A in E. faecalis. The streptogramin B antibiotics quinupristin and linopristin had very little to no effect on growth of the Gram-negative pathogens E. coli and H. influenzae, which is likely due to the inability of these antibiotics to permeate the outer cell membrane; this is in agreement with previous results (43). E. faecalis showed MICs of 64 μg/ml toward streptogramin A antibiotics, likely due to its lsa gene, which encodes a putative ABC transporter leading to streptogramin A efflux (44). In all other bacterial strains tested, flopristin showed higher activity than dalfopristin, and linopristin showed equal or lower activity than quinupristin (Table 1). For all strains, the MIC values for dalfopristin administered in its prehydrolyzed form were within 2-fold of the MIC values of virginiamycin M, which is chemically identical to the hydrolysis product of dalfopristin.
TABLE 1.
Antimicrobial activity of different streptogramin antibioticsa
| Streptogramin | Typeb | MIC (μg/ml) for: |
||||
|---|---|---|---|---|---|---|
| E. coli ATCC 29417 (MRE600) | H. influenzae ATCC 49247 | E. faecalis ATCC 29212 | S. aureus ATCC 29213 | S. pneumoniae ATCC 49619 | ||
| Dalfopristin | A | 4 | 1 | 64 | 4 | 4 |
| Quinupristin | B | >64 | 32 | 8 | 4 | 1 |
| Flopristin | A | 1 | 0.25 | 64 | 1 | 2 |
| Linopristin | B | >64 | >64 | 8 | 32 | 4 |
| Hydrolyzed dalfopristin | A | 4 | 2 | 32 | 4 | 4 |
| Virginiamycin M | A | 4 | 4 | >64 | 8 | 8 |
| Synercid | A+B | 8 | 8 | 4 | 0.25 | 0.25 |
| NXL 103 | A+B | 2 | 0.5 | 1 | 0.125 | 0.125 |
MICs were determined by the broth microdilution technique.
Streptogramin type A or B or combination (A+B). Note that the A+B combinations are formulated in an A-to-B ratio of 70:30 (wt/wt).
We also used a checkerboard assay to evaluate the synergy of Synercid and NXL 103 in E. coli, E. faecalis, and S. aureus. The streptogramin components of Synercid and NXL 103 did not show a synergistic mode of action in E. coli. This finding is expected, since individually, quinupristin and linopristin did not affect bacterial growth even at the highest concentrations tested (Table 1). A synergistic mode of action was observed for Synercid and NXL 103 in E. faecalis, with a lower mean fractional inhibitory concentration (FIC) index for NXL 103 than that for Synercid (Table 2). For Synercid, synergy was observed at three different concentration pairs, whereas for NXL 103, synergy was observed for six different concentration pairs (Table 2). In S. aureus, greater synergy was observed for both Synercid and NXL 103 than for E. faecalis, as judged by their lower mean FIC index values for all combinations that led to synergy. Furthermore, in S. aureus, synergy occurred at lower streptogramin concentrations than those in E. faecalis for both Synercid and NXL 103 (Table 2). Taken together, these results show that NXL 103 generally exhibits a higher degree of synergy than Synercid in the context of a given bacterial strain.
TABLE 2.
Checkerboard analysis to detect synergy
| FIC data | Value(s) for streptogramin combinations for strain: |
|||
|---|---|---|---|---|
|
E. faecalis ATCC 29212 |
S. aureus ATCC 29213 |
|||
| Dalfopristin-quinupristin (Synercid) | Flopristin-linopristin (NXL 103) | Dalfopristin-quinupristin (Synercid) | Flopristin-linopristin (NXL 103) | |
| FIC index range | 0.25 to 4.0 | 0.05 to 4.0 | 0.05 to 4.0 | 0.03 to 1.0 |
| Ø FIC index alla | 0.99 | 0.81 | 0.82 | 0.22 |
| Ø FIC index all synergyb | 0.3 | 0.1 | 0.12 | 0.08 |
| Synergistic combinations (A/B)c | 16/0.5 | 4/0.06 | 0.5/0.06 | 0.06/0.06 |
| 8/1 | 2/0.125 | 0.125/0.125 | 0.06/0.125 | |
| 4/2 | 1/0.25 | 0.125/0.25 | 0.06/0.25 | |
| 0.5/0.5 | 0.06/0.5 | 0.03/0.5 | ||
| 0.25/1 | 0.06/1 | 0.03/1 | ||
| 0.125/2 | 0.06/2 | 0.015/2 | ||
| 0.015/4 | ||||
| 0.015/8 | ||||
Ø FIC index all, mean fractional inhibitory concentration index for all combinations.
