Abstract
Comparison of the transcriptome of Streptococcus pneumoniae strain D39 grown in the presence of either lactose or galactose with that of the strain grown in the presence of glucose revealed the elevated expression of various genes and operons, including the lac gene cluster, which is organized into two operons, i.e., lac operon I (lacABCD) and lac operon II (lacTFEG). Deletion of the DeoR family transcriptional regulator lacR that is present downstream of the lac gene cluster revealed elevated expression of lac operon I even in the absence of lactose. This suggests a function of LacR as a transcriptional repressor of lac operon I, which encodes enzymes involved in the phosphorylated tagatose pathway in the absence of lactose or galactose. Deletion of lacR did not affect the expression of lac operon II, which encodes a lactose-specific phosphotransferase. This finding was further confirmed by β-galactosidase assays with PlacA-lacZ and PlacT-lacZ in the presence of either lactose or glucose as the sole carbon source in the medium. This suggests the involvement of another transcriptional regulator in the regulation of lac operon II, which is the BglG-family transcriptional antiterminator LacT. We demonstrate the role of LacT as a transcriptional activator of lac operon II in the presence of lactose and CcpA-independent regulation of the lac gene cluster in S. pneumoniae.
INTRODUCTION
Carbohydrate metabolism and utilization and their proper regulation play a key role in the survival of prokaryotes, since carbohydrates are the most common sources of energy required to produce essential nucleotides, cofactors, and other metabolites indispensable for growth (1, 2). When encountering multiple sugars and energy sources simultaneously, a cell goes through metabolic assessment and usually prefers a particular energy source, such as glucose, to another (1, 2). Central carbon metabolism in most bacterial species, including the model free-living Gram-positive bacterium Bacillus subtilis, is controlled by a mechanism called carbon catabolite repression (CCR) (1–4). CCR enables a bacterium to select a preferred sugar over a nonpreferred one, aiding the organism with the maintenance of a proper energy balance (5). CCR is mediated by the transcriptional factor CcpA (carbon catabolite protein A) in the presence of a preferred source of energy, such as glucose (2, 5–10). CcpA mediates the repression of genes involved in the utilization of nonpreferred sugars in the presence of the preferred sugar by binding to catabolite repression elements (cre boxes) found in the promoter regions of these genes (11, 12). The strength of binding of CcpA to cre sequences, present in the promoter regions of CcpA targets, is boosted by the histidine phosphoprotein (HPr-Ser-46P) (13). HPr is a central element of the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS), where it usually helps with the transfer of high-energy phosphate from phosphoenolpyruvate to the enzyme II complex during sugar uptake (13, 14).
Low-GC-content bacteria are also able to utilize nonpreferred sugars, like a β-linked disaccharide of β-d-galactose, α/β-d-glucose, or lactose, which are normally found in dairy-rich diets. Galactose is slowly metabolized by bacteria, and in some cases it helps with colonization (9, 10). There are a number of pathways in bacteria that have been shown to be involved in the utilization of lactose found in the environment (15). For instance, Streptococcus salivarius strain 25975 secretes a β-galactosidase enzyme that hydrolyzes extracellular lactose into galactose and glucose, although lactose is normally transported inside the bacterial cell and then gets phosphorylated (lactose-6-phosphate [Lac-6-P]) before being cleaved (16). Lactose and galactose are commonly utilized through the tagatose pathway in streptococci (17, 18). Galactose can also be catabolized by the Leloir pathway (11, 17), which usually involves a multiple-sugar-metabolism (msm) system for galactose transport (12). However, the permease responsible for galactose transport has yet to be identified in Streptococcus pneumoniae (13). The regulatory mechanism of the Leloir pathway has already been studied in Streptococcus mutans (17), Streptococcus gordonii (18), Streptococcus thermophilus (19), and other bacteria, where the transcriptional repressor GalR has been shown to repress the expression of genes involved in the Leloir pathway.
S. pneumoniae is a low-GC-content Gram-positive human pathogen that has the ability to utilize different sources of carbohydrates (3, 20–26), including lactose and galactose. Some strains of Neisseria that are able to utilize lactose have been found in the human nasopharynx (8), suggesting the presence of lactose moieties in the nasopharynx. Unlike various other bacteria, S. pneumoniae possesses a lac gene cluster that is organized into two operons: lac operon I and lac operon II. lac operon I consists of phosphorylated tagatose (tagatose-6-phosphate) pathway genes (lacABCD), and lac operon II consists of a lactose-specific PTS, a β-galactosidase, and a transcriptional antiterminator, lacT. LacT is a member of the BglG/SacY family of proteins (19) and has a coantiterminator (CoAT) RNA-binding domain (20) at its amino terminus. CoAT domains help with binding to ribonucleic antiterminator (RAT) sequences in mRNA transcripts, allowing RNA polymerase to carry out the transcription of downstream genes by preventing the formation of a terminator (27). They are mostly involved in the transcriptional regulation of β-glucoside-specific genes in S. gordonii (18), Escherichia coli (28), Erwinia chrysanthemi (29), Lactococcus lactis (30), Lactobacillus plantarum (31), and B. subtilis (32). Regulation of the lactose utilization operon is under the control of the DeoR family transcriptional repressor LacR in S. mutans (18, 33) and S. gordonii (18). Similarly, LacR regulates lactose and galactose utilization in Lactobacillus helveticus (14) and Streptococcus pyogenes (34). In most studies, they appear to be transcriptional repressors of sugar metabolism. For instance, in B. subtilis, DeoR acts as a transcriptional repressor of the dra-nupC-pdp operon and plays a role in the utilization of deoxyribonucleosides and deoxyribose (35, 36). Similarly, glycerol-3-phosphate (GlpR), l-fucose (FucR), l-ascorbate (UlaR), and deoxyribonucleoside (DeoR) systems are the other examples where DeoR's role has been established to be a transcriptional repressor (37–40). Commonly, phosphorylated intermediates of the pertinent metabolic pathways are the effector molecules for DeoR-type regulators (e.g., besides deoxyribose-5-phosphate, these include fructose-1-phosphate for FruR of Lactococcus lactis [41]). Nevertheless, examples where nonphosphorylated inducers have been shown to play a role are present, e.g., opine for AccR from Agrobacterium tumefaciens (42), fucose for FucR from Bacteroides thetaiotaomicron (43), and likely, N-acetylglucosamine or galactosamine for AgaR from E. coli (44).
