Abstract
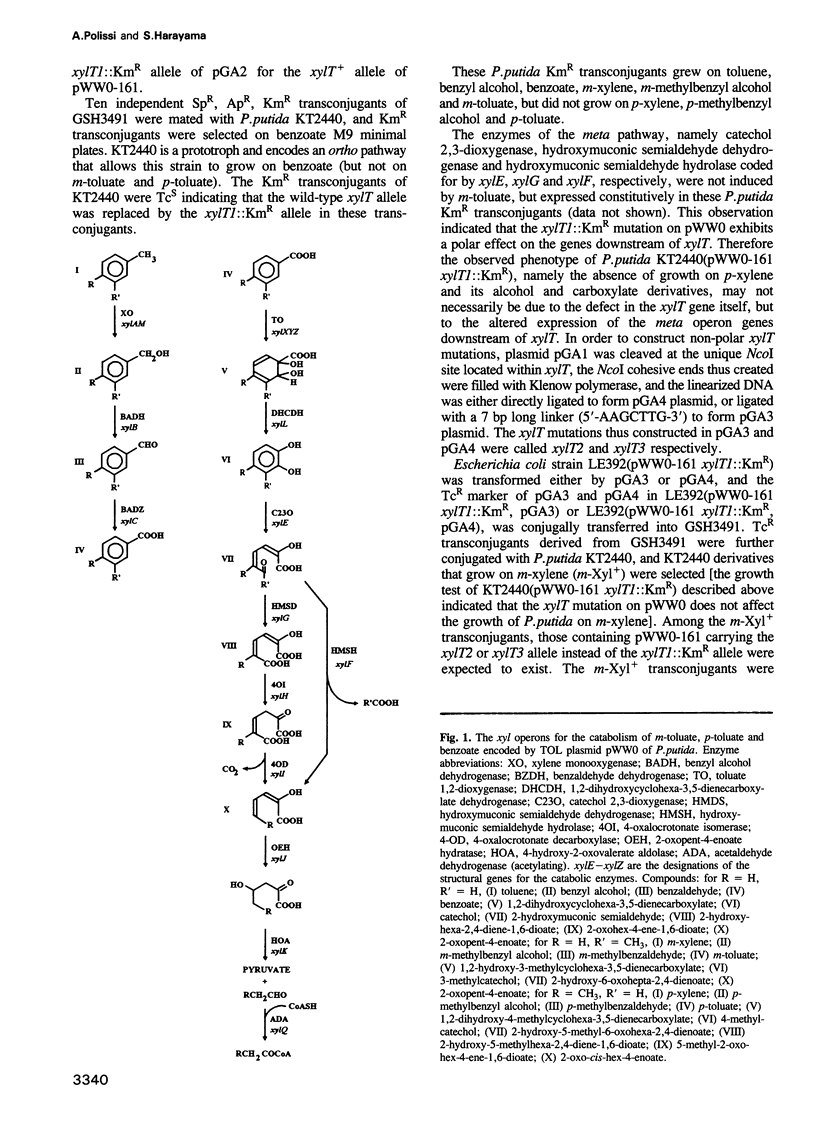
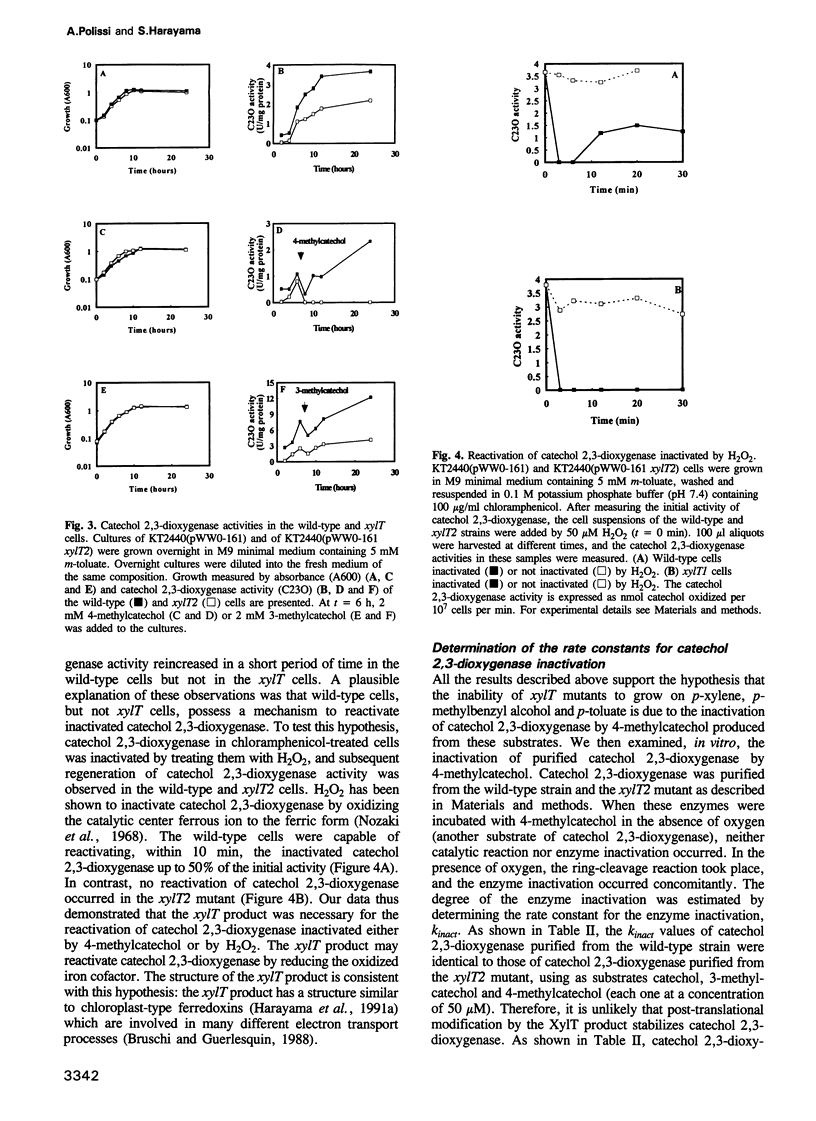
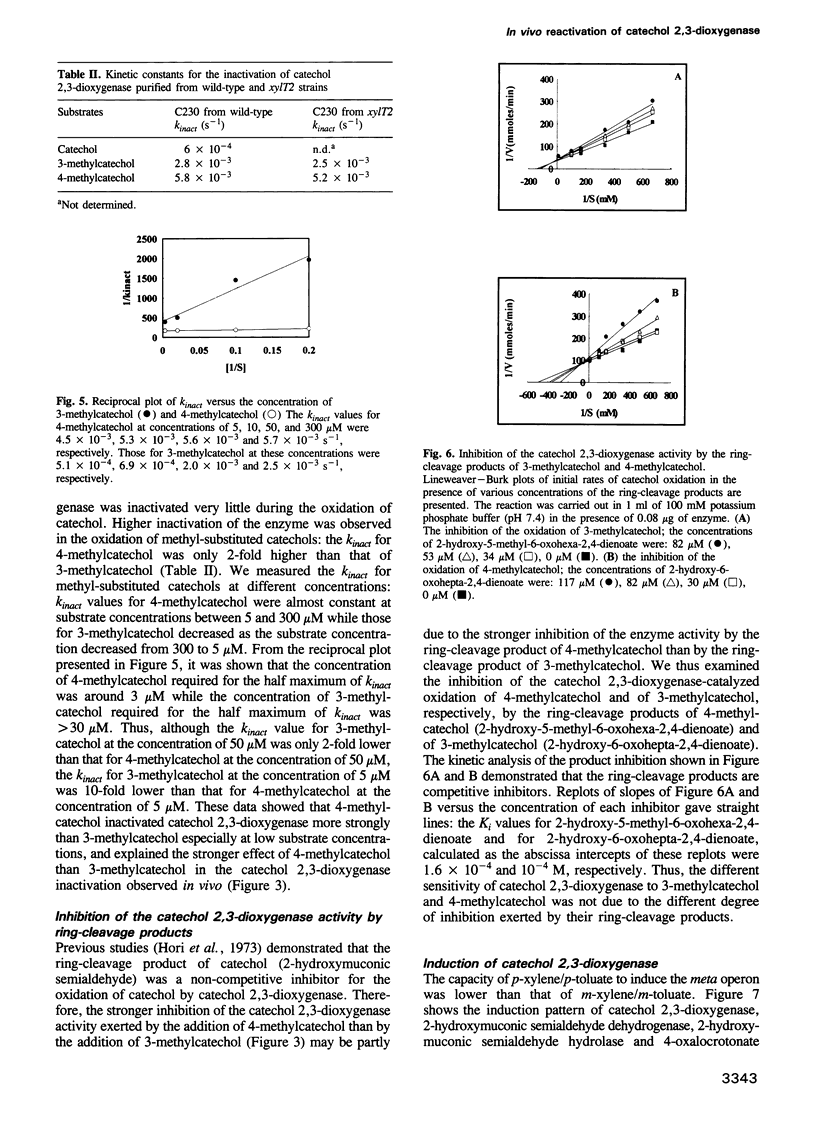
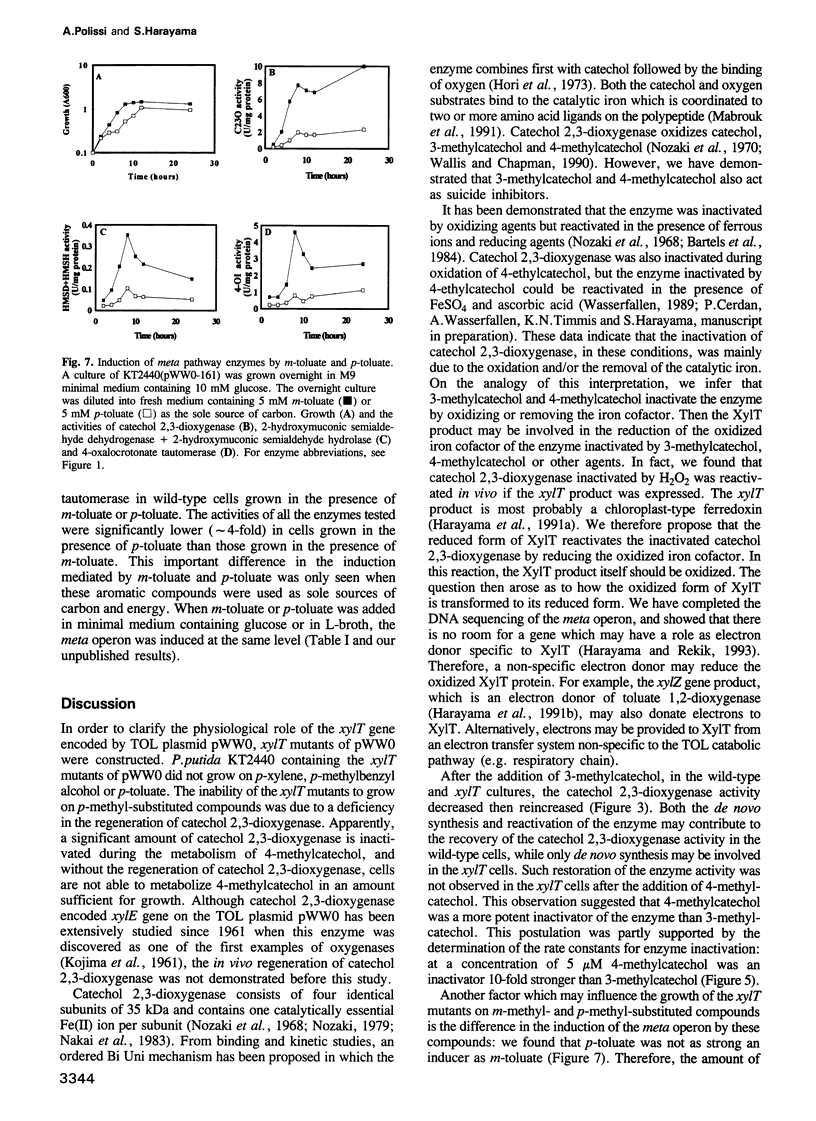
The meta-cleavage operon of the TOL plasmid pWW0 of Pseudomonas putida contains 13 genes responsible for the oxidation of benzoate and toluates to Krebs cycle intermediates via estradiol (meta) cleavage of (methyl)catechol. The functions of all the genes are known with the exception of xylT. We constructed pWW0 mutants defective in the xylT gene, and found that these mutants were not able to grow on p-toluate while they were still capable of growing on benzoate and m-toluate. In the xylT mutants, all the meta-cleavage enzymes were induced by p-toluate with the exception of catechol 2,3-dioxygenase whose activity was 1% of the p-toluate-induced activity in wild-type cells. Addition of 4-methylcatechol to m-toluate-grown wild-type and xylT cells resulted in the inactivation of catechol 2,3-dioxygenase in these cells. In the wild-type strain but not in the xylT mutant, the catechol 2,3-dioxygenase activity was regenerated in a short time. The regeneration of the catechol 2,3-dioxygenase activity was also observed in H2O2-treated wild-type cells, but not in H2O2-treated xylT cells. We concluded that the xylT product is required for the regeneration of catechol 2,3-dioxygenase.