Ø FIC index all synergy, mean fractional inhibitory concentration index for all combinations that lead to synergy.
Concentration of streptogramin A and streptogramin B in μg/ml that lead to synergy.
Effects of streptogramin antibiotics in transcription-coupled translation assays.
We used an E. coli-based transcription-coupled translation assay to determine the inhibitory effects of streptogramin antibiotics on actively translating ribosomes. Although the translation assay employed cell extracts from the Gram-negative organism E. coli and Synercid is indicated for the Gram-positive pathogens S. aureus and S. pyogenes, only one major sequence difference occurs for residues within a radius of 10 Å of the ribosomal binding sites of both streptogramins in a range of Gram-negative and Gram-positive pathogens (Fig. 2A; see also Fig. S5 in the supplemental material). In many Gram-negative pathogenic bacteria, including E. coli, the streptogramin B binding pocket includes a U-U base pair at position 1782–2586, whereas this base pair is a C-C base pair in many Gram-positive pathogenic bacteria, including S. aureus and S. pyogenes (Fig. 2A). Apart from this difference, changes further away from the binding pockets include unpaired U1781, which is found to be a G in B. subtilis, Bacillus anthracis, and Listeria monocytogenes and a C in Clostridium difficile and Propionibacterium avidum, and three canonical W-C base pairs (744–753, 2070–2441, and 2067–2443) that are different canonical W-C base pairs in different pathogens (see also Fig. S5). However, all these changes are located in the second or third shell of the binding site and do not immediately constitute the streptogramin-binding pocket. Notably, the crystal structure of the H. marismortui 50S reveals that a G at position 1781 does not change the geometry of the U1782-U2586 base pair (45). From this alignment, we conclude that the only difference between Gram-negative and Gram-positive pathogens and more specifically between E. coli and S. aureus/S. pyogenes that might impact streptogramin activity is the U1782C U2586C double mutation in the streptogramin B binding site (see Fig. S6 in the supplemental material). In order to account for the rRNA sequence differences between Gram-positive and Gram-negative pathogens, a mutant E. coli strain was constructed containing a C1782-C2586 base pair instead of a U-U base pair in the streptogramin B binding pocket to prepare a transcription-coupled translation extract.
We measured the concentration of the individual streptogramin A and B components that cause 50% inhibition of translation (IC50) in transcription-coupled translation assays in the context of wild-type (U-U) and mutant (C-C) streptogramin-binding pockets. Wild-type and mutant E. coli TT assays were performed with the streptogramin B compounds linopristin and quinupristin and the streptogramin A compounds flopristin, unhydrolyzed dalfopristin, virginiamycin M, and prehydrolyzed dalfopristin. Flopristin has an IC50 of 130 nM and is the compound with the highest activity in wild-type TT assays (Fig. 2B; see also Table S2 in the supplemental material). Notably, dalfopristin preincubated in buffer at pH 7.5 and therefore hydrolyzed to virginiamycin M has an IC50 of 192 nM, nearly identical to that of authentic virginiamycin M (IC50 of 182 nM), as expected. In contrast, unhydrolyzed dalfopristin is about 2.6-fold-less active than hydrolyzed dalfopristin (Fig. 2B; see also Table S2 in the supplemental material). This indicates that the diethylaminoethylsulfonyl group in dalfopristin is detrimental for activity. The activities of all streptogramin A antibiotics were also tested in the context of the Gram-positive streptogramin-binding site using the mutant (C-C) cell extract. The mutant extract yielded comparable activities to that of the wild-type E. coli extract, indicating that the U1782C U2586C double mutation does not affect streptogramin A activity. In contrast, the activity of linopristin increased 1.9-fold in the mutant (C-C) extract compared to in the wild-type (U-U) extract, and the activity of quinupristin increased 1.5-fold. Interestingly, linopristin and quinupristin have more similar activities to each other in the C-C extract than in the U-U extract (Fig. 2B; see also Table S2).