In the current work, we studied the effects of lactose and galactose on global gene expression in S. pneumoniae and characterized the lactose and galactose utilization gene cluster (the lac gene cluster, consisting of lac operons I and II) in S. pneumoniae. Furthermore, we demonstrate that the transcriptional regulator LacR acts as a transcriptional repressor of the tagatose-6-phosphate pathway genes (lac operon I) and LacT acts as a transcriptional activator for genes (lac operon II) encoding the lactose-transporting PTS and a 6-phospho-β-galactosidase. We also demonstrate the CcpA-independent regulation of the lac gene cluster in the presence of lactose, galactose, and glucose.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. M17 broth supplemented with 0.5% (wt/vol) glucose was used for growing S. pneumoniae D39 (45) on blood agar plates supplemented with 1% (vol/vol) defibrinated sheep blood under microaerophilic conditions at 37°C. For β-galactosidase assays, derivatives of the S. pneumoniae D39 strain were grown in M17 medium supplemented with different sugars (arabinose, cellobiose, dextrose, fructose, fucose, glucose, galactose, lactose, maltose, mannitol, mannose, melibiose, sorbitol, trehalose, and xylose) at the concentrations (wt/vol) mentioned in Results. For selection on antibiotics, the medium was supplemented with the following antibiotics at the indicated concentrations: spectinomycin at 150 μg/ml and tetracycline at 2.5 μg/ml for S. pneumoniae and ampicillin at 100 μg/ml for E. coli. All bacterial strains used in this study were stored in 10% (vol/vol) glycerol at −80°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. pneumoniae | ||
| D39 | Serotype 2 strain, cps2 | Laboratory of P. Hermans |
| ΔccpA mutant | D39 ΔccpA; Specr | 3 |
| MA100 | D39 ΔlacR; Specr | This study |
| MA101 | D39 lacT null mutant | This study |
| MA102 | D39 ΔbgaA::PlacA-lacZ; Tetr | This study |
| MA103 | MA100 ΔbgaA::PlacA-lacZ; Tetr | This study |
| MA104 | MA101 ΔbgaA::PlacA-lacZ; Tetr | This study |
| MA105 | D39 ΔbgaA::PlacT-lacZ; Tetr | This study |
| MA106 | MA100 ΔbgaA::PlacT-lacZ; Tetr | This study |
| MA107 | MA101 ΔbgaA::PlacT-lacZ; Tetr | This study |
| MA108 | D39 ΔbgaA::PgalK-lacZ; Tetr | This study |
| MA109 | D39 ΔccpA::PlacT-lacZ; Tetr | This study |
| E. coli EC1000 | Kmr; MC1000 derivative carrying a single copy of the pWV1 repA gene in glgB | Laboratory collection |
| Plasmids | ||
| pPP2 | Ampr Tetr; promoterless lacZ; used for replacement of bgaA with the promoter-lacZ fusion; derivative of pPP1 | 47 |
| pORI280 | Ermr; ori+ ΔrepA; deletion derivative of pWV01; constitutive lacZ expression from the P32 promoter | 71 |
| pORI38 | Specr; ori+ ΔrepA; deletion derivative of pWV01 | 71 |
| pMA101 | pPP2 PlacA-lacZ | This study |
| pMA102 | pPP2 PlacT-lacZ | This study |
| pMA103 | pPP2 PgalK-lacZ | This study |
DNA isolation and manipulation.
All DNA manipulations in this study were done as described before (45). For PCR amplification, chromosomal DNA of the S. pneumoniae D39 strain (25) was used. The primers used in this study are based on the sequence of the D39 genome (25) and are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer name | Nucleotide sequence (5′→3′) | Restriction site |
|---|---|---|
| GalK-Rv | CATGGGATCCTTTGCGAAGAGTTTCAGC | BamHI |
| GalK-Fr | CATGGAATTCAATGTCTTTTAAGGTAGCC | EcoRI |
| LacA-Fr | CATGGAATTCCAAACCTCATCATCTGG | EcoRI |
| LacA-Rv | CATGGGATCCACAAGGTGGAAGTTTTC | BamHI |
| lacR-1 | CCCTCTACTATCTCGGTAACAACAAAC | |
| lacR-2 | GCTATGGCGCGCCTTGTTTGAGCATATTATCACC | AscI |
| lacR-3 | GCTAAGCGGCCGCGTCATCAAGCCTTAATAAAC | NotI |
| lacR-4 | CGTGAAACAACACTTGGAGATCTTG | |
| LacT-Fr | CATGGAATTCATGGAAAGAACGTGTG | EcoRI |
| LacT-Rv | CATGGGATCCGATACATGTCAACCTCC | BamHI |
| lacT-1 | CGATTGCGGCCGCCGCTTGCCAGACTGCTTGG | NotI |
| lacT-2 | CGATACATGTCAACCTCC | |
| lacT-3 | AGGTTGACATGTATCGGATCTATGATGTGATTACGC | |
| lacT-4 | CATGCCATGGCCAACAATCGCTGCTAACAGC | NcoI |
| Spec-R | GCTAAGCGGCCGCACTAAACGAAATAAACGC | NotI |
| Spec-F | GCTATGGCGCGCCCTAATCAAAATAGTGAGGAGG | AscI |
| lacA-1 | CAAACCTCATCATCTGG | |
| lacA-2 | ACAAGGTGGAAGTTTTC | |
| lacG-1 | GCCCTTCTAATCGTGGTTGACG | |
| lacG-2 | GCTTGATAAGCAGCTGTTGCGCC | |
| lacT-1 | ATGGAAAGAACGTGTG | |
| lacT-2 | GATACATGTCAACCTCC |
Construction of lacR and lacT mutants.
A lacR deletion mutant was made by allelic replacement with a spectinomycin resistance marker. Briefly, primers lacR-1/lacR-2 and lacR-3/lacR-4 were used to generate PCR fragments of the left and right flanking regions of lacR, respectively. PCR products of the left and right flanking regions of lacR contain AscI and NotI restriction enzyme sites, respectively. The spectinomycin resistance marker was amplified from plasmid pORI38 with primers Spec-F/Spec-R (46). The spectinomycin resistance marker also contains AscI and NotI restriction enzyme sites on its ends. Then, by restriction and ligation, the left and right flanking regions of lacR were fused to the spectinomycin resistance gene. The resulting ligation product was transformed into the S. pneumoniae D39 wild-type strain, and selection of the lacR mutant strain was done using the appropriate concentration of antibiotic.
To delete lacT, primers lacT-1/lacT-2 and lacT-3/lacT-4 were used to generate PCR fragments of the left and right flanking regions of lacT, respectively. A markerless lacT mutant was constructed using pORI280, as described before (45). Mutants were further examined for the presence of the lacR and lacT deletions by PCR and DNA sequencing.
Construction of promoter-lacZ fusions and β-galactosidase assays.
Chromosomal transcriptional lacZ fusions to the lacA, lacT, and galK promoters were constructed in the integration plasmid pPP2 (47) via double crossover in the bgaA locus with the primer pairs mentioned in Table 2, resulting in pMA101, pMA102, and pMA103, respectively. These constructs were subsequently introduced into the D39 wild type, resulting in strains MA102, MA105, and MA108, respectively. pMA101 was also transformed into the ΔlacR and ΔlacT strains, resulting in strains MA103 and MA104, respectively, and pMA102 was transformed into the ΔlacR and ΔlacT strains, resulting in strains MA106 and MA107, respectively. Similarly, pMA102 was transformed into the ΔccpA strain (3), resulting in strain MA109. All plasmid constructs were checked by PCR and DNA sequencing.
β-Galactosidase assays were performed as described before (45, 48), using cells that were grown in M17 medium with the appropriate sugars, as mentioned in Results. The cells were harvested in their respective mid-exponential phase of growth.
Reverse transcription-PCR.
To confirm that the lac gene cluster transcribes into two transcriptional units, the D39 wild-type strain was grown in 0.5% lactose plus M17 (LM17) medium and total RNA was isolated as described previously (49). The RNA sample was treated with 2 U of RNase-free DNase I (Invitrogen, Paisley, United Kingdom) to remove any DNA contamination. cDNA samples were prepared by using SuperScript III reverse transcriptase and random nanomers at 42°C for 16 h. The intergenic region IR-I was amplified by primer pair lacA-1/lacA-2, intergenic region IR-II was amplified by primer pair lacT-1/lacT-2, and intergenic region IR-III was amplified by primer pair lacG-1/lacG-2. For fair comparison of PCR products, 100 ng of RNA and 20 ng of DNA were used.