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartels I., Knackmuss H. J., Reineke W. Suicide Inactivation of Catechol 2,3-Dioxygenase from Pseudomonas putida mt-2 by 3-Halocatechols. Appl Environ Microbiol. 1984 Mar;47(3):500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartilson M., Nordlund I., Shingler V. Location and organization of the dimethylphenol catabolic genes of Pseudomonas CF600. Mol Gen Genet. 1990 Jan;220(2):294–300. doi: 10.1007/BF00260497. [DOI] [PubMed] [Google Scholar]
- Bruschi M., Guerlesquin F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol Rev. 1988 Apr-Jun;4(2):155–175. doi: 10.1111/j.1574-6968.1988.tb02741.x. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Lehrbach P. R., Lurz R., Rueckert B., Bagdasarian M., Timmis K. N. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J Bacteriol. 1983 May;154(2):676–685. doi: 10.1128/jb.154.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Kok M., Neidle E. L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- Harayama S., Lehrbach P. R., Timmis K. N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J Bacteriol. 1984 Oct;160(1):251–255. doi: 10.1128/jb.160.1.251-255.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Leppik R. A., Rekik M., Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Gene order of the TOL catabolic plasmid upper pathway operon and oxidation of both toluene and benzyl alcohol by the xylA product. J Bacteriol. 1986 Aug;167(2):455–461. doi: 10.1128/jb.167.2.455-461.