To test whether streptogramin antibiotics have synergistic activity on ribosomes translating a natural mRNA in a transcription-coupled translation assay, we determined the IC50 values of the streptogramin A antibiotics in the presence of a streptogramin B antibiotic at its IC50, and vice versa, using a luciferase reporter mRNA. The IC50 values of flopristin and virginiamycin M in the presence of either quinupristin or linopristin at their respective IC50 values decrease only marginally in the wild-type (U-U) extract. In contrast, the IC50 of unhydrolyzed dalfopristin decreases substantially in the presence of quinupristin or linopristin at their IC50 values (Fig. 2C; see also Table S3 in the supplemental material), representing an increase in dalfopristin activity up to 3.7-fold in the presence of linopristin. In the context of the Gram-positive (C-C) streptogramin-binding site, the activity change of either flopristin or virginiamycin M in the presence of either of the streptogramin B compounds is negligible (Fig. 2D; see also Table S3). Furthermore, in the context of the Gram-positive streptogramin-binding site, dalfopristin potency increases only 1.6-fold in the presence of linopristin and only 1.2-fold in the presence of quinupristin at their respective IC50 values, a substantially less pronounced effect than in the Gram-negative system (Fig. 2C and D; see also Table S3).
In the converse experiments, we held the streptogramin A component at its IC50 and varied the concentration of the streptogramin B component. In the context of the U-U streptogramin B binding site, the IC50 of either linopristin or quinupristin in the presence of flopristin or virginiamycin M remained unchanged compared to the IC50 of the respective streptogramin B antibiotic alone (Fig. 2E and F; see also Table S4 in the supplemental material). In contrast, the IC50 of linopristin decreased 4.5-fold in the presence of unhydrolyzed dalfopristin at its IC50 (Fig. 2E; see also Table S4). Furthermore, the IC50 of the other streptogramin B, quinupristin, decreased only 2.1-fold in the presence of unhydrolyzed dalfopristin at its IC50 (Fig. 2F; see also Table S4). In the context of the C-C streptogramin-binding site, the IC50 values of linopristin (Fig. 2G; see also Table S4) and quinupristin (Fig. 2H; see also Table S4) alone remained unchanged compared to their IC50 values in the presence of either flopristin or virginiamycin M. The IC50s of both linopristin and quinupristin decreased in the presence of unhydrolyzed dalfopristin at its IC50 in the context of the C-C streptogramin-binding site, but the decreases were less pronounced compared to those observed with the U-U streptogramin-binding site (Fig. 2G and H versus E and F, respectively; see also Table S4). We also analyzed the effects of prehydrolyzed dalfopristin alone, which is chemically equivalent to virginiamycin M, as well as in combination with either of the two streptogramin B components, quinupristin or linopristin. The results show that prehydrolyzed dalfopristin behaves similarly to virginiamycin M, with slight differences likely due to differences in the antibiotic stock solution compositions (see Materials and Methods).