Microarray analysis.
For DNA microarray analysis in the presence of lactose, the transcriptome of the S. pneumoniae wild-type D39 strain grown in 3 biological replicates in 0.5% glucose plus M17 (GM17) medium was compared to the transcriptome of the same strain grown in 3 biological replicates in LM17 medium. Similarly, for DNA microarray analysis of the response to galactose, the transcriptome of the S. pneumoniae D39 wild-type strain grown in 3 biological replicates in GM17 medium was compared to the transcriptome of the same strain grown in 3 biological replicates in 0.5% galactose plus M17 (GalM17) medium.
To analyze the effect of a lacR deletion on the transcriptome of S. pneumoniae, the D39 wild-type strain and its isogenic lacR mutant were grown in triplicate in GM17 medium and harvested at the mid-exponential phase of growth. To study the impact of the lacT deletion on the transcriptome of S. pneumoniae, the D39 wild type and the ΔlacT mutant were grown in triplicate in LM17 medium and harvested at the mid-exponential growth phase. All other procedures regarding the DNA microarray experiment were performed as described previously (49).
Microarray data analysis.
DNA microarray data were analyzed as previously described (49, 50). For the identification of differentially expressed genes, a Bayesian P value of <0.001 and a fold change cutoff of 3 were applied.
Microarray data accession number.
Microarray data have been submitted to NBCI's GEO database under accession number GSE58184.
RESULTS
Organization and localization of lactose utilization genes in S. pneumoniae D39.
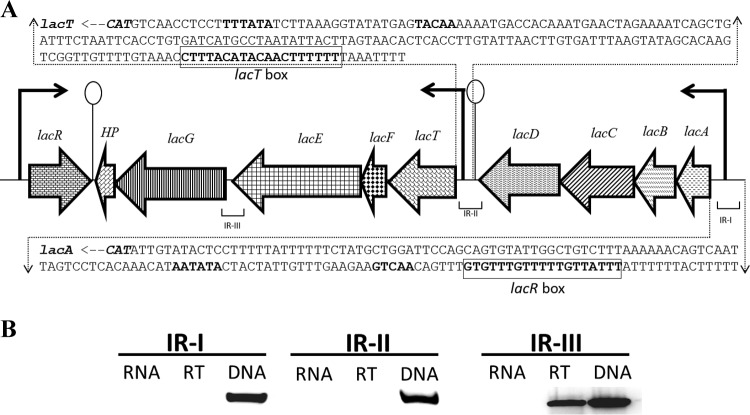
BLAST searches using protein sequences of the lactose utilization operon of S. mutans revealed the presence of a putative lactose utilization gene cluster (lac gene cluster) in the genome of S. pneumoniae D39. Unlike S. mutans (where all these genes are present in one operon [17, 27, 51] and which does not have lacT), the lac gene cluster in S. pneumoniae appears to be organized into two operons that are present next to each other. We named these two operons lac operon I (lacABCD) and lac operon II (lacTFEG) (Fig. 1A). Analysis of the flanking regions of the lac gene cluster identified −10 and −35 promoter sequences in the upstream region of lacA and lacT and possible terminator sequences downstream of lacD and HP (Fig. 1A). Reverse transcription-PCR using all possible intergenic primer sets confirmed that the lac gene cluster is organized into two operons which are transcribed as two transcriptional units (Fig. 1B). Interestingly, a DeoR family transcriptional regulator, lacR, is located downstream of the lac gene cluster, and lacR is transcribed in the opposite direction relative to the direction of transcription of the lac gene cluster. The presence of LacR close to the lac gene cluster may indicate its function as a transcriptional regulator of one or both of the operons in the lac gene cluster.
FIG 1.
(A) Organization of the lac gene cluster in S. pneumoniae D39. Lollipop structures, the transcriptional terminators; black arrows, promoter regions. See the text for further details. Nucleotides in bold indicate the putative core promoter sequences, and bold and boxed nucleotides indicate the putative regulatory consensus sequences. Here, 1 kb is equal to 1.25 cm. (B) Reverse transcription-PCR analysis to confirm the polycistronic nature of the S. pneumoniae lac operons I and II. Reverse transcription-PCR was performed on total RNA isolated from the D39 wild type grown in LM17 medium with reverse transcriptase (RT) and without reverse transcriptase (RNA) treatment using primer pairs specific for the IR-I, IR-II, and IR-III intergenic regions. DNA was used as a positive control.
lac operon I consists of four genes (lacABCD); lacA and lacB encode the A and B subunits of the galactose-6-phosphate isomerase, respectively, whereas lacC encodes the tagatose-6-phosphate kinase and lacD encodes the tagatose-1,6-diphosphate aldolase. lac operon II consists of five genes. These genes are lacF, lacE, lacG, a hypothetical protein (HP), and lacT. lacFE encode the A and BC components of the lactose-specific PTS EII, respectively, lacG encodes the 6-phospho-β-galactosidase, and lacT encodes a BglG-family transcriptional antiterminator. Most likely, in S. pneumoniae, lactose is transported inside the cell by the PEP-dependent lactose-specific PTS (lacFE), as in other Gram-positive bacteria, producing lactose-6-phosphate (Lac-6-P), which is then further hydrolyzed to glucose and galatose-6-phosphate (Gal-6-P) by LacG, and the Gal-6-P is catabolized through the tagatose pathway (52, 53). To further study the role of these genes in lactose utilization, we performed transcriptome analysis in the presence of lactose.
Lactose-dependent gene expression in S. pneumoniae.
To elucidate the transcriptional response of S. pneumoniae to lactose, a comparison of the transcriptome of the D39 wild type grown in LM17 medium with that of D39 grown in GM17 medium was performed. Table 3 summarizes the transcriptome changes observed in S. pneumoniae in the presence of lactose. Lactose is assumed to be an activator of the lac gene cluster, and we expected it to induce activation of the lac cluster. The presence of lactose in the medium has a very profound and specific effect on the tagatose pathway genes (the lac gene cluster, consisting of lac operons I and II), after applying the criteria of a ≥3.0-fold difference and a P value of <0.001. Upregulation of the tagatose pathway gene cluster in the presence of lactose indicates that the tagatose pathway is functional in S. pneumoniae and responds to lactose. β-Galactosidase (SPD_0562) was also unregulated in the presence of lactose. SPD_0562 belongs to glycosyl hydrolase family 2, the members of which have a broad range of enzymatic activity, including β-galactosidase (EC 3.2.1.23), β-glucuronidase (EC 3.2.1.31), and β-mannosidase (EC 3.2.1.25) activities (54). Most β-galactosidases can be induced by lactose (55), and it has been shown that the action of a β-galactosidase increases the rate of lactose transport in S. thermophilus (56).
TABLE 3.
