ABSTRACT
Human immunodeficiency virus type 1 (HIV-1) infection in the central nervous system (CNS) is characterized by replication in macrophages or brain microglia that express low levels of the CD4 receptor and is the cause of HIV-associated dementia and related cognitive and motor disorders that affect 20 to 30% of treatment-naive patients with AIDS. Independent viral envelope evolution in the brain has been reported, with the need for robust replication in resident CD4low cells, as well as CD4-negative cells, such as astrocytes, proposed as a major selective pressure. We previously reported giant-cell encephalitis in subtype B and C R5 simian-human immunodeficiency virus (SHIV)-infected macaques (SHIV-induced encephalitis [SHIVE]) that experienced very high chronic viral loads and progressed rapidly to AIDS, with varying degrees of macrophage or microglia infection and activation of these immune cells, as well as astrocytes, in the CNS. In this study, we characterized envelopes (Env) amplified from the brains of subtype B and C R5 SHIVE macaques. We obtained data in support of an association between severe neuropathological changes, robust macrophage and microglia infection, and evolution to CD4 independence. Moreover, the degree of Env CD4 independence appeared to correlate with the extent of astrocyte infection in vivo. These findings further our knowledge of the CNS viral population phenotypes that are associated with the severity of HIV/SHIV-induced neurological injury and improve our understanding of the mechanism of HIV-1 cellular tropism and persistence in the brain.
IMPORTANCE Human immunodeficiency virus type 1 (HIV-1) infection of astrocytes in the brain has been suggested to be important in HIV persistence and neuropathogenesis but has not been definitively demonstrated in an animal model of HIV-induced encephalitis (HIVE). Here, we describe a new nonhuman primate (NHP) model of R5 simian-human immunodeficiency virus (SHIV)-induced encephalitis (SHIVE) with several classical HIVE features that include astrocyte infection. We further show an association between severe neuropathological changes, robust resident microglia infection, and evolution to CD4 independence of viruses in the central nervous system (CNS), with expansion to infection of truly CD4-negative cells in vivo. These findings support the use of the R5 SHIVE models to study the contribution of the HIV envelope and viral clades to neurovirulence and residual virus replication in the CNS, providing information that should guide efforts to eradicate HIV from the body.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1)-associated dementia (HAD) and related cognitive and motor disorders affect 20 to 30% of treatment-naive patients with advanced immunosuppression or AIDS. Although combination antiretroviral therapy (cART) has markedly diminished the incidence of overt HAD, a milder-to-moderate form of neurocognitive impairment known collectively as HIV-1-associated neurocognitive disorders (HAND) developed in an estimated 50% of treated patients (1–3). Lentiviral neuroinvasion involves the recruitment of infected monocytes/macrophages and/or T cells to the central nervous system (CNS) from circulation, with activation and productive infection of resident microglia and a more restricted/latent infection of astrocytes (4–9). HIV-1 infection of mononuclear phagocytes in the CNS has been suggested to be the critical driving force of HIV-induced encephalitis (HIVE), as well as HAND (10, 11). Infected cells in the brain release neurotoxic viral and host inflammatory proteins that result in neuronal injury and death (6, 12). Identification of the viral and host factors that are associated with neuroinvasion, CNS persistence, and related injury is important, not only for our understanding of HIV-associated neuropathogenesis, but for advancing the design of treatment regimens and knowledge of therapeutic responses.
In this regard, HIV neurotropism is primarily determined by the viral envelope gene (13–16), and compartmentalization of a genetically distinct envelope (Env) population in the CNS from that in the systemic circulation (17–20), as a result of either a founder effect or independent viral adaptive evolution in the brain with long-term HIV infection, has been described (21, 22). Moreover, different env genotypes have been reported in different CNS regions of HIV-1-infected individuals and simian immunodeficiency virus (SIV)-infected macaques (23–26), despite the use of a “clonal” inoculum in the latter (23, 24), suggesting discordant regional virus evolution in this anatomical compartment, as well. The specific pressures that drive the tissue, as well as regional CNS env genetic divergence, remain unclear. Emergence of highly macrophage-tropic viral strains that engage the CD4 and CCR5 receptors differently or have a lower CD4 dependence for entry has been reported in HIV and SIV infection of the brain (27–32). The need for robust replication in macrophages and infection of CNS-resident cells, such as microglia and astrocytes, that express CCR5 but little or no CD4 (33–35), therefore, could be a major selective pressure for independent envelope evolution in the brain that contributes to neurological injury.
Because perivascular macrophages, microglia, and astrocytes in the brain are long lived (36, 37) and many current antiretrovirals fail to reach effective therapeutic levels in the brain (38–42), the CNS is a viral reservoir and a site for drug-resistant viruses to evolve. Appropriate animal models of CNS infection in AIDS, therefore, are critically needed to evaluate the effectiveness of various treatment regimens in crossing the blood brain barrier (BBB) to suppress virus replication and for testing strategies aimed at eradicating viral sanctuaries in the brains of cART-treated HIV-1-infected patients (43). Although studies in macaques infected with neurovirulent SIV strains have led to the critical realization that the brain is a very early target in acute viral infection (44, 45), as well as an important viral reservoir despite antiretroviral therapy (8, 46, 47), the envelope is SIV and not HIV. Moreover, the SIV-induced encephalitis (SIVE) models often require coinoculation of two viruses and/or immune modulation, such as CD8+ T cell depletion, to accelerate neurological disease development (48–53), and infected parenchymal microglia, a key component of HIV neuropathogenesis, is not a consistent feature. Furthermore, while rapid progression to AIDS and development of neurological disease in the SIV-infected macaques is often accompanied by the selection of highly macrophage-tropic, CD4-independent viruses (54, 55), infection of truly CD4-negative cells has not been documented in the brains of these animals.
We recently reported multifocal giant-cell encephalitis in rhesus macaques infected with subtype B and C R5 simian-human immunodeficiency viruses (SHIVs) via intravenous, intrarectal, or intravaginal inoculation in the absence of immune modulation. The neuropathology observed in the SHIV-induced encephalitis (SHIVE) models mirrored that of HIVE in infected patients, with several unique and classical HIVE features that are not commonly seen in other SIVE models, including microglial infection (56, 57). Here, we characterize HIV-1 variants within the brains of R5 SHIVE macaques with varying degrees of neuropathological changes to determine the envelope phenotypes associated with HIV-associated CNS infection and injury. We further explore the relationship between robust macrophage/microglia infection, evolution to CD4 independence (CD4i), and infection of truly CD4-negative cells of viral populations in the brains of the subtype B and C R5 SHIVE animals.
MATERIALS AND METHODS
Cells.
293T, RC49, JC53, and HeLa TZM-bl cells expressing CD4, CCR5, and CXCR4 and containing integrated reporter genes for firefly luciferase and β-galactosidase under the control of the HIV-1 long terminal repeat (LTR) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (complete medium). The Cf2Th canine thymocyte line engineered to express CCR5 without (Cf2Th.CCR5) or with (Cf2Th.CD4hi.CCR5) high levels of the CD4 receptor (58) was maintained in complete medium containing 0.4 mg/ml G418 (for CCR5 selection; Gibco, Life Technology Corporation, Grand Island, NY) and 0.15 mg/ml hygromycin (for CD4 selection; Invitrogen, Carlsbad, CA). Human peripheral blood mononuclear cells (PBMCs) were prepared by Ficoll gradient centrifugation and stimulated with phytohemagglutinin (PHA) (3 μg/ml; Sigma, St. Louis, MO) in RPMI medium containing 10% FCS, penicillin, streptomycin, l-glutamine, and 20 U/ml interleukin-2 (Norvatis, Emeryville, CA). Monocytes were enriched by centrifugation of PBMCs through a 40% Percoll (GE Healthcare Biosciences, Uppsala, Sweden) cushion, followed by plastic adherence, and cultured in RPMI 1640 medium supplemented with 10% FCS, 5% human AB serum, and 25 ng/ml recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) (Invitrogen) for 5 to 7 days to allow differentiation into macrophages.
Detection of antiviral humoral response.
SHIV-specific antibodies in serum samples were measured by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (GS HIV-1/HIV-2 Plus O EIA; Bio-Rad, Redmond, WA). Optical density values at a 1:10 serum dilution that were three times above the cutoff value were considered positive.
Plasmid constructs and pseudotype virus production.
For expression of envelope glycoproteins (Env), viral RNA was prepared from 300 to 500 μl plasma using a commercially available RNA extraction kit (Qiagen, Chatsworth, CA), followed by reverse transcription (RT) with Superscript III reverse transcriptase (Invitrogen) and random hexamer primers (Amersham Pharmacia, Piscataway, NJ). Full-length gp160 coding sequences were amplified from cDNA by single-genome amplification (SGA) or by conventional PCR, since recent studies suggest that bulk sequencing captures a measure of population diversity similar to that determined by SGA (59, 60). For SGA, cDNA was titrated by endpoint dilution, and a single copy was obtained in a two-step nested-PCR procedure using Platinum Taq High Fidelity polymerase (Invitrogen). The primers SH50 (5′-TAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA-3′) and SH51 (5′-TCCAGTCCCCCCTTTTCTTTTATAAAA-3′), and SH43 (5′-AAGACAGAATTCATGAGAGTGAAGGGGATCAGGAAG-3′) and SH44 (5′-AGAGAGGGATCCTTATAGCAAAGCCCTTTCAAAGCCCT-3′) were used for the first and second rounds of PCR, respectively, of the subtype B SHIVE macaques, while the primers SH50 and SH51, and WR40 (5′-AAGACAGAATTCATGAGAGTGAAGGAGAAATATC-3′) and WR41 (5′-AGAGAGGGATCCTCACAAGAGAGTGAGCTCGAGC-3′) were used for nested PCR of the subtype C SHIVE macaque GC98. For conventional PCR, primers SH43 and SH44 were used for the subtype B SHIV-infected animals, with primers WR40 and WR41 in reactions for both subtype C-infected monkeys. Amplicons were subcloned into the pCAGGS vector and sequenced as previously described (61). To generate luciferase reporter viruses capable of only a single round of replication, an envelope trans-complementation assay was used. Briefly, an Env expression plasmid and the NL4.3LucE-R+ vector were cotransfected with polyethylenimine (PEI) (Polyscience, Warrington, PA) into 293T cells. The cell culture supernatants were harvested 72 h later, filtered through 0.45-μm filters, quantified for p24 Gag content (Beckman Coulter, Fullerton, CA), and stored at −80°C.
