ABSTRACT
West Nile virus (WNV) is an enveloped virus with a single-stranded positive-sense RNA genome from the Flaviviridae family. WNV is spread by mosquitoes and able to infect humans, causing encephalitis and meningitis that can be fatal; it therefore presents a significant risk for human health. In insects, innate response to RNA virus infection mostly relies on RNA interference and JAK/SAT pathways; however, some evidence indicates that it can also involve microRNAs (miRNAs). miRNAs are small noncoding RNAs that regulate gene expression at posttranscriptional level and play an important role in a number of processes, including immunity and antiviral response. In this study, we focus on the miRNA-mediated response to WNV in mosquito cells. We demonstrate that in response to WNV infection the expression of a mosquito-specific miRNA, aae-miR-2940, is selectively downregulated in Aedes albopictus cells. This miRNA is known to upregulate the metalloprotease m41 FtsH gene, which we have also shown to be required for efficient WNV replication. Correspondingly, downregulation of aae-miR-2940 reduced the metalloprotease level and restricted WNV replication. Thus, we have identified a novel miRNA-dependent mechanism of antiviral response to WNV in mosquitoes.
IMPORTANCE A detailed understanding of vector-pathogen interactions is essential to address the problems posed by vector-borne diseases. Host and viral miRNAs play an important role in regulating expression of viral and host genes involved in endogenous processes, including antiviral response. There has been no evidence to date for the role of mosquito miRNAs in response to flaviviruses. In this study, we show that downregulation of aae-miR-2940 in mosquito cells acts as a potential antiviral mechanism in the mosquito host to inhibit WNV replication by repressing the expression of the metalloprotease m41 FtsH gene, which is required for efficient WNV replication. This is the first identification of an miRNA-dependent antiviral mechanism in mosquitoes, which inhibits replication of WNV. Our findings should facilitate identification of targets in the mosquito genome that can be utilized to suppress vector population and/or limit WNV replication.
INTRODUCTION
West Nile virus (WNV) is a member of the Japanese encephalitis virus serogroup of the genus Flavivirus. It is an enveloped virus with a single-stranded positive-sense RNA genome of ∼11 kb in length. The WNV genome consists of the coding region and 5′ and 3′ untranslated regions (3′UTRs). The coding region encodes a single polyprotein, which is processed into three structural proteins (C, prM, and E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (1). The 3′UTR of WNV genome is essential for virus replication and is processed into a long noncoding RNA, the subgenomic flaviviral RNA (sfRNA), as well as a microRNA (miRNA), KUN-miR-1 (2, 3). Both of these noncoding RNAs have been previously shown to play an important role in virus-host interactions (2–4). The natural cycle of WNV transmission circulates between mosquitoes from Culex genus and birds; horses and humans are subject to incidental infection (5).
WNV has emerged as a significant viral pathogen, which poses a considerable human health risk across the globe. WNV infection can progress to encephalitis, meningitis, and acute flaccid paralysis and, in some cases, especially in aged and immunocompromised patients, WNV infection can be fatal or develop into serious long-term consequences (6). Among a number of WNV isolates, a strain isolated in New York, WNVNY99, is the most pathogenic, with a mortality rate of >80% in individuals who develop encephalic symptoms. To date, there is no specific treatment or effective vaccine available for WNV (7). The native Australian strain of WNV, Kunjin virus (WNVKUN), is closely related to WNVNY99 (98.5% amino acid homology) but is highly attenuated and not pathogenic for humans (8, 9). The high level of similarity with the WNVNY99 genome and low handling risk has made WNVKUN a popular model for studying WNV replication and virus-host interactions.
The innate immune response to WNV in mammals relies primarily on the induction of interferon (IFN) and related pathways (10). In mosquitoes, WNV also encounters a range of antiviral responses, which are proposed to depend on the Vago-induced JAK-STAT pathway and RNA interference (RNAi) (11, 12). However, there is increasing evidence suggesting that other classes of small noncoding RNAs, such as miRNAs and piwi-interacting RNAs, are also involved in virus-host interactions in insects (13).
miRNAs are short (∼22-nucleotide [nt]) noncoding RNAs that regulate gene expression by binding to a partially complementary region in the target mRNA (14). This typically results in the downregulation of gene expression following mRNA degradation, destabilization, or translational repression. Conversely, there are also instances where miRNAs upregulate the expression of the target genes (3, 14–18, 52). Canonically, miRNAs are produced from longer RNA polymerase II transcripts, which consist of hairpin structures with imperfectly complementary stems named primary miRNAs (pri-miRNAs). These pri-miRNA hairpins are recognized by the nuclear microprocessor complex, consisting of the RNase III-like enzyme, Drosha, and the RNA-binding protein, Pasha. Drosha cleaves the stem of the pri-miRNA, generating ∼70-nt stem-loops with 2- to 3-nt overhangs at the 3′ ends, termed precursor miRNAs (pre-miRNAs), which are exported to the cytoplasm via exportin 5. These pre-miRNAs are recognized by another RNase III-like enzyme, Dicer, which removes the hairpin loop generating imperfectly complementary double-stranded RNAs (dsRNAs). Typically, the more thermodynamically stable strand of the dicer substrate RNA (guide strand) is loaded into the RNA-induced silencing complex (RISC) by binding to the Argonaute protein (Ago), and the second strand (passenger strand) is degraded. These miRNAs direct RISC to the target mRNA in a sequence-specific manner and modulate the expression of the corresponding gene (19).
There are two Dicer and two Ago proteins present in insect cells; Dicer-1 and Ago2 specifically recognize RNA duplexes with perfect complementarity and mediate the RNAi pathway, whereas Dicer-2 and Ago1 interact with imperfectly matched dsRNAs and are involved in the miRNA pathway (20). miRNAs play an important role in a number of cellular processes, including differentiation, development, proliferation, oncogenesis, cell death, and immunity (21, 22). miRNAs are also involved in different types of virus-host interactions.
In addition to hosts, a number of viruses, including WNV (3), bovine leukemia virus (23), Epstein-Barr virus (24), herpesviruses (25), and others (26), encode miRNAs, which regulate host and viral genes to enhance viral replication. Viruses can also recruit cellular miRNAs to facilitate their replication. In particular, hsa-miR-122 is required for normal replication of hepatitis C virus (HCV), where it interacts with the 5′UTR (27). Cellular miRNAs can be also used by the host to inhibit viral replication by guiding RISC to complementary regions in the viral genome or by modulating the expression of antiviral or viral dependent genes. In particular, miRNAs are reported to control the latency of retroviruses restricting their transition to the lytic cycle (28) and to be involved in an IFN-induced response to HCV (29).
Recently, deep-sequencing analysis of HEK-293 cells has revealed that miRNAs are also involved in the response against WNV in mammalian cells (30). WNV infection was reported to induce the expression of three novel miRNAs, and one of them (hsa-miR-6124) inhibited the expression of antiapoptotic factors that promoted the apoptosis of infected cells (30). However, the role of cellular miRNAs in response to WNV in the mosquito host still remains to be elucidated.
To begin filling this gap, we focused on a mosquito-specific miRNA, aae-miR-2940, which was previously reported to inhibit the replication of another flavivirus, dengue virus (DENV) in Aedes aegypti (31). We hypothesized that this miRNA could also be important in WNV replication in mosquitoes. Aae-miR-2940 was first experimentally identified in Aedes albopictus and Culex quinquefasciatus cells, for which no complete genome was published and therefore was mapped to the closely related A. aegypti genome (32). This miRNA is specific to these mosquito species and has no known homologs in either Drosophila or mosquitoes from the Anopheles genus. This miRNA has previously been reported to play a complex role in mosquito-parasite interactions with two different types of parasites: the endosymbiotic bacterium Wolbachia (16) and DENV (31). It was demonstrated that Wolbachia infection induces aae-miR-2940 expression, which in turn facilitates Wolbachia replication by upregulation of the metalloprotease m41 FtsH gene. In coinfection with Wolbachia and DENV, the increased level of aae-miR-2940 induced by Wolbachia inhibited DENV replication.
We used the C6/36 cell line to study the role of aae-miR-2940 in the WNV life cycle and the cellular response to WNV infection. C6/36 cells originated from the A. albopictus mosquito, which is closely related to Culex and is commonly used as a model in flavivirus research. However, unlike Culex cell lines, C6/36 cells are Dicer-1 deficient and do not have a functional RNAi pathway, thus facilitating the study of the miRNA response to WNV infection in isolation (33). We demonstrated that aae-miR-2940 expression is selectively downregulated in mosquito cells in response to WNV infection. We also showed that aae-miR-2940 facilitates virus propagation by upregulating the host metalloprotease m41 FtsH, which is required for efficient WNV replication. Both depletion of aae-miR-2940 prior to infection or metalloprotease knockdown significantly reduced the WNV titer in C6/36 cells. We demonstrate here that the downregulation of aae-miR-2940 in mosquito cells acts as a potential antiviral defense mechanism in the mosquito host that restricts WNV replication by repressing the expression of the metalloprotease m41 FtsH gene.
