Abstract
Artemisinin (ART)-based combination therapy (ACT) is used as the first-line treatment of uncomplicated falciparum malaria worldwide. However, despite high potency and rapid action, there is a high rate of recrudescence associated with ART monotherapy or ACT long before the recent emergence of ART resistance. ART-induced ring-stage dormancy and recovery have been implicated as possible causes of recrudescence; however, little is known about the characteristics of dormant parasites, including whether dormant parasites are metabolically active. We investigated the transcription of 12 genes encoding key enzymes in various metabolic pathways in P. falciparum during dihydroartemisinin (DHA)-induced dormancy and recovery. Transcription analysis showed an immediate downregulation for 10 genes following exposure to DHA but continued transcription of 2 genes encoding apicoplast and mitochondrial proteins. Transcription of several additional genes encoding apicoplast and mitochondrial proteins, particularly of genes encoding enzymes in pyruvate metabolism and fatty acid synthesis pathways, was also maintained. Additions of inhibitors for biotin acetyl-coenzyme A (CoA) carboxylase and enoyl-acyl carrier reductase of the fatty acid synthesis pathways delayed the recovery of dormant parasites by 6 and 4 days, respectively, following DHA treatment. Our results demonstrate that most metabolic pathways are downregulated in DHA-induced dormant parasites. In contrast, fatty acid and pyruvate metabolic pathways remain active. These findings highlight new targets to interrupt recovery of parasites from ART-induced dormancy and to reduce the rate of recrudescence following ART treatment.
INTRODUCTION
Plasmodium falciparum resistance to conventional antimalarial drugs has become a major obstacle in the global effort of malaria control and elimination. To overcome this obstacle, the WHO recommended the use of artemisinin (ART)-based combination therapies (ACTs) as first-line treatment of uncomplicated falciparum malaria in countries where the disease is endemic in 2001 (1). The implementation of ACTs has contributed to the significant reduction in the number of malaria cases and in malaria transmission intensity in many countries over the past decade (2).
ART derivatives have high potency and are fast acting against Plasmodium spp., including parasites that are resistant to conventional antimalarial drugs. However, there is still a high rate of recrudescence (3% to 50%) that is associated with ART monotherapy in nonimmune patients (3). Increasing the treatment duration from 3 to 7 days reduced but did not eliminate recrudescence (4, 5). Combining ART with other antimalarial drugs to form ACTs also reduced the rate of recrudescence.
Several lines of evidence have been developed to explain the observed high rate of recrudescence associated with ART monotherapy and the joint action of ACT in reducing recrudescence. Previous studies demonstrated that ring-stage parasites are arrested within 6 h of exposure to an ART derivative in vitro and that these ring stages transition into a distinctive morphological state and persist without further growth for days followed by recovery and normal development in a dose-dependent manner (6, 7). A mathematical model that incorporates the ring-stage dormancy, recovery rates, and dose dependency of ART-induced dormancy predicts clinical and parasitological failures at rates comparable to those reported in the field with ART monotherapy (8). Dormant parasites similar in morphology to those observed in vitro (7) were also observed in vivo in a rodent malaria model following ART treatment (9). Importantly, transfer of in vivo-derived dormant parasites into new hosts established infection in a dose-dependent manner, demonstrating that these dormant parasites were viable (9). Combined, these results suggest that ART-induced dormancy may be a key factor in P. falciparum malaria treatment failure of ART therapy.
ART-induced dormancy and an arrest of growth at ring stages of development highlight an interesting physiological state of development that has not been fully characterized. As suggested from the model and accumulated data thus far, ART-induced dormant ring stages are likely the source of parasite biomass that recovers to initiate recrudescent infections. Furthermore, ART-induced dormancy has also been shown to be associated with reduced susceptibility to ART (7, 10, 11). Therefore, understanding the metabolism of the parasites during dormancy may lead to novel therapeutic options and provide insight into the mechanism(s) of ART resistance. One of the first issues to be addressed is whether the dormant ring stages remain metabolically active. Interestingly, repeated exposure to dihydroartemisinin (DHA) or 24 h of exposure to mefloquine following a DHA pulse in vitro reduces the overall recovery rate from dormancy by 10-fold (6), suggesting that dormant stages remain partially susceptible to the drugs; these data suggest that the rings may be metabolically active.
To investigate the metabolic activities of DHA-induced P. falciparum dormant parasites, we examined the transcription profiles of genes encoding key enzymes in various metabolic pathways that are important for maintaining parasite viability, growth, and development during the asexual stage of life cycle (12). These include the mitochondrial electron transport chain, glycolysis and tricarboxylic acid (TCA) metabolism, folate synthesis, DNA replication, fatty acid syntheses, and RNA synthesis. Enzyme activity, ATP content, and DNA and protein synthesis were also examined during the dormant recovery period. We found that despite an overall downregulation of most metabolic pathways, two pathways appear to remain active in dormant rings. This finding will have important implications in explaining how companion drugs in ACT work to reduce recrudescence, leading to new approaches to destroy dormant parasites.
MATERIALS AND METHODS
Cultivation of P. falciparum parasites.
Multiple strains of P. falciparum that had not been exposed to DHA prior to this experiment, W2, 3D7, HB3, and S55, were cultivated in vitro in 3% human erythrocytes suspended in RPMI 1640 and 10% human plasma as described by Trager and Jensen (13). Parasite cultures were synchronized at the ring stage by using 5% sorbitol (14) every second day for 2 consecutive life cycles and again immediately before DHA treatment.
Selection of genes encoding key enzymes in parasite metabolic pathways.
Twelve genes encoding key enzymes in several parasite metabolic pathways that have been shown to be active during asexual stage development were selected for transcription analysis, and these included those involved in the electron transport chain, folate synthesis, glycolysis, DNA replication, fatty acid syntheses, and RNA synthesis. Forward and reverse primers were designed to amplify a fragment of each gene. Gene names, abbreviations, genome identifiers (IDs), metabolic pathways, and primer sequences are listed in Table 1. Primers were also designed to amplify an additional six apicoplast genes and three mitochondrial genes. The complete list of genes, abbreviations, gene IDs, and primer sequences is presented in Table 1.
TABLE 1.
