Abstract
Sterile alpha motif- and histidine/aspartic acid domain-containing protein 1 (SAMHD1) limits HIV-1 replication by hydrolyzing deoxynucleoside triphosphates (dNTPs) necessary for reverse transcription. Nucleoside reverse transcriptase inhibitors (NRTIs) are components of anti-HIV therapies. We report here that SAMHD1 cleaves NRTI triphosphates (TPs) at significantly lower rates than dNTPs and that SAMHD1 depletion from monocytic cells affects the susceptibility of HIV-1 infections to NRTIs in complex ways that depend not only on the relative changes in dNTP and NRTI-TP concentrations but also on the NRTI activation pathways.
TEXT
Human immunodeficiency virus type 1 (HIV-1) replicates primarily in activated CD4+ T cells, while showing poor reproductive capacity in monocytes, macrophages, dendritic cells, and resting CD4+ T cells (1–10). Sterile alpha motif- and histidine/aspartic acid domain-containing protein 1 (SAMHD1) is responsible for blocking HIV-1 replication in such cells (5, 11–13), reportedly by acting as a dGTP-stimulated deoxynucleotide triphosphohydrolase that hydrolyzes deoxynucleoside triphosphates (dNTPs), thus decreasing the amounts of dNTPs available for reverse transcription (3, 4, 14–19).
Nucleoside reverse transcriptase inhibitors (NRTIs) are nucleoside analogs and key components of antiretroviral therapies (20–26). They generally lack a 3′-OH group and thus act as chain terminators upon incorporation into viral DNA by reverse transcriptase (RT) (26–29). However, 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) retains a 3′-OH group, acts primarily by blocking RT translocation following incorporation of EFdA monophosphate (MP) into the template-primer, and has picomolar antiviral potency (30–37). NRTIs are administered as nucleosides and are phosphorylated to their active forms by cellular kinases (38). Hence, they compete with dNTPs for activation by cellular kinases, and their incorporation by RT is influenced by the cellular concentrations of dNTPs, which compete with NRTI triphosphates (TPs) at the RT active site (39, 40).
Amie et al. (19) recently reported that SAMHD1 does not significantly hydrolyze dideoxynucleoside triphosphates (ddNTPs) or zidovudine (AZT)-TP and that depletion of SAMHD1 in monocytic THP-1 cells decreased the potency of these NRTIs in a pseudotype-based assay. Strong evidence that the decreased potency of these NRTIs was due to increased amounts of competing dNTPs was presented. Our parallel independent study confirmed their data, extended the number of NRTIs studied, validated the results with fully infectious HIV-1, and found an unexpected disparity in the effects of SAMHD1 on the deoxyribosylthymine (dT) analogs AZT and stavudine (d4T). We demonstrate that this is due to differences in the activation of AZT and d4T, highlighting the importance of distinct metabolic pathways in NRTI activation, in addition to competition with dNTPs.
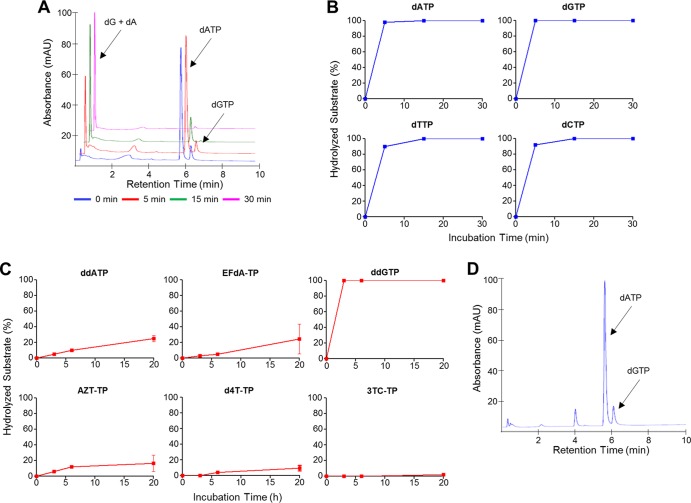
We tested purified Escherichia coli-produced recombinant SAMHD1 for dGTP-regulated NRTI-TP hydrolysis (using dNTPs as a reference) and separated the reaction products by anion-exchange high-performance liquid chromatography (HPLC) (14). A representative chromatogram for dATP hydrolysis by SAMHD1 is shown in Fig. 1A. Notably, NRTI-TP hydrolysis was significantly slower than that of dNTPs, with little hydrolysis after hours of incubation, rather than the minutes required for complete dNTP hydrolysis (Fig. 1B and C). The observed activity was not due to a contaminating phosphatase, as hydrolase activity was ablated by mutating the SAMHD1 active-site residue Asp207 to alanine (Fig. 1D) (14, 18, 41).