MATERIALS AND METHODS
Cell culture and infection.
C6/36 cells were grown in Roswell Park Memorial Institute medium (RPMI) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 μg of penicillin G sodium/ml, and 100 μg of streptomycin sulfate/ml. Aag2 cells were grown in 1:1 mixture of Schneider's and Mitsuhashi & Maramorosch (Sigma) medium supplemented with 10% FBS, 2 mM l-glutamine, 100 μg of penicillin G sodium/ml, and 100 μg of streptomycin sulfate/ml. Insect cells were cultured at 28°C in closed-cap flasks. Vero 76 and BHK-21 cells were grown in Dulbecco modified Eagle medium supplemented with 10% FBS, 2 mM l-glutamine, 100 μg of penicillin G sodium/ml, and 100 μg of streptomycin sulfate/ml. Mammalian cells were cultured in a humidified atmosphere of air and 5% CO2 at 37°C.
Vero 76 cells were infected with West Nile Virus Kunjin isolate (WNVKUN) generated from FLSDX infectious cDNA clone (34) at multiplicity of infection (MOI) of 0.1, and culture fluid was collected at 3, 5, and 7 days postinfection (dpi). The virus titer was determined by plaque assay. Serial dilutions of virus were plated on BHK-21 cells and allowed to absorb for 2 h, followed by overlay with medium containing 0.5% low-melting-point agarose. At 72 h postinfection, the cells were fixed with 4% phosphate-buffered saline (PBS)-buffered formaldehyde, stained with crystal violet, and washed with water. Plaques were then counted, and titers were calculated based on the dilution factor.
All infections were carried out by incubation of cells in low volume of medium containing virus at the indicated MOI, and then the medium was replaced. Infections were performed, and infected cells were grown in medium supplemented with 2% FBS. All cell culture reagents were from Gibco unless specified.
MicroRNA isolation and Northern blotting.
C6/36 cells were infected at an MOI of 1 or left uninfected (mock). Small RNAs were isolated at 3 and 5 dpi using RNAzol RT reagent (Sigma) according to the manufacturer's protocol. A total of 5 μg of small RNAs was mixed with an equal volume of loading buffer II (Ambion) and denatured by heating at 70°C for 5 min. The samples were then loaded onto 15% polyacrylamide Tris-borate-EDTA (TBE)–urea gels (Invitrogen), and electrophoresis was performed in 1× TBE at 180 V for 1 h. The gels were then stained with ethidium bromide, and RNA was electrotransferred to Amersham Hybond-N+ membranes (GE Healthcare) at 35 V for 90 min in 0.5× TBE. The RNA was cross-linked to the membranes by UV cross-linking at 1,200 kDj/cm2, and membranes were prehybridized at 37°C in ExpressHyb hybridization solution (Clontech) for 1 h. The probes were prepared by end labeling 20 pmol of DNA oligonucleotide complementary to the miRNA of interest (Table 1) with [γ-32P]dATP (Perkin-Elmer) using T4 polynucleotide kinase (Roche). Unincorporated nucleotides were then removed by gel filtration on Illustra MicroSpin G-25 columns (GE Healthcare), and purified probes were hybridized to membranes overnight at 37°C in ExpressHyb hybridization solution. After hybridization, the membranes were rinsed with Northern wash buffer (1% sodium dodecyl sulfate, 1× SSC [0.15 M NaCl plus 0.015 M sodium citrate]), washed four time for 15 min each time at 37°C in the same buffer, and exposed overnight to phosphorimager screen (GE Healthcare), which was then scanned on Typhoon 9400 variable mode imager (Amersham Pharmacia Biotech). Northern blot analysis was repeated more than three times for aae-miR-2940 species and twice for other miRNAs.
TABLE 1.
Sequences of DNA oligonucleotides used as probes for miRNA detection
| miRNA | miRBase accession no. | Probe sequence (5′–3′) |
|---|---|---|
| Aae-mir-2940-5p | MIMAT0014283 | GCCTCGACAGATAAGATAAACCA |
| Aae-miR-2940-3p | MIMAT0014284 | TGTGATTTATCTCCCTGTCGTC |
| Pre-aae-miR-2940 | MI0013489 | GATAGTGGTTGCTCACGAA |
| Aae-miR-2a | MIMAT0014225 | TCTTCAAAGCTGGCTGTGATA |
| Aae-miR-998 | MIMAT0014227 | GAGCTGAATCTCATGGTGCTA |
| Aae-miR-184 | MIMAT0014207 | GCCCTTATCAGTTCTCCGTCCA |
| Aae-miR-7 | MIMAT0014290 | AACAACAAAATCACTAGTCTTCCA |
| Aae-bantam-3p | MIMAT0014219 | ATCAGCTTTCAAAATGATCTCA |
Prediction of aae-miR-2940 target sites.
The potential target sites for aae-miR-2940 (miRBase accession no. MIMAT0014283) in WNV genome (GenBank accession no. AY274504) were predicted using RNAHybrid software (35). Hybrids with a minimum fold energy of ≤−22 kcal/mol were considered as potential interactions.
Plasmid construction.
pBS-3′XX-WT construct containing a cDNA fragment of WNV 3′UTR was obtained by cloning a XmaI/XhoI fragment from FLSDX(pro)_HDVr (36) into pBluescript vector (Agilent Technologies) as described previously (2). Mutagenesis of the potential aae-miR-2940-5p seed binding site to generate the pBS-3′ XX-Mut plasmid was performed by site-directed quick change PCR mutagenesis with the primers PK5F 5′-GATACAGTAGGAGATCTTGTCCTCTGCACAACCAGCCACAC-3′ and PK5R 5′-TGTGGCTGGTTGTGCAGAGGACAAGATCTCCTACTGTATC-3′ according to a previously described protocol (37). The presence of the mutation in the pBS-3′XX-Mut plasmid was confirmed by sequencing. Sequencing was performed at the Australian Genome Research Facility (Brisbane, Australia) using an AB3730xl DNA analyzer (Applied Biosystems).
In vitro transcription and transfection of WNV sfRNA.
pBS-3′XX-WT and pBS-3′XX-Mut plasmids that contained cDNA of the wild type (WT) and aae-miR-2940-5p seed mutant (Mut), respectively, under a T7 promoter were linearized by XhoI digestion. The fragments were gel purified using Wizard SV Gel and a PCR Cleanup kit (Promega) and used as the templates for in vitro transcription of WT and Mut WNV sfRNA. The reaction was performed overnight at 37°C using a MEGAscript T7 kit (Ambion). Antisense WT sfRNA was transcribed from NotI-digested pBS3-XX plasmid using a MEGAscript T3 kit (Ambion) under identical reaction conditions and then used as a negative control. FAM-labeled sfRNA was prepared using fluorescein RNA labeling kit (Roche) and T7 RNA polymerase (Promega) according to the manufacturer's recommendations. XhoI-digested pBS-3′XX-WT plasmid was used as a template. All in vitro-transcribed RNAs were DNase I treated, purified by LiCl precipitation, and analyzed by electrophoresis in a 1.5% denaturing agarose gel prior to transfection.
RNA was transfected into C6/36 cells grown in six-well plate to 80% confluence using 5 μg of RNA and 12 μl of Lipofectamine 2000 (Invitrogen) per well. Transfection was carried out according to the manufacturer's instructions. Transfection was performed in RPMI medium supplemented with 2% FBS. At 48 h posttransfection, small RNAs were harvested and subjected to Northern blot analysis as described above. Experiments were performed in duplicate.
To monitor the transfection efficiency and intracellular localization of transfected RNA, FAM-labeled sfRNA was transfected into C6/36 cells grown on coverslips placed into the wells of a 24-well plate. This was done in parallel with the transfection of WT and Mut sfRNAs in the same plates. At 48 h posttransfection, the cells were washed with cold RNase free-PBS and fixed with PBS-buffered 4% paraformaldehyde solution. The cells were then washed three times with RNase-free PBS, counterstained with 4 μM DAPI (4′,6′-diamidino-2-phenylindole) solution for 5 min, and washed three more times. Coverslips were mounted on slides and examined under a Zeiss LSM 510 META confocal microscope.
Actinomycin D treatment of C6/36 cells and miRNA detection by qRT-PCR.
