Abstract
The increasing demand for industrial enzymes and biopharmaceutical proteins relies on robust production hosts with high protein yield and productivity. Being one of the best-studied model organisms and capable of performing posttranslational modifications, the yeast Saccharomyces cerevisiae is widely used as a cell factory for recombinant protein production. However, many recombinant proteins are produced at only 1% (or less) of the theoretical capacity due to the complexity of the secretory pathway, which has not been fully exploited. In this study, we applied the concept of inverse metabolic engineering to identify novel targets for improving protein secretion. Screening that combined UV-random mutagenesis and selection for growth on starch was performed to find mutant strains producing heterologous amylase 5-fold above the level produced by the reference strain. Genomic mutations that could be associated with higher amylase secretion were identified through whole-genome sequencing. Several single-point mutations, including an S196I point mutation in the VTA1 gene coding for a protein involved in vacuolar sorting, were evaluated by introducing these to the starting strain. By applying this modification alone, the amylase secretion could be improved by 35%. As a complement to the identification of genomic variants, transcriptome analysis was also performed in order to understand on a global level the transcriptional changes associated with the improved amylase production caused by UV mutagenesis.
INTRODUCTION
Saccharomyces cerevisiae is one of the best-established host systems for commercial production of biopharmaceutical proteins due to the extensive knowledge of its genome, metabolism, and general physiology (1, 2) and its long history of industrial processing. Many strategies have been applied previously to improve protein production by S. cerevisiae, such as increasing gene expression and optimizing the fermentation process. However, production of secretory proteins is often not improved solely by increasing transcription and translation (3–5), and optimization of the fermentation process is also highly host and protein specific (6). There is, therefore, an increasing focus on host engineering, especially with respect to the protein secretory pathway, as illustrated in a recent study where SNARE proteins were overexpressed, resulting in improved production of two different proteins by S. cerevisiae (7). Since there are many components involved in the secretory pathway and the interactions among them are rather complex, it can be risky to focus on only one or several components. Selective screening following random mutagenesis or adaptive evolution in combination with systems biology analysis (genomics, transcriptomics, etc.) represents another effective approach to identify novel metabolic engineering targets for improving protein production. A similar approach based on adaptive evolution to find new targets for enhancing galactose utilization in yeast has been previously demonstrated (8).
Random mutations can be achieved by different strategies, including induction with chemicals or exposure to physical resources (9), restriction enzyme-mediated integration (10, 11), use of mutation strain collections (12), etc. In each case, it is important to have an efficient screening method and to consider how transcriptional analysis and genomic sequencing can be most efficiently applied in order to identify novel and relevant targets (13, 14). Payne et al. applied chemical mutagenesis and identified chaperone targets (JEM1, SIL1, LHS1, and SCJ1) regulating the activity of Kar2p, overexpression of which significantly improved the production of human albumin, granulocyte-macrophage colony-stimulating factor, and human transferrin in yeast with different degrees and rates of success (15). Wild-type (WT) Pichia pastoris does not secrete β-galactosidase and thus cannot grow on lactose medium. Larsen et al. generated mutation strains that can grow on lactose by random integration of a zeocin-resistant vector to disrupt gene expression. Thereby, a potential Pkc1p ortholog was shown to be involved in regulating secretion efficiency (11). By screening the EUROSCARF yeast deletion library, Kanjou et al. found the target MON2, which is involved in vesicle formation, for engineering and thereby increased the secretion levels of luciferase (12). Lately, novel approaches that combine yeast cDNA overexpression libraries with yeast surface display (e.g., a single-chain T-cell receptor [scTCR] as an indicator) and that allow the rapid flow cytometric (FC) screening of engineered yeast have been developed and new targets for enhancing protein secretion were quickly identified from cDNA overexpression libraries (16–18). Genes identified through this method include the cell wall protein genes (CCW12, CWP2, and SED1) (19), the RPP0 ribosomal subunit gene (19), and the ERO1 thiol oxidase gene (20).
In this report, we present a promising approach that led us to identify new gene targets for enhancing amylase secretion in yeast. The host strain producing heterologous amylase was firstly subjected to random mutagenesis by the use of UV and then selected for improved growth on starch. Strains producing higher levels of amylase were verified by measuring the amylase activity and were then subjected to whole-genome sequencing and genome-wide transcriptome analysis. A list of gene targets for genetic engineering was proposed, and we evaluated the contribution of VTA1S196I to the improvement of protein production in those targets. Surprisingly, this single missense mutation alone was able to improve amylase production in shake flasks by 35%.
MATERIALS AND METHODS
Cultivations of yeast strains.
Yeast extract-peptone-dextrose (YPD) medium was prepared using the following ingredients: 20 g/liter d-glucose, 10 g/liter yeast extract, 20 g/liter peptone. Starch plates were prepared using the following ingredients: 0.04 g/liter d-glucose, 10 g/liter starch, 6.7 g/liter yeast nitrogen base (YNB) without amino acids, and 20 g/liter agar. Delft minimal medium (21) was slightly modified for shake flask cultivation. The detailed composition is described in Text S1 in the supplemental material.
To check amylase production in shake flasks, seed cultures were made by inoculating fresh colonies into 5 ml of YPD medium in 50-ml Falcon tubes with shaking at 200 rpm at 30°C for 24 h. The cultures were diluted in fresh YPD medium to an initial optical density at 600 nm (OD600) of 0.1. The shake flasks were shaken at 200 rpm at 30°C for 60 h, during which cell growth and amylase production were measured.