FIG 1.
SAMHD1 does not efficiently hydrolyze NRTI-TPs. SAMHD1 (5 μM) was incubated at 37°C with NRTI-TP or dNTP (500 μM), in the presence of dGTP (100 μM) and MgCl2 (10 mM). Reactions proceeded for 3, 6, or 20 h for NRTI-TPs and for 5, 15, or 30 min for dNTPs. Reactions were terminated by 10-fold dilution into 25 mM Tris (pH 8.0)-12.5% acetonitrile, and mixtures were analyzed by anion-exchange HPLC (DNAPac PA100 column). (A) Representative chromatograms for dATP hydrolysis. dG, deoxyribosylguanine. (B and C) Data from at least duplicate experiments for dNTP (B) or NRTI-TP (C) hydrolysis, plotted as percent hydrolysis over time (with GraphPad Prism 5). ddATP, dideoxyadenosine triphosphate; ddGTP, dideoxyguanosine triphosphate. (D) Chromatogram for dATP hydrolysis after 30 min of incubation with the SAMHD1 hydrolase active-site D207A mutant, dGTP, and MgCl2.
Next, we assessed whether SAMHD1 affected NRTI potency in the context of HIV-1 infection. We infected parental THP-1 cells and THP-1 cells stably expressing a SAMHD1-targeting short hairpin RNA (shRNA) (THP-1KD-SAMHD1 cells) (42) with infectious HIV-1 carrying a luciferase reporter, in the presence or absence of the previously tested AZT and tenofovir disoproxil fumarate (TDF) (19), as well as d4T, lamivudine (3TC), dideoxyinosine (ddI), and EFdA. The knockdown efficiency of SAMHD1 in the THP-1KD-SAMHD1 cells was >50-fold (see Fig. S1 in the supplemental material) (42). Table 1 lists NRTI 50% effective concentrations (EC50s) determined in four independent experiments. As reported previously, the EC50s of AZT and TDF (dT and deoxyribosyladenine [dA] analogs, respectively) were significantly increased (23- and 18-fold, respectively) in THP-1KD-SAMHD1 cells. Notably, we observed smaller or no increases for other dT, dA, and deoxyribosylcytosine [dC] analogs (∼3.5-fold increase for d4T, 3-fold increase for EFdA, and no increases for 3TC and ddI) (Table 1). The unexpectedly decreased potency of AZT versus d4T in THP-1KD-SAMHD1 cells was not due to a higher rate of hydrolysis of d4T-TP by SAMHD1 (Fig. 1C) (19). It also was not caused by differences in dTTP competition with AZT-TP and d4T-TP at the RT active site, as RT incorporates AZT-TP and d4T-TP with similar efficiencies (43). Thus, we hypothesized that the different effects of SAMHD1 on the potencies of AZT and d4T involved differences in the activation pathways of the two inhibitors.
TABLE 1.
EC50 values for NRTIs in parental THP-1 cells versus THP-1KD-SAMHD1 cells
| Drug | EC50 (nM) fora: |
Fold increase in EC50 | |
|---|---|---|---|
| Parental THP-1 cells | THP-1KD-SAMHD1 cells | ||
| AZT | 1.1 ± 0.5 | 25.3 ± 14.9 | 23 |
| TDF | 0.08 ± 0.01 | 1.44 ± 0.04 | 18 |
| d4T | 47 ± 10 | 166 ± 13 | 3.5 |
| ddI | 44 ± 8 | 42 ± 7 | 1 |
| 3TC | 11 ± 2 | 11 ± 4 | 1 |
| EFdA | 0.04 ± 0.02 | 0.13 ± 0.02 | 3.3 |
Values are the mean ± standard deviation (SD) from four independent experiments and were determined using the one-site competition equation in GraphPad Prism 5.