Description of gene and primer sequences for genes encoding key enzymes in various pathwaysa
| Cellular compartment | Pathway | Gene ID | Gene name (abbreviation) | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|---|---|---|
| Cytosol | Glycolysis | PF3D7_1324900 | l-Lactate dehydrogenase (ldh)* | AGGACAATATGGACACTCCGAT | TTTCAGCTATGGCTTCATCAAA |
| Folate metabolism | PF3D7_0417200 | Dihydrofolate reductase (dhfr)* | ACCTGCGCAGTTCATACACG | TGTTGGGAATGGATAGGGTATTCTGT | |
| Folate metabolism | PF3D7_0810800 | Dihydropteroate synthetase (dhps)* | AGGTATTTTTGTTGAACCTAAACGTGC | AGGACCAGAGGATTCTCCACCT | |
| Aminoacyl tRNA synthesis | PF3D7_0717700 | Seryl-tRNA synthetase (sars)* | AAGTAGCAGGTCATCGTG | CGGCACATTCTTCCATA | |
| Nucleus | DNA replication | PF3D7_0411900 | DNA polymerase alpha (DNA pol α gene)* | GCCAAGCCAACCAACCAAACC | TGTGGCTGTTTGTGGATGCAA |
| Nucleotide metabolism | PF3D7_1127100 | Deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase gene)* | GCCGCCTTGGATAATACTAGTGACC | TCCAAAACCTCCTTCTCCTCTGGA | |
| Apicoplast | Pyruvate metabolism | PF3D7_0530200 | Triose phosphate transporter (itpt) | CACATGCTGTTGCAAGCACAGTTA | AGAGGAACCAAGTCCACCAAGGA |
| Pyruvate metabolism | PF3D7_1037100 | Pyruvate kinase 2 (pykii)* | TGGGTGATATTCAAGGGCCT | ACCCTGTTTTGGTTACCTAATGAA | |
| Pyruvate metabolism | PF3D7_1446400 | Pyruvate dehydrogenase E1 beta subunit (pdh e1β) | TCCGAAGCAGCAAAAGAATTAACGA | CCCAAAACCAGCTGACTCATCCA | |
| Isoprenoids metabolism | PF3D7_1337200 | 1-Deoxy-d-xylulose 5-phosphate synthase (doxp)* | CCCGCTATGTTAGGAGGATCAGG | TGCCATAGCTGCTGCGAAAGT | |
| Fatty acid synthesis | PF3D7_1026900 | Biotin acetyl-CoA carboxylase subunit (acc) | TGAGTATCTCGATTCCACACAACA | CGTTCTTGTAAGTATCTCGTGTTCC | |
| Fatty acid synthesis | PF3D7_1469600 | Biotin carboxylase subunit of acetyl-CoA carboxylase (bc) | TGCAGTATGGCCTGGATGGG | TCCATAACATTACCAGTTGGACCT | |
| Fatty acid synthesis | PF3D7_0615100 | Enoyl-acyl carrier reductase (fabI) | TTTTCGGTATTTGGCCTCCT | CAAAAGAAGCGTCAAAGGGT | |
| Fatty acid synthesis | PF3D7_0920000 | Long-chain fatty acid elongation enzyme (lcfaee)* | CATCAACAAATATTATTGGACACCTCA | TCATGTCCATTTCTTTTCTCTTTTTCA | |
| Lipoic acid metabolism | PF3D7_1344600 | Lipoyl synthase (lipA) | TGCATTTTGGTATCCCATCC | TGTATGAACAGGTTCGTTTCT | |
| Mitochondria | TCA cycle | PF3D7_0618500 | Malate dehydrogenase (mdh)* | AGGGGGCACATCCAGTTGAA | AGTCGAAAGCTTTTTGTGTGTTGCT |
| Electron transport chain | PF3D7_0915000 | NADH:ubiquinone oxidoreductase II (ndh2) | GTTCAGGAAATGTGGACAAGC | ACCACCCCATCCTGAACCTA | |
| Electron transport chain | PF3D7_1034400 | Flavoprotein subunit of succinate dehydrogenase (sdha) | GGTTCAGATTGGCTTGGGGA | TGTTCTTGAAAACGGGAGTCCA | |
| Electron transport chain | PF3D7_1439400 | Ubiquinnol-cytochrome c reductase iron-sulfur subunit (uqcr) | ACCTAGGTTGTGTTCCAGCTC | AGGTGCAGGTCCTTGTCTGA | |
| Electron transport chain | mal_mito_3 | Cytochrome b (cyt b)* | AGCAAGTCGATATACACCAGATGTT | GAGAAGCACCTGTTGCGTGC | |
| Electron transport chain | PF3D7_1430900 | Cytochrome c oxidase subunit II (coxii)* | GCTATTCCGGGGCGCTTACA | TGCCTCTGGTGAGACAGCCT |
Asterisks indicate genes included in the initial screening. ID, identifier.
Dynamics of parasitemia and proportions of dormant parasites after DHA treatment.
Synchronized W2 ring-stage parasites (at 2% parasitemia) were treated with 200 ng/ml DHA for 6 h and then washed with culture medium (6). A parallel parasite culture was treated with 100 mM sodium azide for 24 h and used as a negative control. All treated parasite cultures were passed through a magnetic column (magnetic cell separator [MACS] column; Miltenyi Biotec) at 24, 48, and 72 h posttreatment to remove any growing parasites (6). Equal volumes of parasite culture were collected each day before (day 0) and after (days 2, 3, 4, 5, 6, 8, 10, and 12) DHA treatment for making blood smears and for RNA isolation. Proportions of dormant rings and dead and other developmental stages of live parasites were determined as described earlier (7, 10). Briefly, dormant ring-stage parasites have condensed chromatin surrounded by a small amount of cytoplasm; dead parasites have diffuse or degraded chromatin and cytoplasm, whereas viable parasites are present with the normal morphology of the ring, trophozoite, and schizont stages. The entire experiment was repeated three times using three biological replicates.
Reverse transcription and real-time qRT-PCR.
Total RNA was isolated from parasite samples using a NucleoSpin RNA II kit (Macherey-Nagel). rRNasin (RNase inhibitor; Promega) was added to each RNA sample before storage at −80°C. cDNA was synthesized using Superscript III reverse transcriptase (RT) (Invitrogen) and gene-specific primers following the manufacturer's instructions. Real-time quantitative RT-PCR (qRT-PCR) was carried out using Brilliant II SYBR green QPCR master mix (Stratagene) on a Stratagene MX4000 QPCR Thermal Cycler in triplicate. The entire experiment was repeated three times using three biological replicates.
Analysis of qRT-PCR data.
For each gene examined at each time point, triplicate samples from three different cultures (n = 3 × 3 = 9) were analyzed and the average quantification cycle (Cq) values were calculated. These values were normalized against the parasite density (including dormant, dead, and normal parasites) of the sample determined by microscopy. Values representing the relative change of transcripts at each time point were then calculated using the relative transcription level before treatment (day 0) as the baseline. Confidence intervals were determined from 3 independent experiments.
Comparison of transcription levels in different strains.
Synchronized ring-stage parasite cultures of W2, 3D7, HB3, and S55 with 2% parasitemia were treated with DHA (200 ng/ml) for 6 h. A sample was collected from each culture 48 h posttreatment. The transcription levels of seryl-tRNA synthetase (sars) and biotin carboxylase subunit of acetyl-coenzyme A (CoA) carboxylase (bc) were measured by qRT-PCR. The experiment was repeated 3 times.
LDH assay and cellular ATP assay.
A P. falciparum lactate dehydrogenase (LDH) enzyme activity assay was performed in triplicate using an LDH-based In Vitro Toxicology Assay kit (Sigma-Aldrich) following the manufacturer's instructions. Cellular ATP content was detected in triplicate using a CellTiter-Glo Luminescent Cell Viability Assay kit (Promega) following the manufacturer's instructions.
Detection of DNA and protein synthesis.