To measure invertase production in shake flasks, the same seed cultures were used and inoculated in YPD medium at an initial OD600 of 0.05. Cells were cultured under the same conditions as before. Invertase activity assay was performed as described in reference 22. The presence of glucose hydrolyzed from invertase was determined using a d-glucose (GOPOD) kit (Megazyme K-Cera, Wicklow, Ireland). Cell-bound invertase activity was defined as follows: 1 unit of invertase releases 1 mmol glucose in 1 min at 30°C and pH 5.
To compare strain performances in Delft medium supplemented with bovine serum albumin (BSA), seed cultures were prepared in 5 ml of Delft medium, cultured as described above, and transferred into 5 ml of the media in question, which were Delft, Delft–1 g/liter BSA, and Delft–2 g/liter BSA, respectively, with an initial OD600 of 0.05 in 50-ml Falcon tubes. The tubes were shaken at 200 rpm at 30°C for 60 h, during which cell growth and α-amylase production were measured.
For growth in bioreactors, seed cultures were grown overnight before being inoculated into the fermenters with an initial OD600 of 0.01. All fermentations were performed in DasGip 1.0-liter stirrer-pro vessels (Eppendorf, Jülich, Germany) with a working volume of 500 ml YPD medium at 30°C and 600-rpm agitation. Aerobic conditions were controlled by keeping a flow of 1 vvm (volume of flow per working volume per min) of air during fermentation. One drop of antifoam 204 (Sigma) was added to each fermentation. Dissolved oxygen was measured using a polarographic oxygen electrode (Mettler Toledo, Switzerland). The pH was monitored by the use of a pH sensor (Mettler Toledo, Switzerland) and maintained at 6.0 using 2 M KOH. All fermentations were done in biological triplicates.
Induction of mutagenesis by UV.
A single colony was picked from the YPD plate and transferred into 5 ml of YPD medium and incubated overnight at 30°C. A 1-μl volume of the culture was used to achieve a 1,000× dilution in 1 ml, and 10 μl was spread on starch agar plates. These plates were immediately exposed to ultraviolet light (UV λ = 254 nm) with doses of 4, 5, 6, 7, 8, 9, 10, and 11 mJ/cm2 and were subsequently incubated in darkness at 30°C for 7 to 10 days.
Analytical methods.
A 1-ml volume of the culture was centrifuged at 4,000 × g for 5 min. A total of 8 volumes of the supernatant was added to 1 volume of 5.5 μM sodium azide and stored at 4°C until measurement. Concentrations of glucose, glycerol, ethanol, and acetate were analyzed by the use of a Dionex Ultimate 3000 high-performance liquid chromatography (HPLC) system (Dionex Softron GmbH, Germany) with an Aminex HPX-87H column (Bio-Rad) at 65°C using 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 ml/min. The activity of α-amylase was measured using a Ceralpha kit (Megazyme, Ireland) and α-amylase from Aspergillus oryzae (Sigma) as the standard. The dry cell weight (DCW) was acquired by filtering the cell culture through a 0.45-μm-pore-size filter membrane (Sartorius Stedim, Germany) that had been dried for 24 h in a desiccator and measuring the increased weight.
Genome-wide transcription data analysis.
RNA for a DNA microarray analysis was isolated using an RNeasy minikit (Qiagen) and processed to biotin-labeled amplified RNA (aRNA; sometimes called complementary RNA [cRNA]) using a Genechip 3′ IVT Express kit (Affymetrix) and hybridized/scanned on a Yeast Genome 2.0 array (Affymetrix). CEL files were normalized using Plier normalization with perfect-match probes only. Statistical analysis using the moderated t-statistic and reporter analysis were performed using the Platform for Integrated Analysis of Omics (PIANO) data package for R (23).
Whole-genome Illumina sequencing and data analysis.
Whole-genome sequencing of the two UV mutagenized strains (M715 and M1052) and the wild-type CEN.PK113-7D strain was performed using an Illumina Hiseq 2000 system. The reads were checked for quality, and reads with an average Phred quality score of less than 28 were filtered out (a score of 30 means 1 error in 1,000 nucleotides [nt]). The reads were aligned to the recently published genome sequence of CEN.PK113-7D (24) using Stampy version 1.0.17 (25). On average, 83% of the reads could be mapped to the reference sequence. Point mutations in each of the sequenced genomes, compared to the reference sequence, were identified using Atlas-SNP2 version 1.0 (26), and indels were detected using Atlas-Indel2 version 1.0 (27). Identification of a single-nucleotide point mutation was considered highly confident by the Atlas-SNP2 algorithm if it was found on both strands, if the calculated posterior probability was greater than 0.95, and if the coverage (number of aligned reads) at that position was at least 8 (26, 27). Indels were detected using the default parameters of Atlas-Indel2, and indels that were located in regions where the reference sequence contained at least one unknown base (N) were filtered out.
Single nucleotide variants (SNVs) and indels which were detected as having the same variant in all three strains (the mutants and the WT) were considered to represent genetic differences between the background strain and the published reference sequence and therefore were not further investigated. Mutations detected in exons and upstream regions (0 to 1,000 bp upstream of the exon start) in the M715 and M1052 strains could have been of interest and were therefore investigated further by verifying transcription level changes of those genes.
Significant statistic tests.
The significant statistic tests were done with a one-tailed homoscedastic (equal-variance) t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
RESULTS AND DISCUSSION
Random mutagenesis of the amylase-producing yeast.