To study the activation of AZT and d4T, we treated phorbol myristate acetate (PMA)-differentiated THP-1 and THP-1KD-SAMHD1 cells with radiolabeled [14C]AZT or [14C]d4T, incubated the cells for 24 h at 37°C, lysed the cells, separated the NRTI metabolites by HPLC, and analyzed the metabolites with a liquid scintillation counter. We found an ∼10-fold decrease in the amount of AZT-TP recovered from THP-1KD-SAMHD1 cells, in comparison with THP-1 cells, but no significant difference in the amounts of d4T-TP recovered from the two cell lines (Fig. 2). These results are consistent with the observation that AZT experiences a larger EC50 increase than does d4T upon SAMHD1 knockdown, suggesting that AZT does not compete as well as d4T for phosphorylation by cellular kinases when there are increased levels of dNTPs and therefore it experiences a larger change in EC50.
FIG 2.
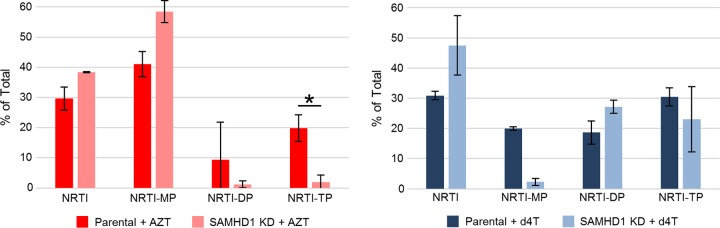
Knockdown of SAMHD1 has differential effects on NRTI activation. PMA-differentiated parental and THP-1KD-SAMHD1 cells were treated with 1.5 μM (0.5 μCi) [2-14C]AZT or [4-14C]d4T. After 24 h at 37°C, the cells were lysed, the dNTP and NRTI metabolites were separated by anion-exchange HPLC (DNAPac PA100 column), and the NRTI metabolites in the collected fractions were quantified with a liquid scintillation counter. Data represent the mean ± standard deviation (SD) from 2 independent experiments. *, P < 0.05.
We directly explored the impact of increased cellular dTTP levels on the inhibitory potential of AZT and d4T by exogenously adding thymidine. We treated TZM-bl cells with phosphate-buffered saline (PBS) or 100 μM dT or dC (as a control as a noncompeting nucleoside) and infected the cells with HIV-1NL4-3 (multiplicity of infection [MOI], 0.02) in the presence of increasing inhibitor concentrations. At 48 h postinfection, cells were lysed and luciferase activity was measured. As expected, exogenous dT increased the EC50s for HIV-1NL4-3 inhibition by AZT and d4T. Whereas the EC50 for AZT increased >100-fold upon addition of exogenous dT, the EC50 for d4T appeared to increase significantly less, although the exact EC50s could not be estimated because we could not reach extremely high NRTI concentrations (Fig. 3). These data agree with our observation that SAMHD1 knockdown has a greater effect on AZT than on d4T. Notably, addition of 100 μM dA did not affect the ddI EC50 (Fig. 3), consistent with reported differences in the ddI and dA activation mechanisms (39, 44–48) and also with the lack of differences in EC50 values for ddI in THP-1 versus THP-1KD-SAMHD1 cells (Table 1). While addition of 100 μM dC blocked HIV inhibition by 3TC (Fig. 3), the unchanged 3TC EC50 values in THP-1 and THP-1KD-SAMHD1 cells may be partly attributed to the findings that SAMHD1 depletion had the smallest effect on the concentration of dCTP, compared to other dNTPs (19), and that 3TC-TP was a poorer substrate for SAMHD1 (Fig. 1).
FIG 3.