Synchronized ring-stage parasites of W2 were treated with 20 ng/ml DHA to induce dormancy (6). Parasite samples were collected before treatment (day 0) and after treatment (day 3). For measuring DNA synthesis, [3H]hypoxanthine was added to the parasite culture 24 h posttreatment, and the culture was incubated for 72 h, harvested, and analyzed (15). For measuring protein synthesis, [3H]isoleucine (0.25mCi) was added to the parasite culture, which was then maintained in isoleucine-free media and incubated for 72 h. Parasites were then harvested and analyzed on SDS-PAGE.
Effect of inhibitors on dormancy recovery.
Three sets of experiments were carried out to assess the effect of inhibitors on dormancy recovery. In each set of experiments, parallel synchronized ring-stage parasite cultures of W2 (2% parasitemia) were treated with one of four treatment options: (i) sham (untreated); (ii) DHA (200 ng/ml for 6 h); (iii) an inhibitor (50% inhibitory concentration [c] for 48 h); or (iv) DHA (200 ng/ml for 6 h) followed by an inhibitor (IC50 for 48 h). Haloxyfob (Sigma-Aldrich), an inhibitor of biotin acetyl-CoA carboxylase (ACC), and triclosan (Sigma-Aldrich), an inhibitor of enoyl-acyl carrier reductase (FABI) (16), were used as the inhibitors in the first and second sets of experiments, respectively, while pyrimethamine was used in the third experiment as a control. The IC50s of these inhibitors were determined by inhibition of [3H]hypoxanthine uptake as described by Desjardins et al. (15). Parasitemias in treated cultures were estimated daily by microscopy until the parasitemia reached 10%. The effect of inhibitors on the recovery of dormant parasites was quantified by comparing the numbers of days required to reach 10% parasitemia in cultures with and without inhibitors. Each assay was carried out in triplicate.
RESULTS
Dynamics of parasite density and proportion of dormant parasites following 6-h DHA treatment of W2 P. falciparum parasites.
Parasite density and parasite classification (dormant, dead, and viable) were determined microscopically by examining smears made before (day 0) and after (days 2 to 12) DHA treatment in three independent experiments. Parasites were highly synchronized rings on day 0. On day 2 after DHA treatment, the proportion of parasites classified as dormant averaged 51.2%, with the remaining parasites classified as dead based on their morphology. The average proportion of dormant parasites decreased from day 2, with only 13.6% being dormant rings on day 8 (Fig. 1). The proportion of parasites classified as dead increased as the proportion of dormant parasites decreased. Therefore, the parasite population collected on day 2 had the highest prevalence of dormant yet still viable parasites for investigating gene transcription. The average parasite density, including both dormant and dead parasites, remained stable until day 6. A small number of healthy growing parasites was observed on day 8, and their numbers increased rapidly on days 10 and 12 (Fig. 1). In control cultures where parasites were treated with sodium azide, all parasites were dead on day 2 (not shown), and cultures lysed by day 6 without recovery of parasite growth.
FIG 1.
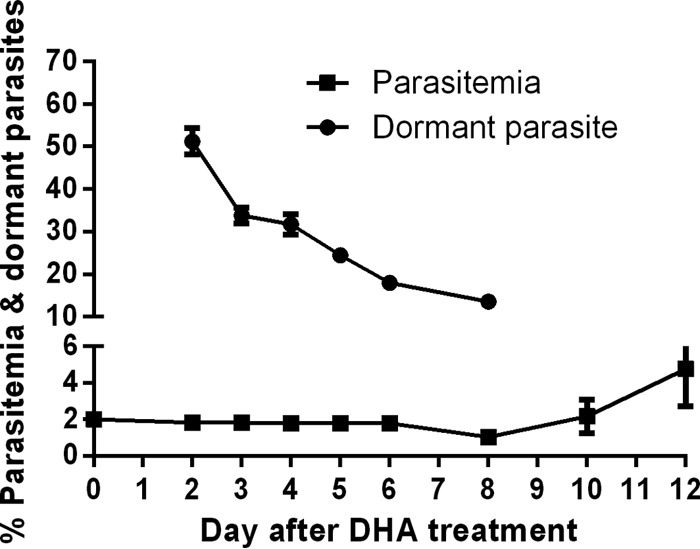
Percentages of parasite density (including normal, dormant, and dead parasites) and dormant rings prior to and post-DHA treatment. Data (means with 95% confidence intervals) were obtained from 3 independent experiments.
Transcription of genes encoding key enzymes in major metabolic pathways during dormancy and recovery.
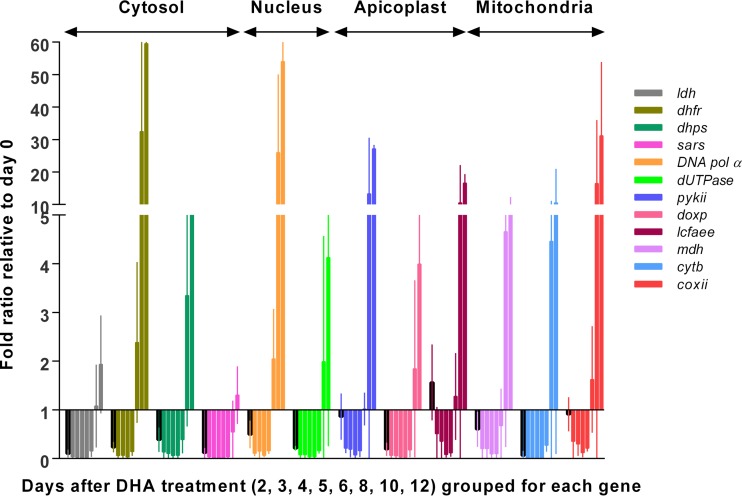
Transcription levels of 12 genes in major metabolic pathways were determined in dormant parasites and compared to the pretreatment levels (day 0) (Fig. 2). On day 2 (48 h posttreatment), transcription levels in 7 genes, including cytochrome b (cytb), ldh, sars, 1-deoxy-d-xylulose 5-phosphate synthase (doxp), deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase), dihydrofolate reductase (dhfr), and dihydropteroate synthetase (dhps), decreased to below 40% of the day 0 transcription levels, suggesting a marked downregulation in the electron transport chain, glycolysis, and DNA, tRNA, and folate synthesis pathways. The transcription levels of two genes, DNA polymerase alpha (DNA pol α) and malate dehydrogenase (mdh), were moderately downregulated to 50% to 60% of day 0 levels (Fig. 2). Interestingly, genes encoding two apicoplast enzymes, pyruvate kinase 2 (pykii) and long-chain fatty acid elongation enzyme (lcfaee), and cytochrome c oxidase subunit II (coxii) of the mitochondrial electron transport chain maintained transcription levels comparable to or higher than that of untreated ring parasites (day 0). This suggests that the apicoplast and mitochondrial pathways are active in dormant parasites of P. falciparum 48 h after DHA treatment. In contrast, the transcription levels of all 12 selected genes in the azide-treated control decreased to 0% to 0.01% of pretreatment levels on day 2 and to below detectable levels by day 6.
FIG 2.