Phylogenetic analysis of viral env sequences.
Envelope DNA sequences were aligned by Clustal W (62) using the parental subtype B HIV-1SF162 or subtype C ZM109F.PB4 strain as a reference. A codon-based alignment was also performed in order to remove sequencing errors, gaps, and homopolymeric regions. Neighbor-joining phylogenetic trees were generated by PhyML 3.0 (63) using the Jukes-Cantor model of evolution (64), with a gamma distribution of site-to-site rate variation, as estimated by the FindModel tool from the Los Alamos National Laboratory (LANL) HIV Database (http://hiv.lanl.gov). Compartmentalization of viral sequences was assessed visually using the Highlighter tool at the LANL HIV Database, as well as by quantitative evaluation using the parametric bootstrapping Slatkin-Maddison approach.
Envelope characterization.
For assessment of Env entry efficiency and CD4 dependency, 7 × 103 RC49, JC53, Cf2Th.CD4hi.CCR5, and Cf2Th.CCR5 cells were seeded in 96-well plates 24 h before use and infected in duplicate with the indicated pseudotype viruses. The infected cells were cultured for 72 h at 37°C, at which time the cells were harvested, lysed, and processed for luciferase activity according to the manufacturer's instructions (luciferase assay system; Promega, Madison, WI). Entry, as quantified by luciferase activity, was measured with an MLX microtiter plate luminometer (Dynex Technologies, Inc., Chantilly, VA). For infection of primary cells, 105 and 106 human PBMCs and monocyte-derived macrophages (MDMs), respectively, were infected and processed 72 h later for luciferase activity. To control for differences in Env entry efficiencies, infectivity in MDMs was expressed as a ratio of the infectivity for these cells compared to the infectivity in PBMCs from the same donor. Similarly, infectivity in RC49 and Cf2Th.CCR5 cells was expressed as a percentage of that achieved in JC53 and Cf2Th.CD4hi.CCR5 cells, respectively.
Immunophenotyping of SHIV-infected cells.
Identification of SHIV-infected and activated cells in the CNS was accomplished by immunohistochemical staining for SIVgag (1:400) (SIVp27, clone 183–H12-5C; NIH AIDS Reagent Program), Iba-1 (1:1,000) (polyclonal, 019-19741; Wako Chemicals, Richmond, VA), CD68 (1:410) (clone KP-1; Dako, Carpinteria, CA), CD163 (1:100) (clone 10D6; ThermoScientific, Waltham, MA), MRP14 (clone Mac387, IgG1;ThermoScientific, Waltham, MA), and glial fibrillary acidic protein (GFAP) (1:340) (polyclonal; Dako). Double-label in situ hybridization (ISH) for SIVmac and immunohistochemistry (IHC) for CD68, the endothelial marker CD31 (1:50) (clone JC70A; Dako), and GFAP (polyclonal; Dako) were also performed. SIV ISH was performed on Ventana Medical Systems' (Tucson, AZ) Discovery Ultra platform. Briefly, the tissue sections were pretreated with reagents from the RiboMap kit (Ventana Medical Systems) and digested with protease 3 (Ventana Medical Systems) for 4 min at 37°C. Tissue sections were prehybridized with RiboHyb (Ventana Medical Systems) and hybridized with digoxigenin (Dig)-labeled probe for 6 h at 55°C. Stringency washes were done using 1.0× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (Ventana Medical Systems) at 60°C. The bound probe was detected with rabbit anti-Dig (1:20,000; Sigma-Aldrich, St. Louis, MO) and UMap anti-rabbit alkaline phosphatase conjugate (Ventana Medical Systems). On the bench, the sections were then blocked for endogenous peroxidase activity with 3% H2O2 in phosphate-buffered saline (PBS). No pretreatment was necessary for GFAP, with pretreatment for CD31 involving a 5-min incubation with proteinase K (Dako). The sections were then blocked for endogenous biotin with an avidin-biotin block, followed by incubation with the primary antibody and the secondary antibody biotinylated horse anti-mouse (CD31; Vector Laboratories) or secondary antibody biotinylated goat anti-rabbit (GFAP; Vector Laboratories). Sections were detected using a standard avidin-biotin peroxidase complex technique (Vectastain ABC Elite; Vector Laboratories) and DAB chromogen (Dako) and counterstained with Nuclear Fast Red (Vector Laboratories). Full-length SIVmac ISH probe was used for maximum sensitivity in detecting viral RNA and DNA. Isotype-matched irrelevant controls were included for all runs.
RESULTS
Characteristics of R5 SHIV-infected macaques with encephalitis.
In a previous study, we documented progression to AIDS in 64% (26/43) of Indian rhesus monkeys (Macaca mulatta) infected with the late-stage subtype B R5 SHIVSF162P3N by the intravenous (i.v.) (n = 9), intrarectal (i.r.) (n = 17), or intravaginal (i.vg.) (n = 17) route (65) (see Table S1 in the supplemental material). Of the 26 immunodeficient animals, 27% (7/26) developed SHIVE at terminal disease (57). Three of 13 macaques (23%) infected intrarectally with the lineage-related subtype C R5 SHIVC109P3 (n = 7) and SHIVC109P3N (n = 6) also progressed to AIDS (see Table S1 in the supplemental material), with severe SHIVE documented in two of the three animals with advanced immunodeficiency (56). Table 1 shows the age, MHC class I genotype, route, dose, time to clinical immunodeficiency, coreceptor switch, and development of SHIV binding antibody in the seven subtype B and the two subtype C SHIV-infected rhesus macaques with neurological disease. The SHIVE macaques ranged from 6 to 10 years of age at the time of virus exposure and were infected via various routes and inoculum doses. With the exception of macaque GC98, which tested positive for Mamu-A*01, none expressed this as well as the class I Mamu-B*08 and Manu-B*17 alleles reported to be associated with spontaneous virologic control of pathogenic SIVmac239 replication (66–68). Consistent with studies showing that rapid disease progression is the best correlate of the development of SIV encephalitis (69), a rapid-progressor (RP) phenotype characterized by high levels of virus replication and weak or undetectable antiviral antibody response was evident in 77.8% (7/9) of the AIDS-defining animals with SHIVE, with a switch to CXCR4 preference dictated by amino acid changes in the V3 loop documented in 55.5% (5/9) of the animals with neurological disease.
TABLE 1.
Characteristics of subtype B R5 SHIVSF162P3N-, subtype C SHIVC109P3-, and SHIVC109P3N-infected macaques with SHIVEa
| Virus | Animal | Age (yr) |
Mamu alleleb |
Route | Dose (TCID50)c | Time to AIDS (wk) | Coreceptor switch (X4 V3 sequence)e | SHIV binding antibody | ||
|---|---|---|---|---|---|---|---|---|---|---|
| A*01 | B*08 | B*17 | ||||||||
| SHIVSF162P3N | DE86 | 5.69 | − | − | − | I.v. | 3,000 | 12d | Yes (GHI insertion) | Transient |
| T799 | 9.90 | − | − | − | I.v. | 3,000 | 224 | Yes (S11 K/R) | High, persistent | |
| DG07 | 6.71 | − | − | − | I.r. | 10,000 | 7d | No | Transient | |
| DG08 | 6.98 | − | − | − | I.r. | 10,000 | 20d | Yes (HI/HR insertion, ΔATGD) | Not detected | |
| DN57 | 5.84 | − | − | − | I.r. | 10,000 | 30d | No | Weak, persistent | |
| ET94 | 6.50 | − | − | − | I.r. | 10,000 | 39 | Yes (HR insertion) | Weak, persistent | |
| DG17 | 8.55 | − | − | − | I.vg. | 1,000 | 22d | No | Not detected | |
| SHIVC109P3 | GC98 | 7.21 | + | − | − | I.r. | 5,000 | 20d | No | Not detected |
| SHIVC109P3N | FI08 | 7.62 | − | − | − | I.r. | 5,000 | 10d | Yes (ΔIGDI) | Not detected |
The age at the time of virus challenge, status of restrictive MHC class I genotype, inoculation route, dose of the virus inoculum and time to AIDS of the SHIVE macaques are shown. Presence of coreceptor switch, V3 sequence dictating the switch to CXCR4 usage, and status of SHIV binding antibody in the animals are also indicated.
−, absent; +, present.
TCID50, 50% tissue culture infectious dose.
Rapid progressor.
Δ denotes deletion in the V3 loop.
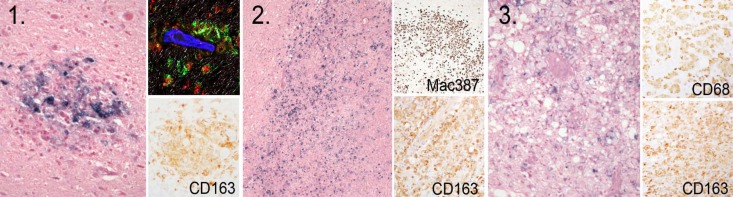
As reported recently (57), CNS lesions in the subtype B R5 SHIVSF162P3N-infected SHIVE monkeys could be divided into three main histopathological patterns that are consistent with descriptions of HIVE in humans in the pre-cART era (5, 70–72). The first type (type I) represents classical perivascular lesions comprised of CD68+ CD163+ SHIV-infected macrophages and multinucleated giant cells (MNGCs) (Fig. 1). The second type (type II) is characterized by very large, expansive lesions extending deep into the gray and white matter parenchyma, with marked activation of surrounding microglia and astroglia. The third type of lesion (type III) is comprised of chronic “burnt-out” lesions centered on vessels surrounded by large numbers of vacuolated macrophages/microglia that contain little virus. All three types of lesions were detected in macaques ET94 and DG17, with varying degrees of type I and II lesions seen in DN57, DG08, and DE86 and only mild type I lesions observed in T799 and DG07 (Table 2). Furthermore, immunohistochemistry staining of brain tissues for SIVp27 and the CD68/CD163 and the GFAP markers for macrophage/microglia and astroglial activation, respectively, indicated a difference among the seven subtype B SHIVE animals in CNS viral burdens, cellular targets, and neuropathological changes, as well. Staining for SIVp27 and reactivity with CD68/CD163 and GFAP were strongest for ET94 and DG17, with mild to moderate staining and reactivity for the SIV gag antigen and the three activation markers demonstrable in DN57 and DG08. Further decreases in staining for SIVp27 and the macrophage/microglia and astrocyte markers were noted in DE86 and T799, with DG07 displaying little or no reactivity. Moreover, two patterns of SIVp27 antigen-staining cells were observed in ET94 and DG17 (57). One is in macrophage nodules, as seen in DE86, DN57, DG08, and T799, and the other is a pattern of scattered cells distant from the perivascular lesion center with the appearance of microglia. Robust microglia or macrophage infection and activation were also documented in both subtype C SHIVE macaques (56), with more vacuolar leukoencephalopathy in white matter tracts but fewer observed multinucleated giant cells in the two subtype C- than in the subtype B-infected macaques with severe SHIVE.