Comparison of transcriptomes of the S. pneumoniae D39 wild-type strain grown in LM17 and GM17
| D39 taga | Functionb | Ratioc |
|---|---|---|
| SPD_0562 | β-Galactosidase | 4.1 |
| SPD_1044 | Lactose phosphotransferase system repressor (LacR) | 1.9 |
| SPD_1046 | 6-Phospho-β-galactosidase (LacG) | 5.5 |
| SPD_1047 | PTS system, lactose-specific IIBC components (LacE) | 6.0 |
| SPD_1048 | PTS system, lactose-specific IIA component (LacF) | 5.7 |
| SPD_1049 | Transcription antiterminator (LacT) | 4.4 |
| SPD_1050 | Tagatose-1,6-diphosphate aldolase (LacD) | 27.1 |
| SPD_1051 | Tagatose-6-phosphate kinase (LacC) | 30.5 |
| SPD_1052 | Galactose-6-phosphate isomerase (LacB subunit) | 28.5 |
| SPD_1053 | Galactose-6-phosphate isomerase (LacA subunit) | 16.2 |
The expression of some other genes and operons was also affected in the presence of lactose (see Table S1 in the supplemental material). To find out why the expression of these genes was affected in our microarray analysis, we further analyzed the promoter regions of these genes/operons and found out that these genes/operons have putative CcpA binding sites (cre box) in their promoter regions (see Table S1 in the supplemental material). The CcpA repression of these genes was most likely relieved in the absence of glucose. These findings are also supported by the findings from a previous study of Carvalho et al. (3). Interestingly, S. pneumoniae also harbors genes involved in the Leloir pathway, i.e., galKTE. galK encodes the galactokinase, galT encodes the galactose-1-phosphate uridylyltransferase, and galE encodes the UDP-glucose-4 epimerase. However, no change in the expression of these genes was observed in the presence of lactose. Therefore, we decided to also perform a microarray analysis in the presence of galactose to study the expression/regulation of genes involved in the Leloir pathway.
Galactose-dependent gene expression in S. pneumoniae.
To elucidate the transcriptomic response of S. pneumoniae to galactose, microarray analyses of the D39 wild type were performed in GalM17 medium to compare the transcriptome of S. pneumoniae strain D39 grown in GalM17 medium with that of the strain grown in GM17 medium. Table 4 lists the transcriptome changes incurred in S. pneumoniae D39 in the presence of galactose. The presence of galactose in the medium seems to have a very profound and specific effect on the tagatose pathway genes when the criteria of a ≥3.0-fold difference and a P value of <0.001 were used. The tagatose pathway genes were highly upregulated in the presence of galactose, suggesting that galactose can also be metabolized through the tagatose pathway. However, no effect on the expression of genes encoding the Leloir pathway enzymes was observed.
TABLE 4.
Comparison of transcriptomes of the S. pneumoniae D39 wild-type strain grown in GalM17 and GM17
| D39 taga | Functionb | Ratioc |
|---|---|---|
| SPD_0562 | β-Galactosidase | 3.2 |
| SPD_0264 | PTS system, mannose-specific IIAB components | −3.0 |
| SPD_1046 | 6-Phospho-β-galactosidase (LacG) | 10.0 |
| SPD_1047 | PTS system, lactose-specific IIBC components (LacE) | 4.0 |
| SPD_1048 | PTS system, lactose-specific IIA component (LacF) | 3.3 |
| SPD_1049 | Transcription antiterminator (LacT) | 4.5 |
| SPD_1050 | Tagatose-1,6-diphosphate aldolase (LacD) | 114.0 |
| SPD_1051 | Tagatose-6-phosphate kinase (LacC) | 112.2 |
| SPD_1052 | Galactose-6-phosphate isomerase (LacB subunit) | 91.5 |
| SPD_1053 | Galactose-6-phosphate isomerase (LacA subunit) | 111.3 |
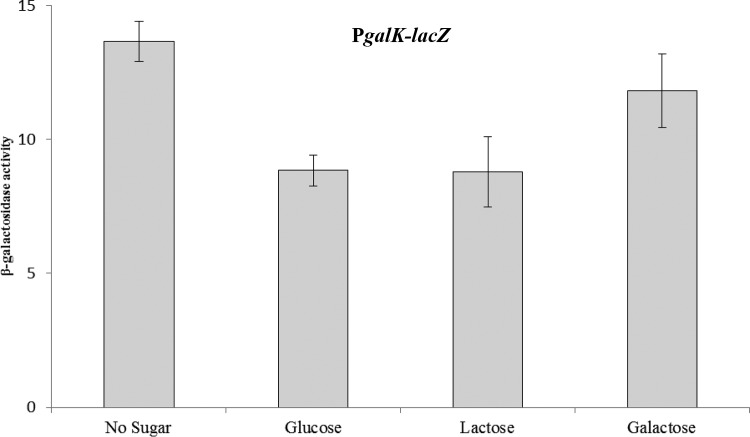
To confirm this finding further, we made a promoter-lacZ fusion of galK and transformed it into the D39 wild-type strain and checked the expression of PgalK-lacZ in the presence of galactose through β-galactosidase assays. We did not see any activation of PgalK-lacZ in response to galactose, confirming our microarray results in the presence of galactose (Fig. 2). These data further suggest the involvement of another regulator that represses the expression of genes involved in the Leloir pathway in the presence of glucose, lactose, and galactose. To solve this mystery of another regulator, we analyzed the promoter region of galK and found a cre box (5′-AAGAAAACGATTACAC-3′) in the promoter region of galK. The presence of a cre box in the promoter region of galK suggests that CcpA strongly represses this operon (galKT) in the presence of glucose and galactose (3).
FIG 2.
Expression levels (in Miller units) of PgalK-lacZ in the D39 wild type grown in M17 (without any sugar), GM17, LM17, and GalM17 media. The standard deviations from three independent experiments or replicates are indicated by bars.
Lactose induces, while glucose represses, the expression of the lac gene cluster.
To confirm our transcriptome results in response to lactose and galactose, we made a transcriptional lacZ fusion of PlacA, transformed it into the D39 wild-type strain, and checked the promoter activity in the presence of various sugars (Table 5). The expression of PlacA-lacZ was significantly higher in the presence of galactose and lactose in the medium than in the presence of other sugars. These results suggest that the lac gene cluster is activated in the presence of galactose or lactose, while it is repressed in the presence of other sugars, including glucose. Moreover, these results are also in accordance with our microarray data mentioned above.
TABLE 5.
Expression levels of PlacA-lacZ transcriptional fusion in D39 wild type grown in M17 medium with different added sugarsa
| Sugar | β-Galactosidase activity (Miller units) of wild-type PlacA-lacZb |
|---|---|
| No sugar | 209 (2) |
| Arabinose | 301 (30) |
| Cellobiose | 198 (6) |
| Dextrose | 171 (5) |
| Fructose | 157 (17) |
| Fucose | 353 (8) |
| Galactose | 1078 (36) |
| Glucose | 141 (6) |
| Lactose | 502 (7) |
| Maltose | 165 (6) |
| Mannitol | 322 (2) |
| Mannose | 173 (7) |
| Melibiose | 358 (30) |
| Sorbitol | 339 (7) |
| Trehalose | 342 (7) |
| Xylose | 297 (10) |
Sugars were present at 0.5% (wt/vol).
The standard deviations from three independent experiments are given in parentheses.
LacR acts as a transcriptional repressor of lac operon I, while LacT acts as a transcriptional activator of lac operon II.
LacR, a DeoR-family transcriptional regulator, is present downstream of the lac gene cluster. To study whether lacR is involved in the regulation of the lac gene cluster, we constructed a lacR isogenic mutant by replacing lacR with a spectinomycin resistance marker and transformed the PlacA-lacZ and PlacT-lacZ transcriptional fusions into the ΔlacR strain. β-Galactosidase assays were performed with the strains containing these transcriptional lacZ fusions grown in M17, GM17, and LM17 media. The β-galactosidase assay data showed that the deletion of lacR leads to a high level of expression of PlacA-lacZ even in the presence of glucose (Fig. 3A). However, the lacR deletion had no effect on the expression of PlacT-lacZ (Fig. 3B), which suggests a putative role of another transcriptional regulator in the regulation of lac operon II.