Transcription levels of 12 genes encoding enzymes of several metabolic pathways in DHA-treated rings relative to pretreatment rings (fold ratio ± 95% confidence intervals). Relative transcription levels measured for each of the 12 genes are represented with different color bars. For each gene, relative transcription levels measured on days 2 (outlined in black), 3, 4, 5, 6, 8, 10, and 12 post-DHA treatment (normalized against overall parasitemia, including dormant, dead, and normal parasites) are grouped together from left to right. The transcription level measured on day 0 (pretreatment) is set as 1 for each gene. Data (means with 95% confidence intervals) were obtained from the results determined for samples in triplicate and three independent experiments. Note: lcfaee could also be active in the endoplasmic reticulum.
Transcription levels of all 12 genes were reduced from day 3 onwards and reached the lowest levels on days 5 and 6. From day 8 onward, transcription levels of all 12 genes began to increase during recovery of dormant parasites and the subsequent increase in parasitemia. The transcription levels of five genes (dhfr, DNA pol α, pykii, coxii, and lcfaee) recovered more quickly, with levels at or above the baseline level by day 8. These 5 genes were transcribed at much higher levels in the recovered parasites on days 10 and 12 than in the pretreatment parasites after normalizing to parasite density (Fig. 2). The overexpression of these 5 genes after recovery was likely a result of the presence of a mixture of rings, trophozoites, and schizonts in cultures on days 10 and 12 and higher transcription levels of these genes in trophozoites and schizonts as shown in PlasmoDB (17).
LDH activity during dormancy and recovery.
On day 1 (24 h after DHA treatment), LDH enzyme activity decreased to only 10% of the pretreatment enzyme activity. LDH activity was below detectable levels between day 2 and day 8 after DHA treatment but increased on days 10 and 12 (Fig. 3). This result is in agreement with the ldh transcription profile, suggesting that glycolysis pathway was severely downregulated in dormant parasites and recovered when normal parasite growth resumed.
FIG 3.
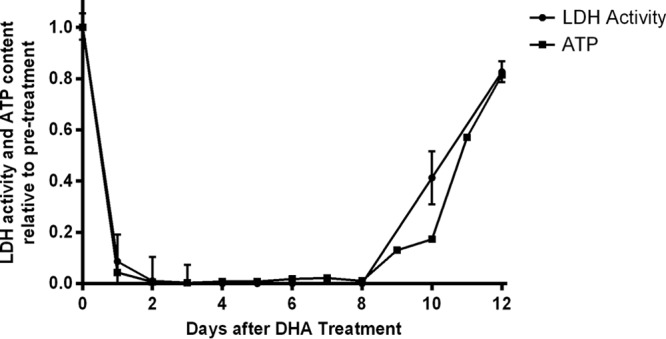
LDH activity and cellular ATP level relative to pretreatment levels during DHA-induced dormancy recovery period. Pretreatment levels are set as 1. Data (means and standard errors) were obtained from the results determined for triplicate samples.
Cellular ATP levels during dormancy and recovery.
ATP levels mirrored the trend of LDH levels (Fig. 3). Since glycolysis is the main source of ATP production in blood stages of malaria parasites (18), reductions in ATP levels are expected when glycolysis has been suspended.
Transcription of genes encoding apicoplast pyruvate metabolism and fatty acid synthesis pathways (FASII) during dormancy and recovery.
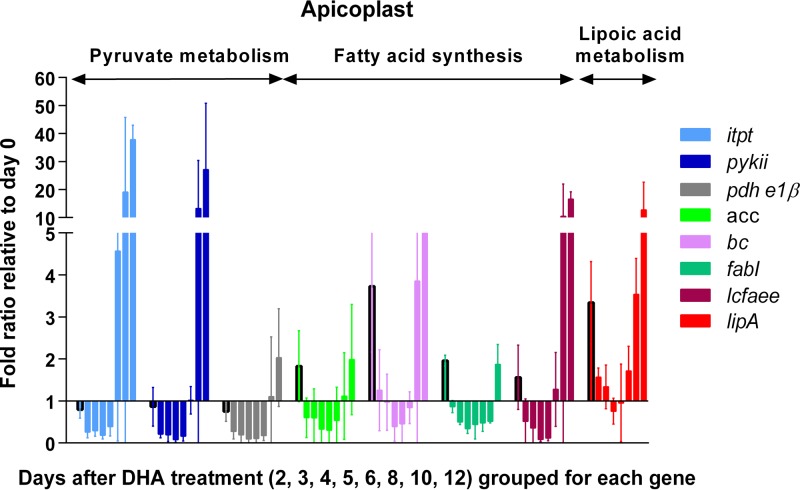
We found that two enzyme-encoding genes (pykii and lcfaee) in the apicoplast maintained high levels of transcription during DHA-induced dormancy. To further investigate metabolism in the apicoplast during dormancy and recovery, transcription of six additional genes encoding enzymes in pyruvate metabolism and fatty acid synthase II (FASII) pathways was examined (Fig. 4). Similarly to pykii, triose phosphate transporter (itpt) and pyruvate dehydrogenase (PDH) E1 beta subunit (pdh e1β) of the pyruvate metabolism pathway maintained their baseline transcription levels on day 2, suggesting that this pathway is active in dormant parasites following DHA treatment.
FIG 4.
Transcription levels of 8 genes encoding enzymes of apicoplast pyruvate metabolism and fatty acid synthesis pathways in DHA-treated rings relative to pretreatment rings (fold ratio ± 95% confidence intervals). Relative transcription levels measured for each of the 8 genes are represented with different color bars. For each gene, relative transcription levels measured on days 2 (outlined in black), 3, 4, 5, 6, 8, 10, and 12 post-DHA treatment are grouped together from left to right. The transcription level measured on day 0 (pretreatment) is set as 1 for each gene. Data (means with 95% confidence intervals) were obtained from the results determined for samples in triplicate and three independent experiments. Note: lcfaee could also be active in the endoplasmic reticulum.
Transcription of genes encoding enzymes in the FASII pathway and lipoyl synthesis pathway downstream of FASII, including acc, fabi, and lipoyl synthase (lipA) was also examined. Both subunits of ACC, biotin acetyl-CoA carboxylase (acc) and bc, had transcription levels on day 2 that were 1.83- and 3.72-fold higher, respectively, than those seen with untreated rings (Fig. 4). Their transcription levels remained above 30% of the baseline level throughout the dormancy recovery experiment. Transcriptions of fabi and lipA were also upregulated on day 2 by approximately 2- and 3-fold, respectively, compared to the baseline (Fig. 4). Interestingly, transcription of lipA was maintained at levels comparable to, or higher than, those of untreated rings throughout the dormancy recovery period. It is worth noting that lipoic acid produced by the lipoyl metabolism pathway is a cofactor of the E2 subunit of pyruvate dehydrogenase (PDH) (19); thus, it may have a positive-feedback effect on PDH in the upstream pyruvate metabolism pathway. The transcription levels of lcfaee in the apicoplast fatty acid synthesis pathway on day 2 were equivalent to those of pretreatment parasites (Fig. 2). In contrast to these results, transcription of doxp, a gene encoding a key enzyme of the isoprenoid metabolism pathway, was markedly downregulated in dormant parasites (Fig. 2). These results suggest that the apicoplast fatty acid synthesis pathway is important for survival of dormant parasites and that other metabolic pathways in the apicoplast such as isoprenoid metabolism are not.
Effect of fatty acid synthesis inhibitors on dormancy recovery.