The starting strain subjected to random mutagenesis by UV light is called AAC, a CEN.PK530-1C [MATa URA3HIS3 LEU2 TRP1 SUC2 MAL2-8c tpi1(41-707)::loxP-KanMX4-loxP]-based strain expressing fungal α-amylase on the CPOTud plasmid (28). The amylase expression cassette is comprised of the codon-optimized amylase gene from A. oryzae and the yeast alpha factor leader (28). The POT1 gene from Schizosaccharomyces pombe (a triose phosphate isomerase gene from a different species to avoid homologous recombination) is used as the marker for selection in the TPI1 deletion strain. Expression of the protection of telomeres (POT)-based plasmid can reach up to 18 copies (29) through which the amylase gene can be highly expressed.
The AAC strain was spread on starch plates and exposed to different doses (from 4 to 11 mJ/cm2) of UV light (λ = 254 nm). The mortality rate under these conditions was between 60% and 90%. A total of 591 colonies were selected from starch plates after UV treatment and cultivation for further analysis. When performing mutagenesis experiments, trade-offs should be considered; i.e., improvement of fitness in certain circumstances is often accompanied by reduced fitness under other conditions (30). This can be reflected by the phenomenon that enhanced recombinant protein production (RPP) usually results in slower growth and reduced biomass yield. To minimize the problem, in the first round of selection, we grew the mutated colonies on starch plates; selected colonies generated a bigger activity halo until full growth was achieved. The growth rate was not considered at this stage. In the second round, 591 selected strains were cultivated in YPD medium in test tubes and further verified in shake flask and bioreactors. Two strains with the highest amylase titers were selected, namely, M715 and M1052 (“7” and “10” stand for 7 or 10 mJ/cm2 UV dose, and 15 and 52 are identification numbers within the series; Table 1]).
TABLE 1.
Strains and plasmids used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| NC | CEN.PK530-1C with TPI1 promoter (2 μm POT1) | 31 |
| AAC | CEN.PK530-1C with pTPI1-alpha factor leader amylase (2 μm POT1) | 31 |
| M715 | UV-mutated AAC strain exposed to 7 mJ/cm2 | This study |
| M1052 | UV-mutated AAC strain exposed to 10 mJ/cm2 | This study |
| M715n | CEN.PK 530-1C with plasmid extracted from M715 | This study |
| M1052n | CEN.PK 530-1C with plasmid extracted from M1052 | This study |
Characterization of strains M715 and M1052.
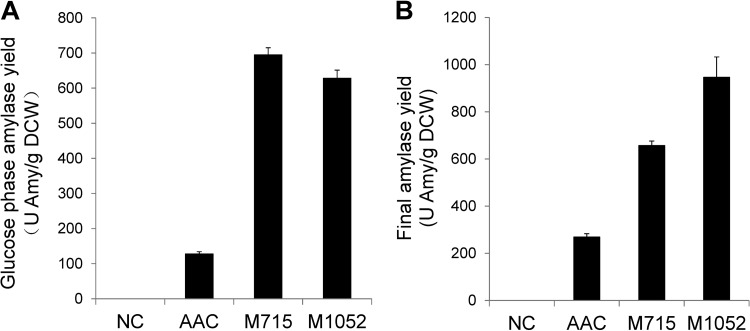
Strains M715 and M1052 were evaluated in batch cultivations and compared to the reference amylase-producing AAC strain (mother strain for M715 and M1052) and the nonproducing NC strain (background identical to that of AAC but harboring an empty CPOTud vector) (28). The improvement in amylase production was particularly significant during growth on glucose, where the fermentative metabolism happens. Compared to AAC results, the amylase yields on glucose increased 5.4-fold and 4.9-fold, respectively, for strains M715 and M1052 during the glucose phase (Fig. 1A) and increased 2.4-fold and 3.5-fold, respectively, at the end of the ethanol phase (Fig. 1B). The glycerol production rates for all amylase-producing strains (AAC, M715, and M1052) were higher than that seen with the NC strain, while the specific growth rates were lower (Table 2). Both phenomena are usually seen when recombinant proteins are produced in S. cerevisiae (7, 32). The reduced specific-growth rate might have come from shifting of amino acid use and the translational machinery to heterologous protein production rather than biomass formation, and the increased glycerol formation rate might have been a response to neutralize surplus NADH produced from amino acid biosynthesis (33). Synthesis of 1 mole of glycerol results in the consumption of 1 mole of NADH. The oxygen consumption rate for protein production is usually high (34). If the oxygen supply is insufficient during oxidative protein folding, the cell might behave anaerobically and produce glycerol as the only way to neutralize cytosolic NADH (35).
FIG 1.
Protein yield during batch fermentations. (A) Amylase yield in cell mass during the exponential phase. (B) Final amylase yield.
TABLE 2.
Physiological characterization of the strainsa
| Strain | Yield (g/g glucose) |
μmax(h−1)b | Final biomass (g/liter) | |||
|---|---|---|---|---|---|---|
| Biomass | Glycerol | Ethanol | Acetate | |||
| NC | 0.20 ± 0.02 | 0.05 ± 0.01 | 0.25 ± 0.02 | 0.033 ± 0.007 | 0.40 ± 0.01 | 6.5 ± 0.2 |
| AAC | 0.23 ± 0.02 | 0.15 ± 0.01 | 0.20 ± 0.01 | 0.034 ± 0.001 | 0.38 ± 0.01 | 6.7 ± 0.1 |
| M715 | 0.18 ± 0.01 | 0.15 ± 0.01 | 0.25 ± 0.02 | 0.036 ± 0.001 | 0.31 ± 0.01 | 6.6 ± 0.1 |
| M1052 | 0.17 ± 0.01 | 0.14 ± 0.01 | 0.28 ± 0.02 | 0.045 ± 0.005 | 0.24 ± 0.01 | 5.6 ± 0.1 |
Yield data calculated here only consider the exponential phase and the total consumed substrate. The data represent results from biological triplicates.