FIG 3.
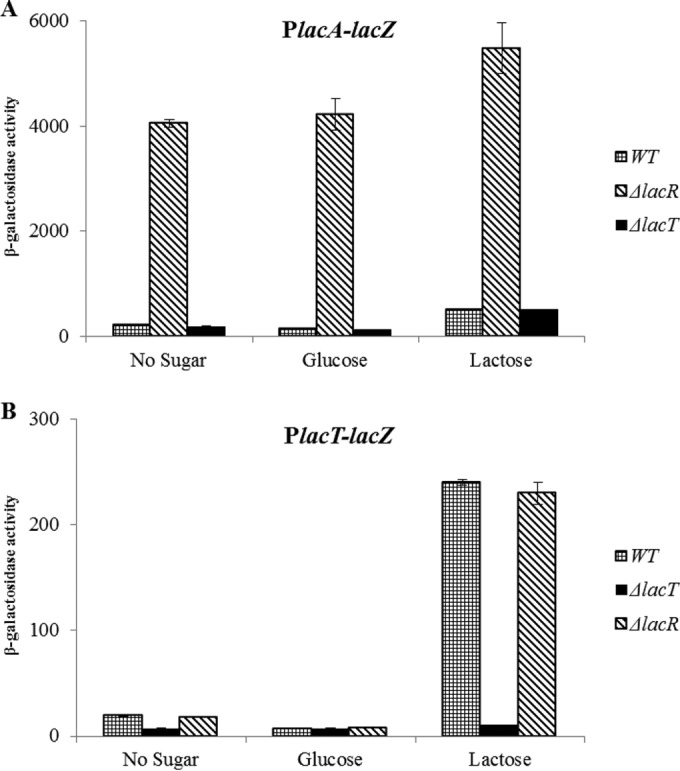
Levels (in Miller units) of PlacA-lacZ (A) and PlacT-lacZ (B) expression in the D39 wild-type, D39 ΔlacR, and D39 ΔlacT strains grown in M17 (without any sugar), GM17, and LM17 media. The standard deviations from three independent experiments or replicates are indicated by bars.
lac operon II consists of a lactose-specific PTS and a 6-phospho-β-galactosidase. It also encodes a BglG-family transcriptional antiterminator, LacT. The presence of LacT in lac operon II indicates the putative role of LacT in the regulation of lac operon II. Therefore, we decided to further investigate the role of LacT in the regulation of lac operon II. As lacT is the first gene of lac operon II (Fig. 1), we decided to make a clean knockout of the lacT gene to avoid a polar effect of the lacT deletion on the rest of the genes present in lac operon II. To study the effect of the lacT deletion on the regulation of lac operon II, we transformed a PlacT-lacZ transcriptional fusion into both the ΔlacT and D39 wild-type strains. β-Galactosidase assays were performed with the strains containing PlacT-lacZ grown in M17, GM17, and LM17 media. The activity of PlacT-lacZ was abolished in the ΔlacT strain in the presence of lactose, whereas it was retained in the wild-type strain (Fig. 3B), suggesting a role for LacT as a transcriptional activator of lac operon II.
To further investigate the role of LacT in the regulation of lac operon I, we transformed PlacA-lacZ into the ΔlacT strain. β-Galactosidase assays were performed with the strain containing this transcriptional lacZ fusion grown in LM17 medium. No difference in the activity of PlacA-lacZ in the ΔlacT strain from that in the wild type was observed in the presence of lactose and glucose, indicating that LacT has no role in the regulation of lac operon I (Fig. 3A).
DNA microarray analysis of the ΔlacR strain.
To elucidate the effect of the lacR deletion on the gene expression of S. pneumoniae, DNA microarray analyses were performed with the D39 wild type, and the results were compared with those obtained with its isogenic lacR mutant grown in GM17 medium. GM17 medium was used, as LacR represses the expression of its target genes in the presence of glucose (shown above). Table 6 provides the results of transcriptome changes in S. pneumoniae caused by the deletion of lacR. The deletion of lacR did not have a broad effect on the transcriptome of S. pneumoniae. According to the criteria of a ≥3.0-fold difference as the threshold change and a P value of <0.001, lac operon I was the only operon that was significantly upregulated in the ΔlacR strain, suggesting that lac operon I is the only target of LacR and confirming the role of LacR as a negative transcriptional regulator of lac operon I. No effect on the expression of lac operon II was observed in the absence of lacR. These data are also in accordance with those from the β-galactosidase assays mentioned above.
TABLE 6.
Comparison of transcriptomes of the S. pneumoniae D39 ΔlacR and D39 wild-type strains grown in GM17
| D39 taga | Functionb | Ratioc |
|---|---|---|
| SPD_0562 | β-Galactosidase | 4.9 |
| SPD_1044 | Lactose phosphotransferase system repressor (LacR) | −27.3 |
| SPD_1046 | 6-Phospho-β-galactosidase (LacG) | 1.0 |
| SPD_1047 | PTS system, lactose-specific IIBC components (LacE) | 1.6 |
| SPD_1048 | PTS system, lactose-specific IIA component (LacF) | −1.3 |
| SPD_1049 | Transcription antiterminator (LacT) | −2.0 |
| SPD_1050 | Tagatose-1,6-diphosphate aldolase (LacD) | 18.5 |
| SPD_1051 | Tagatose-6-phosphate kinase (LacC) | 10.5 |
| SPD_1052 | Galactose-6-phosphate isomerase (LacB subunit) | 33.0 |
| SPD_1053 | Galactose-6-phosphate isomerase (LacA subunit) | 15.5 |
lacT acts as a transcriptional activator of lac operon II.
To find more targets of LacT, we decided to perform microarray analyses of the S. pneumoniae ΔlacT strain with the D39 wild-type strain in LM17 medium. LM17 medium was used because our β-galactosidase assays showed that LacT activates its targets in the presence of lactose. The results of the microarray analyses are summarized in Table 7. The lacT mutation did not have broader effects on the transcriptome of S. pneumoniae. lac operon II was the only operon that was downregulated in the ΔlacT strain in the presence of lactose. The downregulation of lac operon II in the ΔlacT strain not only confirms the findings of our β-galactosidase assays with PlacT-lacZ but also demonstrates the role of LacT as a transcriptional activator of lac operon II in the presence of lactose.
TABLE 7.
Comparison of transcriptomes of the S. pneumoniae D39 wild-type and ΔlacT strains grown in LM17
Role of CcpA in regulation of lac operons I and II.
CcpA is a global transcriptional regulator that represses the expression of genes involved in the utilization of nonpreferred sugars in the presence of a preferred one (3). To study the role of CcpA in the regulation of lac operons I and II, we analyzed the promoter regions of lacA and lacT for the presence of cre boxes. Interestingly, a putative cre box (5′-ATGTAAAGGTTTACAA-3′) was present only in the lacT promoter region, suggesting the putative role of CcpA in the LacT-dependent regulation of lac operon II. However, no cre box was found in the lacA promoter region, suggesting the CcpA-independent regulation of lac operon I by transcriptional repressor LacR.