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Mermod N., Rekik M., Lehrbach P. R., Timmis K. N. Roles of the divergent branches of the meta-cleavage pathway in the degradation of benzoate and substituted benzoates. J Bacteriol. 1987 Feb;169(2):558–564. doi: 10.1128/jb.169.2.558-564.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Polissi A., Rekik M. Divergent evolution of chloroplast-type ferredoxins. FEBS Lett. 1991 Jul 8;285(1):85–88. doi: 10.1016/0014-5793(91)80730-q. [DOI] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Bairoch A., Neidle E. L., Ornston L. N. Potential DNA slippage structures acquired during evolutionary divergence of Acinetobacter calcoaceticus chromosomal benABC and Pseudomonas putida TOL pWW0 plasmid xylXYZ, genes encoding benzoate dioxygenases. J Bacteriol. 1991 Dec;173(23):7540–7548. doi: 10.1128/jb.173.23.7540-7548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Rekik M. The meta cleavage operon of TOL degradative plasmid pWW0 comprises 13 genes. Mol Gen Genet. 1990 Mar;221(1):113–120. doi: 10.1007/BF00280375. [DOI] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Wubbolts M., Rose K., Leppik R. A., Timmis K. N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989 Sep;171(9):5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Hashimoto T., Nozaki M. Kinetic studies on the reaction mechanism of dioxygenases. J Biochem. 