FIG 1.
Types of neuropathologic lesions in the brains of subtype B R5 SHIVSF162P3N-infected macaques. Shown are SIV+ cells as revealed by in situ hybridization in cortical perivascular histolytic lesions with MNGCs (1), thalamic expansive lesions with white matter damage (2), and vacuolated chronic burnt-out lesions (3). Subsets of CD163+, Mac387+, and CD68+ macrophages are demonstrated by IHC in the insets, with SIV (green), CD68 (red), and glut-1 (blue) shown in the confocal inset.
TABLE 2.
Histopathologic lesions and staining characteristics in the brains of subtype B R5 SHIVSF162P3N-infected macaques with SHIVE
| Animal | Time to AIDS (wk) | Histopathologic lesion type(s)a | Immunohistochemical stainingb |
||
|---|---|---|---|---|---|
| SIVp27 (morphology) | CD68/CD163 | GFAP | |||
| DG07 | 7 | I | +/− | +/− | ND |
| T799 | 224 | I | + (Mϕ) | ++ | ++ |
| DE86 | 12 | I, II | ++ (Mϕ) | ++ | + |
| DG08 | 20 | I, II | +++ (Mϕ) | +++ | + |
| DN57 | 30 | I, II | +++ (Mϕ) | +++ | + |
| ET94 | 39 | I, II, III | ++++ (Mϕ; microglia) | +++ | ++ |
| DG17 | 22 | I, II, III | ++++ (Mϕ; microglia) | ++++ | ++ |
Three main types of lesions were observed: type I, perivascular histolytic with MNGCs; type II, severe parenchymal with white matter damage; type III, chronic burnt out.
Subjective semiquantitative scoring (from −, indicating no change over control brain sections, to ++++, indicating most severe) for the presence of SIVp27 and reactivity with the macrophage/microglia (CD68/CD163) and GFAP astrocyte marker by immunohistochemistry. Mϕ, macrophage; ND, not performed.
Anatomical and regional env compartmentalization in the brains of subtype B R5 SHIVE macaques.
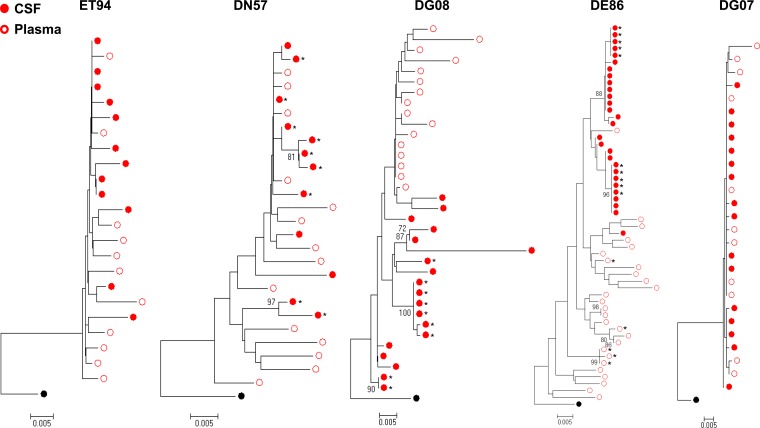
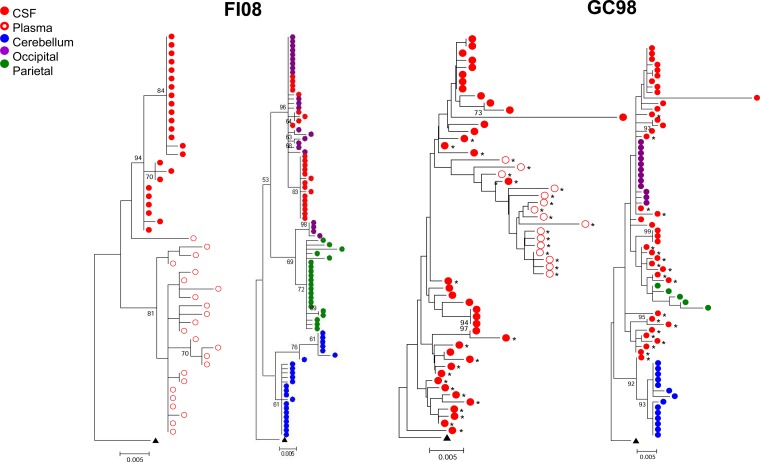
Compartmentalization of viral variants in the cerebrospinal fluid (CSF) can be detected in the course of HIV and SIV infections, the degree of which had been reported to be a strong indicator of HIV neuropathogenesis (18, 73, 74). CSF and plasma samples were available from five (ET94, DN57, DG08, DE86, and DG07) of the seven subtype B R5 SHIVE macaques at terminal disease, allowing us to examine whether there was compartmentalization of viral genomes. Sequence analysis of CSF env variants showed the absence of signature CXCR4-determining V3 sequences in ET94, DG08, and DE86 (data not shown), even though these animals had X4 viruses in the blood and in at least one peripheral lymph node (LN) at the time of necropsy (75, 76). Phylogenetic analysis of the env V3-to-V5 region (660 bp) showed clustering of Envs from the CSF in a lineage that is separated from the plasma in macaques DG08 and DE86. CSF and plasma compartmentalization in these two animals was confirmed by Slatkin-Maddison analysis, consistent with local virus expansion and adaptation to the CNS (Fig. 2). In contrast, env V3-to-V5 sequences from the CSF and plasma of ET94, DN57, and DG07 were well equilibrated, with low genetic diversity in DG07 that was confirmed by tree analysis of full-length gp120s from the plasma and CSF of the animal (data not shown). ET94 and DN57 were infected for longer periods (39 and 30 weeks, respectively) than DG08 and DE86 (20 and 12 weeks, respectively), with the shortest infection (7 weeks) in DG07. Additionally, as shown in Table 2, CNS lesions and degrees of macrophage or microglia infection and activation were more robust in ET94 and DN57 than in the other macaques. Thus, it is tempting to speculate that, similar to reports in HIV-infected patients and SIV-infected monkeys (21, 22, 50), compartmentalization in SHIV-infected macaques requires a period of time for variants to adapt within the CNS (Table 3) but may dissipate with increased time of infection and continuous or repeated viral invasion of the CNS that drives neuronal damage and dysfunction of the blood brain barrier.
FIG 2.
Phylogenetic tree analysis of CSF and blood env sequences in subtype B R5 SHIVE macaques. Neighbor-joining phylogenetic trees of env V3-to-V5 sequences from the CSF and blood plasma rooted to HIV-1SF162 are shown, with bootstrap values of >70% indicated at the appropriate nodes. Genetic-distance scales representing the number of nucleotide substitutions per site between env sequences (0.005) are shown at the bottom of each tree. *, sequence obtained by conventional PCR.
TABLE 3.
Relationship between the duration of infection, CNS histopathological lesions, viral burden, cellular targets, and anatomical compartmentalization in subtype B SHIVE macaquesa
| Animal | Time to AIDS | Histopathologic lesion type(s) | SIVp27 staining (morphology) | Plasma and CSF compartmentalization |
|---|---|---|---|---|
| DG07 | 7 | I | +/− | No |
| DE86 | 12 | I, II | ++ (Mϕ) | Yes |
| DG08 | 20 | I, II | +++ (Mϕ) | Yes |
| DN57 | 30 | I, II | +++ (Mϕ) | No |
| ET94 | 39 | I, II, III | ++++ (Mϕ; microglia) | No |
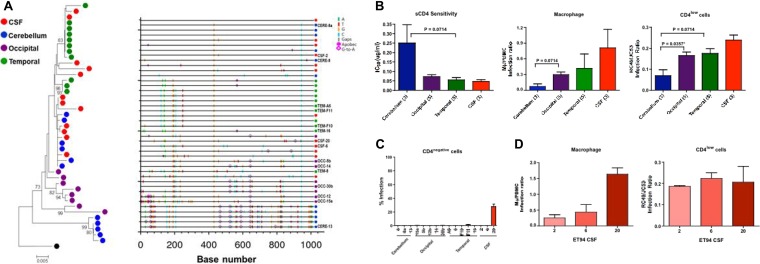
In addition to anatomical compartmentalization, regional compartmentalization of env genotypes in the brains of HIV-infected individuals and SIV-infected macaques has also been reported (23–26). For most of the SHIVE macaques (DN57, DG08, DE86, and DG07), only tissues from the gray and white matter of the brain were collected at the time of necropsy, with no specific indication of the regions of the brain being sampled. In contrast, tissues from different parts of the brain of ET94 were collected, providing an opportunity to investigate whether they harbored different Env populations. SGA of the full-length env gene was performed with cDNA templates from the CSF, the cerebellum, and the occipital and temporal regions of the brain of the animal. A total of 42 env V1-to-V5 sequences (1,047 bp) were analyzed, with an average of 10 (range, 8 to 12) from each regional compartment. Phylogenetic and Highlighter plot analyses showed that the cerebellum viral population was not uniformly equilibrated with the CSF, occipital, or temporal subpopulation (Fig. 3A), and this was confirmed by performing the Slatkin-Maddison test. To determine if there are phenotypic properties associated with the regional genotypic differences, single-round replication-incompetent luciferase reporter viruses were generated with Envs from the CSF and brain tissues of ET94 and examined for their sensitivity to CD4-IgG2, a tetrameric soluble-CD4 (sCD4) construct based on IgG, with this measurement serving as a surrogate marker for CD4 utilization efficiency. The abilities of the reporter viruses to infect CD4low cells, such as primary macrophages and RC49 cells, were also determined, since emergence of highly macrophage-tropic viral strains has been reported in HIV and SIV infection of the brain. The results showed that Env variants in the cerebellum (n = 3) were more resistant to neutralization with sCD4 than those in the occipital (n = 5), temporal (n = 5), and CSF (n = 3) compartments, with the last being the most sensitive (Fig. 3B). Moreover, compared to Envs from the other regions, those from the cerebellum infected primary macrophages and the CD4low RC49 cells less efficiently, consistent with reports that efficient infection of macrophages in vitro correlates with the capacity to use low CD4 levels and with increased sensitivity to sCD4 (29, 32, 77–79).