C6/36 cells grown in six-well plates to 80% confluence were incubated in media containing 8 μg of actinomycin D/ml or without the drug (control). After 12, 24, and 48 h of incubation, the cells were lysed in 1 ml of RNAzol RT reagent, and small RNA fractions were isolated. Prior to isolation lysates were spiked with nonspecific miRNA (5′-UGGUGCGAGAACACAGGAUCU-3′) at 10 pmol/ml, which served as an internal control for normalization. The expression of aae-miR-2940 species was then determined by Northern blotting and quantitative stem-loop reverse transcription-PCR (qRT-PCR) (38). An RT reaction was performed using TaqMan microRNA reverse transcription kit (Applied Biosystems) according to the manufacturer's recommendations. Each reaction mixture contained 10 ng of RNA and 1 pmol of each of the following miRNA-specific stem-loop RT primers: miR-2940-5p-RT (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCTCG-3′), miR-2940-3p-RT (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGTGAT-3′), and NSmiR-RT (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGATCC-3′). qRT-PCR was carried out using common reverse primer (5′-CCAGTGCAGGGTCCGAGGTA-3′) and specific forward primer aae-miR-2940-5p-F (5′-GTCCGCCGCCTGGTTTATCTTA-3′), aae-miR-2940-3p-F (GAGAAGTCGACAGGGAGATAA), or NS-miR-F (5′-GACAGGCGATGGTGCGAGAAC-3′). A PCR was performed on a ViiA 7 real-time PCR instrument (Applied Biosystems) using SYBR green PCR master mix (Applied Biosystems), and gene expression was determined by the ΔΔCT method with normalization to the level of nonspecific control miRNA. The cycling conditions were the same for all primers: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 30 s, and melting-curve analysis.
Analysis of WNV replication and metalloprotease expression in cells transfected with aae-miR-2940 mimic and inhibitor.
Aae-miR-2940-5p inhibitor (5′-GCCUCGACAGAUAAGAUAAACCA-3′), aae-miR-2940-3p inhibitor (5′-AGUGAUUUAUCUCCCUGUCGAC-3′), nonspecific miRNA inhibitor (5′-CAGUACUUUUGUGUAGUACAA-3′), aae-miR-2940 mimic dsRNA (5′-UGUUUAUCUUAUCUGUCGAGGC-3′ and 5′-CUCGACAGAUAAGAUAAACCAUU-3′), nonspecific miRNA mimic dsRNA (5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′), and FAM-labeled nonspecific miRNA mimic and inhibitor were synthesized by Shanghai GenePharma Co., Ltd. C6/36 cells were transfected with 90 pmol of miRNA mimics or inhibitor and 12 μl of Lipofectamine 2000 (Invitrogen) per well of a six-well plate. At 24 h posttransfection, the cells were infected with WNVKUN at an MOI of 1, and at 72 h postinfection transfection was repeated. Culture fluids were harvested from cells at 0, 1, 2, 3, 4, and 5 dpi, and WNV titers were determined by plaque assay on BHK-21 cells. Transfection experiments were performed three times, and plaque assays were performed in duplicate for each sample.
Detection of metalloprotease by RT-PCR.
C6/36 cells were infected with WNVKUN at an MOI of 1, and total RNA was isolated at 3, 5, and 7 dpi using an SV total RNA isolation system (Promega). RNA from mock-infected cells was used as a control. RNA (1 μg) was subjected to amplification with primers specific to metalloprotease m41 FtsH (MetP Forward, 5′-CCCGGACCAAGTCCTAGTA-3′; MetP Reverse, 5′-CAACTCTTTCGGCACGTAA-3′). A fragment of ribosomal protein L11 (RPL 11) mRNA was amplified from the same samples and used as a reference. Two primers were used for RPL11: RPL11 Forward (5′-CGGTCTGGACTTCTACGTTGTGCT-3′) and RPL11 Reverse (5′-CCACTTCATGGCGTCTTCCTTGG-3′). RT-PCR was performed using a SuperScript III One-Step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen) according to the manufacturer's instructions. The cycling conditions were as follows: 55°C for 30 min, 94°C for 2 min, and 30 cycles of 94°C for 15 s, 56°C for 30 s, and 68°C for 30 s, followed by a 5-min extension at 68°C.
RNAi-mediated depletion of metalloprotease.
dsRNA fragments of metalloprotease m41 (MetP) and green fluorescent protein (GFP) mRNA were prepared by in vitro transcription as described previously (16). dsRNAs (5 μg/well) were transfected into C6/36 cells with Lipofectamine 2000 according to the manufacturer's recommendations. At 48 h posttransfection, the cells were infected with WNVKUN at an MOI of 1, and at 24 h postinfection the cells were transfected again. At 72 h postinfection, culture fluids were harvested, virus titers were determined by plaque assay, and WNVKUN genomic RNA levels were measured by qRT-PCR as described below. To assess the knockdown efficiency, RNA was isolated from cells and subjected to qRT-PCR with primers specific to MetP as described below.
Quantitative RT-PCR.
To determine viral RNA levels in culture fluids, viral RNA was isolated from 150 μl of fluids spiked with 4 μg of total human RNA using a NucleoSpin RNA virus kit (Macherey-Nagel). First-strand cDNA was synthesized from 1 μg of viral RNA with a qScript cDNA synthesis kit (Quanta Biosciences) according to the manufacturer's instructions and subjected to qPCR with primers specific to WNV NS3 gene (qWNV Forward, 5′-GCAGAGGAAGGGCATCGGAAAGA-3′; qWNV Reverse, 5′-CAGAAGGAGGGAAAGCAGCAGCATT-3′) and human TATA-binding protein 1 (TBP1) gene (TBP Forward, 5′-GCTGGCCCATAGTGATCTTT-3′; TBP Reverse, 5′-CTTCACACGCCAAGAAACAGT-3′). The cycling conditions were as follows: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by melting-curve analysis.
For the analysis of MetP gene expression, total RNA was isolated from C6/36 cells using an SV total RNA isolation system (Promega) according to the manufacturer's instructions, and 2 μg of RNA was used for cDNA synthesis as described above. Then, 3 μl of cDNA was used as a template in a PCR with primers specific to MetP (MetP Forward, 5′-GACTGGGGTATGTCGGAAAAGGTTG-3′; MetP Reverse, 5′-TGGCGTGCTGCTTGAGAATGG) and RPL11 genes. Amplification was performed under the following conditions: 50°C for 5 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s, 57°C for 30 s, and 68°C for 15 s, followed by melting-curve analysis.
All qRT-PCR analyses were performed on ViiA 7 real-time PCR instrument (Applied Biosystems) using SYBR green PCR master mix (Applied Biosystems) in a reaction volume of 20 μl with 10 pmol of each primer. Gene expression analysis was performed by ΔΔCT method. The expression of WNV RNA was normalized to the TBP mRNA level in mock-treated samples; the expression of MetP was normalized to the RPL11 mRNA level in mock-infected samples. For each experiment, RNA from three biological replicates was used, an RT reaction was performed in triplicate for each mRNA sample, and PCR amplification of each cDNA sample was performed in triplicate. “No-RT” controls were included for each RNA sample, and “no-template” controls were included for each primer pair on each plate.
Statistic analysis.
Data were analyzed by an unpaired two-tailed t test with the Welch's correction or by two-way analysis of variance (ANOVA). The data on miRNA decay were analyzed by linear regression analysis and two-way ANOVA. Analysis was performed using GraphPad Prism software.
RESULTS
The expression of aae-miR-2940-5p but not aae-miR-2940-3p is highly repressed in WNV-infected C6/36 cells.
The biogenesis of most miRNAs involves the cleavage of pre-miRNA hairpin by endonuclease III-like enzyme Dicer with the generation of short dsRNA, followed by the incorporation of one strand of the molecule (guide strand) into RISC and degradation of the complementary (passenger) strand. The incorporation of a guide or passenger strand is made based on the thermodynamic stability of the ends. However, the processing of some pre-miRNAs, including pre-aae-miR-2940, results in both strands (aae-miR-2940-5p and aae-miR-2940-3p in the case of aae-miR-2940; Fig. 1A) being incorporated into RISC and functionally active (39). Aae-miR-2940-5p was recently shown to be functional in facilitating Wolbachia infection of A. aegypti (16). In order to examine the effect of WNV infection on expression of both aae-miR-2940 strands, C6/36 cells were infected at an MOI of 1 PFU/cell, small RNA samples were collected at 3 and 5 dpi, and the miRNA expression was examined by Northern blot analysis. In parallel, small RNAs were extracted from uninfected cells and used for comparison. The blot was first probed with DNA oligonucleotide complementary to aae-miR-2940-5p. The experiment demonstrated that WNV-infected C6/36 cells contained less aae-miR-2940-5p than mock-infected cells (Fig. 1B, left panel). Expression of this miRNA in infected cells decreased in time and was highly suppressed at 5 dpi. Thus, either the production of aae-miR-2940-5p is inhibited in cells upon infection, or it is subjected to degradation. To clarify this matter, we assessed the expression of the corresponding miRNA precursor and aae-miR-2940-3p strand in WNV-infected cells. Surprisingly, Northern blot analysis of small RNAs from uninfected C6/36 cells and cells at 3 and 5 dpi using the probe complementary to aae-miR-2940-3p (Fig. 1A) demonstrated that WNV infection does not change the level of this miRNA strand (Fig. 1B, middle panel). To elucidate whether the observed phenomenon results from selective destabilization/degradation of aae-miR-2940-5p or decreased production of the corresponding pre-miRNA, the blot was also probed with the oligonucleotide complementary to the loop region of pre-aae-miR-2940 (Fig. 1A), which selectively detects pre-aae-miR-2940. The experiment demonstrated that at 5 dpi pre-aae-miR-2940 becomes undetectable (Fig. 1B, right panel), suggesting that the expression of this pre-miRNA is notably downregulated in WNV-infected cells.