Specific growth rate.
To ensure that the improved amylase production was caused by mutations on the chromosome but not by the amylase expression plasmid, the plasmids from both mutant strains (M715 and M1052) were extracted and sequenced. No mutations were found on either plasmid. The extracted plasmids were then transformed into the standard empty strain (CEN.PK530-1C). Shake flask cultivations of these two strains showed no improvement in amylase production compared with the AAC strain (see Fig. S1A in the supplemental material), confirming that the improvements in amylase production in the mutant strains were caused by mutations in the host chromosome.
Genome-wide transcriptional responses in mutant strains M715 and M1052.
Genome-wide transcription analysis of the two mutant strains (M715 and M1052) and the two control strains (NC and AAC) was carried out during the exponential-growth phase on glucose. The expression of 63 genes changed significantly (false-discovery rate [FDR] < 0.05) in M715 in comparison to AAC, and the expression of 1,452 genes changed significantly (FDR < 0.05) in M1052 in comparison to to AAC (see Fig. S2A in the supplemental material). There were an additional 129 genes with expression changes that occurred in both strains. The transcriptional responses (including adjusted P values and log fold change) of all the genes are summarized in Dataset S1.
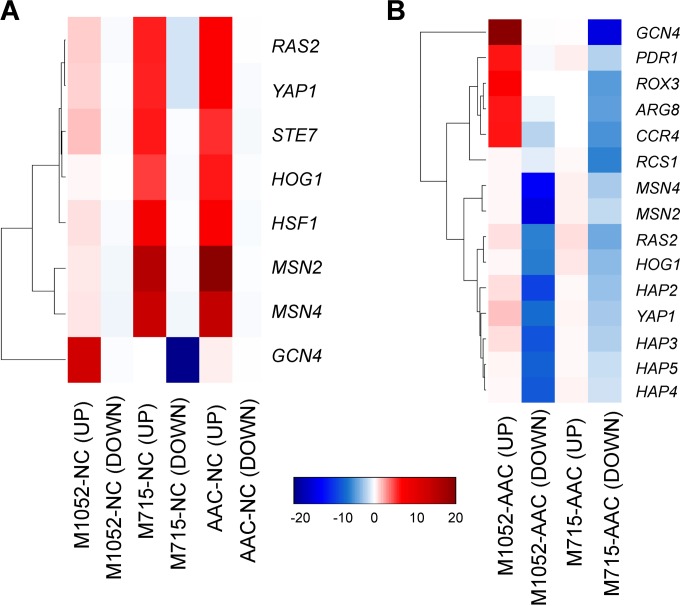
Cells can regulate their functions in many different ways, and the regulatory mechanisms are not always understood. One type of regulation is transcriptional regulation, where a transcription factor (TF) can regulate the transcription of its target genes. Potential TF-DNA regulatory interactions for S. cerevisiae have been identified (36, 37). In many cases, the TF gene itself does not change in expression but the expression of the regulatory target genes is altered as a response to a certain change in conditions. We therefore applied the reporter TF algorithm (31, 38) using the TF-DNA interaction network (37) to identify key TFs that could play an important role in the regulation of RPP. As shown in Fig. 2, most genes related to stress responses, such as genes regulated by oxidative stress (transcriptionally regulated by Yap1p), osmotic stress (regulated by Hog1p), and general stress (regulated by Msn2p and Msn4p), were upregulated in a RPP-dependent manner (as seen in comparisons of the amylase-producing AAC, M715, and M1052 strains to the nonproducing NC strain; Fig. 2A), indicating that increased protein production results in the induction of cellular stress responses. In contrast, the same classes of genes were downregulated in a mutation-dependent manner (as seen in comparisons of mutant strains M715 and M1052 to the nonmutated amylase-producing AAC strain; Fig. 2B), suggesting that the mutations existing in the mutant strains contributed to a reduction of cellular stress induced by RPP.
FIG 2.
Reporter TF analysis. (A) Many stress-related transcription factors were identified as key transcription factors in comparisons of mRNA levels of the amylase-producing strains with those of strain NC (reporter P value < 10−6). (B) The reporter TF results of comparisons of UV mutation strains with strain AAC. Red indicates that the genes regulated by this transcription factor were upregulated, and blue indicates they were downregulated. Reporter P value < 10−6. The color key shows significance levels from the reporter TF analysis [−log(reporter P value)]. Red indicates that the genes regulated by the specific TF were upregulated in comparisons of the transcriptomes of two strains, and blue indicates downregulation in transcription of the genes regulated by the TF.
Interestingly, genes regulated by Gcn4p, which regulates amino acid biosynthesis during amino acid starvation, were regulated differently in the two mutant strains; i.e., they were significantly upregulated in M1052 but downregulated in M715 in comparison to both NC and AAC. A strain producing recombinant protein at high levels has an increasing demand for amino acids. The upregulation of amino acid biosynthesis genes in M1052 may be due to a response to a certain level of amino acid starvation. Downregulation of amino acid biosynthesis has been noticed in HAC1 deletion strains producing recombinant amylase and an insulin precursor where the unfolded-protein-response (UPR)-ensured efficient protein folding was affected (34). We cannot deduce exactly why amino acid biosynthesis in M715 was downregulated, but one possibility could be that the increased amylase production in the glucose phase (Fig. 1A) caused perturbation in the UPR and thus generated responses similar to those observed in a HAC1 deletion strain.
Whole-genome sequencing of the M715 and M1052 mutant strains.