FIG 3.
ET94 Env analysis. (A) Phylogenetic tree and Highlighter plot analyses of the env sequences from the CSF, cerebellum, and occipital and temporal regions of ET94. A neighbor-joining tree of the env V1-to-V5 region rooted to HIV-1SF162 is shown, with bootstrap values of >70% indicated at the appropriate nodes. All sequences were obtained by SGA, and the genetic distance, representing the number of nucleotide substitutions per site between env sequences, is scaled at the bottom (0.005). A Highlighter plot was generated at www.hiv.lanl.gov, and the sequence identifiers for the Env clones used in functional studies are indicated on the y axis. The positions of nucleotide base transitions and transversions in the Highlighter plots are indicated by short colored-coded bars. Tics that are bracketed represent G-to-A changes, while those in circles represent G-to-A changes in a sequence consistent with an APOBEC signature. (B and C) sCD4 sensitivity and infection of primary macrophages and CD4low (B) and CD4-negative (C) cells of pseudoviruses expressing Envs from the different brain regions and the CSF compartment. (B)The numbers in parentheses indicate the number of clones analyzed per compartment. (C) The level of infection of CD4-negative cells is expressed as a percentage of that of cells expressing both CD4 and CCR5. P values by Mann-Whitney t tests that approach significance (<0.1) are shown, with asterisks indicating those that reached statistical significance (P < 0.05). (D) Comparison of the abilities of the three individual CSF Env clones of ET94 to mediate infection of primary macrophages and CD4low cells. The error bars in panels B, C, and D indicate means and standard deviations (SD) of 2 or 3 independent experiments.
Variability in sCD4 neutralization and infection of macrophages suggests that the viruses in different brain regions of ET94 evolve with different CD4 requirements that may expand tropism. Indeed, microglia in the brain of ET94 were infected (Table 2) (57), and these CNS-resident cells expressed little or no CD4 (33–35). We therefore investigated the abilities of the Envs originating from different brain compartments and the CSF of ET94 to mediate entry independently of CD4. The results showed that only one of the three Env clones from the CSF compartment tested (clone 20) mediated entry into the Cf2Th canine cells that expressed CCR5, but not CD4, with close to 30% efficiency of the signal seen in cells expressing both CD4 and CCR5 (Fig. 3C). Clone 20 was more efficient at infecting primary macrophages and CD4low cells than the other two CSF clones analyzed (Fig. 3D), supporting the evolution of highly macrophage-tropic viruses to CD4 independence, which may have contributed to the severe neuropathological changes seen in ET94.
Evolution of CNS viruses to CD4 independence is associated with increased neuropathology in subtype B R5 SHIVE macaques.
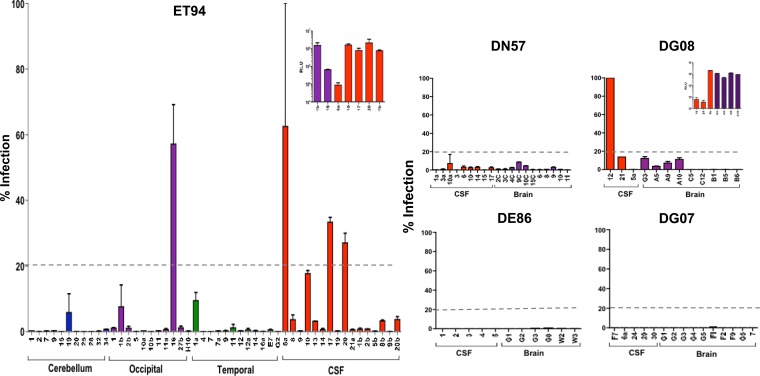
To further examine the association between evolution to CD4i and the severity of SHIV-induced encephalitis, we characterized Env populations from the brains and the CSF compartments of DN57, DG08, DE86, and DG07 for the ability to mediate infection of CD4-negative cells, expanding the panel of variants obtained from the CSF and brain tissues of ET94 for comparison (Fig. 4). As described above and shown in Table 2, there is a defined gradation of CNS lesion severity, perivascular macrophage infection, and resident cell activation among these five subtype B R5 SHIVE macaques, in the ascending rank order of DG07, DE86, DG08, DN57, and ET94. Furthermore, microglia infection was detected only in ET94. Full-length gp160s were amplified from the CSF and gray and white matter of the brains of DN57, DG08, DE86, and DG07 and from the CSF compartment and different regions of the brain of ET94. The results confirmed that Envs from the cerebellum (n = 11) of ET94 were largely CD4 dependent, but several Envs in the occipital (n = 11) and temporal (n = 12) regions of the brain of this rhesus monkey mediated CD4-independent entry (Fig. 4). Entry into CD4-negative cells, however, was generally low, with ≤10% of the efficiency seen when CD4 and CCR5 are coexpressed. The exception was clone 16 from the occipital region, which mediated infection of cells expressing CCR5 alone at 60% of the signal observed when both CD4 and CCR5 are present. In comparison, a greater proportion of Envs amplified from the CSF of ET94 (n = 16) mediated CD4-independent entry with varying efficiencies, with one (clone 6a) reaching the same level as clone 16 from the occipital region. However, these highly CD4i Envs are attenuated, mediating entry into cells expressing both CD4 and CCR5 at 1- to 2-log-unit-lower efficiency than those that are more CD4 dependent (e.g., clones 10, 17, and 20 from the CSF and clone 1b from the occipital region) (Fig. 4, inset). CD4-independent Envs were also found in the brains and CSF compartments of DN57 and DG08, but not in DE86 or DG07. Entry into CD4-negative cells mediated by Envs from DN57 was negligible, with <5% efficiency compared to entry when CD4 was present. In contrast, CD4-independent entry appeared to be more robust for Envs from DG08, with clone 12 from the CSF mediating CCR5-dependent, CD4-independent entry as efficiently as when CD4 was present. However, similar to findings in ET94, this highly CD4-independent Env in DG08 was defective (Fig. 4, inset). Acquisition of high CD4 independence (≥50% efficiency) by subtype B R5 SHIVSF162P3N, therefore, appears to come at a cost to viral fitness. Nonetheless, our finding that fully functional Envs that displayed a moderate degree (≥20%) of CD4-independent entry efficiency were detected only in the brain of ET94, but not in DN57, DG08, DE86, and DG07, suggests an association between emergence of CNS viruses with reduced CD4 affinity, microglia infection in vivo, and severity of neuropathological changes.
FIG 4.
CD4-independent Envs in the brains and CSF of R5 SHIVSF162P3N SHIVE macaques. Infection mediated by Envs amplified from the brains and CSF of the five SHIVSF162P3N SHIVE macaques in Cf2Th.CCR5 was assessed, with the level of infection in CD4-negative cells expressed as a percentage of that of Cf2Th.CD4hi.CCR5 cells that express both receptors. The dashed lines delineate 20% CD4-independent entry efficiency, and the insets show absolute relative light units (RLU) in Cf2Th.CD4hi.CCR5 cells of select CD4i Env populations from ET94 and DG08. The error bars indicate means and SD of 2 or 3 independent experiments.
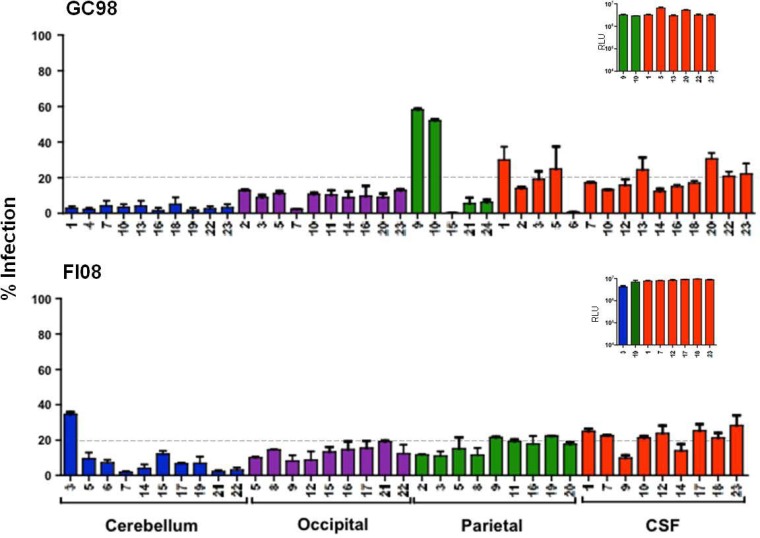
Emergence of CD4-independent Envs in the brains of subtype C R5 SHIVE macaques.
To further investigate the relationship between emergence of CD4-independent viruses, macrophage or microglia infection in vivo, and SHIVE severity, we determined the abilities of Envs recovered from the brains and the CSF compartments of the two subtype C R5 SHIVE macaques to mediate entry in the absence of CD4. Severe neuropathological changes with robust macrophage or microglia infection were previously documented in the brains of both macaques (56). Phylogenetic tree analyses and Slatkin-Maddison tests of the env V3-to-V5 sequences of GC98 and FI08 showed plasma and CSF compartmentalization, more so in FI08 than in GC98, where the CSF Envs were not uniformly compartmentalized (Fig. 5). Distinct clustering of sequences from the cerebellum region of the brain in both animals was also observed, consistent with findings in the subtype B SHIVE macaque ET94. Furthermore, similar to the subtype B SHIVE animals with expansion to CXCR4 use, V3 signature sequence associated with the coreceptor switch in FI08 was not detected in the brain (data not shown), even though X4 viruses could be found in the blood and peripheral LNs of the animal at end-stage disease (56). When CD4-independent infection was assessed using pseudovirions expressing Envs from GC98, a good proportion of Envs (9 of 40 clones tested) were found to mediate ≥20% entry into CD4-negative cells, with two clones from the parietal region achieving close to 50% of the CD4-independent infection efficiency seen in the presence of CD4 (Fig. 6). Furthermore, in contrast to the findings for the subtype B SHIVE macaques, the highly CD4i Envs are fully functional (Fig. 6, inset). Similar observations were made in FI08, with a higher proportion of Envs (12 of 37 clones tested), particularly those from the CSF and parietal region, mediating entry into CD4-negative cells at 20 to 30% of the efficiency of the signal seen in cells coexpressing CD4 and CCR5. None, however, achieved the high degree of CD4 independence seen in the two GC98 parietal Env clones, perhaps reflecting the shorter period of virus infection and replication in this macaque (10 weeks compared to 20 weeks for GC98). However, in agreement with findings in GC98, the CD4i Envs are fully functional. The differences in the frequency, functionality, and efficiency of CD4i Envs in the subtype B and subtype C R5 SHIVE animals cannot be explained by differences in the viral burdens (data not shown), suggesting that the barrier to subtype B R5 SHIVSF162P3N acquiring CD4 independence is greater than that of the two lineage-related SHIVC109 viruses.