To confirm the importance of FASII in the survival of DHA-induced dormant parasites, the effects of haloxyfop and triclosan on the dormancy recovery profile in W2 were examined. Haloxyfob is an inhibitor of ACC, while triclosan is an inhibitor of FABI (16). As shown in Fig. 5A, haloxyfob treatment (IC50 = 440 μM for W2) alone for 48 h delayed parasite growth by 2 days, while the same treatment of haloxyfob after 6 h treatment with DHA delayed dormant parasite recovery to 10% parasitemia by 6 days compared to DHA treatment alone (day 20 compared to day 14). Triclosan (IC50 = 5 μM for W2) treatment delayed both parasite growth and dormant parasite recovery to 10% parasitemia by 4 days (Fig. 5B). In contrast, treatment with pyrimethamine (IC50 = 27 μM for W2), a DHFR inhibitor, delayed parasite growth by 2 days when it was used alone but had no effect on the recovery of dormant parasites (Fig. 5C). These results demonstrate that both haloxyfob and triclosan had an inhibitory effect on dormant parasites and were in good agreement with findings of the transcription component of this study, where acc and fabi were found to be upregulated whereas dhfr was downregulated in dormant parasites. As both haloxyfob and triclosan are inhibitors of enzymes in the apicoplast FASII pathway, the results suggest that FASII is active during DHA-induced dormancy and is important for the survival and recovery of dormant parasites.
FIG 5.
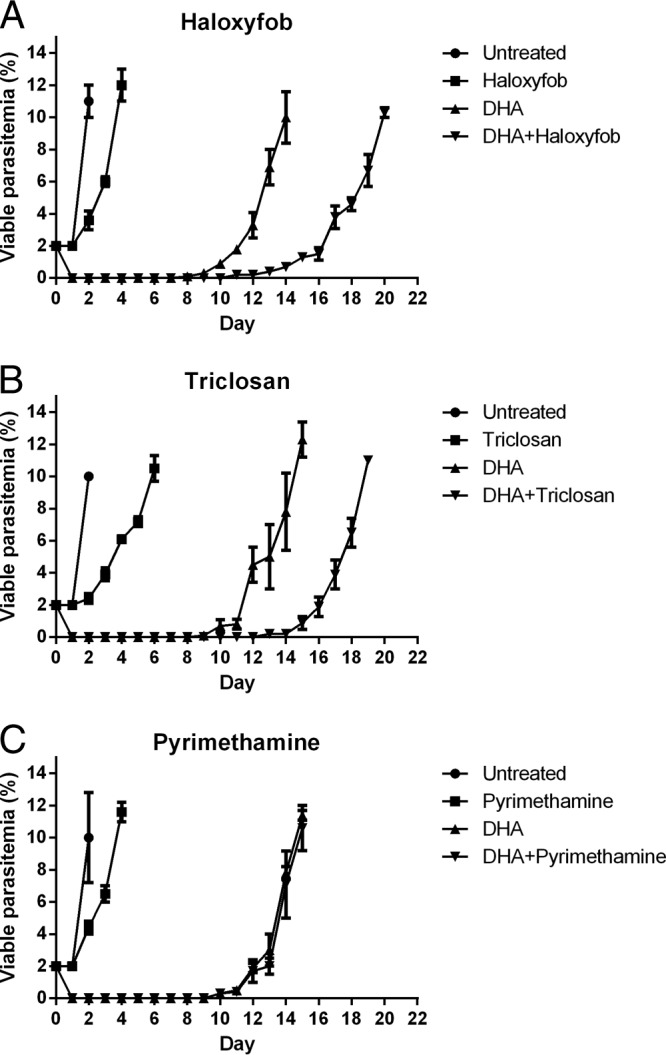
Effect of haloxyfob (A), triclosan (B), and pyrimethamine (C) on the recovery of DHA-induced dormant parasites. Parasitemia of viable parasites on various days is shown in the figure. Data (means with 95% confidence intervals) were obtained from the results determined for samples in triplicate and three independent experiments.
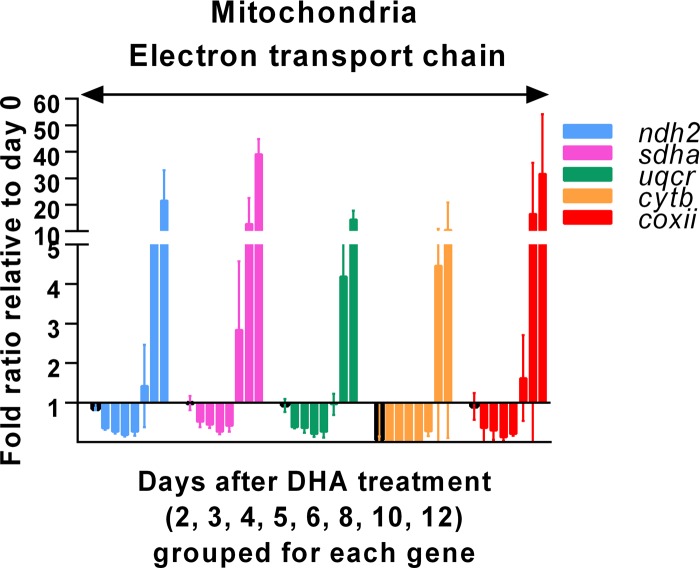
Transcription of genes encoding the mitochondrial electron transport chain during dormancy and recovery.
Transcription of two genes encoding proteins in the mitochondrial electron transport chain, cytb and coxii, was included in the study of the initial 12 genes. While transcription of cytb (complex iii) was significantly reduced in dormant parasites on day 2, coxii (complex iv) maintained its transcription at a level comparable to the baseline level (Fig. 2). To further investigate the activity of the electron transport chain in dormant parasites, transcription of genes encoding NADH-ubiquinone oxidoreductase II (ndh2, complex i), flavoprotein subunit of succinate dehydrogenase (sdha, complex ii), and ubiquinol-cytochrome c reductase iron-sulfur subunit (uqcr, complex iii) in this transport chain was investigated. Similarly to coxii, the three additional genes maintained their transcription at baseline levels on day 2 (Fig. 6). All except cytb (encoded by the mitochondrial genome) are nuclear genes encoding enzymes in the mitochondria. It is possible that transcription of these nuclear genes accumulates, preparing for recovery from dormancy, and that cytb serves as a control point of the pathway. As soon as cytb transcription in mitochondria is resumed, the pathway is immediately fully functional.
FIG 6.
Transcription levels of 5 genes encoding enzymes of the mitochondrial electron transport chain in DHA-treated rings relative to pretreatment rings (ratio ± confidence intervals). Relative transcription levels measured for each of the 5 genes are represented with different color bars. For each gene, relative transcription levels measured on days 2 (outlined in black), 3, 4, 5, 6, 8, 10, and 12 post-DHA treatment are grouped together from left to right. The transcription level measured on day 0 (pretreatment) is set as 1 for each gene. Data (means with 95% confidence intervals) were obtained from the results determined for samples in triplicate and three independent experiments.
Comparison of transcription levels in different parasite strains.