Taken together, these results reveal that neither of the streptogramin A compounds flopristin and virginiamycin M or the streptogramin B compounds quinupristin and linopristin influence their respective IC50 values in either the Gram-negative or Gram-positive streptogramin-binding site contexts. However, the IC50 of dalfopristin is significantly lower in the presence of either linopristin or quinupristin than by itself, especially in the context of the Gram-negative (U-U) streptogramin-binding site (Fig. 2C and D; see also Table S3 in the supplemental material). The IC50 values of linopristin and quinupristin also drop in the presence of dalfopristin (Fig. 2E and F; see also Table S4 in the supplemental material). The effects in the context of the Gram-positive (C-C) streptogramin-binding site are not as strong but are still observed (Fig. 2G and H; see also Table S4). Dalfopristin and virginiamycin M are chemically identical except for the presence of the additional sulfonyl group in dalfopristin. The results described here indicate that this bulky sulfonyl group in dalfopristin is detrimental to dalfopristin activity, as evidenced by the reduced activity of dalfopristin compared to that of virginiamycin M. However, the sulfonyl group leads to some degree of synergy between dalfopristin in the presence of a streptogramin B component on actively translating ribosomes.
Binding affinities of streptogramin antibiotics to vacant 70S ribosomes.
We also investigated whether the dissociation constant of streptogramin B to the isolated E. coli vacant 70S ribosome is affected in the presence of saturating concentrations of streptogramin A. We determined the dissociation constant of either quinupristin or linopristin alone or in the presence of dalfopristin or flopristin, respectively, using isothermal titration calorimetry (ITC). Quinupristin alone binds to the 70S ribosome with a Kd of 44 ± 21 nM, and its affinity for the ribosome increases about 6-fold in the presence of dalfopristin (Table 3; see also Fig. S7 and Table S5 in the supplemental material). In these experiments, performed at pH 7.5, dalfopristin was present in its hydrolyzed form. Similarly, the affinity of linopristin for the ribosome increases >3-fold when the ribosome was preincubated with flopristin (Table 3; see also Fig. S7 and Table S5). These results show that the affinity of the streptogramin B component increases in the presence of the streptogramin A component, as observed previously (20, 21). The crystallographic structures of the complexes of either dalfopristin or flopristin alone bound to the E. coli ribosome reveal that the base of A2062 has already moved by 2 Å to stack on the amide of the streptogramin A component, and that subsequent binding of streptogramin B results in only a small shift of A2062 in the plane of its nucleobase to maximize stacking interactions (see Fig. S3 in the supplemental material). Since large areas of the streptogramin B component do not interact with the ribosome directly, the prepositioning of A2062 due to streptogramin A binding likely leads to a more defined and rigid binding pocket for subsequent streptogramin B binding (22, 24).
TABLE 3.
Determination of streptogramin affinity for 70S ribosomes by isothermal titration calorimetry
| Cell content | Injectant | Dissociation constant (Kd)a |
|---|---|---|
| 70S | Quinupristin | 44 ± 21 |
| 70S + dalfopristinb | Quinupristin | 7 ± 5 |
| 70S | Linopristin | 40 ± 6 |
| 70S + flopristinb | Linopristin | 12 ± 8 |
Values are given in nM and are the average of the results from three independent experiments, plus or minus their standard deviations.
70S ribosomes were prebound to either dalfopristin or flopristin.
DISCUSSION
Here, we assessed the activities of streptogramin combinations and their individual components in a transcription-coupled translation assay and determined the binding mode of both streptogramin combinations in a common system, the Gram-negative E. coli ribosome. Prior studies demonstrated that NXL 103 displays higher antimicrobial activity than that of Synercid; it further showed that flopristin, the individual streptogramin A component of NXL 103, showed higher activity than that of the corresponding streptogramin A in Synercid, dalfopristin. In contrast, the activity of linopristin, the streptogramin B component of NXL 103, showed varied activity compared to that of quinupristin, the corresponding streptogramin B in Synercid. From these studies, it was concluded that the higher activity of NXL 103 versus that of Synercid is based on its streptogramin A component, flopristin (6–9, 38). Our antimicrobial and biochemical investigations confirm that flopristin is consistently more active than virginiamycin M or prehydrolyzed dalfopristin, whereas linopristin either displays similar or reduced activity compared to that of quinupristin.