1973 Aug;74(2):375–384. [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Köhler T., Rekik M., Harayama S. Growth-phase-dependent expression of the Pseudomonas putida TOL plasmid pWW0 catabolic genes. J Bacteriol. 1990 Dec;172(12):6651–6660. doi: 10.1128/jb.172.12.6651-6660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOJIMA Y., ITADA N., HAYAISHI O. Metapyrocatachase: a new catechol-cleaving enzyme. J Biol Chem. 1961 Aug;236:2223–2228. [PubMed] [Google Scholar]
- Lehrbach P. R., Zeyer J., Reineke W., Knackmuss H. J., Timmis K. N. Enzyme recruitment in vitro: use of cloned genes to extend the range of haloaromatics degraded by Pseudomonas sp. strain B13. J Bacteriol. 1984 Jun;158(3):1025–1032. doi: 10.1128/jb.158.3.1025-1032.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai C., Kagamiyama H., Nozaki M., Nakazawa T., Inouye S., Ebina Y., Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983 Mar 10;258(5):2923–2928. [PubMed] [Google Scholar]
- Nakazawa T., Inouye S., Nakazawa A. Physical and functional mapping of RP4-TOL plasmid recombinants: analysis of insertion and deletion mutants. J Bacteriol. 1980 Oct;144(1):222–231. doi: 10.1128/jb.144.1.222-231.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund I., Powlowski J., Shingler V. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990 Dec;172(12):6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki M., Kotani S., Ono K., Seno S. Metapyrocatechase. 3. Substrate specificity and mode of ring fission. Biochim Biophys Acta. 1970 Nov 11;220(2):213–223. doi: 10.1016/0005-2744(70)90007-0. [DOI] [PubMed] [Google Scholar]