FIG 5.
Phylogenetic tree analysis of env sequences in the blood and brains of subtype C SHIVE macaques. Neighbor-joining phylogenetic trees of env V3-to-V5 sequences from the plasma, CSF, cerebellum, and occipital and parietal regions of the brains of SHIVC-infected macaques GC98 and FI08 rooted to ZM109F.PB4 are shown, with bootstrap values of >70% indicated at the appropriate nodes. Genetic distances representing the number of nucleotide substitutions per site between env sequences (0.005) are indicated at the bottom of each tree. *, sequence obtained by SGA.
FIG 6.
CD4-independent Envs in the brains of subtype C SHIVE macaques GC98 and FI08. Env clones amplified from the cerebellum (blue) and occipital (purple) and parietal (green) regions and from the CSF (red) were assessed for the ability to enter Cf2th.CCR5 cells. Infectivity in these CD4-negative cells is expressed as a percentage of that of cells expressing both receptors. The dashed lines delineate 20% CD4-independent entry efficiency, and the insets show absolute RLU in Cf2Th.CD4hi.CCR5 cells of pseudovirions expressing representative Env populations with >20% CD4i entry efficiency. The data are the means and SD of 2 or 3 independent experiments.
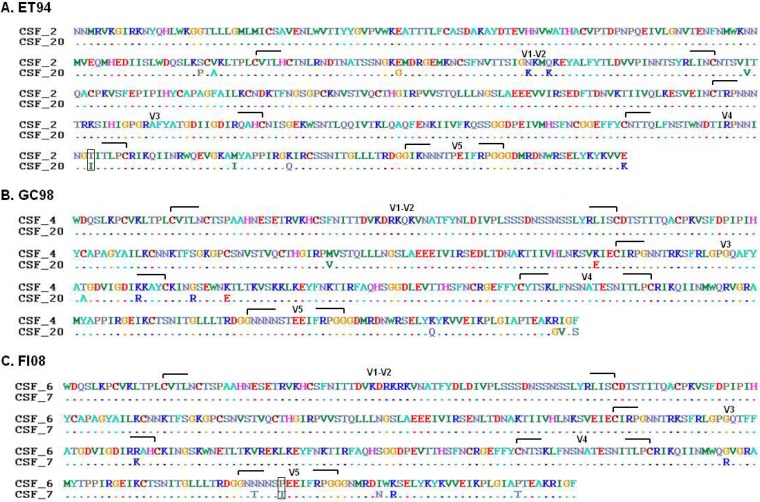
Amino acid changes in gp120 V1/V2, V3, C2, C3, and V4/C4, as well as an altered cytoplasmic tail, have been reported to be sufficient to render HIV-1 (58, 80–85) and SIV (32, 54, 55, 86) CD4 independent. Comparison of the gp120 amino acid sequences of the CD4-dependent clone 2 and the CD4-independent clone 20 from the CSF of subtype B SHIVE macaque ET94 (Fig. 2) showed changes in the V1/V2 loop structure, as well as in V4/C4, with the change in V4 resulting in the loss of an N-linked glycosylation site (Fig. 7A). For the subtype C SHIVE macaque GC98 (Fig. 7B), comparison of the V1-to-V5 gp120 region of the CD4-dependent clone 4 and the CD4-independent clone 20 CSF Envs revealed no changes in the V1/V2 or V4 region but differences in the C2, V3/C3, and C5 domains. Similar lack of amino acid changes in V1/V2 and V4 was also observed between the CD4-dependent clone 6 and the CD4-independent clone 7 from the CSF of the other subtype C SHIVE macaque, FI08 (Fig. 7C). The differences were mainly in the V5 and C5 structures, with one of the changes in the former involving the addition of an N-linked glycosylation site. Further genetic sequence analyses of gp120, as well as gp41, combined with mutagenesis studies on these and additional CD4-dependent and CD4-independent Env clones from the subtype B and C SHIVE macaques will be required to delineate whether the determinants of and barriers to CD4 independence differed between the two subtype R5 SHIVs.
FIG 7.
Amino acid sequence alignment of CD4-dependent and CD4i envelope clones from the CSF of ET94 (A), GC98 (B), and FI08 (C). The full-length gp120 of the CD4-dependent clone 2 and the CD4i clone 20 of ET94, as well as the V1-to-V5 gp120 of CD4-dependent clones 4 and 6 and CD4i clones 20 and 7 from GC98 and FI08, respectively, are shown. The dashes indicate amino acid identity, and changes altering potential sites of N-linked glycosylation are boxed.
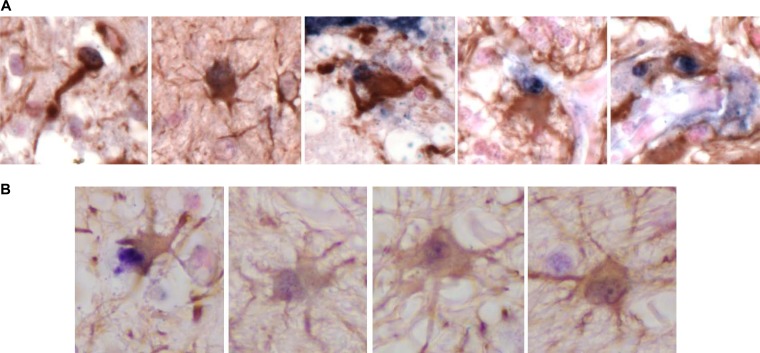
Reduced CD4 affinity correlates with expanded tropism for truly CD4-negative cells in the brain.
To determine if the reduced CD4 dependence of the fully functional subtype C R5 SHIVE Envs correlates with expanded tropism for truly CD4-negative cells in vivo, double-label in situ hybridization for SIVmac RNA and DNA and immunohistochemistry for the endothelial cell marker CD31 or the astrocyte marker GFAP were conducted on brain tissues from GC98 and FI08, with tissues from the subtype B SHIVE macaque ET94 examined as well for comparison. The results showed that, in contrast to ET94, SIV+ cells that also stained for GFAP but not CD31 could be detected in brain tissues of GC98 (Fig. 8A) and FI08 (Fig. 8B), supporting infection of astrocytes and not endothelial cells in the CNS of the subtype C SHIV-infected animals. The SIV signal in astrocytes appeared to be stronger in GC98 than FI08, consistent with an overall more efficient CD4-negative cell infection mediated by Envs in the brain of the former than the latter in vitro.
FIG 8.
Astrocyte infection in the brains of subtype C SHIVE macaque GC98 (A) and FI08 (B). Immunohistochemistry and in situ hybridization staining of brain sections were carried out (SIV ISH and IHC for GFAP).
DISCUSSION
The CNS and long-lived resident cells, such as perivascular macrophages, microglia, and astrocytes, are tissue and cellular reservoirs that can sequester replication-competent HIV-1, constituting a site for viral persistence and for drug-resident viruses to evolve, since many antiviral drugs access the brain poorly (87). Studies in relevant nonhuman primate models of CNS infection could help to further delineate the viral mechanisms that maintain HIV persistence in the brain and to identify the nature of the reservoir in the brain in the presence of cART and upon its removal, as well as to develop and test therapeutic agents to eliminate latently infected cells in the brain. In this study, we used the R5 SHIV model of HIV/AIDS, which recapitulates the features of HIVE and accounts for the role of the HIV-1 envelope in neuropathogenesis, to identify viral phenotypes that alter the spectrum of infected CNS-resident cells and impact CNS disease severity.
In a cross-sectional study of five subtype B R5 SHIV-infected macaques with neurological AIDS, we show that the severity of neuropathological changes is associated with the duration of the infection and robust macrophage/microglia infection in the CNS (Table 2), consistent with a model of neuropathogenesis in which enhanced infection and activation of resident phagocytic cells lead to increased release of neurotoxic viral and inflammatory proteins and neuronal damage. The degree of env genetic compartmentalization in the CSF and blood also appeared to be related to the infection time and extent of neurological damage (Fig. 2 and Table 3). For example, CSF compartmentalization in DE86 and DG08, the two macaques infected for 12 and 20 weeks, respectively, but not in DG07, which was infected for only 7 weeks, is consistent with the notion that the short period of infection in the last animal did not provide sufficient time for env evolution and adaptation to the CNS. Discordant viral evolution in the brain was also clearly evident in the two subtype C SHIVE macaques that were infected for 10 and 20 weeks (Fig. 5). Conversely, the equilibration of CSF and plasma sequences in ET94 and DN57, the two macaques infected for the longest time and with more severe CNS pathology, could be the consequence of dysfunction of BBB integrity. In most cases, the envelope sequences analyzed were obtained by SGA, including those from the plasma and CSF of DG07, which showed low genetic diversity and lack of anatomical compartmentalization (Fig. 2). The sequence homogeneity observed in this animal, therefore, is likely the consequence of the outgrowth of a dominant variant during the short infection time (7 weeks) and in the absence of antiviral immunity. Sets of identical sequences were also frequently seen in macaques DG08, DE86, FI08, and GC98 (Fig. 2 and 5), all of which were infected for 20 weeks or less in the absence of persistent seroconversion. Moreover, identical sequences were present in SGA, as well as conventional PCR V3-to-V5 amplicons of DG08, DE86, and GC98. For FI08, however (Fig. 5), because the sequences were all obtained by conventional PCR, the possibility that the discordant CSF/plasma viral evolution and frequent occurrence of identical sequences in what are expected to be variable and informative regions of gp120 are the result of potential artifacts, such as preferential amplification and/or template resampling associated with conventional PCR, cannot be excluded.