FIG 1.

Detection of aae-miR-2940 species in small RNA fractions from C6/36 A. albopictus cells. (A) Sequence and secondary structure of pre-aae-miR-2940 hairpin obtained from miRBase with the location of probes used for detection of aae-miR-2940-5p (5p-probe), aae-mir-2940-3p (3p-probe), and pre-aae-miR-2940 (pre-miRNA-probe). (B) Northern blot analysis with probes in panel A of small RNAs from C6/36 cells infected with WNVKUN at 3 and 5 dpi or mock infected. The experiments were repeated five times, and the blots shown are representative of these experiments. The levels of 5S rRNA and tRNA detected by ethidium bromide staining of the gels prior to transfer (lower panels) serve as loading controls and evidence for RNA integrity. M, mock; dpi, days postinfection; MW, molecular weight marker (GeneRuller Ultra-Low-Range DNA Ladder; Thermo Scientific).
Downregulation of aae-miR-2940-5p expression in WNV-infected C6/36 cells is selective and achieved by a mechanism other than inhibition of miRNA-processing enzymes or miRNA sponging by sfRNA.
There are several possible mechanisms that may be responsible for virus-induced downregulation of miRNA expression, including transcriptional silencing of the corresponding gene (29), inhibition of miRNA processing enzymes (4), and sponging of the miRNA by long noncoding RNAs (40, 54, 55). To determine the mechanism of downregulation of aae-miR-2940 expression in WNV-infected cells, we examined the ability of WNV to inhibit general miRNA biogenesis pathways and to specifically absorb aae-miR-2940-5p.
It was previously demonstrated that flaviviruses could inhibit Dicer activity in mammalian cells (4, 41). This may result in alteration of the entire cellular miRNA profile since the production of most cellular miRNAs is suppressed when the activity of the miRNA-processing enzyme is inhibited. To examine whether WNV infection can also suppress general miRNA processing machinery in mosquito cells, the expression of several cellular miRNAs after infection was assessed. For this purpose, identical small RNA samples used to detect aae-miR-2940 species were analyzed by Northern blotting with probes specific to five unrelated cellular miRNAs. To ensure that the results were not biased against certain types of miRNAs, the RNAs for the analysis were chosen to represent 5p- and 3p-derived species, as well as abundant (aae-miR-184, aae-miR-998, and aae-miR-2a) and moderately expressed (aae-miR-7 and aae-bantam-3p) miRNAs based on previously published deep-sequencing data (32). Northern blot analysis showed that the expression of these miRNAs was not altered in infected cells (Fig. 2), indicating that the activity of the general miRNA processing machinery is not inhibited by WNV. Thus, downregulation of aae-miR-2940 expression in WNV-infected C6/36 cells is achieved by a mechanism selective to this particular miRNA rather than by modification of the overall miRNA biogenesis pathway.
FIG 2.

Northern blot analysis of the expression of selected A. albopictus miRNAs in WNVKUN-infected and uninfected C6/36 cells. Blots were probed with DNA probes complementary to the sequences of corresponding miRNAs, as published in miRBase. Each experiment was repeated in duplicate using RNA samples from independently infected cells. The levels of 5S rRNA and tRNA detected by ethidium bromide staining of the gels prior to transfer (lower panels) serve as loading controls and evidence for RNA integrity. M, mock; dpi, days postinfection; MW, molecular weight marker (GeneRuller Ultra Low Range DNA Ladder).
Recently, evidence has begun to accumulate indicating that the expression of certain miRNAs can be selectively inhibited by long noncoding RNAs containing corresponding miRNA binding sites that can act as miRNA sponges (40, 42). Interestingly, WNV is known to produce a noncoding RNA ∼500 nt in length named subgenomic flaviviral RNA (sfRNA). This RNA derives from the highly conservative 3′UTR region of the genome, is highly abundant in infected cells, and is required for viral pathogenicity (2). To determine whether viral subgenomic or genomic RNAs can act as sponges for aae-miR-2940, we performed computational prediction of the potential aae-miR-2940-5p and aae-miR-2940-3p target sites in WNV genome. This analysis with RNAHybrid software resulted in prediction of a potential aae-miR-2940-5p target site at the position from nt 10567 to nt 10594 of the viral genome (Fig. 3A). This site is located in the 3′UTR and is contained in both genomic RNA and sfRNA.
FIG 3.
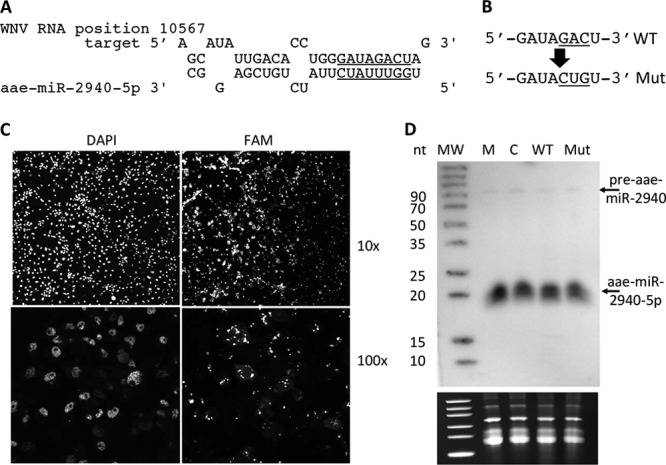
Effect of WNV subgenomic RNA (sfRNA) on aae-miR-2940-5p levels in C6/36 cells. (A) The predicted target of aae-miR-2940-5p in WNV 3′UTR and the seed region is underlined. (B) Mutated predicted target site of aae-miR-2940-5p in WNV 3′UTR. (C) Detection of FAM-labeled in vitro-transcribed sfRNA in transfected C6/36 cells. (D) Northern blot analysis of small RNAs from C6/36 cells transfected with in vitro-transcribed wild-type WNV sfRNA (WT) and sfRNA bearing a mutated aae-miR-2940 target site (Mut). RNA from cells transfected with in vitro-transcribed RNA complementary to sfRNA (NC) and from mock-transfected cells (M) serve as a negative control. A blot was probed with DNA probe complementary to aae-miR-2940-5p. In vitro transcription, transfection, and detection were repeated twice. The levels of 5S rRNA and tRNA detected by ethidium bromide staining of the gels prior to transfer (lower panel) serve as loading controls and evidence for RNA integrity.
To experimentally validate this predicted interaction and the role of this site in aae-miR-2940 absorption, C6/36 cells were transfected with in vitro-transcribed WT sfRNA, sfRNA bearing the mutations in the potential aae-miR-2940-5p seed-interacting region (Mut sfRNA, Fig. 3B) or control RNA. In order to assess the transfection efficiency and to confirm that transfected sfRNA has an intracellular distribution similar to that of an infection, C6/36 cells were transfected with FAM-labeled sfRNA and at 2 dpi examined under confocal microscope. Fluorescence microscopy of transfected cells demonstrated that more than 90% of cells were transfected and in vitro delivered sfRNA localized in cells similar to the previously shown localization of sfRNA in infected cells (2), mostly segregated to the perinuclear foci where viral replication usually occurs (Fig. 3C). To assess aae-miR-2940 levels, small RNAs from cells transfected with WT sfRNA, Mut sfRNA, control RNA and mock-transfected cells were extracted at 2 dpi and subjected to Northern blot hybridization with the probe specific to aae-miR-2940-5p. The analysis indicated that C6/36 cells transfected with sfRNA had the same level of aae-miR-2940-5p and pre-aae-miR-2940 as those transfected with control RNA (Fig. 3D). There were no differences in aae-miR-2940 species between cells transfected with Mut and WT sfRNA (Fig. 3D). These data clearly indicate that sfRNA does not act as a sponge for aae-miR-2940 regardless of the presence of the potential target site for this miRNA and thus is unlikely to contribute to downregulation of aae-miR-2940 expression in WNV-infected mosquito cells.