Transcription levels of two genes that were either upregulated (bc) and downregulated (sars) in the W2 strain following DHA treatment were compared in 3 additional P. falciparum strains with different genetic backgrounds (in samples collected on day 2 following DHA exposure). The transcription levels of sars in all strains were low, ranging from 3.6% to 8.6% of those in pretreatment rings. While the transcription levels of bc in 3D7, HB3, and S55 strains were not upregulated as in strain W2, they remained above 43% of those in untreated rings and were considerably higher than those of sars (data not shown).
DNA and protein synthesis in dormant parasites.
[3H]hypoxanthine uptake in dormant parasites was analyzed and compared with the levels in normal ring-stage parasites as well as in red blood cell controls. There was no detectable [3H]hypoxanthine uptake in dormant parasites, indicating that DNA synthesis was suspended (data not shown). Similarly, [3H]isoleucine uptake was not detected in dormant parasites (data not shown), suggesting that protein synthesis was suspended also or was at levels below detection in dormant parasites after DHA treatment.
DISCUSSION
A proportion of ring-stage P. falciparum parasites are capable of arresting their development after a short exposure to DHA in vitro and resuming growth several days later. This DHA-induced dormancy phenomenon has similarities to environmentally induced microbial persistence that causes recurrent infections after therapy (reviewed by Cohen et al.) (20). To better understand the mechanisms underlying this phenomenon, it is important to first determine whether the arrested parasites are metabolically active. In a previous study, we investigated DHA-induced dormancy in P. falciparum in vitro and observed that treatment of dormant parasites with mefloquine or repeating DHA treatment delayed parasite recovery markedly (6). The observation suggested that DHA-induced dormant parasites were susceptible to other drugs, meaning that some metabolic pathways were active.
In the current study, we investigated metabolism in DHA-induced dormant parasites by using several different approaches, including quantifying transcription levels for genes encoding key enzymes, measuring LDH activity, ATP content, and DNA/protein syntheses, and examining the effect of enzyme inhibitors during dormancy recovery. The outcome of these investigations provides direct and indirect evidence for metabolic activities in dormant parasites. Overall, dormant parasites appear to be metabolically inactive, as DNA and protein synthesis was not detected and LDH activity and ATP content were also not detectable. These results indicate that glycolysis, the main ATP-producing pathway, is suspended in dormant parasites; thus, most cellular activities that require energy would be expected to be reduced significantly. However, ATP can also be produced within the apicoplast by the pyruvate metabolism pathway (19) and within mitochondria by the electron transport chain (18). Our transcription analysis demonstrated upregulation of the pyruvate pathway, suggesting that ATP may be produced by the pyruvate metabolism pathway to power other metabolic pathways within the apicoplast and mitochondria (e.g., FASII and electron transport chain). If ATP is used as it is produced, the overall ATP level would remain in the steady state in dormant parasites and could be measured as low or undetectable.
Gene transcription activities are important biomarkers for metabolism. The results of qRT-PCR indicate that all 12 genes selected from major metabolic pathways were still transcribed at some level in dormant parasites (day 2 after DHA treatment). Among these genes, those encoding enzymes in pyruvate and fatty acid metabolism pathways in the apicoplast (pykii and lcfaee) were most interesting, as their transcription in dormant parasites was maintained at a level comparable to or even higher than that in corresponding untreated ring-stage parasites. Further examination of transcription of other nuclear genes encoding enzymes in pyruvate metabolism, fatty acid synthesis (FASII), and lipoyl metabolism in apicoplast demonstrates that they also maintained comparable or elevated transcription levels. In the apicoplast, the pyruvate metabolism pathway is upstream of the fatty acid synthesis pathway, while lipoyl metabolism is downstream (19). Pyruvate metabolism produces ATP and acetyl-CoA for fatty acid synthesis (19, 21), whereas the lipoyl metabolism pathway provides a cofactor for PDH to facilitate pyruvate metabolism. It is very interesting to see that all the examined genes encoding enzymes on these pathways were transcriptionally active. Therefore, transcription analysis suggests that this extended fatty acid synthesis pathway in the apicoplast is still active in dormant parasites. It should be noted that, although the 12 genes selected for transcription analysis encode key enzymes in several important metabolic pathways, they may not reflect all metabolic activities of dormant parasites. A comprehensive transcriptional profiling and metabolomics analysis of dormant parasites may identify additional active pathways.
The steady state in the transcription level is generally determined by the balance between gene transcription and degradation of mRNA molecules. Therefore, mRNA with a shorter half-life has a lower mRNA level when gene transcription suspends. In the genes we examined, mRNAs of ldh and sars have been shown to have a relatively long half-lives of 17 and 21.4 h, respectively, at the ring stage (PlasmoDB [22]), while lcfaee, pykii, and coxii have relatively shorter half-lives of 7.4 h, 12.2 h, and 9.1 h at the ring stage, respectively (PlasmoDB [22]). Since three genes with shorter half-lives had transcription levels detected in dormant rings that were higher than those detected for the two genes with longer half-lives, we believe that the differences in the transcription levels of the genes examined were mainly due to the differences in the levels of gene transcription rather than to the speed of mRNA degradation. It is also unlikely that the upregulation of genes in two pathways was due to parasites arrested at the ring stage, leading to accumulation of transcripts, because only 3 of the 12 genes examined were found to be upregulated whereas the remaining 9 genes were found to be downregulated.
Transcription profiles for genes of interest are usually normalized against the transcription of a housekeeping gene. For P. falciparum, sars is one of the housekeeping genes that is transcribed consistently throughout blood-stage schizogony and is often used as a normalizer (23–25). However, in the current study, the total parasitemia of each sample was used as a normalizer because transcription of sars could be affected during DHA-induced dormancy. Indeed, transcription of sars was severely downregulated from the beginning of dormancy, and sars was among the genes whose transcription was slowest to return to the baseline level. To minimize the effect of dead parasites, the transcription level for each gene was primarily assessed on day 2 after treatment, when over 50% of parasites were dormant, so that the effects of dead parasites were similar for all genes measured. Importantly, the relative proportions of dormant and dead parasites were identical for each gene measured at the same time point for each gene transcript. In addition, since our main finding was the upregulation of genes encoding enzymes in two pathways, it is likely that transcription levels of these genes were underestimated due to the effect of the presence of dead parasites.
Further evidence for the fatty acid synthesis pathway being active in dormant rings came from inhibitor experiments where the effect of inhibitors of these pathways, haloxyfob (inhibitor of ACC) and triclosan (inhibitor of FABI), on parasite recovery from dormancy was assessed. Both inhibitors significantly delayed the recovery of DHA-induced dormant parasites when the inhibitors were added after DHA treatment for 48 h, while pyrimethamine, added as a negative control to inhibit the downregulated folate synthesis pathway, failed to do so. This suggests that ACC and FABI were active in dormant rings and that the recovery of dormant parasites could be impacted by blocking their activities for a period of 48 h during dormancy. In contrast, DHFR was not active in dormant parasites, judging from a strong downregulation of its transcription level and the cessation of DNA synthesis; therefore, it was not unexpected that dormant parasites were not susceptible to pyrimethamine, making it a good negative control. Haloxyfob is a known inhibitor of plant plastid ACC (26) and has been shown to inhibit ACC of Toxoplasma gondii (27). In the current study, it is likely that haloxyfob inhibited ACC of dormant parasites and the downstream fatty acid synthesis pathway, delaying their recovery by 6 days. Triclosan is an inhibitor of FABI in bacteria (28, 29) and was initially shown to inhibit P. falciparum growth (30, 31). However, triclosan also has an unknown off-target effect(s) in P. falciparum since parasites with a genetic knockout of FabI remain susceptible to triclosan (32). Therefore, we could not rule out an off-target effect of triclosan as impacting the recovery of dormant parasites.