Regional env compartmentalization was documented in ET94 (Fig. 3), with viral populations in the cerebellum of ET94 being genotypically distinct from those present in the CSF compartment and the occipital and temporal regions of the brain and less able to infect target cells, such as primary macrophages that express low levels of CD4. Interestingly, genotypically distinct env viral variants were also seen in the cerebella of the two subtype C R5 SHIVE macaques (Fig. 5), suggestive of differential selective constraints within the brain. Since ET94, GC98, and FI08 all failed to mount or sustain a strong anti-SHIV antibody response and the brain is a largely immunologically privileged site, the development of specific Env populations in the cerebellum is unlikely to be driven by the immune system. Anatomically, the cerebellum has the appearance of a separate structure attached to the bottom of the brain and is a region that plays an important role in motor control and contains more neurons than the rest of the brain. This, together with the finding that Env variants in the cerebellum of ET94 are less able to infect CD4low cells, suggests that the reduced availability of resident microglia, as well as the apparent physical separation of the cerebellum, which hinders virus trafficking from other regions of the brain, could be selective factors. Further studies are needed to address these possibilities, as well as to determine if the engagement of the envelope gp120 with both CD4 and CCR5 is altered for the cerebellum Env variants compared to those from other regions of the brain.
Notably, an association between the severity of neuropathological changes, robust microglia infection, evolution to CD4 indepedence, and expanded tropism for truly CD4-negative cells in vivo was observed. Envs with ≥20% CD4i entry efficiency were more readily detected in ET94 than in the other subtype B R5 SHIVE macaques with less severe CNS lesions, and microglia infection was documented only in this macaque, as well as in the two subtype C R5 SHIVE macaques with emergence of CD4i Envs. Moreover, the degree of CD4 independence is higher and functionality is greater for the subtype C than the subtype B Envs, and astrocyte infection could be detected in the brains of the subtype C, but not the subtype B, SHIVE monkeys. Collectively, the data support virus evolution over time in the brain to require less CD4, expanding its tropism to targets, such as microglia and astrocytes, that express little or no CD4. Activated microglia trigger brain inflammation, and recent studies have revealed that, in addition to their phagocytic function, these immune cells are also required for the healthy production of myelin and in shoring up the junctions between neurons (88). Similarly, astrocytes perform vital functions to regulate myelination and maintain brain homeostasis, releasing gliotransmitters to modify neuronal functions (89). Astrocytes are the most abundant cell type in the brain, and integrated viral DNA has been detected within postmortem brain astrocytes of HIV-1-infected individuals with encephalitis (8, 90). Thus, akin to HIVE, infection and activation of microglia and astrocytes are likely to play pivotal roles in SHIV-induced neuropathogenesis.
Neurological disorders appear late in HIV/SIV infection, in association with immunosuppression. Furthermore, isolates from the brain have been shown to be sensitive to neutralizing antibodies (91, 92), and highly neutralization-sensitive CD4-independent envelopes are found primarily in SIV-infected macaques with weak virus-specific antibody responses (32, 54, 55), suggesting that the evolution to a low CD4 requirement or CD4 independence is only possible in the absence of a neutralizing antibody response, in immunologically privileged sites such as the brain, or when the immune constraints decline toward end-stage disease. Our finding that five of the seven subtype B and both subtype C SHIVE animals are rapid progressors that failed to mount or sustain an anti-SHIV antibody response supports this requirement and is in agreement with reports of a correlation between the presence of SIV-induced encephalitis and rapid disease progression (69, 93). Examination of the CD4 independence of envelope clones derived from the plasma of ET94, GC98, and FI08, however, indicated the presence of CD4i envelope variants only in the periphery of the latter (data not shown). Since both subtype C SHIVE macaques (FI08 and GC98) failed to mount plasma antibody responses, the finding of CD4i Envs in the plasma of FI08, but not GC98, implies that while the absence of antiviral antibodies is required for CD4i evolution, it is by itself insufficient to favor CD4i presence in the periphery.
The absence of antibody selective pressure is also conducive to switching or expansion to CXCR4 usage in the R5 SHIV models (56, 61). Indeed, coreceptor switching was documented in three of the five subtype B, as well as in one of the two subtype C, SHIVE macaques studied here, with X4 viruses readily detected by clonal sequence analysis in blood and several lymphoid organs at terminal disease. This contrasts with our findings in this study, where only Envs bearing CCR5-using wild-type sequences could be amplified in the CNS. Because PCRs that distinguish between wild-type and CXCR4 signature V3 sequences had been developed for ET94, DG08, and DE86 (75), we employed these assays, which have a detection sensitivity of 1 X4 variant copy among 104 to 105 R5 targets, to probe the CNS. However, CXCR4-using variants remained undetectable (data not shown). X4 viruses are generally less macrophage tropic than R5 viruses, which may impose constraints on their ability to invade and establish infection in the brain. Because longitudinal CSF and brain tissues were not sampled for the animals with a coreceptor switch, the possibility that X4 virus did penetrate the CNS during the disease course to activate resident cells and contributed to the development and/or severity of the neurological disorder cannot be excluded. Nonetheless, the finding that encephalitis developed in macaques with and without a coreceptor switch, with varying degrees of lesion severity seen in those that harbored X4 viruses, indicates that switching from CCR5 to CXCR4 does not invariably exacerbate the immune activation and pathogenesis of neuroAIDS.
In summary, the SHIVE models on which we based our work are established by infection with two different subtypes of R5 SHIVs via biologically relevant routes (e.g., intrarectal and intravaginal); do not require immunologic manipulations, like CD8 or CD4 depletion; and result in coreceptor switching. Moreover, because there is infection of microglia as well as astrocytes, CNS-resident cells that are less often infected by SIV and that are likely important for viral persistence and neuropathogenesis, these models hold promise for studying the contributions of the HIV envelope and viral clades to neurovirulence and residual virus replication in the CNS, providing information that should guide efforts toward eradicating HIV from the body.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joseph Sodroski for the canine thymocyte Cf2Th cell lines and David Kabat for the RC49 and JC53 cells. TZM-bl cells (catalog no. 8129, from John Kappes, Xiaoyun Wu, and Tranzyme, Inc.) and HIV-1 p24 hybridoma (catalog no. 1513, from Bruce Chesebro) were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported by National Institutes of Health grant R01AI084765 and by Primate Center Base grant P51-OD011104-51.
Footnotes
Published ahead of print 14 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01237-14.
REFERENCES
- 1.Heaton RK, Clifford DB, Franklin DR, Jr, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, CHARTER Group 2010. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75:2087–2096. 10.1212/WNL.0b013e318200d727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McArthur JC, Steiner J, Sacktor N, Nath A. 2010. Human immunodeficiency virus-associated neurocognitive disorders: mind the gap. Ann. Neurol. 67:699–714. 10.1002/ana.22053 [DOI] [PubMed] [Google Scholar]
- 3.Liner KJ, II, Hall CD, Robertson KR. 2008. Effects of antiretroviral therapy on cognitive impairment. Curr. HIV/AIDS Rep. 5:64–71. 10.1007/s11904-008-0011-7 [DOI] [PubMed] [Google Scholar]
- 4.Bagasra O, Lavi E, Bobroski L, Khalili K, Pestaner JP, Tawadros R, Pomerantz RJ. 1996. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS 10:573–585. 10.1097/00002030-199606000-00002 [DOI] [PubMed] [Google Scholar]
- 5.Wiley CA, Schrier RD, Nelson JA, Lampert PW, Oldstone MB. 1986. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc. Natl. Acad. Sci. U. S. A. 83:7089–7093. 10.1073/pnas.83.18.7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gendelman HE, Lipton SA, Tardieu M, Bukrinsky MI, Nottet HS. 1994. The neuropathogenesis of HIV-1 infection. J. Leukoc. Biol. 56:389–398 [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Scarano F, Martin-Garcia J. 2005. The neuropathogenesis of AIDS. Nat. Rev. Immunol. 5:69–81. 10.1038/nri1527 [DOI] [PubMed] [Google Scholar]
- 8.Churchill MJ, Wesselingh SL, Cowley D, Pardo CA, McArthur JC, Brew BJ, Gorry PR. 2009. Extensive astrocyte infection is prominent in human immunodeficiency virus-associated dementia. Ann. Neurol. 66:253–258. 10.1002/ana.21697 [DOI] [PubMed] [Google Scholar]
- 9.Williams R, Bokhari S, Silverstein P, Pinson D, Kumar A, Buch S. 2008. Nonhuman primate models of NeuroAIDS. J. Neurovirol. 14:292–300. 10.1080/13550280802074539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiley CA, Soontornniyomkij V, Radhakrishnan L, Masliah E, Mellors J, Hermann SA, Dailey P, Achim CL. 1998. Distribution of brain HIV load in AIDS. Brain Pathol. 8:277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelman BB, Lisinicchia JG, Morgello S, Masliah E, Commins D, Achim CL, Fox HS, Kolson DL, Grant I, Singer E, Yiannoutsos CT, Sherman S, Gensler G, Moore DJ, Chen T, Soukup VM. 2013. Neurovirological correlation with HIV-associated neurocognitive disorders and encephalitis in a HAART-era cohort. J. Acquir. Immune Defic. Syndr. 62:487–495. 10.1097/QAI.0b013e31827f1bdb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams KC, Hickey WF. 2002. Central nervous system damage, monocytes and macrophages, and neurological disorders in AIDS. Annu. Rev. Neurosci. 25:537–562. 10.1146/annurev.neuro.25.112701.142822 [DOI] [PubMed] [Google Scholar]