Collectively, the evidence of specificity of aae-miR-2940 downregulation and the inability of viral RNA to sequester this miRNA suggest that the mechanism responsible for this effect is independent of the alteration of miRNA biogenesis pathways or RNA sponging and may involve transcriptional repression of the corresponding miRNA gene. Only the level of aae-miR-2940-5p decreases upon infection, whereas the level of the corresponding -3p strand remains unchanged, this could be explained if we assume that these molecules have different stabilities. If this is true and the -5p strand is less stable than the -3p strand, when gene transcription is silenced and new pre-miRNA is no longer produced, we might detect decreased levels of aae-miR-2940-5p, whereas the level of aae-miR-2940-3p would remain unchanged.
Aae-miR-2940-5p is less stable than aae-miR-2940-3p.
Typically, miRNAs are relatively stable RNA molecules, and the average half-life of the miRNAs is known to be ∼5 days (43). However, the stability of individual miRNAs can vary depending on their thermodynamic properties and presence of posttranscriptional modifications such as 2′-O-methylation, uridylation, and adenylation of 3′ ends (43, 44). To elucidate whether the stability of aae-miR-2940-5p differs from the stability of the -3p strand, we performed actinomycin D treatment of C6/36 cells to inhibit transcription of new RNA molecules. These cells were maintained in medium containing 8 μg of actinomycin D/ml for up to 48 h; after this time, we observed significant cell death due to the toxicity of actinomycin D. The concentration of actinomycin D used in this experiment was previously reported to efficiently inhibit transcription in C6/36 cells while having a relatively moderate toxic effect on cells (45). RNA was harvested at 0, 12, 24, and 48 h and subjected to Northern blot analysis to determine the expression of aae-miR-2940-5p and aae-miR-2940-3p. The intensity of the band corresponding to aae-miR-2940-5p was slightly decreased at 48 h compared to the untreated control (0 h), whereas the aae-miR-2940-3p band density was unchanged (Fig. 4A), indicating that aae-miR-2940-3p is more stable than aae-miR-2940-5p.
FIG 4.
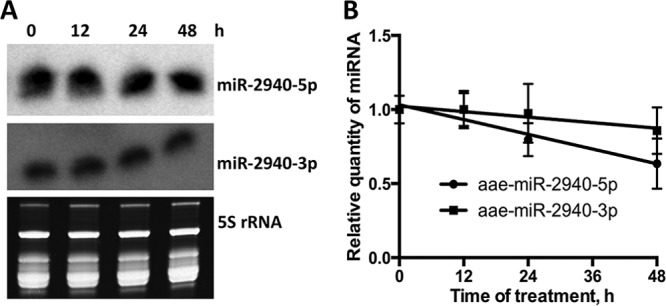
Effect of actinomycin D treatment on levels of aae-miR-2940-5p and aae-miR-2940-3p in C6/36 cells. (A) Northern blot analysis with probes for aae-miR-2940-5p and aae-miR-2940-3p of small RNAs from actinomycin D-treated or untreated C6/36 cells. The experiment was repeated three times, and the blots are representative of these experiments. The levels of 5S rRNA and tRNA detected by ethidium bromide staining serve as loading controls and evidence for RNA integrity. (B) qRT-PCR analysis of aae-miR-2940-5p and aae-miR-2940-3p stability in actinomycin D-treated C6/36 cells. The quantity of miRNA was determined by the ΔΔCT method with normalization to the nonspecific miRNA added to the cell lysates prior to RNA extraction. Each RT reaction and PCR of cDNA samples was performed in triplicate. The data on the graph represent the means of triplicate independent treatment experiments with the standard deviations.
To verify the results of Northern blotting and to quantitatively estimate the level of miRNAs in treated cells, qRT-PCR analysis of the same RNA samples was performed. The results of qRT-PCR demonstrated that the level of aae-miR-2940-5p decreased by 37% at 48 h of treatment, while only 10% decrease of the -3p strand level was observed, further suggesting that the -5p strand is less stable (Fig. 4B). Linear regression analysis identified the significant nonzero slope of the aae-miR-2940 decay curve (P = 0.0017, R2 = 0.6428), but not of the aae-miR-2940-3p curve (P = 0.1679, R2 = 0.181), confirming the observation that the level of the -5p miRNA, but not of the -3p miRNA, was significantly decreased at 48 h after transcriptional repression. However, ANOVA did not identify the difference between aae-miR-2940-5p and aae-miR-2940-3p decay dynamics as statistically significant; however, the prominent decline in the aae-miR-2940-5p degradation curves but not in the aae-miR-2940-3p degradation curves leads us to presume that the difference will become more pronounced at 3 to 5 days after transcriptional repression.
After actinomycin D treatment we demonstrated that the -5p strand of aae-miR-2940 is less stable than the -3p strand. Although we could not analyze the levels of these miRNA species more than 48 h after transcriptional repression due to technical limitations of the method, we demonstrated that degradation of aae-miR-2940-5p occurs more rapidly than aae-miR-2940-3p with 37% lower expression at 48 h postinhibition. We assume that the phenomenon observed at 5 days after infection, when only the level of aae-miR-2940-5p strand is decreased and pre-miRNA is undetectable, is explained by transcriptional repression of the corresponding gene with subsequent degradation of relatively unstable aae-miR-2940-5p.
Aae-miR-2940-5p facilitates WNV replication.
To elucidate whether downregulation of aae-miR-2940 in WNV-infected cells is part of a host-controlled antiviral response or results from WNV induction and is beneficial for the virus, we assessed the effect of aae-miR-2940 species on WNV replication in C6/36 cells. We used synthetic miRNA inhibitors complementary to aae-miR-2940-5p or aae-miR-2940-3p to inhibit functions of the corresponding miRNAs and aae-miR-2940 mimics to artificially increase the levels of these miRNAs in cells. The inhibitors were designed to strongly bind the miRNAs and prevent their interactions with target mRNAs abolishing their functionality. MicroRNA mimics were RNA duplexes designed to specifically load mature aae-miR-2940 strands into RISC and thus compensate for the loss of aae-miR-2940 expression in infected cells. C6/36 cells were transfected with aae-miR-2940-5p inhibitor, aae-miR-2940-3p inhibitor, aae-miR-2940 mimic or nonspecific miRNA inhibitor and mimic.
At 24 h posttransfection, the cells were infected with WNV at an MOI of 1 and at 72 h after infection, the transfection was repeated again to ensure high levels of transfected RNAs during the entire time course. At 1, 2, 3, 4, and 5 dpi, the production of infectious particles in transfected cells was determined by plaque assay on BHK-21 cells. The results demonstrated that the WNV growth kinetics in cells transfected with aae-miR-2940 mimic and aae-miR-2940-5p inhibitor significantly differs from those of cells transfected with nonspecific RNA oligonucleotides (Fig. 5). Transfection with aae-miR-2940-5p inhibitor resulted in significant repression of WNV replication at 3, 4, and 5 dpi, with the highest effect (∼10-fold difference) observed at 3 and 4 dpi. In contrast, aae-miR-2940-3p inhibitor had no significant effect on WNV replication in transfected cells. These data indicate that aae-miR-2940-5p miRNA facilitates efficient propagation of the virus, whereas aae-miR-2940-3p does not. Transfection with aae-miR-2940 mimic resulted in a drastic increase in WNV titers starting at 1 dpi (∼16-fold), and from 2 dpi the difference became statistically significant as determined by ANOVA. At 4 dpi, virus production by cells transfected with nonspecific mimics nearly reached the level observed in transfection with aae-miR-2940 mimics at 2 dpi (∼107 PFU/ml). This indicates that aae-miR-2940 has a pronounced positive effect on WNV replication. Thus, we demonstrated that aae-miR-2940-5p facilitates WNV replication in mosquito cells and hypothesized that the observed downregulation of aae-miR-2940 expression in WNV-infected cells is likely to be the result of the cellular response to infection aimed to restrict virus propagation.
FIG 5.