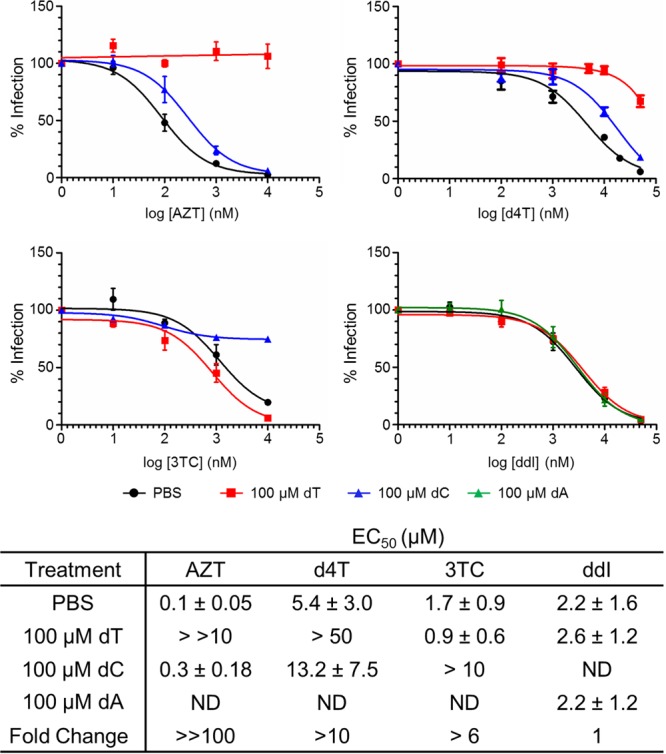
Exogenously added dT, but not dC, affects AZT and d4T potencies. TZM-bl cells were treated with PBS, 100 μM dT, or 100 μM dC or 100 µM dA (as controls for noncompeting nucleosides) and infected with HIV-1NL4-3 at an MOI of 0.02, in the presence of increasing concentrations of inhibitor (AZT or d4T). At 48 h postinfection, cells were lysed and luciferase activity was detected. Luciferase activity at various drug concentrations was plotted using the one-site competition equation in GraphPad Prism 5, and data were normalized to the no-nucleoside control results. Data represent the mean ± SD from at least three independent experiments. Data in the table represent the mean ± SD from at least three independent experiments. Shown also are the fold changes in the EC50 of NRTI in the presence or absence of cognate nucleoside, which indicate change in sensitivity to AZT/d4T, 3TC, or ddI, in the presence of dT, dC, or dA, respectively. ND, not determined.
We have demonstrated that SAMHD1 downregulation affects not only dNTP concentrations (3, 4, 14–19) but also the concentrations of AZT and d4T metabolites (Fig. 2). Our data are consistent with previous reports noting that the rate-limiting step in activation is the second phosphorylation step, catalyzed by thymidylate kinase, for AZT but the first phosphorylation step, catalyzed by thymidine kinase, for d4T (39, 49–54), as shown by the accumulation of AZT-MP and d4T in treated THP-1 and THP-1KD-SAMHD1 cells (Fig. 3). d4T diphosphate (DP) is more readily phosphorylated to the triphosphate form by nucleoside diphosphate kinase, the final kinase in the activation pathway for d4T and AZT, as well as other analogs and deoxynucleoside diphosphates (dNDPs) (39, 55–58), than is AZT-DP, addressing why the potency of AZT is affected more than that of d4T with SAMHD1 depletion and increased dNTP concentrations.
In conclusion, the presence of SAMHD1, or its depletion, as occurs for lentiviruses that encode the Vpx accessory protein (12, 59–63), can affect NRTI susceptibility in multiple ways that depend not only on the relative changes in the concentrations of dNTPs and NRTI-TPs but also on the activation pathways of NRTIs. Our study highlights the importance of the metabolic pathways for activation of different NRTIs to NRTI-TPs, especially in cells in which dNTP concentrations are low and competition with NRTI-TPs does not mask the effects of differential NRTI activation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH (grants AI076119, AI100890, AI099284, AI112417, and GM103368 to S.G.S., grant AI079801 to Michael A. Parniak, and grant AI058864 to N.R.L.), Mizzou Advantage, and the Ministry of Knowledge and Economy, Bilateral International Collaborative R&D Program, Republic of Korea.
Footnotes
Published ahead of print 27 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02745-14.