- 13.Anderson MG, Hauer D, Sharma DP, Joag SV, Narayan O, Zink MC, Clements JE. 1993. Analysis of envelope changes acquired by SIVmac239 during neuroadaption in rhesus macaques. Virology 195:616–626. 10.1006/viro.1993.1413 [DOI] [PubMed] [Google Scholar]
- 14.Babas T, Munoz D, Mankowski JL, Tarwater PM, Clements JE, Zink MC. 2003. Role of microglial cells in selective replication of simian immunodeficiency virus genotypes in the brain. J. Virol. 77:208–216. 10.1128/JVI.77.1.208-216.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Power C, McArthur JC, Johnson RT, Griffin DE, Glass JD, Perryman S, Chesebro B. 1994. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J. Virol. 68:4643–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zink MC, Amedee AM, Mankowski JL, Craig L, Didier P, Carter DL, Munoz A, Murphey-Corb M, Clements JE. 1997. Pathogenesis of SIV encephalitis. Selection and replication of neurovirulent SIV. Am. J. Pathol. 151:793–803 [PMC free article] [PubMed] [Google Scholar]
- 17.Brown RJ, Peters PJ, Caron C, Gonzalez-Perez MP, Stones L, Ankghuambom C, Pondei K, McClure CP, Alemnji G, Taylor S, Sharp PM, Clapham PR, Ball JK. 2011. Intercompartmental recombination of HIV-1 contributes to env intrahost diversity and modulates viral tropism and sensitivity to entry inhibitors. J. Virol. 85:6024–6037. 10.1128/JVI.00131-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohagen A, Devitt A, Kunstman KJ, Gorry PR, Rose PP, Korber B, Taylor J, Levy R, Murphy RL, Wolinsky SM, Gabuzda D. 2003. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J. Virol. 77:12336–12345. 10.1128/JVI.77.22.12336-12345.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H, Schols D, De Clercq E, Kunstman K, Wolinsky SM, Gabuzda D. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073–10089. 10.1128/JVI.75.21.10073-10089.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salemi M, Lamers SL, Yu S, de Oliveira T, Fitch WM, McGrath MS. 2005. Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J. Virol. 79:11343–11352. 10.1128/JVI.79.17.11343-11352.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnell G, Price RW, Swanstrom R, Spudich S. 2010. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J. Virol. 84:2395–2407. 10.1128/JVI.01863-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sturdevant CB, Dow A, Jabara CB, Joseph SB, Schnell G, Takamune N, Mallewa M, Heyderman RS, Van Rie A, Swanstrom R. 2012. Central nervous system compartmentalization of HIV-1 subtype C variants early and late in infection in young children. PLoS Pathog. 8:e1003094. 10.1371/journal.ppat.1003094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen MF, Westmoreland S, Ryzhova EV, Martin-Garcia J, Soldan SS, Lackner A, Gonzalez-Scarano F. 2006. Simian immunodeficiency virus envelope compartmentalizes in brain regions independent of neuropathology. J. Neurovirol. 12:73–89. 10.1080/13550280600654565 [DOI] [PubMed] [Google Scholar]
- 24.Reeve AB, Pearce NC, Patel K, Augustus KV, Novembre FJ. 2010. Neuropathogenic SIVsmmFGb genetic diversity and selection-induced tissue-specific compartmentalization during chronic infection and temporal evolution of viral genes in lymphoid tissues and regions of the central nervous system. AIDS Res. Hum. Retroviruses 26:663–679. 10.1089/aid.2009.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shapshak P, Segal DM, Crandall KA, Fujimura RK, Zhang BT, Xin KQ, Okuda K, Petito CK, Eisdorfer C, Goodkin K. 1999. Independent evolution of HIV type 1 in different brain regions. AIDS Res. Hum. Retroviruses 15:811–820. 10.1089/088922299310719 [DOI] [PubMed] [Google Scholar]
- 26.Smit TK, Wang B, Ng T, Osborne R, Brew B, Saksena NK. 2001. Varied tropism of HIV-1 isolates derived from different regions of adult brain cortex discriminate between patients with and without AIDS dementia complex (ADC): evidence for neurotropic HIV variants. Virology 279:509–526. 10.1006/viro.2000.0681 [DOI] [PubMed] [Google Scholar]
- 27.Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J. Virol. 78:6915–6926. 10.1128/JVI.78.13.6915-6926.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F. 2006. HIV-1 tropism for the central nervous system: brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology 346:169–179. 10.1016/j.virol.2005.10.031 [DOI] [PubMed] [Google Scholar]
- 29.Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D. 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360:105–119. 10.1016/j.virol.2006.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salimi H, Roche M, Webb N, Gray LR, Chikere K, Sterjovski J, Ellett A, Wesselingh SL, Ramsland PA, Lee B, Churchill MJ, Gorry PR. 2013. Macrophage-tropic HIV-1 variants from brain demonstrate alterations in the way gp120 engages both CD4 and CCR5. J. Leukoc. Biol. 93:113–126. 10.1189/jlb.0612308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277–6292. 10.1128/JVI.76.12.6277-6292.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, Reitter J, Watry DD, Fox HS, Desrosiers RC, Doms RW. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595–2605. 10.1128/JVI.76.6.2595-2605.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westmoreland SV, Rottman JB, Williams KC, Lackner AA, Sasseville VG. 1998. Chemokine receptor expression on resident and inflammatory cells in the brain of macaques with simian immunodeficiency virus encephalitis. Am. J. Pathol. 152:659–665 [PMC free article] [PubMed] [Google Scholar]
- 34.Lewin SR, Sonza S, Irving LB, McDonald CF, Mills J, Crowe SM. 1996. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res. Hum. Retroviruses 12:877–883. 10.1089/aid.1996.12.877 [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Crawford K, Yuan M, Wang H, Gorry PR, Gabuzda D. 2002. Regulation of CC chemokine receptor 5 and CD4 expression and human immunodeficiency virus type 1 replication in human macrophages and microglia by T helper type 2 cytokines. J. Infect. Dis. 185:885–897. 10.1086/339522 [DOI] [PubMed] [Google Scholar]
- 36.Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R. 2009. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog. 5:e1000395. 10.1371/journal.ppat.1000395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burdo TH, Soulas C, Orzechowski K, Button J, Krishnan A, Sugimoto C, Alvarez X, Kuroda MJ, Williams KC. 2010. Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS Pathog. 6:e1000842. 10.1371/journal.ppat.1000842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sacktor N, Tarwater PM, Skolasky RL, McArthur JC, Selnes OA, Becker J, Cohen B, Miller EN, Multicenter for AIDS Cohort Study (ACS) 2001. CSF antiretroviral drug penetrance and the treatment of HIV-associated psychomotor slowing. Neurology 57:542–544. 10.1212/WNL.57.3.542 [DOI] [PubMed] [Google Scholar]
- 39.Staprans S, Marlowe N, Glidden D, Novakovic-Agopian T, Grant RM, Heyes M, Aweeka F, Deeks S, Price RW. 1999. Time course of cerebrospinal fluid responses to antiretroviral therapy: evidence for variable compartmentalization of infection. AIDS 13:1051–1061. 10.1097/00002030-199906180-00008 [DOI] [PubMed] [Google Scholar]
- 40.Gunthard HF, Havlir DV, Fiscus S, Zhang ZQ, Eron J, Mellors J, Gulick R, Frost SD, Brown AJ, Schleif W, Valentine F, Jonas L, Meibohm A, Ignacio CC, Isaacs R, Gamagami R, Emini E, Haase A, Richman DD, Wong JK. 2001. Residual human immunodeficiency virus (HIV) type 1 RNA and DNA in lymph nodes and HIV RNA in genital secretions and in cerebrospinal fluid after suppression of viremia for 2 years. J. Infect. Dis. 183:1318–1327. 10.1086/319864 [DOI] [PubMed] [Google Scholar]
- 41.Kim RB, Fromm MF, Wandel C, Leake B, Wood AJ, Roden DM, Wilkinson GR. 1998. The drug transporter P-glycoprotein limits oral absorption and brain entry of HIV-1 protease inhibitors. J. Clin. Invest. 101:289–294. 10.1172/JCI1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynn HE, Brundage RC, Fletcher CV. 2002. Clinical implications of CNS penetration of antiretroviral drugs. CNS Drugs 16:595–609. 10.2165/00023210-200216090-00002 [DOI] [PubMed] [Google Scholar]
- 43.Deeks SG, Lewin SR, Havlir DV. 2013. The end of AIDS: HIV infection as a chronic disease. Lancet 382:1525–1533. 10.1016/S0140-6736(13)61809-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane JH, Sasseville VG, Smith MO, Vogel P, Pauley DR, Heyes MP, Lackner AA. 1996. Neuroinvasion by simian immunodeficiency virus coincides with increased numbers of perivascular macrophages/microglia and intrathecal immune activation. J. Neurovirol. 2:423–432. 10.3109/13550289609146909 [DOI] [PubMed] [Google Scholar]
- 45.Sasseville VG, Lane JH, Walsh D, Ringler DJ, Lackner AA. 1995. VCAM-1 expression and leukocyte trafficking to the CNS occur early in infection with pathogenic isolates of SIV. J. Med. Primatol. 24:123–131. 10.1111/j.1600-0684.1995.tb00157.x [DOI] [PubMed] [Google Scholar]
- 46.Clements JE, Li M, Gama L, Bullock B, Carruth LM, Mankowski JL, Zink MC. 2005. The central nervous system is a viral reservoir in simian immunodeficiency virus-infected macaques on combined antiretroviral therapy: a model for human immunodeficiency virus patients on highly active antiretroviral therapy. J. Neurovirol. 11:180–189. 10.1080/13550280590922748-1 [DOI] [PubMed] [Google Scholar]
- 47.Dinoso JB, Rabi SA, Blankson JN, Gama L, Mankowski JL, Siliciano RF, Zink MC, Clements JE. 2009. A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J. Virol. 83:9247–9257. 10.1128/JVI.00840-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mankowski JL, Clements JE, Zink MC. 2002. Searching for clues: tracking the pathogenesis of human immunodeficiency virus central nervous system disease by use of an accelerated, consistent simian immunodeficiency virus macaque model. J. Infect. Dis. 186(Suppl 2):S199–S208. 10.1086/344938 [DOI] [PubMed] [Google Scholar]
- 49.Madden LJ, Zandonatti MA, Flynn CT, Taffe MA, Marcondes MC, Schmitz JE, Reimann KA, Henriksen SJ, Fox HS. 2004. CD8+ cell depletion amplifies the acute retroviral syndrome. J. Neurovirol. 10(Suppl 1):S58–S66. 10.1080/13550280490268269 [DOI] [PubMed] [Google Scholar]
- 50.Babas T, Dewitt JB, Mankowski JL, Tarwater PM, Clements JE, Zink MC. 2006. Progressive selection for neurovirulent genotypes in the brain of SIV-infected macaques. AIDS 20:197–205. 10.1097/01.aids.0000198078.24584.21 [DOI] [PubMed] [Google Scholar]