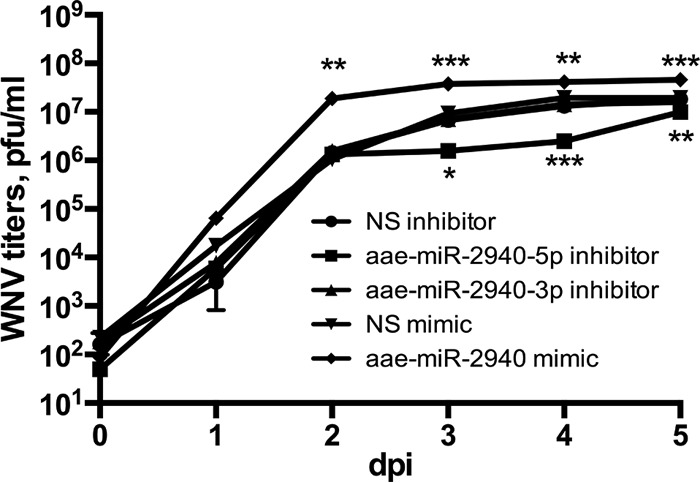
Effect of aae-miR-2940 mimic and inhibitor on WNV replication in infected C6/36 cells. Virus growth kinetics were assessed in C6/36 cells transfected with aae-miR-2940 mimic or inhibitors and infected with WNVKUN at an MOI of 1. Titer was determined by plaque assay on BHK-21 cells. WNV titer in culture fluids from cells transfected with nonspecific miRNA mimic and inhibitor were used for comparison. NS, nonspecific. Each transfection experiment was repeated three times, and plaque assays were performed in duplicate for each sample. Data on the graph are means of triplicates, with error bars indicating minimum and maximum values. Statistical analysis was performed using two-way ANOVA (***, P < 0.001; **, P < 0.01; *, P < 0.1).
Infection-induced downregulation of aae-miR-2940-5p leads to the repression of metalloprotease m41 FtsH expression.
It was recently demonstrated that aae-miR-2940 can bind to a complementary region in metalloprotease m41 FtsH (MetP) mRNA and upregulate its transcript levels (16). Therefore, we decided to investigate whether MetP expression is affected by WNV infection and whether MetP influences WNV replication. Analysis of the metalloprotease expression in infected cells at different time points (3, 5, and 7 dpi) showed that MetP mRNA levels progressively declined over time (Fig. 6B), which correlated well with the decrease in aae-miR-2940-5p expression (Fig. 1B). Similarly, qRT-PCR analysis of MetP mRNA at 4 dpi showed significantly lower MetP mRNA levels in infected cells (compare “Mock” and “No RNA” data in Fig. 6B). To further confirm that MetP m41 downregulation in infected cells results from a decreased aae-miR-2940-5p level, we assessed the ability of aae-miR-2940 mimic to restore MetP transcript levels in WNV-infected cells. C6/36 cells were infected with WNV and subsequently transfected with aae-miR-2940 mimic or a nonspecific miRNA mimic at 24 h after infection. At 3 dpi, MetP transcript level was determined by qRT-PCR. Transfection with aae-miR-2940 mimic resulted in significant recovery of MetP transcript levels relative to cells transfected with the nonspecific miRNA mimic (Fig. 6B), indicating that the effect of WNV infection on the transcript level of this gene was primarily due to the downregulation of aae-miR-2940-5p miRNA expression.
FIG 6.
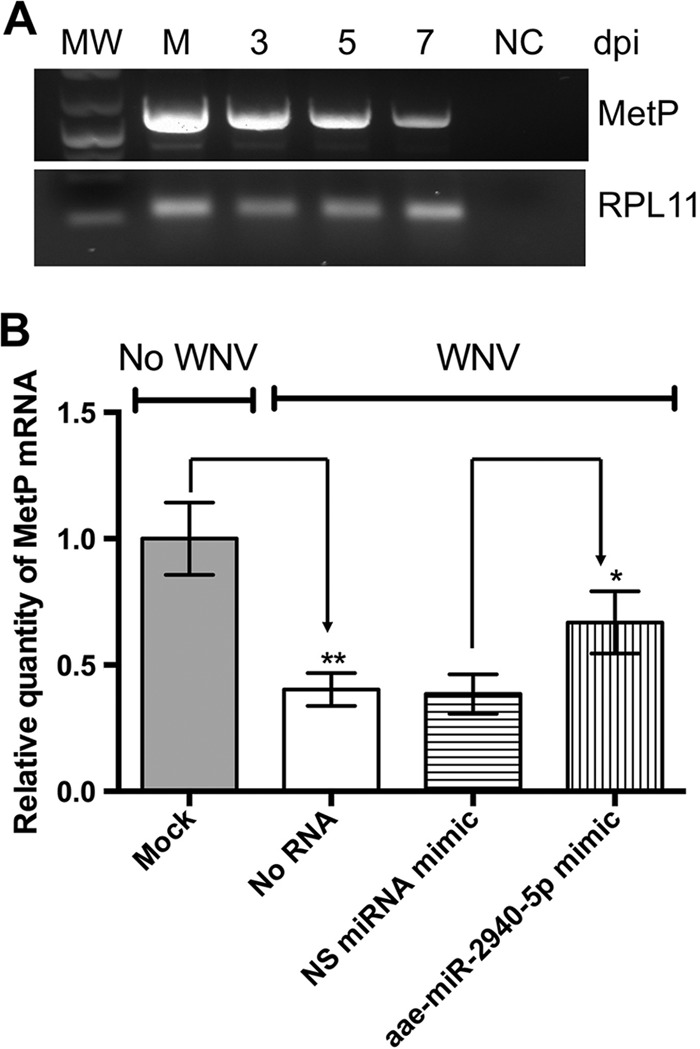
Effect of WNV-induced downregulation of aae-miR-2940-5p expression on MetP mRNA level. (A) Detection of MetP expression in uninfected and infected C6/36 cells at different time points by RT-PCR. Amplification of RPL11 is shown as a control. (B) qRT-PCR analysis of MetP expression in WNV-infected C6/36 cells at 4 dpi versus uninfected cells and in infected C6/36 cells transfected with aae-miR-2940 inhibitor compared or nonspecific (NS) miRNA inhibitor. Relative quantity of MetP mRNA was determined by ΔΔCT method with normalization to ribosomal protein L11 (RPL11) mRNA level. The data are presented as means of triplicates with standard deviations (**, P = 0.0085 [compared to uninfected samples]; *, P = 0.037 [compared to samples transfected with nonspecific miRNA inhibitor]).
Depletion of metalloprotease m41 FtsH restricts WNV replication.
To determine whether MetP is indeed required for efficient WNV propagation, we depleted MetP transcripts using RNAi. In this experiment, we used the RNAi-competent Aag2 cell line derived from A. aegypti. Aag2 cells were transfected with dsRNA fragments specific to the coding region of MetP mRNA or to GFP, which was used as a control. The knockdown efficiency was confirmed by qRT-PCR (Fig. 7A). At 24 h after transfection the cells were infected with WNVKUN at an MOI of 1, and at 24 h postinfection transfection with dsRNA was repeated. At 3 dpi, culture media were collected and used to determine WNV titers and gRNA levels. The WNV titers produced in MetP-depleted cells were significantly lower compared to those detected in control, GFP-depleted cells (Fig. 7B). Quantification of WNV gRNA by qRT-PCR with WNV-specific primers demonstrated that the level of viral RNA was also significantly reduced in culture media from MetP-depleted cells (Fig. 7C), thus supporting the conclusion that MetP facilitates WNV replication.
FIG 7.
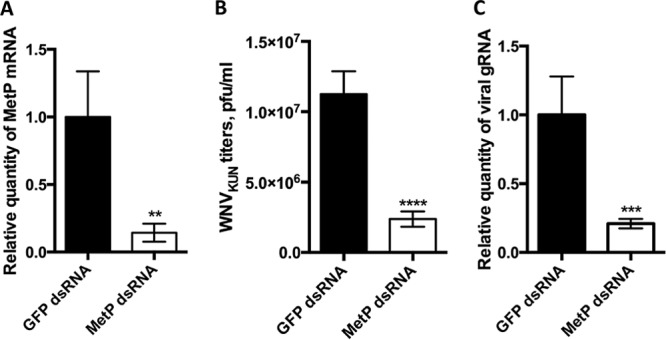
Effect of MetP m41 knockdown on WNV replication in C6/36 cells. To knock down MetP1 expression, C6/36 cells were transfected with double-stranded MetP m41 RNA. Cells transfected with GFP dsRNA served as a negative control. (A) Knockdown efficiency was assessed by qRT-PCR, and the relative quantity of MetP m41 mRNA was determined by the ΔΔCT method with normalization to the ribosomal protein L11 (RPL11) mRNA level. (B) WNV titers in 3pi culture fluid of infected cells depleted for MetP m41 and GFP(control). (C) Detection of WNV gRNA in culture fluids from MetP-depleted Aag2 cells at 3 days after infection at an MOI of 1. Detection was performed by qRT-PCR. Prior to RNA isolation, the samples were spiked with mammalian total RNA, and the level of viral RNA was determined by the ΔΔCT method with normalization to the level of mammalian TBP mRNA. All data are presented as the means of three independent experiments with standard deviations (**, P < 0.05; ***, P < 0.005; ****, P < 0.0005).