The rapid hydrolysis of the diethylaminoethylsulfonyl group in dalfopristin to virginiamycin M at physiological pH, with a half-life of 11 min, indicates that this group is not the basis for the decreased antimicrobial activity of dalfopristin compared to that of flopristin, due to the duration of the experiment. However, our studies reveal that hydrolyzed dalfopristin or virginiamycin M bound to the ribosome is structurally essentially identical to flopristin and that neither the fluorine in flopristin nor the carbonyl group present in hydrolyzed dalfopristin are positioned in such a way so as to interact with the ribosome. Thus, differences in the biochemical and antimicrobial activities of flopristin (Fig. 2 and Table 1; see also Table S1 in the supplemental material) compared to those of virginiamycin M are not structurally observable but likely depend on energetic and kinetic parameters of translation.
Since Synercid is used to treat infections caused by Gram-positive species, we investigated differences in the constitution of both the streptogramin A and B binding pockets of the Gram-negative E. coli compared to those of pathogens treated by Synercid and to that of B. subtilis. The sequence alignment of all 23S rRNA residues within 10 Å of the bound streptogramin components reveals one major difference: in Gram-negative pathogens, a U-U base pair is found at position 1782–2586, whereas it is a C-C base pair in most Gram-positive pathogens. This indicates that a U1782C U2586C double mutation in the E. coli system would mimic a Gram-positive-like system in terms of its streptogramin-binding site. While the activities of streptogramin A components are identical in wild-type and mutant translation extracts, the activities of linopristin and quinupristin increased in the context of the Gram-positive versus the Gram-negative binding site. The increased activity of streptogramin B components seems surprising at first, since the base pairing geometry and the hydrogen bonding pattern in a C-C base pair are retained with respect to a U-U base pair (see Fig. S8 in the supplemental material). However, more favorable dipole-dipole interactions between the streptogramin B component and the C-C base pair compared to a U-U base pair may rationalize the observed increased activity of streptogramin B components to ribosomes with a C-C pair at position 1782–2586 (46). The streptogramin B component positions its amide group at position 17, directly under the nucleobase of nucleotide 2586. This amide group has a dipole moment that is positioned at about 50° with respect to the dipole moment of the uracil base of 2586. However, a cytosine base at position 2586 results in a dipole moment at about 130° with respect to the dipole moment of the streptogramin B amide group. Further, cytosine bases have a larger dipole moment compared to uracil bases (see Fig. S8 in the supplemental material) (46), which further favors positive attractions between the streptogramin B component and a cytidine at 2586. These findings may explain the increased activities of streptogramin B components observed in transcription-coupled translation assays utilizing cell extract from an E. coli strain bearing a C-C base pair at position 1782–2586, which mimics the streptogramin-binding site in Gram-positive pathogens.
Streptogramin antibiotics are known for their synergistic mode of action, as in vitro and in vivo studies revealed that the activity of a streptogramin combination exceeds the sum of the activities of the individual components (13–15). Our in vitro studies showed the synergistic effect of NXL 103 and Synercid in Gram-positive pathogens (Table 1), with a higher degree of synergy for NXL 103 versus Synercid in S. aureus and E. faecalis, as judged by their mean FIC indices (Table 2). The increased binding affinities of streptogramin B in the presence of streptogramin A compared to those of streptogramin B alone were previously suggested to confer a synergistic effect on a molecular level (20, 21). However, these studies used purified empty ribosomes, which may not reflect actively translating ribosomes. Furthermore, the binding affinity of antibiotics to their target may not be directly correlated with their activity.
Only one study has been published that demonstrated some degree of synergy for the combination of virginiamycin M and virginiamycin S using a translation system derived from B. subtilis (19). Using transcription-coupled translation assays with cell extract from either a wild-type E. coli strain or an E. coli mutant strain bearing the U1782C U2586C double mutant, we found that no synergy occurs for any streptogramin combination in either extract, with the exception of combinations that include dalfopristin. Therefore, since no general synergistic effects were observed in either U-U- or C-C-based transcription-coupled translation systems but were observed in antimicrobial assays, the synergistic effect of streptogramin components likely occurs independent of protein synthesis. Future experiments will be required to dissect the basis for streptogramin synergy in cells.