To determine the functionality of the cre box present in the lacT promoter region, we transformed PlacT-lacZ into the ΔccpA strain. β-Galactosidase assays showed that the ccpA deletion has no effect on the expression of lac operon II even in the presence of glucose (data not shown here). These results suggest that the cre box present in PlacT is most likely not functional and CcpA has no role in the regulation of the lac gene cluster. These findings are also consistent with the previous findings of Carvalho et al. (3).
DISCUSSION
S. pneumoniae, like many other bacteria, utilizes glucose as a preferred carbon/energy source (23). However, it also has the ability to utilize other carbon sources, if glucose is not available in the environment, which is also evident from the presence of several other sugar-specific systems in S. pneumoniae (23, 25, 57). The presence of such systems is a representation of a pattern of the self-regulating evolution of the regulatory and metabolic genes in S. pneumoniae (58). The regulation of many of these systems dedicated to sugars, including sucrose, maltose, raffinose, cellobiose, and others, has been studied extensively in S. pneumoniae (3, 22, 24, 26, 59). However, lactose- and galactose-dependent systems have not yet been explored in S. pneumoniae. Lactose and galactose are usually metabolized by the tagatose and Leloir pathways, respectively, and regulation of these pathways has already been studied in various bacteria. BLAST searches showed that S. pneumoniae also possesses a gene cluster (the lac gene cluster) that encodes enzymes required for the functionality of the tagatose and Leloir pathways. In this study, we have studied the effect of lactose and galactose on the transcriptome of S. pneumoniae and characterized the role of two transcriptional factors (LacR and lacT) that are required for the regulation of the lac gene cluster.
The lac gene cluster (consisting of two operons, lac operon I and lac operon II) of S. pneumoniae shares high sequence homology with the lac operon in S. mutans and the lac gene cluster in S. gordonii. In S. mutans, the lac genes are organized in one operon (33), whereas in S. gordonii, the lac genes are organized in two operons (18), as they are in S. pneumoniae. Moreover, S. mutans lacks the BglG-family transcriptional antiterminator LacT in the lac operon. S. gordonii and S. pneumoniae both have a gene for LacT. The S. mutans lac (lacSM) operon is regulated by the single regulator LacR (33), whereas the S. pneumoniae lac (lacSP) gene cluster is regulated by two different transcriptional regulators; i.e., LacR acts as a repressor for lac operon I in the presence of glucose and LacT activates lac operon II in the presence of lactose/galactose. In S. pneumoniae, LacT regulates the putative lactose transport part, while LacR regulates the lactose utilization part. Similarly, the S. gordonii lac (lacSG) gene cluster has both regulators (LacR and LacT) (18, 33). However, the role of LacT has not yet been explored in S. gordonii.
The PEP-dependent PTSs are the primary carbohydrate uptake systems in all bacteria. PTSs phosphorylate substrates during uptake and play a key role in the regulation of metabolic activities (57, 60–62). lac operon II of S. pneumoniae encodes a lactose-dependent PTS (LacFE) that is probably involved in the transport and phosphorylation of lactose inside the cell and a 6-phospho-β-galactosidase (LacG) that putatively breaks Lac-6-P down into glucose and Gal-6-P (33). The lacTFEG genes (lac operon II) of S. pneumoniae also show 90 to 95% sequence homology to those of S. gordonii, Streptococcus mitis, Streptococcus infantis, and Streptococcus oralis. The lacABCD genes are organized on lac operon I of S. pneumoniae and encode enzymes involved in the tagatose pathway that metabolize Gal-6-P. They have >90% sequence homology to their counterparts in S. gordonii, S. mitis, S. infantis, and S. oralis.
Our data show that LacR, a DeoR-type regulator present downstream of tagatose pathway genes (lac operon I), acts as a transcriptional repressor of lac operon I in the absence of lactose/galactose. DeoR-type regulators have been shown to be transcriptional repressors of sugar-specific genes involved in the uptake and metabolism of different sugars: lactose (L. lactis [63], Staphylococcus aureus [64]), fructose (Lactococcus lactis [41], Streptococcus gordonii [65]), and sorbose (Lactobacillus casei [66]). These DeoR-type repressors have in common the characteristics that in most cases they regulate neighboring genes and act as transcriptional repressors in sugar metabolism (39, 40, 67). LacR in S. pneumoniae also shares >80% sequence homology with its counterparts in S. mitis, S. infantis, and S. oralis. However, some streptococci like S. gordonii possess two copies of the tagatose pathway genes. Therefore, we looked further for a second copy of tagatose pathway genes in the D39 strain of S. pneumoniae. Interestingly, the S. pneumoniae D39 strain does not have the second copy of the tagatose pathway genes like S. gordonii does.
LacT in S. pneumoniae activates the expression of lac operon II. LacT is also present in S. gordonii, S. mitis, S. infantis, and S. oralis and shares high sequence homology (∼90%) with these species, but it is missing in S. mutans. LacT belongs to the BglG family of transcriptional antiterminators and possesses PTS regulatory domains (PRDs) and a CoAT RNA-binding domain. Usually, these PRDs have conserved histidine residues that require phosphorylation, one by one, of the certain carbohydrate-specific PTS components (68). On the basis of models for the PTS-dependent regulation of antitermination available in the literature (69), it can be assumed that when the PTS permease for lactose is involved in sugar transport, the PRD in LacT would be dephosphorylated, allowing the antitermination of the expression of lac operon II. To find the putative LacT site in the promoter region of lacT, we looked into the RegPrecise database (70) and propose a putative LacT binding site spanning 19 bp (5′-AAAAAAGTTGTATGTAAAG-3′) based on the already predicted binding sites for BglG-type regulators.
Lactose and most of the galactose are usually utilized through the tagatose pathway (LacABCD), but galactose can also be utilized by the Leloir pathway (33). Galactose enters the cell through an unknown permease in S. mutans and gets phosphorylated by a galactokinase (GalK) to produce galactose-1-phosphate, which is then transformed into glucose-1-phosphate by hexose-1-phosphate uridyltransferase (GalT) and UPD-glucose epimerase (GalE) (13). The glucose produced in this process enters the glycolytic pathway. No significant change in the expression of genes encoding Leloir pathway enzymes was detected in our microarray studies. However, tagatose pathway genes were upregulated in the presence of galactose under our tested conditions. The repression of Leloir pathway genes in the presence of glucose and galactose is due to CcpA (carbon catabolite protein A), as CcpA causes the repression of certain genes that have cre boxes in their promoter regions (3). Also, no strong change in the expression of lacFE was seen in our galactose microarray results, suggesting that galactose is not fully transported through this PTS and there must be some other transport system for galactose.
CcpA is the master regulator that regulates genes involved in sugar metabolism (3, 4, 6). There are many other systems specified for nonpreferred sugars that are regulated independently of CcpA, like CelR, in S. pneumoniae (59). In this study, we could not see an effect of CcpA on the regulation of the lac gene cluster of S. pneumoniae, though there is a putative cre box in the promoter region of lacT. Similar results were found in a recent transcriptome-wide analysis of a ΔccpA mutant with glucose and galactose, where ccpA deletion had no effect on the expression of the lac gene cluster (3). This suggests that expression of the lac gene cluster is independent of CcpA and that most likely the putative cre box present in PlacT is not functional, probably because it is not located properly or due to the missing important central CG in the putative cre box.