DISCUSSION
Antiviral response in insects involves a number of pathways, including autophagy, the Janus kinase-signal transducer and activator of transcription (JAK-STAT) pathway, Toll and immunodeficiency (IMD) pathways, and RNAi response (46). The RNAi pathway is thought to be the major pathway of innate immune response to RNA viruses in insects (47). However, recent studies indicate that other types of small RNAs, particularly miRNAs, also play an important role in the antiviral response in insects and mammals (12, 28, 42, 43, 45).
Recently, the role of miRNAs in response to flaviviral infection has been reported. Specifically, WNV infection of human cells was shown to induce the expression of miRNA hsa-miR-154, which downregulates the expression of antiapoptotic proteins CCCTC-binding factor and EGFR-coamplified and overexpressed protein, thus inducing apoptosis in infected cells (30). A number of differentially expressed miRNAs have been identified in DENV-infected A. aegypti cells (48). Targets of these miRNAs include genes responsible for transcription regulation, mitochondrial function, intracellular transport, chromatin modification, and signal transduction that are known to be required for viral replication and dissemination (48). In the present study, we identified a novel mechanism of antiviral response in insects that is dependent on a host miRNA. We demonstrated that aae-miR-2940 is downregulated in mosquito cells in response to WNV infection which leads to restriction of viral replication via suppression of metalloprotease m41 FtsH (MetP) transcript levels.
The mosquito-specific miRNA, aae-miR-2940, has been recently reported to play an important role in mosquito host-parasite interactions. It was demonstrated that aae-miR-2940 is highly upregulated in A. aegypti cells and mosquitoes infected with the endosymbiotic bacteria Wolbachia pipientis and is essential for Wolbachia maintenance. MetP was identified as a target of aae-miR-2940 in Wolbachia-infected cells, and the ability of this gene to facilitate replication of the endosymbiont was demonstrated (16). Interestingly, unlike most miRNA targets, the MetP transcript level is upregulated by aae-miR-2940. Furthermore, it was demonstrated that Wolbachia-induced overexpression of aae-miR-2940 in mosquito cells also contributes to the suppression of DENV replication in the case of coinfection (31). Based on these findings, we hypothesized that aae-miR-2940 could also be involved in interactions between mosquito hosts and other flaviviruses, particularly WNV.
In order to investigate the role of aae-miR-2940 in response to WNV, we compared the expression of aae-miR-2940-related miRNA species in infected versus uninfected A. albopictus C6/36 cells, analyzed their effect on WNV replication, and assessed the expression of the known aae-miR-2940 target, MetP, in WNV-infected cells and its effect on viral replication. We demonstrated that WNV infection leads to a significant decrease in the production of aae-miR-2940-5p and the corresponding pre-miRNA and addressed the question of how this effect is achieved.
Previously, it has been demonstrated that flaviviruses are capable of impairing the general siRNA and miRNA biogenesis pathways (4, 41). Particularly, DENV nonstructural protein NS4B could directly bind to Dicer, suppressing its activity and thus downregulating the production of all cellular miRNAs (41). WNV was shown to be capable of inhibiting mammalian Dicer by using a different mechanism, which involves binding of the enzyme to the double-stranded structures in sfRNA (4), as well as demonstrating that WNV exhibits sfRNA-mediated miRNA and RNAi suppressor activity in insect cells, but direct interaction of sfRNA with either of insect Dicers was not shown (4). The molecular mechanisms of these inhibitions are still unclear and may occur either on the level of miRNA/siRNA production or RISC activity. To investigate whether the decreased aae-miR-2940 level, observed in WNV-infected mosquito cells, results from the general repression of small RNA biogenesis after inhibition of the corresponding processing enzymes or by another mechanism specific to this miRNA, we assessed the expression of several random cellular miRNAs in infected and uninfected C6/36 cells. We demonstrated that the expression of other cellular miRNAs assessed in the experiment was not influenced by WNV infection. Hence, WNV does not appear to inhibit the general miRNA biogenesis pathways in mosquito cells, and downregulation of aae-miR-2940-5p by WNV is clearly achieved by a mechanism specific for this miRNA.
To date, there are two known mechanisms thought to play major roles in specific regulation of miRNA expression: transcriptional regulation of the corresponding genes (14) and depletion of miRNA due to specific binding to long noncoding or circular RNAs that act as molecular sponges (40, 49, 50). We first assessed the ability of viral genomic and subgenomic RNA to act as the sponges for aae-miR-2940 and showed that despite the presence of a potential aae-imR-2940-5p binding site in the 3′UTR of WNV genome, viral RNAs do not neutralize aae-miR-2940. Taking into account the corresponding decrease in the levels of pre-aee-miR-2940, we propose that the miRNA downregulation is likely to be due to the transcriptional repression of the aee-miR-2940 gene in response to WNV infection, since the level of aae-miR-2940-5p, which is less stable, is decreased in WNV-infected cells, whereas the level of aae-miR-2940-3p, which has a longer half-life, remains uninfluenced at the chosen time points.
Unlike the inhibitory effect of aae-miR-2940 on DENV replication (31), aae-miR-2940 in our experiments facilitated WNV replication. This conclusion was made primarily based on results obtained with an inhibitor of aae-miR-2940-5p, the addition of which significantly repressed WNV replication. In addition, the restoration of aae-miR-2940 levels in infected cells by transfecting aae-miR-2940 mimic also led to a significant increase in virus replication, especially at early time points. Hence, the downregulation of aae-miR-2940 levels observed in infected cells is likely to restrict WNV replication and therefore can be considered an antiviral response mechanism of the host. On the other hand, it is intriguing that aae-miR-2940 has different effects on replication of different flaviviruses. However, taking into account that no changes in aae-miR-2940 expression were found in DENV-infected mosquito cells examined using a deep-sequencing approach (48), it is likely that aae-miR-2940 is not involved in DENV-mosquito host interactions in the absence of coinfection with Wolbachia. Therefore, the downregulation of aae-miR-2940 expression observed in our experiments is likely to be attributed to a specific antiviral response against WNV.
We hypothesized that cellular targets of this miRNA mediate the observed repressive effect on virus replication by the aae-miR-2940-5p inhibitor. In order to elucidate this matter, we assessed the transcript levels of the previously identified aae-miR-2940 target, MetP in infected cells and the effect of this metalloprotease on WNV replication. We demonstrated that in WNV-infected cells the expression of MetP is significantly downregulated compared to uninfected cells, but this effect can be partially reversed by transfecting synthetic aae-miR-2940 mimic. These data indicate that downregulation of aae-miR-2940 expression in response to WNV infection also results in the suppression of MetP transcript levels. To elucidate whether the observed downregulation of MetP can mediate the effect of aae-miR-2940 on WNV replication, we tested whether the metalloprotease is required for WNV propagation by assessing viral replication efficiency in MetP-depleted cells. WNV replication was significantly reduced in cells with depleted MetP, demonstrating that MetP is indeed required for efficient WNV replication in mosquito cells.
Metalloprotease m41 FtsH belongs to the ancient and evolutionary conservative family of ATP-dependent metalloproteases named FtsH (filamentation temperature sensitive H). FtsH proteases are the integral membrane proteins that are localized to the inner mitochondrial membranes and primarily responsible for the quality control of membrane protein folding by degrading the improperly folded molecules (51). The potential roles of these enzymes in the life cycle of viruses and antiviral response have not been investigated. Interestingly, it has been recently reported that knockdown of metalloprotease FtsH YME1L in mammalian cells HEK293 results in the accumulation of nonassembled and oxidized proteins in the inner mitochondrial membrane, abnormal cristae morphology, impaired cell proliferation, and high susceptibility to apoptotic stimuli (51). If a deficiency of metalloprotease FtsH has a similar influence on resistance to apoptosis in insects, this could perhaps explain the antiviral effect of MetP depletion observed in our experiments; however, further research is clearly required to verify this assumption.
In conclusion, we have shown that the expression of aae-miR-2940 in mosquito cells is downregulated in response to infection with WNV. This results in the suppression of MetP transcript levels, which is normally upregulated by aae-miR-2940-5p. WNV replicates less efficiently in the absence of this metalloprotease, and thus a virus-induced and aae-miR-2940-mediated decline in MetP transcript levels is likely to act as an antiviral response mechanism in mosquito cells. To our knowledge, this is the first identification of an antiviral mechanism restricting the replication of WNV in a mosquito host by using the miRNA-regulated pathway.
ACKNOWLEDGMENTS
The study was funded by the NHMRC grant APP1027110 to A.A.K. and A.S. A.A.K. is the research fellow with NHMRC.
We acknowledge Brian Clarke (RVL, UQ) for critical readings of the manuscript and all members of A.A.K.'s group for helpful discussions.