In order to identify random UV mutagenesis-generated mutations that led to higher amylase production, the genomes of the M715 and M1052 mutant amylase-producing strains and our wild-type CEN.PK113-7D strain (8) were sequenced by Illumina HiSeq (Materials and Methods; see also Table S1 in the supplemental material), aligned, and compared to the reference genome sequence of CEN.PK113-7D from a different study (24). In total, 1,713 putative mutations were identified in either of the two protein-producing strains. Compared to the reference sequence of CEN.PK113-7D (24), mutations that present in all three strains (M715, M1052, and our resequenced CEN.PK113-7D [8]) at the same position and with the same variant were considered to be background mutations found in the original host strain and were therefore filtered out in this first stage. This step reduced the number of unique mutations to 496 (the combined total for the two mutant strains), 328 of which were single-nucleotide point mutations and 84 of which were insertions or deletions (indels). Of 328 single-nucleotide point mutations, 271 were found in noncoding regions and the remaining 57 were found in coding regions. Of the 57 mutations in the coding regions, 1 was a nonsense mutation (with the codon changed to a STOP codon), 22 were same-sense mutations (with no change in the amino acid sequence in the protein), and 34 were missense mutations (with an introduction of an amino acid change in the protein). All mutations identified in the coding and upstream regions in all strains are presented in Datasets S2 to S4 in the supplemental material. Same-sense mutations and mutations in the upstream regions of genes that were not transcriptionally changed (FDR < 0.05) (i.e., the mutation did not alter the gene expression level) were filtered out in the next step. This resulted in the identification of mutations in 39 unique genes in the M715 strain and of mutations in 35 unique genes in the M1052 strain. The nonsynonymous single-nucleotide point mutations in the coding regions found in the mutation strains are summarized in Table 3.
TABLE 3.
Nonsynonymous SNVs found in the coding regions of strains M715 and M1052a
| Strain and ORF | Gene | SNV | Description |
|---|---|---|---|
| M715 | |||
| YBR018C | GAL7 | GAL7S177F | Galactose-1-phosphate uridyl transferase; galactose catabolism |
| YBR026C | ETR1 | ETR1E82G | 2-Enoyl thioester reductase; probable role in fatty acid synthesis |
| YBR120C | CBP6 | CBP6P133S | Mitochondrial translational activator of the COB mRNA; phosphorylated |
| YBR295W | PCA1 | PCA1S431L | Cadmium-transporting P-type ATPase; may have a role in copper and iron homeostasis |
| YKR105C | NA | “NA”K333N | NA |
| YDL194W | SNF3 | SNF3S97F | Plasma membrane low-glucose sensor that regulates glucose transport |
| YDR050C | TPI1 | TPI1N245D | Triose phosphate isomerase |
| YGR047C | TFC4 | TFC4L273F | Subunit of the RNA polymerase III transcription initiation factor complex (TFIIIC) |
| YGR083C | GCD2 | GCD2S2R | Delta subunit of the translation initiation factor eIF2B |
| YHL038C | CBP2 | CBP2M385T | Mitochondrial protein required for splicing of the group I intron aI5 of the COB pre-mRNA |
| YKL148C | SDH1 | SDH1S121F | Flavoprotein subunit of succinate dehydrogenase, which couples the oxidation of succinate to the transfer of electrons to ubiquinone as part of the TCA cycle and the mitochondrial respiratory chain |
| YKL087C | CYT2 | CYT2*54Y | Cytochrome c1 heme lyase; involved in maturation of cytochrome c1, which is a subunit of the mitochondrial ubiquinol-cytochrome c reductase |
| YLL051C | FRE6 | FRE6S421F | Putative ferric reductase with similarity to Fre2p |
| YMR076C | PDS5 | PDS5S289F | Protein required for establishment and maintenance of sister chromatid condensation and cohesion |
| YOR009W | TIR4 | TIR4S178P | Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins |
| YPL189W | GUP2 | GUP2*133K | Probable membrane protein with a possible role in proton symport of glycerol |
| M1052 | |||
| YAL016W | TPD3 | TPD3S138F | Regulatory subunit A of the heterotrimeric protein phosphatase 2A (PP2A) |
| YBL018C | POP8 | POP8H65Y | Subunit of both RNase MRP |
| YCR020C | PET18 | PET18A150V | Protein of unknown function |
| YDR472W | TRS31 | TRS31Q220H | One of 10 subunits of the transport protein particle (TRAPP) complex of the cis-Golgi which mediates vesicle docking and fusion |
| YDR495C | VPS3 | VPS3S521L | Component of CORVET complex |
| YDR505C | PSP1 | PSP1W761* | Asn- and Gln-rich protein of unknown function |
| YER176W | ECM32 | ECM32E530K | DNA-dependent ATPase/DNA helicase belonging to the Dna2p- and Nam7p-like family of helicases that is involved in modulating translation termination |
| YHL009W-B | NA | “NA”G539S | Retrotransposon TYA Gag and TYB Pol genes |
| YLR181C | VTA1 | VTA1S196I | Multivesicular body (MVB) protein involved in endosomal protein sorting |
| YML080W | DUS1 | DUS1S364L | Dihydrouridine synthase |
| YOR019W | NA | “NA” S693L | Protein of unknown function that may interact with ribosomes |
| YOR191W | ULS1 | ULS1K341N | RING finger protein involved in proteolytic control of sumoylated substrates |
| YPL009C | NA | “NA”E744K | Protein of unknown function |
ORF, open reading frame; TCA, tricarboxylic acid; CORVET, class C core vacuole/endosome tethering.