REFERENCES
- 1.Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. 2004. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279:51545–51553. 10.1074/jbc.M408573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy EM, Gavegnano C, Nguyen L, Slater R, Lucas A, Fromentin E, Schinazi RF, Kim B. 2010. Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285:39380–39391. 10.1074/jbc.M110.178582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13:223–228. 10.1038/ni.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. 2012. SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18:1682–1687. 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. 2012. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9:87. 10.1186/1742-4690-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St. Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. 2012. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9:105. 10.1186/1742-4690-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123–126. 10.1038/373123a0 [DOI] [PubMed] [Google Scholar]
- 8.Coleman CM, Wu L. 2009. HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology 6:51. 10.1186/1742-4690-6-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR. 2010. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467:214–217. 10.1038/nature09337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergamaschi A, Pancino G. 2010. Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology 7:31. 10.1186/1742-4690-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E. 2011. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7:e1002425. 10.1371/journal.ppat.1002425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480:379–382. 10.1038/nature10623 [DOI] [PubMed] [Google Scholar]
- 15.Powell RD, Holland PJ, Hollis T, Perrino FW. 2011. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286:43596–43600. 10.1074/jbc.C111.317628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F. 2013. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436:81–90. 10.1016/j.virol.2012.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. 2012. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287:21570–21574. 10.1074/jbc.C112.374843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. 2013. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 288:8101–8110. 10.1074/jbc.M112.431148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amie SM, Daly MB, Noble E, Schinazi RF, Bambara RA, Kim B. 2013. Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. J. Biol. Chem. 288:20683–20691. 10.1074/jbc.M113.472159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer SM, Saag MS, Schechter M, Montaner JS, Schooley RT, Jacobsen DM, Thompson MA, Carpenter CC, Fischl MA, Gazzard BG, Gatell JM, Hirsch MS, Katzenstein DA, Richman DD, Vella S, Yeni PG, Volberding PA, International AIDS Society-USA Panel 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society-USA panel. JAMA 296:827–843. 10.1001/jama.296.7.827 [DOI] [PubMed] [Google Scholar]
- 21.Schinazi RF, Hernandez-Santiago BI, Hurwitz SJ. 2006. Pharmacology of current and promising nucleosides for the treatment of human immunodeficiency viruses. Antiviral Res. 71:322–334. 10.1016/j.antiviral.2006.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parniak MA, Sluis-Cremer N. 2000. Inhibitors of HIV-1 reverse transcriptase. Adv. Pharmacol. 49:67–109. 10.1016/S1054-3589(00)49024-1 [DOI] [PubMed] [Google Scholar]
- 23.De Clercq E. 2007. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Verh. K. Acad. Geneeskd. Belg. 69:81–104. 10.1016/j.ijantimicag.2008.10.010 [DOI] [PubMed] [Google Scholar]
- 24.Deval J. 2009. Antimicrobial strategies: inhibition of viral polymerases by 3′-hydroxyl nucleosides. Drugs 69:151–166. 10.2165/00003495-200969020-00002 [DOI] [PubMed] [Google Scholar]
- 25.Ilina T, Parniak MA. 2008. Inhibitors of HIV-1 reverse transcriptase. Adv. Pharmacol. 56:121–167. 10.1016/S1054-3589(07)56005-9 [DOI] [PubMed] [Google Scholar]
- 26.Singh K, Marchand B, Kirby KA, Michailidis E, Sarafianos SG. 2010. Structural aspects of drug resistance and inhibition of HIV-1 reverse transcriptase. Viruses 2:606–638. 10.3390/v2020606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693–713. 10.1016/j.jmb.2008.10.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menendez-Arias L. 2002. Targeting HIV: antiretroviral therapy and development of drug resistance. Trends Pharmacol. Sci. 23:381–388. 10.1016/S0165-6147(02)02054-0 [DOI] [PubMed] [Google Scholar]
- 29.Menendez-Arias L. 2008. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 134:124–146. 10.1016/j.virusres.2007.12.015 [DOI] [PubMed] [Google Scholar]