To find the putative LacR binding site in the promoter region of lacA, we looked in the RegPrecise database (70) for already predicted sites and found a stretch of DNA spanning 18 bp (5′-AAATAACAAAACAAACAC-3′). To explore whether there are more putative LacR binding sites in the D39 genome, we conducted a genome-wide search with the putative pneumococcal LacR operator site mentioned above. The putative LacR operator site was exclusively found in the promoter region of lac operon I, confirming that lac operon I is the only target of LacR in S. pneumoniae. This predicted LacR operator site is also found to be highly conserved in other streptococci as well (70), suggesting a similar function of LacR in other streptococci.
Supplementary Material
ACKNOWLEDGMENT
M.A. is supported by the GC University, Faisalabad, Pakistan, under the faculty development program of HEC Pakistan.
Footnotes
Published ahead of print 20 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01370-14.
REFERENCES
- 1.Stulke J, Hillen W. 1998. Coupling physiology and gene regulation in bacteria: the phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften 85:583–592. 10.1007/s001140050555 [DOI] [PubMed] [Google Scholar]
- 2.Titgemeyer F, Hillen W. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82:59–71. 10.1023/A:1020628909429 [DOI] [PubMed] [Google Scholar]
- 3.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. 2011. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae D39. PLoS One 6:e26707. 10.1371/journal.pone.0026707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lulko AT, Buist G, Kok J, Kuipers OP. 2007. Transcriptome analysis of temporal regulation of carbon metabolism by CcpA in Bacillus subtilis reveals additional target genes. J. Mol. Microbiol. Biotechnol. 12:82–95. 10.1159/000096463 [DOI] [PubMed] [Google Scholar]
- 5.Gorke B, Stulke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- 6.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:1366–1381. 10.1128/JB.01013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93. 10.1016/j.mib.2008.02.007; 10.1016/j.mib.2008.02.007 [DOI] [PubMed] [Google Scholar]
- 8.Hollis DG, Wiggins GL, Weaver RE. 1969. Neisseria lactamicus sp. n., a lactose-fermenting species resembling Neisseria meningitidis. Appl. Microbiol. 17:71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King SJ. 2010. Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol. Oral Microbiol. 25:15–24. 10.1111/j.2041-1014.2009.00564.x [DOI] [PubMed] [Google Scholar]
- 10.Yesilkaya H, Manco S, Kadioglu A, Terra VS, Andrew PW. 2008. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett. 278:231–235. 10.1111/j.1574-6968.2007.01003.x [DOI] [PubMed] [Google Scholar]
- 11.Fridovich-Keil JL. 2006. Galactosemia: the good, the bad, and the unknown. J. Cell. Physiol. 209:701–705. 10.1002/jcp.20820 [DOI] [PubMed] [Google Scholar]
- 12.Russell RR, Aduse-Opoku J, Sutcliffe IC, Tao L, Ferretti JJ. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 267:4631–4637 [PubMed] [Google Scholar]
- 13.Abranches J, Chen YY, Burne RA. 2004. Galactose metabolism by Streptococcus mutans. Appl. Environ. Microbiol. 70:6047–6052. 10.1128/AEM.70.10.6047-6052.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortina MG, Ricci G, Mora D, Guglielmetti S, Manachini PL. 2003. Unusual organization for lactose and galactose gene clusters in Lactobacillus helveticus. Appl. Environ. Microbiol. 69:3238–3243. 10.1128/AEM.69.6.3238-3243.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frostell G, Keyes PH, Larson RH. 1967. Effect of various sugars and sugar substitutes on dental caries in hamsters and rats. J. Nutr. 93:65–76 [DOI] [PubMed] [Google Scholar]
- 16.de Vos WM, Vaughan EE. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:217–237. 10.1016/0168-6445(94)90114-7 [DOI] [PubMed] [Google Scholar]
- 17.Ajdic D, Sutcliffe IC, Russell RR, Ferretti JJ. 1996. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene 180:137–144. 10.1016/S0378-1119(96)00434-9 [DOI] [PubMed] [Google Scholar]
- 18.Zeng L, Martino NC, Burne RA. 2012. Two gene clusters coordinate galactose and lactose metabolism in Streptococcus gordonii. Appl. Environ. Microbiol. 78:5597–5605. 10.1128/AEM.01393-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Declerck N, Vincent F, Hoh F, Aymerich S, van Tilbeurgh H. 1999. RNA recognition by transcriptional antiterminators of the BglG/SacY family: functional and structural comparison of the CAT domain from SacY and LicT. J. Mol. Biol. 294:389–402. 10.1006/jmbi.1999.3256 [DOI] [PubMed] [Google Scholar]
- 20.Langbein I, Bachem S, Stulke J. 1999. Specific interaction of the RNA-binding domain of the Bacillus subtilis transcriptional antiterminator GlcT with its RNA target, RAT. J. Mol. Biol. 293:795–805. 10.1006/jmbi.1999.3176 [DOI] [PubMed] [Google Scholar]
- 21.Iyer R, Baliga NS, Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340–8349. 10.1128/JB.187.24.8340-8349.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giammarinaro P, Paton JC. 2002. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect. Immun. 70:5454–5461. 10.1128/IAI.70.10.5454-5461.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins J, Alborn WE, Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR, Jr, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709–5717. 10.1128/JB.183.19.5709-5717.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer R, Camilli A. 2007. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol. Microbiol. 66:1–13. 10.1111/j.1365-2958.2007.05878.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38–51. 10.1128/JB.01148-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shafeeq S, Kuipers OP, Kloosterman TG. 2013. Cellobiose-mediated gene expression in Streptococcus pneumoniae: a repressor function of the novel GntR-type regulator BguR. PLoS One 8:e57586. 10.1371/journal.pone.0057586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosey EL, Stewart GC. 1992. Nucleotide and deduced amino acid sequences of the lacR, lacABCD, and lacFE genes encoding the repressor, tagatose 6-phosphate gene cluster, and sugar-specific phosphotransferase system components of the lactose operon of Streptococcus mutans. J. Bacteriol. 174:6159–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall BG, Xu L. 1992. Nucleotide sequence, function, activation, and evolution of the cryptic asc operon of Escherichia coli K12. Mol. Biol. Evol. 9:688–706 [DOI] [PubMed] [Google Scholar]
- 29.el Hassouni M, Chippaux M, Barras F. 1990. Analysis of the Erwinia chrysanthemi arb genes, which mediate metabolism of aromatic beta-glucosides. J. Bacteriol. 172:6261–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bardowski J, Ehrlich SD, Chopin A. 1994. BglR protein, which belongs to the BglG family of transcriptional antiterminators, is involved in beta-glucoside utilization in Lactococcus lactis. J. Bacteriol. 176:5681–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marasco R, Salatiello I, De Felice M, Sacco M. 2000. A physical and functional analysis of the newly-identified bglGPT operon of Lactobacillus plantarum. FEMS Microbiol. Lett. 186:269–273. 10.1111/j.1574-6968.2000.tb09116.x [DOI] [PubMed] [Google Scholar]
- 32.Tobisch S, Glaser P, Kruger S, Hecker M. 1997. Identification and characterization of a new beta-glucoside utilization system in Bacillus subtilis. J. Bacteriol. 179:496–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng L, Das S, Burne RA. 2010. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression J. Bacteriol. 192:2434–2444. 10.1128/JB.01624-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogaert D, de Groot R, Hermans PW. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4:144–154. 10.1016/S1473-3099(04)00938-7 [DOI] [PubMed] [Google Scholar]