Variations in the upstream gene sequence might also be of interest since there could be mutations in a regulatory (transcriptional and translational) element. Even though no mutations were found in the coding regions of the stress-related genes in either strain, we found single-nucleotide point mutations in the upstream regions (0 to 1,000 bp) of several cellular stress-related genes (CCS1, SRX1, UBC4, and GPD1) in M1052 which may contribute to the downregulation of these genes (Fig. 2B). The availabilities of foldases, chaperones, redox-equivalents, glycans, trafficking vesicles, etc., are tuned in response to the expression level of the recombinant protein and the load with respect to the secretory pathway. Layers of quality control corresponding to protein folding and secretion are involved and must be well coordinated in order to avoid cellular stresses that may cause reductions of cell growth and protein secretion (41, 42) or even apoptosis and cell death (43). No mutations were found in stress-related genes in M715, suggesting that the stress response may be regulated by a mechanism different from that seen with the M1052 strain.
In addition to the stress-responsive genes, we also found single-point mutations in the coding (TRS31, VPS3, and VTA1) and upstream (VPS35) regions of several genes involved in protein trafficking in strain M1052. These mutations might be direct targets for enhancing protein secretion, as some studies demonstrated a good effect of overexpression of SNARE proteins, including those expressed by the SEC1 and SSO1 genes, to increase protein secretions in S. cerevisiae (7, 44).
Role of VTA1S196I in protein secretion.
To identify the effects of some mutations on amylase production, we selected a few gene targets in the secretory pathway found in M1052 and reconstructed single-nucleotide point mutations individually using site-directed mutagenesis (see Fig. S3 in the supplemental material), including TRS31Q220H (Trs31p; involved in endoplasmic reticulum [ER]-to-Golgi trafficking) and VTA1S196I (Vta1p, involved in the late endosome sorting in endocytosis). We found that TRS31Q220H did not influence amylase production (data not shown).
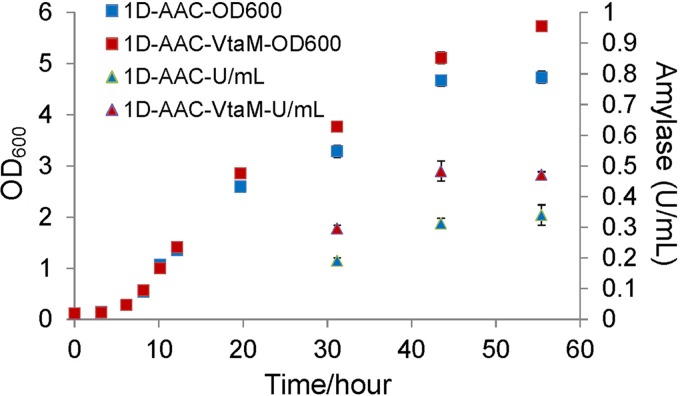
Vta1p, encoded by VTA1, is a regulatory protein which interacts with several vacuolar-protein-sorting proteins, including Vps4p, Vps60p, and Vps46p, in the multivesicular body (MVB) sorting pathway during late endosome sorting (45). The crystal structure of Vta1p has been intensively studied in recent years, with a special focus on its interaction with Vps4, an AAA ATPase that is required for dissociation of the endosomal sorting complexes required for transport (ESCRT) from endosomes (45–47). Secondary-structure prediction by circular-dichroism spectral analysis suggested that the linker region between Vta1NTD and Vta1CTD (residues 168 to 279) contains few regular ordered secondary structures (45). The linker region was recently reported to autoinhibit its regulation of the Vps4 ATP hydrolysis (48). From the genomic sequencing results, we found that the Ser196 in Vta1p has been changed to Ile in the M1052 strain (Table 3), which showed a significant enhancement of α-amylase yield compared to the AAC strain. For evaluation of this single-nucleotide point mutation, CEN.PK530-1D (49) was transformed with the pAlphaAmyCPOT α-amylase overexpression vector (28), giving rise to strain 1D-AAC, which was then used as the starting strain to introduce the point mutation (the only difference between 1D-AAC and AAC is that the URA3 gene has been deleted in 1D-AAC, as is used as a standard selective marker). After replacing the WT VTA1 with VTA1S196I1, the strain named 1D-AAC-VtaM was obtained. Compared to the 1D-AAC strain, the 1D-AAC-VtaM strain showed a 35% improvement in amylase production and a 21% increase in cell growth when cultivated in YPD medium (Fig. 3). Low levels of glucose are required for SUC2 transcription (50). Yeast periplasmic invertase (encoded by SUC2) production in the same medium (YPD) was also increased (14%) in the 1D-AAC-VtaM strain (see Fig. S4 in the supplemental material), suggesting that the efficiency of the secretory pathway in the 1D-AAC-VtaM strain had generally increased.
FIG 3.
Effect of the Vta1S196I mutation on α-amylase production in shake flask fermentation.