In translation extracts, the only synergy that we were able to observe occurred with streptogramin combinations in which one of the components is dalfopristin containing its diethylaminoethylsulfonyl group. However, the diethylaminoethylsulfonyl group reduces the activity of dalfopristin by ≥2.8-fold compared to that of virginiamycin M, the hydrolysis product of dalfopristin. As the transcription-coupled translation assay is run at pH 7.4 for 30 min, during which time extensive hydrolysis of dalfopristin will have occurred, it is likely that the activity for completely unhydrolyzed dalfopristin is even lower. Thus, while the detrimental effect of the diethylaminoethylsulfonyl group of dalfopristin can be overcome by synergy in the presence of a streptogramin B compound, it will likely be necessary to employ an analogue of dalfopristin with a nonhydrolyzable group instead of the diethylaminoethylsulfonyl group, so long as the size of this added group does not severely impact dalfopristin activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Wilson for technical assistance with isothermal titration calorimetry experiments, G. Meigs and J. Holton for assistance at beamline 8.3.1 at the Advanced Light Source (ALS), and T. Doukov for assistance at beamline 11-1 at the Stanford Synchrotron Radiation Laboratory (SSRL). We also thank S. Blanchard (Weill Cornell Medical College) for providing the pKK3535 ribosome plasmid and E. coli strain SCB 53. We also acknowledge helpful discussions by members of the Cate laboratory, and we acknowledge the assistance and consulting of S. Mills in establishing a productive collaboration between UC Berkeley and AstraZeneca.
This work was supported by NIH grant R01-GM65050 to J.H.D.C., by the NIH project MINOS grant R01GM105404 for the Structural Integrated Biology for Life Sciences (SIBYLS) and 8.3.1 beamlines at the ALS, and by the U.S. Department of Energy (grants DEAC02-05CH11231 for the SIBYLS and 8.3.1 beamlines at the ALS). J.N. was funded by a Human Frontiers in Science Program long-term postdoctoral fellowship.
Footnotes
Published ahead of print 23 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03389-14.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Threat report 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/ [Google Scholar]
- 2.Donadio S, Maffioli S, Monciardini P, Sosio M, Jabes D. 2010. Antibiotic discovery in the twenty-first century: current trends and future perspectives. J. Antibiot. (Tokyo) 63:423–430. 10.1038/ja.2010.62 [DOI] [PubMed] [Google Scholar]
- 3.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12:35–48. 10.1038/nrmicro3155 [DOI] [PubMed] [Google Scholar]
- 4.Yates JD, Schaible PJ. 1962. Virginiamycin as an antibiotic for poultry feeds. Nature 194:183–184. 10.1038/194183b0 [DOI] [PubMed] [Google Scholar]
- 5.Pfizer. 2013. Synercid I.V. (quinupristin and dalfopristin for injection), Pfizer Inc., New York, NY: http://labeling.pfizer.com/ShowLabeling.aspx?id=712 [Google Scholar]
- 6.Dupuis M, Leclercq R. 2006. Activity of a new oral streptogramin, XRP2868, against Gram-positive cocci harboring various mechanisms of resistance to streptogramins. Antimicrob. Agents Chemother. 50:237–242. 10.1128/AAC.50.1.237-242.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eliopoulos GM, Ferraro MJ, Wennersten CB, Moellering RC., Jr 2005. In vitro activity of an oral streptogramin antimicrobial, XRP2868, against Gram-positive bacteria. Antimicrob. Agents Chemother. 49:3034–3039. 10.1128/AAC.49.7.3034-3039.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein EJ, Citron DM, Merriam CV, Warren YA, Tyrrell KL, Fernandez HT, Bryskier A. 2005. Comparative in vitro activities of XRP 2868, pristinamycin, quinupristin-dalfopristin, vancomycin, daptomycin, linezolid, clarithromycin, telithromycin, clindamycin, and ampicillin against anaerobic Gram-positive species, actinomycetes, and lactobacilli. Antimicrob. Agents Chemother. 49:408–413. 10.1128/AAC.49.1.408-413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mabe S, Champney WS. 2005. A comparison of a new oral streptogramin XRP 2868 with quinupristin-dalfopristin against antibiotic-resistant strains of Haemophilus influenzae, Staphylococcus aureus, and Streptococcus pneumoniae. Curr. Microbiol. 51:363–366. 10.1007/s00284-005-0027-9 [DOI] [PubMed] [Google Scholar]