- Nozaki M., Ono K., Nakazawa T., Kotani S., Hayaishi O. Metapyrocatechase. II. The role of iron and sulfhydryl groups. J Biol Chem. 1968 May 25;243(10):2682–2690. [PubMed] [Google Scholar]
- Nozaki M. Oxygenases and dioxygenases. Top Curr Chem. 1979;78:145–186. doi: 10.1007/BFb0048193. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Shingler V., Powlowski J., Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992 Feb;174(3):711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M. B., Lynch G. P., Paroczay E., Norton S. Influence of rapeseed meal, whole rapeseed, and soybean meal on fatty acid composition and cholesterol content of muscle and adipose tissue from ram lambs. J Anim Sci. 1991 Oct;69(10):4055–4061. doi: 10.2527/1991.69104055x. [DOI] [PubMed] [Google Scholar]
- Wallis M. G., Chapman S. K. Isolation and partial characterization of an extradiol non-haem iron dioxygenase which preferentially cleaves 3-methylcatechol. Biochem J. 1990 Mar 1;266(2):605–609. [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You I. S., Ghosal D., Gunsalus I. C. Nucleotide sequence analysis of the Pseudomonas putida PpG7 salicylate hydroxylase gene (nahG) and its 3'-flanking region. Biochemistry. 1991 Feb 12;30(6):1635–1641. doi: 10.1021/bi00220a028. [DOI] [PubMed] [Google Scholar]