- 30.Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, Mitsuya H, Arnold E, Matsuoka M. 2008. 2′-Deoxy-4′-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int. J. Biochem. Cell Biol. 40:2410–2420. 10.1016/j.biocel.2008.04.007 [DOI] [PubMed] [Google Scholar]
- 31.Michailidis E, Marchand B, Kodama EN, Singh K, Matsuoka M, Kirby KA, Ryan EM, Sawani AM, Nagy E, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. 2009. Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-ethynyl-2-fluoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. J. Biol. Chem. 284:35681–35691. 10.1074/jbc.M109.036616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michailidis E, Ryan EM, Hachiya A, Kirby KA, Marchand B, Leslie MD, Huber AD, Ong YT, Jackson JC, Singh K, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Hypersusceptibility mechanism of tenofovir-resistant HIV to EFdA. Retrovirology 10:65. 10.1186/1742-4690-10-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hachiya A, Reeve AB, Marchand B, Michailidis E, Ong YT, Kirby KA, Leslie MD, Oka S, Kodama EN, Rohan LC, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Evaluation of combinations of 4′-ethynyl-2-fluoro-2′-deoxyadenosine with clinically used antiretroviral drugs. Antimicrob. Agents Chemother. 57:4554–4558. 10.1128/AAC.00283-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michailidis E, Singh K, Ryan EM, Hachiya A, Ong YT, Kirby KA, Marchand B, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2012. Effect of translocation defective reverse transcriptase inhibitors on the activity of N348I, a connection subdomain drug resistant HIV-1 reverse transcriptase mutant. Cell. Mol. Biol. (Noisy-le-grand) 58:187–195 [PMC free article] [PubMed] [Google Scholar]
- 35.Kirby KA, Singh K, Michailidis E, Marchand B, Kodama EN, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. 2011. The sugar ring conformation of 4′-ethynyl-2-fluoro-2′-deoxyadenosine and its recognition by the polymerase active site of HIV reverse transcriptase. Cell. Mol. Biol. (Noisy-le-grand) 57:40–46 [PMC free article] [PubMed] [Google Scholar]
- 36.Murphey-Corb M, Rajakumar P, Michael H, Nyaundi J, Didier PJ, Reeve AB, Mitsuya H, Sarafianos SG, Parniak MA. 2012. Response of simian immunodeficiency virus to the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine in vitro and in vivo. Antimicrob. Agents Chemother. 56:4707–4712. 10.1128/AAC.00723-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirby KA, Michailidis E, Fetterly TL, Steinbach MA, Singh K, Marchand B, Leslie MD, Hagedorn AN, Kodama EN, Marquez VE, Hughes SH, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Effects of substitutions at the 4′ and 2 positions on the bioactivity of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob. Agents Chemother. 57:6254–6264. 10.1128/AAC.01703-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perno CF, Yarchoan R, Cooney DA, Hartman NR, Gartner S, Popovic M, Hao Z, Gerrard TL, Wilson YA, Johns DG, Broder S. 1988. Inhibition of human immunodeficiency virus (HIV-1/HTLV-IIIBa-L) replication in fresh and cultured human peripheral blood monocytes/macrophages by azidothymidine and related 2′,3′-dideoxynucleosides. J. Exp. Med. 168:1111–1125. 10.1084/jem.168.3.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein DS, Moore KH. 2001. Phosphorylation of nucleoside analog antiretrovirals: a review for clinicians. Pharmacotherapy 21:11–34. 10.1592/phco.21.1.11.34439 [DOI] [PubMed] [Google Scholar]
- 40.Ray AS. 2005. Intracellular interactions between nucleos(t)ide inhibitors of HIV reverse transcriptase. AIDS Rev. 7:113–125 [PubMed] [Google Scholar]
- 41.Welbourn S, Miyagi E, White TE, Diaz-Griffero F, Strebel K. 2012. Identification and characterization of naturally occurring splice variants of SAMHD1. Retrovirology 9:86. 10.1186/1742-4690-9-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gramberg T, Kahle T, Bloch N, Wittmann S, Mullers E, Daddacha W, Hofmann H, Kim B, Lindemann D, Landau NR. 2013. Restriction of diverse retroviruses by SAMHD1. Retrovirology 10:26. 10.1186/1742-4690-10-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer P, Schneider B, Sarfati S, Deville-Bonne D, Guerreiro C, Boretto J, Janin J, Veron M, Canard B. 2000. Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase. EMBO J. 19:3520–3529. 10.1093/emboj/19.14.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson PL, Kakuda TN, Lichtenstein KA. 2004. The cellular pharmacology of nucleoside- and nucleotide-analogue reverse-transcriptase inhibitors and its relationship to clinical toxicities. Clin. Infect. Dis. 38:743–753. 10.1086/381678 [DOI] [PubMed] [Google Scholar]