- 10.Politano AD, Sawyer RG. 2010. NXL-103, a combination of flopristin and linopristin, for the potential treatment of bacterial infections including community-acquired pneumonia and MRSA. Curr. Opin. Investig. Drugs 11:225–236 [PMC free article] [PubMed] [Google Scholar]
- 11.Delgado G, Jr, Neuhauser MM, Bearden DT, Danziger LH. 2000. Quinupristin-dalfopristin: an overview. Pharmacotherapy 20:1469–1485. 10.1592/phco.20.19.1469.34858 [DOI] [PubMed] [Google Scholar]
- 12.Vazquez D. 1966. Studies on the mode of action of the streptogramin antibiotics. J. Gen. Microbiol. 42:93–106. 10.1099/00221287-42-1-93 [DOI] [PubMed] [Google Scholar]
- 13.Bouanchaud DH. 1992. In-vitro and in-vivo synergic activity and fractional inhibitory concentration (FIC) of the components of a semisynthetic streptogramin, RP 59500. J. Antimicrob. Chemother. 30(Suppl A):95–99 [DOI] [PubMed] [Google Scholar]
- 14.Cocito C. 1979. Antibiotics of the virginiamycin family, inhibitors which contain synergistic components. Microbiol. Rev. 43:145–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neu HC, Chin NX, Gu JW. 1992. The in-vitro activity of new streptogramins, RP 59500, RP 57669 and RP 54476, alone and in combination. J. Antimicrob. Chemother. 30(Suppl A):83–94 [DOI] [PubMed] [Google Scholar]
- 16.Chinali G, Nyssen E, Di Giambattista M, Cocito C. 1988. Action of erythromycin and virginiamycin S on polypeptide synthesis in cell-free systems. Biochim. Biophys. Acta 951:42–52. 10.1016/0167-4781(88)90023-1 [DOI] [PubMed] [Google Scholar]
- 17.Chinali G, Nyssen E, Di Giambattista M, Cocito C. 1988. Inhibition of polypeptide synthesis in cell-free systems by virginiamycin S and erythromycin. Evidence for a common mode of action of type B synergimycins and 14-membered macrolides. Biochim. Biophys. Acta 949:71–78 [DOI] [PubMed] [Google Scholar]
- 18.Pestka S. 1977. Molecular mechanisms of protein biosynthesis. Academic Press, New York, NY [Google Scholar]
- 19.Cocito C, Vanlinden F. 1983. Inhibitory action of virginiamycin components on cell-free systems for polypeptide formation from Bacillus subtilis. Arch. Microbiol. 135:8–11. 10.1007/BF00419474 [DOI] [PubMed] [Google Scholar]
- 20.Contreras A, Vázquez D. 1977. Synergistic interaction of the streptogramins with the ribosome. Eur. J. Biochem. 74:549–551. 10.1111/j.1432-1033.1977.tb11423.x [DOI] [PubMed] [Google Scholar]
- 21.Parfait R, de Béthune MP, Cocito C. 1978. A spectrofluorimetric study of the interaction between virginiamycin S and bacterial ribosomes. Mol. Gen. Genet. 166:45–51. 10.1007/BF00379728 [DOI] [PubMed] [Google Scholar]
- 22.Hansen JL, Moore PB, Steitz TA. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061–1075. 10.1016/S0022-2836(03)00668-5 [DOI] [PubMed] [Google Scholar]
- 23.Harms JM, Schlünzen F, Fucini P, Bartels H, Yonath A. 2004. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2:4. 10.1186/1741-7007-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu D, Blaha G, Moore PB, Steitz TA. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257–270. 10.1016/j.cell.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 25.Hsieh MH, Yu CM, Yu VL, Chow JW. 1993. Synergy assessed by checkerboard. A critical analysis. Diagn. Microbiol. Infect. Dis. 16:343–349. 10.1016/0732-8893(93)90087-N [DOI] [PubMed] [Google Scholar]