- 35.Zeng X, Saxild HH, Switzer RL. 2000. Purification and characterization of the DeoR repressor of Bacillus subtilis. J. Bacteriol. 182:1916–1922. 10.1128/JB.182.7.1916-1922.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortensen L, Dandanell G, Hammer K. 1989. Purification and characterization of the deoR repressor of Escherichia coli. EMBO J. 8:325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbier CS, Short SA. 1985. Studies on deo operon regulation in Escherichia coli: cloning and expression of the cytR structural gene. Gene 36:37–44. 10.1016/0378-1119(85)90067-8 [DOI] [PubMed] [Google Scholar]
- 38.Campos E, Baldoma L, Aguilar J, Badia J. 2004. Regulation of expression of the divergent ulaG and ulaABCDEF operons involved in l-ascorbate dissimilation in Escherichia coli. J. Bacteriol. 186:1720–1728. 10.1128/JB.186.6.1720-1728.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Z, Lin EC. 1989. The nucleotide sequence of Escherichia coli genes for l-fucose dissimilation. Nucleic Acids Res. 17:4883–4884. 10.1093/nar/17.12.4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valentin-Hansen P, Hojrup P, Short S. 1985. The primary structure of the DeoR repressor from Escherichia coli K-12. Nucleic Acids Res. 13:5927–5936. 10.1093/nar/13.16.5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barriere C, Veiga-da-Cunha M, Pons N, Guedon E, van Hijum SA, Kok J, Kuipers OP, Ehrlich DS, Renault P. 2005. Fructose utilization in Lactococcus lactis as a model for low-GC gram-positive bacteria: its regulator, signal, and DNA-binding site. J. Bacteriol. 187:3752–3761. 10.1128/JB.187.11.3752-3761.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beck von Bodman S, Hayman GT, Farrand SK. 1992. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl. Acad. Sci. U. S. A. 89:643–647. 10.1073/pnas.89.2.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. U. S. A. 96:9833–9838. 10.1073/pnas.96.17.9833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray WK, Larson TJ. 2004. Application of AgaR repressor and dominant repressor variants for verification of a gene cluster involved in N-acetylgalactosamine metabolism in Escherichia coli K-12. Mol. Microbiol. 51:813–826. 10.1046/j.1365-2958.2003.03868.x [DOI] [PubMed] [Google Scholar]
- 45.Kloosterman TG, Bijlsma JJE, Kok J, Kuipers OP. 2006. To have neighbour's fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiology 152:351–359. 10.1099/mic.0.28521-0 [DOI] [PubMed] [Google Scholar]
- 46.Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217–224. 10.1007/s004380050315 [DOI] [PubMed] [Google Scholar]
- 47.Halfmann A, Hakenbeck R, Bruckner R. 2007. A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268:217–224. 10.1111/j.1574-6968.2006.00584.x [DOI] [PubMed] [Google Scholar]
- 48.Israelsen H, Madsen SM, Vrang A, Hansen EB, Johansen E. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, Kuipers OP, Morrissey JA. 2011. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81:1255–1270. 10.1111/j.1365-2958.2011.07758.x [DOI] [PubMed] [Google Scholar]
- 50.Shafeeq S, Kloosterman TG, Kuipers OP. 2011. Transcriptional response of Streptococcus pneumoniae to Zn2+ limitation and the repressor/activator function of AdcR. Metallomics 3:609–618. 10.1039/c1mt00030f [DOI] [PubMed] [Google Scholar]
- 51.Jagusztyn-Krynicka EK, Hansen JB, Crow VL, Thomas TD, Honeyman AL, Curtiss R., III 1992. Streptococcus mutans serotype c tagatose 6-phosphate pathway gene cluster. J. Bacteriol. 174:6152–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Vos WM, Vaughan EE. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:217–237 [DOI] [PubMed] [Google Scholar]
- 53.Morse ML, Hill KL, Egan JB, Hengstenberg W. 1968. Metabolism of lactose by Staphylococcus aureus and its genetic basis. J. Bacteriol. 95:2270–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terra VS, Homer KA, Rao SG, Andrew PW, Yesilkaya H. 2010. Characterization of novel β-galactosidase activity that contributes to glycoprotein degradation and virulence in Streptococcus pneumoniae. Infect. Immun. 78:348–357. 10.1128/IAI.00721-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zahner D, Hakenbeck R. 2000. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 182:5919–5921. 10.1128/JB.182.20.5919-5921.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geertsma ER, Duurkens RH, Poolman B. 2005. The activity of the lactose transporter from Streptococcus thermophilus is increased by phosphorylated IIA and the action of β-galactosidase. Biochemistry 44:15889–15897. 10.1021/bi051638w [DOI] [PubMed] [Google Scholar]
- 57.Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. 2012. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7:e33320. 10.1371/journal.pone.0033320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nieto C, Espinosa M, Puyet A. 1997. The maltose/maltodextrin regulon of Streptococcus pneumoniae. Differential promoter regulation by the transcriptional repressor MalR. J. Biol. Chem. 272:30860–30865 [DOI] [PubMed] [Google Scholar]
- 59.Shafeeq S, Kloosterman TG, Kuipers OP. 2011. CelR-mediated activation of the cellobiose-utilization gene cluster in Streptococcus pneumoniae. Microbiology 157:2854–2861. 10.1099/mic.0.051359-0 [DOI] [PubMed] [Google Scholar]
- 60.Barabote RD, Saier MH., Jr 2005. Comparative genomic analyses of the bacterial phosphotransferase system Microbiol. Mol. Biol. Rev. 69:608–634. 10.1128/MMBR.69.4.608-634.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031. 10.1128/MMBR.00024-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lengeler JW, Jahreis K. 2009. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib. Microbiol. 16:65–87. 10.1159/000219373 [DOI] [PubMed] [Google Scholar]
- 63.van Rooijen RJ, de Vos WM. 1990. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J. Biol. Chem. 265:18499–18503 [PubMed] [Google Scholar]
- 64.Oskouian B, Stewart GC. 1990. Repression and catabolite repression of the lactose operon of Staphylococcus aureus. J. Bacteriol. 172:3804–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:6241–6254. 10.1128/JB.185.21.6241-6254.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yebra MJ, Veyrat A, Santos MA, Perez-Martinez G. 2000. Genetics of l-sorbose transport and metabolism in Lactobacillus casei. J. Bacteriol. 182:155–163. 10.1128/JB.182.1.155-163.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada M, Saier MH., Jr 1988. Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J. Mol. Biol. 203:569–583. 10.1016/0022-2836(88)90193-3 [DOI] [PubMed] [Google Scholar]
- 68.van Tilbeurgh H, Declerck N. 2001. Structural insights into the regulation of bacterial signalling proteins containing PRDs. Curr. Opin. Struct. Biol. 11:685–693. 10.1016/S0959-440X(01)00267-6 [DOI] [PubMed] [Google Scholar]
- 69.Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73:245–259. 10.1271/bbb.80479 [DOI] [PubMed] [Google Scholar]
- 70.Novichkov PS, Laikova ON, Novichkova ES, Gelfand MS, Arkin AP, Dubchak I, Rodionov DA. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118. 10.1093/nar/gkp894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leenhouts K, Venema G, Kok J. 1998. A lactococcal pWV01 based integration toolbox for bacteria. Methods Cell Sci. 20:35–50. 10.1023/A:1009862119114 [DOI] [Google Scholar]
- 72.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. 10.1126/science.1061217 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.