The FASII pathway is present in all bacteria and in the plastid of plants, algae, and apicomplexan parasites. Its presence in P. falciparum was discovered in the late 1990s (33, 34), and it was targeted for antimalarial drug development. Recent knockout studies show that FASII is essential for the development of the liver stage of the parasite but not for the development of blood-stage parasites in vitro (32, 35). This is in agreement with earlier reports that blood-stage parasites could scavenge fatty acid (36, 37) and lipoic acid (38) from the host. However, FASII may play a role in vivo and when parasites are under stress. Indeed, genes encoding FASII enzymes were reported to have been upregulated in 43 patients in Senegal (39), especially among a cluster of parasite isolates with transcription profiles matching starvation response. Exposure to artemisinin drugs such as DHA also results in strong stress to malaria ring-stage parasites as evidenced by the abrupt arrest of parasite development and the immediate killing effect. Although the mechanisms underlying dormancy recovery remain to be elucidated, the apicoplast fatty acid synthesis pathway is likely important for the survival and/or recovery of dormant parasites.
Drug-induced persistence is common in microbes. Its underlying mechanism had been linked to metabolic dormancy because antibiotics kill growing microbes by interrupting various metabolic pathways. However, increasing evidence suggests that persistence does not represent passive dormancy, as some metabolic pathways, such as those corresponding to global regulators, amino acid synthesis, DNA repair (40), and nucleotide metabolism (41), are active in microbe persisters. Recently, it was proposed that persistence is an active process that microbes use for survival (42). Our current report on DHA-induced dormancy provides supporting evidence for this mechanism in P. falciparum. Our results show that parasites at the dormant ring stage downregulated gene transcription in the glycolysis, TCA, DNA, and protein synthesis pathways after exposure to DHA. These dormant parasites are not metabolically quiescent, and yet they have stopped growing and dividing. Genes in the pyruvate and FASII pathways continue to be transcribed in dormant parasites; furthermore, inhibitors of enzymes of the FASII pathway delayed the recovery of dormant parasites. The results suggest that the extended FASII pathway is active during dormancy and is important for the survival and recovery of dormant parasites. In summary, these data highlight new avenues to interrupt dormancy recovery that will help to reduce the rate of recrudescence following ART treatment. Successful reduction of the survival of dormant parasites will reduce the reliance and consequential pressure on companion drugs in ACT, especially where resistance to the companion drug already exists.
These observations shed new light on the biology of ART-induced dormant ring stages of P. falciparum, and yet the precise role of dormancy as it relates to the emergence of ART resistance remains to be elucidated. Importantly, the metabolic data obtained in this study were from parasites that expressed the previously described, morphologically distinct dormant rings (6, 7) rather than from parasites in the quiescent stage with normal ring-stage morphology (11). Our current understanding of the biology of dormant ring stages is that ART-induced dormancy is a phenotype expressed in both susceptible and ART-resistant P. falciparum (6, 7) and therefore could confound the underlying biology of emerging ART resistance. Also, a proportion of dormant parasites remain viable, and this viability is linked to upregulation of two key pathways, fatty acid synthesis and pyruvate metabolism. On the basis of accumulated evidence, ART-induced dormancy therefore enhances survival of parasites following treatment with ART therapy and is a likely mechanism for the high rate of recrudescence observed even prior to the emergence of ART resistance. The proportion of ring stages that survive dormancy following treatment should enhance the potential acquisition of secondary resistance traits that would allow the parasite to continue to develop in the presence of increasing drug concentrations (43). More-detailed studies are required to assess on a global scale if additional pathways are important in the biology of dormant parasites and to determine if these same pathways are upregulated in ART-resistant parasites following exposure to drug.
ACKNOWLEDGMENTS
This work was partially funded by a NIH grant (RO1AI058973) and a NHMRC grant (APP1021273).
We thank the Australian Red Cross Blood Service in Brisbane for providing human erythrocytes and serum used for in vitro cultivation of P. falciparum. We thank Daren Krause for performing SDS-PAGE.
We do not have a commercial or other association that might pose a conflict of interest.
The opinions expressed herein are ours and do not necessarily reflect those of the Australian Defence Force Joint Health Command or the U.S. Department of Defense.
Footnotes
Published ahead of print 9 June 2014
REFERENCES
- 1.WHO. 2001. Antimalarial drug combination therapy. Report of a WHO technical consultation. World Health Organization, Geneva, Switzerland [Google Scholar]
- 2.WHO. 2011. World malaria report: 2010. World Health Organization, Geneva, Switzerland [Google Scholar]
- 3.Meshnick SR, Taylor TE, Kamchonwongpaisan S. 1996. Artemisinin and the antimalarial endoperoxides: from herbal remedy to targeted chemotherapy. Microbiol. Rev. 60:301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIntosh HM, Olliaro P. 2000. Artemisinin derivatives for treating uncomplicated malaria. Cochrane Database Syst. Rev. 2:CD000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giao PT, Binh TQ, Kager PA, Long HP, Van Thang N, Van Nam N, de Vries PJ. 2001. Artemisinin for treatment of uncomplicated falciparum malaria: is there a place for monotherapy? Am. J. Trop. Med. Hyg. 65:690–695 [DOI] [PubMed] [Google Scholar]
- 6.Teuscher F, Gatton ML, Chen N, Peters J, Kyle DE, Cheng Q. 2010. Artemisinin-induced dormancy in Plasmodium falciparum: duration, recovery rates, and implications in treatment failure. J. Infect. Dis. 202:1362–1368. 10.1086/656476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tucker MS, Mutka T, Sparks K, Patel J, Kyle DE. 2012. Phenotypic and genotypic analysis of in vitro selected artemisinin-resistant progeny of Plasmodium falciparum. Antimicrob. Agents Chemother. 56:302–314. 10.1128/AAC.05540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Codd A, Teuscher F, Kyle DE, Cheng Q, Gatton ML. 2011. Artemisinin-induced parasite dormancy: a plausible mechanism for treatment failure. Malar. J. 10:56. 10.1186/1475-2875-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaCrue AN, Scheel M, Kennedy K, Kumar N, Kyle DE. 2011. Effects of artesunate on parasite recrudescence and dormancy in the rodent malaria model Plasmodium vinckei. PLoS One 6:e26689. 10.1371/journal.pone.0026689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teuscher F, Chen N, Kyle DE, Gatton ML, Cheng Q. 2012. Phenotypic changes in artemisinin-resistant Plasmodium falciparum lines in vitro: evidence for decreased sensitivity to dormancy and growth inhibition. Antimicrob. Agents Chemother. 56:428–431. 10.1128/AAC.05456-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witkowski B, Lelievre J, Barragan MJ, Laurent V, Su XZ, Berry A, Benoit-Vical F. 2010. Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob. Agents Chemother. 54:1872–1877. 10.1128/AAC.01636-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen A, Dumetre A, Azas N. 2013. A decade of Plasmodium falciparum metabolic pathways of therapeutic interest to develop new selective antimalarial drugs. Mini Rev. Med. Chem. 13:1340–1347. 10.2174/13895575113139990060 [DOI] [PubMed] [Google Scholar]