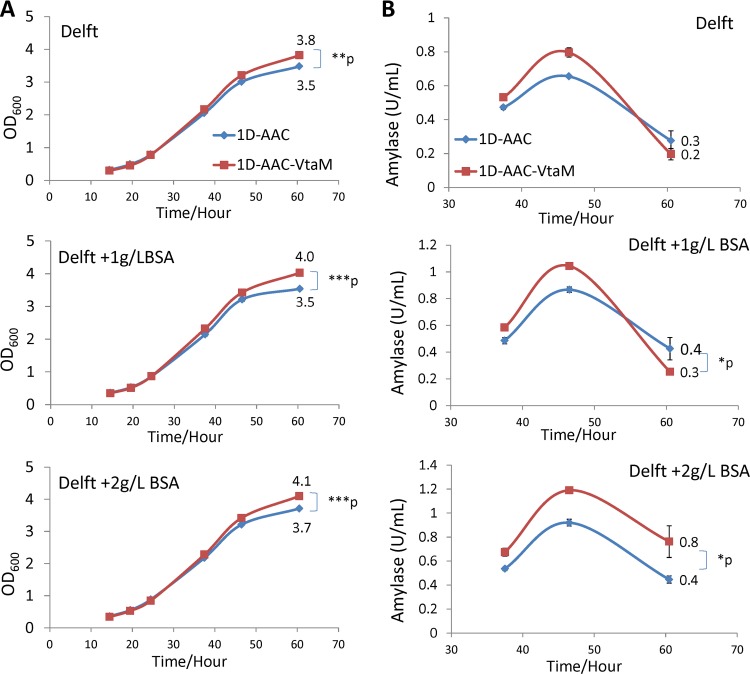
To understand what effect the VTA1S196I mutation may have on endocytosis, we added different concentrations of bovine serum albumin (BSA) into the Delft medium, which lacks peptone and nutrient-rich yeast nitrogen source compared to the YPD medium. Both cell growth and the production of amylase were monitored. As shown in Fig. 4A, strains 1D-AAC and 1D-AAC-VtaM grew (as indicated by optical density at 600 nm [OD600]) in all three media (Delft, Delft–1 g/liter BSA, and Delft–2 g/liter BSA) following the same trend as seen in the YPD medium (Fig. 3) in that 1D-AAC-VtaM grew slightly faster than 1D-AAC. In media with no BSA or a lower concentration of BSA (1 g/liter) (Fig. 4B), amylase production in the two strains exhibited trends similar to those in the YPD medium (Fig. 3) in that 1D-AAC-VtaM produced more amylase than 1D-AAC until up to around 55 h. It has been reported previously that yeast can take up amylase through endocytosis (51, 52). After that point (55 h), the main nutrients supporting cell growth as well as amylase production, the carbon source (glucose) and the added BSA, were depleted, so it was possible that the yeast cells started taking up the produced amylase through endocytosis. It is likely that the lower amylase activity measured in 1D-AAC-VtaM than in 1D-AAC after the 55 h was due to the fact that endocytosis is more efficient in this strain under starvation conditions (no and 1 g/liter BSA). With more BSA (2 g/liter) present, amylase uptake was reduced, as indicated by the higher amylase amount (0.8 U/ml) in 1D-AAC-VtaM than 1D-AAC (0.4 U/ml) after 60 h (Fig. 4B). This experiment suggests that the endocytosis process is enhanced by the VTA1S196I point mutation in starvation. There is another possibility: that proteolysis caused the reductions of amylase activity. S. cerevisiae naturally produces very small amount of extracellular proteins, including proteases. We have observed that the amylase activity is rather stable in yeast culture supernatants at room temperatures. The basal expression of protease(s) should be at similar levels in 1D-AAC and 1D-AAC-VtaM, as the only difference between these two strains is the VTA1S196I mutation. If the presence of more BSA prevented protease degradation with amylase, there should have been more amylase left in the Delft–2 g/liter BSA medium than in the Delft–1 g/liter BSA medium for both strains. However, we saw this phenomenon only in strain 1D-AAC-VtaM (0.3 U/liter in Delft–1 g/liter BSA and 0.8 U/liter in Delft–2 g/liter BSA). The amylase activity levels remaining in 1D-AAC were similar (0.4 U/liter) in Delft–1 g/liter BSA and Delft–2 g/liter BSA. Based on this analysis, we are confident that the reduction in amylase activity mainly results from endocytosis and not proteolysis.
FIG 4.
Effect of the Vta1S196I mutation on amylase production in Delft medium supplemented with BSA. (A) Growth profiles. (B) Amylase production levels.
The relevance of the VTA1S196I point mutation to boost endocytosis could lie in the structural stabilization of this protein. There are a series of neutral or polar residues (Asp192, His193, Gln194, Thr195, Ser196, and Asp197) in the linker region of Vta1p. Mutation from a polar uncharged serine to a nonpolar hydrophobic isoleucine could change the linker structure and create a more favorable configuration for Vta1p's interaction with the client proteins, especially Vps4p, therefore enhancing the efficiency of the late endosome sorting. In contrast to the exocytic pathway where proteins and lipids are carried from the ER through Golgi trafficking and eventually to the plasma membrane, the endocytic pathway internalizes cargo from the environment, or the plasma membrane, through a set of endosomes to the vacuole for degradation (53). A faster recycling of the nutrients through subsequent degradation in the vacuole (53) is hence expected for amino acid biosynthesis (for protein) as well as for cell growth as seen in Fig. 3. A group of genes related to protein secretion, including SEC72 in ER translocation, ERO1 in protein folding, ERI1 and BST1 in glycosylphosphatidylinositol (GPI) biosynthesis and anchoring, TRS20, SFB2, TRS120, and SFB3 in ER-to-Golgi vesicle trafficking, SEC3, SEC8, and EXO84 as low-density secretory vesicles, etc. (identified in reference 54), were found upregulated in M1052. The upregulation of these genes indicates that the secretory pathway in M1052 was more actively turned on, and VTA1S196I may have contributed to this process through the hypothesis discussed above. Several genes (VPS20, VPS33, and PEP5) involved in the late endosome-to-vacuole transport were also significantly upregulated, further supporting our hypothesis.