Footnotes
Published ahead of print 14 May 2014
REFERENCES
- 1.Roby JA, Funk A, Khromykh AA. 2012. Flavivirus replication and assembly, p 21–49 In Shipy I. (ed), Molecular virology and control of flaviviruses. Caister Academic Press, Wymondham, United Kingdom [Google Scholar]
- 2.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi P-Y, Hall RA, Khromykh AA. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591. 10.1016/j.chom.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 3.Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, Pijlman GP, Khromykh AA, Asgari S. 2012. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which upregulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 40:2210–2223. 10.1093/nar/gkr848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. 2012. Noncoding flavivirus RNA displays RNAi suppressor activity in insect and mammalian cells. J. Virol. 86:13486–13500. 10.1128/JVI.01104-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kramer LD, Li J, Shi P. 2007. West Nile virus. Lancet Neurol. 6:171–181. 10.1016/S1474-4422(07)70030-3 [DOI] [PubMed] [Google Scholar]
- 6.Brinton MA. 2002. The molecular biology of West Nile virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371–402. 10.1146/annurev.micro.56.012302.160654 [DOI] [PubMed] [Google Scholar]
- 7.Hall RA, Khromykh AA. 2004. West Nile virus vaccines. Expert Opin. Biol. Ther. 4:1295–1305. 10.1517/14712598.4.8.1295 [DOI] [PubMed] [Google Scholar]
- 8.Audsley M, Edmonds J, Liu W, Mokhonov V, Mokhonova E, Melian EB, Prow N, Hall RA, Khromykh AA. 2011. Virulence determinants between New York 99 and Kunjin strains of West Nile virus. Virology 414:63–73. 10.1016/j.virol.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall RA, Broom AK, Smith DW, Mackenzie JS. 2002. The ecology and epidemiology of Kunjin virus. Curr. Top. Microbiol. Immunol. 267:253–269 [DOI] [PubMed] [Google Scholar]
- 10.Suthar MS, Diamond MS, Gale M. 2013. West Nile virus infection and immunity. Nat. Rev. Microbiol. 11:115–128. 10.1038/nrmicro2950 [DOI] [PubMed] [Google Scholar]
- 11.Paradkar PN, Trinidad L, Voysey R, Duchemin J-B, Walker PJ. 2012. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. U. S. A. 109:18915–18920. 10.1073/pnas.1205231109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain M, Asgari S. 2010. Functional analysis of a cellular microRNA in insect host-ascovirus interaction. J. Virol. 84:612–620. 10.1128/JVI.01794-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vodovar N, Saleh M. 2012. Of insects and viruses: the role of small RNAs in insect defence, p 1–36 In Jockusch EL. (ed), Advances in insect physiology. Elsevier, Ltd, London, England [Google Scholar]
- 14.Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambros V. 2001. MicroRNAs: tiny regulators with great potential. Cell 107:823–826. 10.1016/S0092-8674(01)00616-X [DOI] [PubMed] [Google Scholar]
- 16.Hussain M, Frentiu FD, Moreira LA, O'Neill SL, Asgari S. 2011. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 108:9250–9255. 10.1073/pnas.1105469108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rusk N. 2008. When microRNAs activate translation. Nat. Methods 5:122–123. 10.1038/nmeth0208-122a [DOI] [Google Scholar]
- 18.Vasudevan S. 2012. Posttranscriptional upregulation by microRNAs. Wiley Interdiscipl. Rev. RNA 3:311–330. 10.1002/wrna.121 [DOI] [PubMed] [Google Scholar]
- 19.Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 10:126–139. 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- 20.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117:69–81. 10.1016/S0092-8674(04)00261-2 [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asgari S. 2013. MicroRNA functions in insects. Insect Biochem. Mol. Biol. 43:388–397. 10.1016/j.ibmb.2012.10.005 [DOI] [PubMed] [Google Scholar]
- 23.Kincaid RP, Burke JM, Sullivan CS. 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. U. S. A. 109:3077–3082. 10.1073/pnas.1116107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H-J, Huang T-J, Yang C-F, Peng L-X, Liu R-Y, Yang G-D, Chu Q-Q, Huang J-L, Liu N, Huang H-B, Zhu Z-Y, Qian C-N, Huang B-J. 2013. Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells. Virol. J. 10:314–329. 10.1186/1743-422X-10-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang X-J, Coen DM. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 80:5499–5508. 10.1128/JVI.00200-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kincaid R, Sullivan C. 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 8:e1003018. 10.1371/journal.ppat.1003018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimakami T, Yamane D, Welsch C, Hensley L, Jangra RK, Lemon SM. 2012. Base-pairing between hepatitis C virus RNA and miR-122 3′ of its seed sequence is essential for genome stabilization and production of infectious virus. J. Virol. 86:7372–7385. 10.1128/JVI.00513-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Detsika MG, Psarris A, Paraskevis D. 2012. MicroRNAs and HIV latency: a complex and promising relationship. AIDS Rev. 14:188–194 [PubMed] [Google Scholar]
- 29.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922. 10.1038/nature06205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith JL, Grey FE, Uhrlaub JL, Nikolich-Zugich J, Hirsch AJ. 2012. Induction of the cellular microRNA, Hs_154, by West Nile virus contributes to virus-mediated apoptosis through repression of antiapoptotic factors. J. Virol. 86:5278–5287. 10.1128/JVI.06883-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, Hussain M. 2013. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 110:10276–10281. 10.1073/pnas.1303603110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skalsky R, Vanlandingham D. 2010. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics 11:119–135. 10.1186/1471-2164-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD. 2010. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl. Trop. Dis. 4:e856. 10.1371/journal.pntd.0000856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khromykh AA, Kenney MT, Edwin G, Westaway EG. 1998. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270–7279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA. 10:1507–1517. 10.1261/rna.5248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu WJ, Chen HB, Khromykh AA. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804–7813. 10.1128/JVI.77.14.7804-7813.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi P-Y, Khromykh AA. 2010. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 84:11407–11417. 10.1128/JVI.01159-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varkonyi-Gasic E, Hellens RP. 2011. Quantitative stem-loop RT-PCR for detection of microRNAs. Methods Mol. Biol. 744:145–157. 10.1007/978-1-61779-123-9_10 [DOI] [PubMed] [Google Scholar]
- 39.Okamura K, Phillips MD, Tyler DM, Duan H, Chou Y, Lai EC. 2008. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′UTR evolution. Nat. Struct. Mol. Biol. 15:354–363. 10.1038/nsmb.1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kallen AN, Zhou X-B, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi J-S, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. 2013. The imprinted H19 lncRNA antagonizes Let-7 microRNAs. Mol. Cell 52:101–112. 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kakumani PK, Ponia SS, Rajgokul KS, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, Bhatnagar RK, S RK. 2013. Role of RNAi in Dengue viral replication and identification of NS4B as a RNAi suppressor. J. Virol. 87:8870–8883. 10.1128/JVI.02774-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369. 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, Williams BRG. 2011. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 39:5692–5703. 10.1093/nar/gkr148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kai ZS, Pasquinelli AE. 2010. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat. Struct. Mol. Biol. 17:5–10. 10.1038/nsmb.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Condreay LD, Adams RH, Edwards J, Brown DT. 1988. Effect of actinomycin D and cycloheximide on replication of Sindbis virus in Aedes albopictus effect of actinomycin D and cycloheximide on replication of Sindbis virus in Aedes albopictus (mosquito) cells. J. Virol. 62:2629–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Cherry S. 2014. Viruses and antiviral immunity in Drosophila. Dev. Comp. Immunol. 42:67–84. 10.1016/j.dci.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blair CD. 2011. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 6:265–277. 10.2217/fmb.11.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell CL, Harrison T, Hess AM, Ebel GD. 2014. MicroRNA levels are modulated in Aedes aegypti after exposure to Dengue-2. Insect Mol. Biol. 23:132–139. 10.1111/imb.12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cullen B. 2013. How do viruses avoid inhibition by endogenous cellular microRNAs? PLoS Pathog. 9:12–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucas K, Raikhel AS. 2012. Insect MicroRNAs: biogenesis, expression profiling, and biological functions. Insect Biochem. Mol. Biol. 43:1–15. 10.1016/j.ibmb.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiburek L, Cesnekova J, Kostkova O, Fornuskova D, Vinsova K, Wenchich L, Houstek J, Zeman J. 2012. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell 23:1010–1023. 10.1091/mbc.E11-08-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasudevan S, Tong Y, Steitz JA. 2007. Switching from repression to activation: microRNAs can upregulate translation. Science 318:1931–1934. 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 53. Reference deleted.
- 54.Ebert MS, Neilson JR, Sharp PA. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4:721–726. 10.1038/NMETH1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bak RO, Hollensen AK, Primo MN, Sørensen CD, Mikkelsen JG. 2013. Potent microRNA suppression by RNA Pol II-transcribed “tough decoy” inhibitors. RNA 19:280–293. 10.1261/rna.034850.112 [DOI] [PMC free article] [PubMed] [Google Scholar]