- 13.Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. 10.1126/science.781840 [DOI] [PubMed] [Google Scholar]
- 14.Lambros C, Vanderberg JP. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418–420. 10.2307/3280287 [DOI] [PubMed] [Google Scholar]
- 15.Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16:710–718. 10.1128/AAC.16.6.710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramya TN, Mishra S, Karmodiya K, Surolia N, Surolia A. 2007. Inhibitors of nonhousekeeping functions of the apicoplast defy delayed death in Plasmodium falciparum. Antimicrob. Agents Chemother. 51:307–316. 10.1128/AAC.00808-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. 2006. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 34:1166–1173. 10.1093/nar/gkj517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaidya AB, Mather MW. 2009. Mitochondrial evolution and functions in malaria parasites. Annu. Rev. Microbiol. 63:249–267. 10.1146/annurev.micro.091208.073424 [DOI] [PubMed] [Google Scholar]
- 19.Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. 2004. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2:203–216. 10.1038/nrmicro843 [DOI] [PubMed] [Google Scholar]
- 20.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. 10.1016/j.chom.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim L, Linka M, Mullin KA, Weber AP, McFadden GI. 2010. The carbon and energy sources of the non-photosynthetic plastid in the malaria parasite. FEBS Lett. 584:549–554. 10.1016/j.febslet.2009.11.097 [DOI] [PubMed] [Google Scholar]
- 22.Shock J, Fischer KF, Derisi JL. 2007. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biol. 8:R134. 10.1186/gb-2007-8-7-r134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, Hviid L, Theander TG. 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 49:179–191. 10.1046/j.1365-2958.2003.03570.x [DOI] [PubMed] [Google Scholar]
- 24.Peters JM, Fowler EV, Krause DR, Cheng Q, Gatton ML. 2007. Differential changes in Plasmodium falciparum var transcription during adaptation to culture. J. Infect. Dis. 195:748–755. 10.1086/511436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker J, Gatton ML, Peters J, Ho MF, McCarthy JS, Cheng Q. 2011. Transcription and expression of Plasmodium falciparum histidine-rich proteins in different stages and strains: implications for rapid diagnostic tests. PLoS One 6:e22593. 10.1371/journal.pone.0022593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuther E, Johnson JJ, Haselkorn R, McLeod R, Gornicki P. 1999. Growth of Toxoplasma gondii is inhibited by aryloxyphenoxypropionate herbicides targeting acetyl-CoA carboxylase. Proc. Natl. Acad. Sci. U. S. A. 96:13387–13392. 10.1073/pnas.96.23.13387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisanz C, Bastien O, Grando D, Jouhet J, Marechal E, Cesbron-Delauw MF. 2006. Toxoplasma gondii acyl-lipid metabolism: de novo synthesis from apicoplast-generated fatty acids versus scavenging of host cell precursors. Biochem. J. 394:197–205. 10.1042/BJ20050609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMurry LM, Oethinger M, Levy SB. 1998. Triclosan targets lipid synthesis. Nature 394:531–532. 10.1038/28970 [DOI] [PubMed] [Google Scholar]
- 29.Heath RJ, Yu YT, Shapiro MA, Olson E, Rock CO. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 273:30316–30320. 10.1074/jbc.273.46.30316 [DOI] [PubMed] [Google Scholar]
- 30.Surolia N, Surolia A. 2001. Triclosan offers protection against blood stages of malaria by inhibiting enoyl-ACP reductase of Plasmodium falciparum. Nat. Med. 7:167–173. 10.1038/84612 [DOI] [PubMed] [Google Scholar]
- 31.McLeod R, Muench SP, Rafferty JB, Kyle DE, Mui EJ, Kirisits MJ, Mack DG, Roberts CW, Samuel BU, Lyons RE, Dorris M, Milhous WK, Rice DW. 2001. Triclosan inhibits the growth of Plasmodium falciparum and Toxoplasma gondii by inhibition of apicomplexan Fab I. Int. J. Parasitol. 31:109–113. 10.1016/S0020-7519(01)00111-4 [DOI] [PubMed] [Google Scholar]
- 32.Yu M, Kumar TR, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, Kelly BJ, Moura PA, Lakshmanan V, Freundlich JS, Valderramos JC, Vilcheze C, Siedner M, Tsai JH, Falkard B, Sidhu AB, Purcell LA, Gratraud P, Kremer L, Waters AP, Schiehser G, Jacobus DP, Janse CJ, Ager A, Jacobs WR, Jr, Sacchettini JC, Heussler V, Sinnis P, Fidock DA. 2008. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe 4:567–578. 10.1016/j.chom.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waller RF, Keeling PJ, Donald RG, Striepen B, Handman E, Lang UN, Cowman AF, Besra GS, Roos DS, McFadden GI. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 95:12352–12357. 10.1073/pnas.95.21.12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waller RF, Reed MB, Cowman AF, McFadden GI. 2000. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 19:1794–1802. 10.1093/emboj/19.8.1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, Cowman AF, Kappe SH. 2009. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell. Microbiol. 11:506–520. 10.1111/j.1462-5822.2008.01270.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holz GG., Jr 1977. Lipids and the malarial parasite. Bull. World Health Organ. 55:237–248 [PMC free article] [PubMed] [Google Scholar]
- 37.Vial HJ, Ancelin ML, Philippot JR, Thuet MJ. 1990. Biosynthesis and dynamics of lipids in Plasmodium-infected mature mammalian erythrocytes. Blood Cells 16:531–555 [PubMed] [Google Scholar]
- 38.Allary M, Lu JZ, Zhu L, Prigge ST. 2007. Scavenging of the cofactor lipoate is essential for the survival of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 63:1331–1344. 10.1111/j.1365-2958.2007.05592.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, Kamal M, Sarr O, Mboup S, Ndir O, Wypij D, Levasseur K, Thomas E, Tamayo P, Dong C, Zhou Y, Lander ES, Ndiaye D, Wirth D, Winzeler EA, Mesirov JP, Regev A. 2007. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 450:1091–1095. 10.1038/nature06311 [DOI] [PubMed] [Google Scholar]
- 40.De Groote VN, Verstraeten N, Fauvart M, Kint CI, Verbeeck AM, Beullens S, Cornelis P, Michiels J. 2009. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 297:73–79. 10.1111/j.1574-6968.2009.01657.x [DOI] [PubMed] [Google Scholar]
- 41.Hansen S, Lewis K, Vulic M. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52:2718–2726. 10.1128/AAC.00144-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng Q, Kyle DE, Gatton ML. 2012. Artemisinin resistance in Plasmodium falciparum: a process linked to dormancy? Int. J. Parasitol. Drugs Drug Resist. 2:249–255. 10.1016/j.ijpddr.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]