- 51.Bissel SJ, Wang G, Trichel AM, Murphey-Corb M, Wiley CA. 2006. Longitudinal analysis of monocyte/macrophage infection in simian immunodeficiency virus-infected, CD8+ T-cell-depleted macaques that develop lentiviral encephalitis. Am. J. Pathol. 168:1553–1569. 10.2353/ajpath.2006.050240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clements JE, Mankowski JL, Gama L, Zink MC. 2008. The accelerated simian immunodeficiency virus macaque model of human immunodeficiency virus-associated neurological disease: from mechanism to treatment. J. Neurovirol. 14:309–317. 10.1080/13550280802132832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zink MC, Clements JE. 2002. A novel simian immunodeficiency virus model that provides insight into mechanisms of human immunodeficiency virus central nervous system disease. J. Neurovirol. 8(Suppl 2):S42–S48. 10.1080/13550280290101076 [DOI] [PubMed] [Google Scholar]
- 54.Ryzhova E, Whitbeck JC, Canziani G, Westmoreland SV, Cohen GH, Eisenberg RJ, Lackner A, Gonzalez-Scarano F. 2002. Rapid progression to simian AIDS can be accompanied by selection of CD4-independent gp120 variants with impaired ability to bind CD4. J. Virol. 76:7903–7909. 10.1128/JVI.76.15.7903-7909.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dehghani H, Puffer BA, Doms RW, Hirsch VM. 2003. Unique pattern of convergent envelope evolution in simian immunodeficiency virus-infected rapid progressor macaques: association with CD4-independent usage of CCR5. J. Virol. 77:6405–6418. 10.1128/JVI.77.11.6405-6418.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren W, Mumbauer A, Gettie A, Seaman MS, Russell-Lodrigue K, Blanchard J, Westmoreland S, Cheng-Mayer C. 2013. Generation of lineage-related, mucosally transmissible subtype C R5 simian-human immunodeficiency viruses capable of AIDS development, induction of neurological disease, and coreceptor switching in rhesus macaques. J. Virol. 87:6137–6149. 10.1128/JVI.00178-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harbison C, Zhuang K, Gettie A, Blanchard J, Knight H, Didier P, Cheng-Mayer C, Westmoreland S. 2014. Giant cell encephalitis and microglial infection with mucosally transmitted simian-human immunodeficiency virus SHIVSF162P3N in rhesus macaques. J. Neurovirol. 20:62–72. 10.1007/s13365-013-0229-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney LJ, Choe H, Sodroski J. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jordan MR, Kearney M, Palmer S, Shao W, Maldarelli F, Coakley EP, Chappey C, Wanke C, Coffin JM. 2010. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J. Virol. Methods 168:114–120. 10.1016/j.jviromet.2010.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai L, Tasovski I, Leda AR, Chin M, Cheng-Mayer C. 2014. The number and genetic relatedness of transmitted/founder virus impact clinical outcome in vaginal R5 SHIVSF162P3N infection. Retrovirology 11:22. 10.1186/1742-4690-11-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho SH, Tasca S, Shek L, Li A, Gettie A, Blanchard J, Boden D, Cheng-Mayer C. 2007. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 81:8621–8633. 10.1128/JVI.00759-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 64.Jukes TH, Cantor CR. 1969. Evolution of protien molecules, p 21–132 In Mammalian protein metabolism. Academy Press, New York, NY [Google Scholar]
- 65.Shakirzyanova M, Tsai L, Ren W, Gettie A, Blanchard J, Cheng-Mayer C. 2012. Pathogenic consequences of vaginal infection with CCR5-tropic simian-human immunodeficiency virus SHIVSF162P3N. J. Virol. 86:9432–9442. 10.1128/JVI.00852-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mothe BR, Weinfurter J, Wang C, Rehrauer W, Wilson N, Allen TM, Allison DB, Watkins DI. 2003. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 77:2736–2740. 10.1128/JVI.77.4.2736-2740.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 80:5074–5077. 10.1128/JVI.80.10.5074-5077.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 81:8827–8832. 10.1128/JVI.00895-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westmoreland SV, Halpern E, Lackner AA. 1998. Simian immunodeficiency virus encephalitis in rhesus macaques is associated with rapid disease progression. J. Neurovirol. 4:260–268. 10.3109/13550289809114527 [DOI] [PubMed] [Google Scholar]
- 70.Budka H, Costanzi G, Cristina S, Lechi A, Parravicini C, Trabattoni R, Vago L. 1987. Brain pathology induced by infection with the human immunodeficiency virus (HIV). A histological, immunocytochemical, and electron microscopical study of 100 autopsy cases. Acta Neuropathol. 75:185–198 [DOI] [PubMed] [Google Scholar]
- 71.Michaels J, Sharer LR, Epstein LG. 1988. Human immunodeficiency virus type 1 (HIV-1) infection of the nervous system: a review. Immunodefic. Rev. 1:71–104 [PubMed] [Google Scholar]
- 72.Sharer LR. 1992. Pathology of HIV-1 infection of the central nervous system. A review. J. Neuropathol Exp. Neurol. 51:3–11. 10.1097/00005072-199201000-00002 [DOI] [PubMed] [Google Scholar]
- 73.Ritola K, Robertson K, Fiscus SA, Hall C, Swanstrom R. 2005. Increased human immunodeficiency virus type 1 (HIV-1) env compartmentalization in the presence of HIV-1-associated dementia. J. Virol. 79:10830–10834. 10.1128/JVI.79.16.10830-10834.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harrington PR, Schnell G, Letendre SL, Ritola K, Robertson K, Hall C, Burch CL, Jabara CB, Moore DT, Ellis RJ, Price RW, Swanstrom R. 2009. Cross-sectional characterization of HIV-1 env compartmentalization in cerebrospinal fluid over the full disease course. AIDS 23:907–915. 10.1097/QAD.0b013e3283299129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ren W, Tasca S, Zhuang K, Gettie A, Blanchard J, Cheng-Mayer C. 2010. Different tempo and anatomic location of dual-tropic and X4 virus emergence in a model of R5 simian-human immunodeficiency virus infection. J. Virol. 84:340–351. 10.1128/JVI.01865-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shakirzyanova M, Ren W, Zhuang K, Tasca S, Cheng-Mayer C. 2010. Fitness disadvantage of transitional intermediates contributes to dynamic change in the infecting-virus population during coreceptor switch in R5 simian/human immunodeficiency virus-infected macaques. J. Virol. 84:12862–12871. 10.1128/JVI.01478-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bannert N, Schenten D, Craig S, Sodroski J. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984–10993. 10.1128/JVI.74.23.10984-10993.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walter BL, Wehrly K, Swanstrom R, Platt E, Kabat D, Chesebro B. 2005. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J. Virol. 79:4828–4837. 10.1128/JVI.79.8.4828-4837.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peters PJ, Duenas-Decamp MJ, Sullivan WM, Brown R, Ankghuambom C, Luzuriaga K, Robinson J, Burton DR, Bell J, Simmonds P, Ball J, Clapham PR. 2008. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology 5:5. 10.1186/1742-4690-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. 1998. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J. Virol. 72:512–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.LaBranche CC, Hoffman TL, Romano J, Haggarty BS, Edwards TG, Matthews TJ, Doms RW, Hoxie JA. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 73:10310–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. U. S. A. 96:6359–6364. 10.1073/pnas.96.11.6359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edwards TG, Hoffman TL, Baribaud F, Wyss S, LaBranche CC, Romano J, Adkinson J, Sharron M, Hoxie JA, Doms RW. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J. Virol. 75:5230–5239. 10.1128/JVI.75.11.5230-5239.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, Baribaud F. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683–2691. 10.1128/JVI.76.6.2683-2691.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taylor BM, Foulke JS, Flinko R, Heredia A, DeVico A, Reitz M. 2008. An alteration of human immunodeficiency virus gp41 leads to reduced CCR5 dependence and CD4 independence. J. Virol. 82:5460–5471. 10.1128/JVI.01049-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pohlmann S, Davis C, Meister S, Leslie GJ, Otto C, Reeves JD, Puffer BA, Papkalla A, Krumbiegel M, Marzi A, Lorenz S, Munch J, Doms RW, Kirchhoff F. 2004. Amino acid 324 in the simian immunodeficiency virus SIVmac V3 loop can confer CD4 independence and modulate the interaction with CCR5 and alternative coreceptors. J. Virol. 78:3223–3232. 10.1128/JVI.78.7.3223-3232.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spudich S, Gonzalez-Scarano F. 2012. HIV-1-related central nervous system disease: current issues in pathogenesis, diagnosis, and treatment. Cold Spring Harbor Perspect. Med. 2:a007120. 10.1101/cshperspect.a007120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tremblay ME, Stevens B, Sierra A, Wake H, Bessis A, Nimmerjahn A. 2011. The role of microglia in the healthy brain. J. Neurosci. 31:16064–16069. 10.1523/JNEUROSCI.4158-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clarke LE, Barres BA. 2013. Emerging roles of astrocytes in neural circuit development. Nat. Rev. Neurosci. 14:311–321. 10.1038/nrn3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trillo-Pazos G, Diamanturos A, Rislove L, Menza T, Chao W, Belem P, Sadiq S, Morgello S, Sharer L, Volsky DJ. 2003. Detection of HIV-1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV-1 encephalitis by laser capture microdissection. Brain Pathol. 13:144–154. 10.1111/j.1750-3639.2003.tb00014.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Marle G, Rourke SB, Zhang K, Silva C, Ethier J, Gill MJ, Power C. 2002. HIV dementia patients exhibit reduced viral neutralization and increased envelope sequence diversity in blood and brain. AIDS 16:1905–1914. 10.1097/00002030-200209270-00007 [DOI] [PubMed] [Google Scholar]
- 92.Dunfee RL, Thomas ER, Gabuzda D. 2009. Enhanced macrophage tropism of HIV in brain and lymphoid tissues is associated with sensitivity to the broadly neutralizing CD4 binding site antibody b12. Retrovirology 20:69. 10.1186/1742-4690-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baskin GB, Murphey-Corb M, Roberts ED, Didier PJ, Martin LN. 1992. Correlates of SIV encephalitis in rhesus monkeys. J. Med. Primatol. 21:59–63 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.