- 26.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310:827–834. 10.1126/science.1117230 [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Dunkle JA, Cate JH. 2009. Structures of the ribosome in intermediate states of ratcheting. Science 325:1014–1017. 10.1126/science.1175275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch W. 2010. XDS. Acta Cryst. 66:125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 31.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—9th ed. CLSI document M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 32.CLSI. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S23 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 33.Pillai SK, Moellering RC, Jr, Eliopoulos GM. 2005. Antimicrobial combinations, p 365–440 In Lorian V. (ed), Antibiotics in laboratory medicine, 5th ed. Lipincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 34.Meletiadis J, Pournaras S, Roilides E, Walsh TJ. 2010. Defining fractional inhibitory concentration index cutoffs for additive interactions based on self-drug additive combinations, Monte Carlo simulation analysis, and in vitro-in vivo correlation data for antifungal drug combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 54:602–609. 10.1128/AAC.00999-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52:1. 10.1093/jac/dkg301 [DOI] [PubMed] [Google Scholar]
- 36.Brosius J, Ullrich A, Raker MA, Gray A, Dull TJ, Gutell RR, Noller HF. 1981. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6:112–118. 10.1016/0147-619X(81)90058-5 [DOI] [PubMed] [Google Scholar]
- 37.Buurman ET, Foulk MA, Gao N, Laganas VA, McKinney DC, Moustakas DT, Rose JA, Shapiro AB, Fleming PR. 2012. Novel rapidly diversifiable antimicrobial RNA polymerase switch region inhibitors with confirmed mode of action in Haemophilus influenzae. J. Bacteriol. 194:5504–5512. 10.1128/JB.01103-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pankuch GA, Kelly LM, Lin G, Bryskier A, Couturier C, Jacobs MR, Appelbaum PC. 2003. Activities of a new oral streptogramin, XRP 2868, compared to those of other agents against Streptococcus pneumoniae and Haemophilus species. Antimicrob. Agents Chemother. 47:3270–3274. 10.1128/AAC.47.10.3270-3274.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhtar TA, Wright GD. 2005. Streptogramins, oxazolidinones, and other inhibitors of bacterial protein synthesis. Chem. Rev. 105:529–542. 10.1021/cr030110z [DOI] [PubMed] [Google Scholar]
- 40.Depardieu F, Courvalin P. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319–323. 10.1128/AAC.45.1.319-323.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G, Taylor DE. 1998. Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics. Antimicrob. Agents Chemother. 42:1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigmund CD, Ettayebi M, Morgan EA. 1984. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 12:4653–4663. 10.1093/nar/12.11.4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cocito C, Di Giambattista M, Nyssen E, Vannuffel P. 1997. Inhibition of protein synthesis by streptogramins and related antibiotics. J. Antimicrob. Chemother. 39(Suppl A):7–13 [DOI] [PubMed] [Google Scholar]
- 44.Singh KV, Weinstock GM, Murray BE. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 46:1845–1850. 10.1128/AAC.46.6.1845-1850.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905–920. 10.1126/science.289.5481.905 [DOI] [PubMed] [Google Scholar]
- 46.Churchill CD, Navarro-Whyte L, Rutledge LR, Wetmore SD. 2009. Effects of the biological backbone on DNA-protein stacking interactions: the interplay between the backbone···π and π···π components. Phys. Chem. Chem. Phys. 11:10657–10670. 10.1039/b910747a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.