- 45.Samuels DC. 2006. Mitochondrial AZT metabolism. IUBMB Life 58:403–408. 10.1080/15216540600791571 [DOI] [PubMed] [Google Scholar]
- 46.Johnson MA, Fridland A. 1989. Phosphorylation of 2′,3′-dideoxyinosine by cytosolic 5′-nucleotidase of human lymphoid cells. Mol. Pharmacol. 36:291–295 [PubMed] [Google Scholar]
- 47.Bradshaw PC, Samuels DC. 2005. A computational model of mitochondrial deoxynucleotide metabolism and DNA replication. Am. J. Physiol. Cell Physiol. 288:C989–C1002. 10.1152/ajpcell.00530.2004 [DOI] [PubMed] [Google Scholar]
- 48.Eriksson S, Munch-Petersen B, Johansson K, Eklund H. 2002. Structure and function of cellular deoxyribonucleoside kinases. Cell. Mol. Life Sci. 59:1327–1346. 10.1007/s00018-002-8511-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tornevik Y, Ullman B, Balzarini J, Wahren B, Eriksson S. 1995. Cytotoxicity of 3′-azido-3′-deoxythymidine correlates with 3′-azidothymidine-5′-monophosphate (AZTMP) levels, whereas anti-human immunodeficiency virus (HIV) activity correlates with 3′-azidothymidine-5′-triphosphate (AZTTP) levels in cultured CEM T-lymphoblastoid cells. Biochem. Pharmacol. 49:829–837. 10.1016/0006-2952(94)00453-S [DOI] [PubMed] [Google Scholar]
- 50.Qian M, Bui T, Ho RJ, Unadkat JD. 1994. Metabolism of 3′-azido-3′-deoxythymidine (AZT) in human placental trophoblasts and Hofbauer cells. Biochem. Pharmacol. 48:383–389. 10.1016/0091-3057(94)90542-8, [DOI] [PubMed] [Google Scholar]
- 51.Lavie A, Schlichting I, Vetter IR, Konrad M, Reinstein J, Goody RS. 1997. The bottleneck in AZT activation. Nat. Med. 3:922–924. 10.1038/nm0897-922 [DOI] [PubMed] [Google Scholar]
- 52.Gao WY, Agbaria R, Driscoll JS, Mitsuya H. 1994. Divergent anti-human immunodeficiency virus activity and anabolic phosphorylation of 2′,3′-dideoxynucleoside analogs in resting and activated human cells. J. Biol. Chem. 269:12633–12638 [PubMed] [Google Scholar]
- 53.Furman PA, Fyfe JA, St. Clair MH, Weinhold K, Rideout JL, Freeman GA, Lehrman SN, Bolognesi DP, Broder S, Mitsuya H, Barry DW. 1986. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 83:8333–8337. 10.1073/pnas.83.21.8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balzarini J, Herdewijn P, De Clercq E. 1989. Differential patterns of intracellular metabolism of 2′,3′-didehydro-2′,3′-dideoxythymidine and 3′-azido-2′,3′-dideoxythymidine, two potent anti-human immunodeficiency virus compounds. J. Biol. Chem. 264:6127–6133 [PubMed] [Google Scholar]
- 55.Hernandez-Santiago BI, Mathew JS, Rapp KL, Grier JP, Schinazi RF. 2007. Antiviral and cellular metabolism interactions between dexelvucitabine and lamivudine. Antimicrob. Agents Chemother. 51:2130–2135. 10.1128/AAC.01543-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider B, Xu YW, Sellam O, Sarfati R, Janin J, Veron M, Deville-Bonne D. 1998. Pre-steady state of reaction of nucleoside diphosphate kinase with anti-HIV nucleotides. J. Biol. Chem. 273:11491–11497. 10.1074/jbc.273.19.11491 [DOI] [PubMed] [Google Scholar]
- 57.Bourdais J, Biondi R, Sarfati S, Guerreiro C, Lascu I, Janin J, Veron M. 1996. Cellular phosphorylation of anti-HIV nucleosides: role of nucleoside diphosphate kinase. J. Biol. Chem. 271:7887–7890 [DOI] [PubMed] [Google Scholar]
- 58.Schneider B, Biondi R, Sarfati R, Agou F, Guerreiro C, Deville-Bonne D, Veron M. 2000. The mechanism of phosphorylation of anti-HIV D4T by nucleoside diphosphate kinase. Mol. Pharmacol. 57:948–953 [PubMed] [Google Scholar]
- 59.Ayinde D, Maudet C, Transy C, Margottin-Goguet F. 2010. Limelight on two HIV/SIV accessory proteins in macrophage infection: is Vpx overshadowing Vpr? Retrovirology 7:35. 10.1186/1742-4690-7-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berger G, Durand S, Goujon C, Nguyen XN, Cordeil S, Darlix JL, Cimarelli A. 2011. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat. Protoc. 6:806–816. 10.1038/nprot.2011.327 [DOI] [PubMed] [Google Scholar]
- 61.Goujon C, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A. 2006. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIVMAC. Gene Ther. 13:991–994. 10.1038/sj.gt.3302753 [DOI] [PubMed] [Google Scholar]
- 62.Goujon C, Riviere L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A. 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4:2. 10.1186/1742-4690-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Munch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, Delsuc F, Telenti A, Benkirane M. 2012. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11:205–217. 10.1016/j.chom.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.