With the advances in systems biology, it has become convenient to identify gene targets from strains developed by random mutagenesis or evolution. Whole-genome sequencing enables identification of nonsynonymous variations that directly change the amino acid and may subsequently affect the expression level or the activity of the protein. Transcriptome analysis is usually applied in combination with other approaches, such as proteomics (55), genomics and intracellular metabolite measurements (8, 56), etc., to identify targets for strain improvement. The transcriptome is very sensitive, and a global response may be triggered by a variety of factors, such as changes to the cultivation conditions or the expression level of a transcription factor. In many cases, transcriptional change does not necessarily affect the protein expression level and vice versa. In our study, the Vta1p function may have been affected by the S196I mutation; however, the gene transcriptional level per se was not changed and therefore the functional mutation could not have been detected by transcriptome analysis alone.
In conclusion, we demonstrated how random mutagenesis and screening enabled us to identify strains with improved production of recombinant protein. We further demonstrated how a combination of systems biology characterization and genome sequencing enabled us to identify key mechanisms altered in the improved strains. Finally, we identified from the genome-sequencing analysis a novel target for improvement of heterologous protein production by S. cerevisiae, namely, S196I in VTA1. By introducing a single-nucleotide point mutation in this gene, we could improve production of amylase by 35%, most likely due to increased endocytosis resulting from the mutation in this gene. We found not only that the secretion of amylase is increased but also that invertase, a protein naturally secreted by yeast, has increased secretion. We are hereby suggesting that our approach demonstrated here may be used for further improvement of recombinant protein production by yeast and not only that better production strains can be obtained in the future but that our approach may also result in new insight into the complex secretory pathway of yeast.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by European Research Council ERC project INSYSBIO (grant no. 247013), the Novo Nordisk Foundation, and the Chalmers Foundation.
Footnotes
Published ahead of print 27 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00712-14.
REFERENCES
- 1.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- 2.Petranovic D, Tyo K, Vemuri GN, Nielsen J. 2010. Prospects of yeast systems biology for human health: integrating lipid, protein and energy metabolism. FEMS Yeast Res. 10:1046–1059. 10.1111/j.1567-1364.2010.00689.x [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Tyo KEJ, Martínez JL, Petranovic D, Nielsen J. 2012. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol. Bioeng. 10.1002/bit.24409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porro D, Sauer M, Branduardi P, Mattanovich D. 2005. Recombinant protein production in yeasts. Mol. Biotechnol. 31:245–259. 10.1385/MB:31:3:245 [DOI] [PubMed] [Google Scholar]
- 5.Schröder M. 2007. The cellular response to protein unfolding stress, p 117–139 Exploitation of fungi, vol 26 Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 6.Idiris A, Tohda H, Kumagai H, Takegawa K. 2010. Engineering of protein secretion in yeast: strategies and impact on protein production. Appl. Microbiol. Biotechnol. 86:403–417. 10.1007/s00253-010-2447-0 [DOI] [PubMed] [Google Scholar]
- 7.Hou J, Tyo K, Liu Z, Petranovic D, Nielsen J. 2012. Engineering of vesicle trafficking improves heterologous protein secretion in Saccharomyces cerevisiae. Metab. Eng. 14:120–127. 10.1016/j.ymben.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 8.Hong KK, Vongsangnak W, Vemuri GN, Nielsen J. 2011. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc. Natl. Acad. Sci. U. S. A. 108:12179–12184. 10.1073/pnas.1103219108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durnev AD. 2008. Methodological aspects of studies of chemical mutagenesis modification. Bull. Exp. Biol. Med. 146:307–312. 10.1007/s10517-008-0273-5 [DOI] [PubMed] [Google Scholar]
- 10.Schroder LA, Glick BS, Dunn WA., Jr 2007. Identification of pexophagy genes by restriction enzyme-mediated integration, p 203–218 Pichia protocols. Springer, New York, NY: [DOI] [PubMed] [Google Scholar]
- 11.Larsen S, Weaver J, de Sa Campos K, Bulahan R, Nguyen J, Grove H, Huang A, Low L, Tran N, Gomez S. 2013. Mutant strains of Pichia pastoris with enhanced secretion of recombinant proteins. Biotechnol. Lett. 35:1925–1935. 10.1007/s10529-013-1290-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanjou N, Nagao A, Ohmiya Y, Ohgiya S. 2007. Yeast mutant with efficient secretion identified by a novel secretory reporter, Cluc. Biochem. Biophys. Res. Commun. 358:429–434. 10.1016/j.bbrc.2007.04.140 [DOI] [PubMed] [Google Scholar]
- 13.Desai TA, Rodionov DA, Gelfand MS, Alm EJ, Rao CV. 2009. Engineering transcription factors with novel DNA-binding specificity using comparative genomics. Nucleic Acids Res. 37:2493–2503. 10.1093/nar/gkp079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. 2009. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 10:252–263. 10.1038/nrg2538 [DOI] [PubMed] [Google Scholar]
- 15.Payne T, Finnis C, Evans LR, Mead DJ, Avery SV, Archer DB, Sleep D. 2008. Modulation of chaperone gene expression in mutagenized Saccharomyces cerevisiae strains developed for recombinant human albumin production results in increased production of multiple heterologous proteins. Appl. Environ. Microbiol. 74:7759–7766. 10.1128/AEM.01178-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. 1999. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J. Mol. Biol. 292:949–956. 10.1006/jmbi.1999.3130 [DOI] [PubMed] [Google Scholar]
- 17.Wentz AE, Shusta EV. 2007. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl. Environ. Microbiol. 73:1189–1198. 10.1128/AEM.02427-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gai SA, Wittrup KD. 2007. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 17:467–473. 10.1016/j.sbi.2007.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wentz AE, Shusta EV. 2008. Enhanced secretion of heterologous proteins from yeast by overexpression of ribosomal subunit RPP0. Biotechnol. Prog. 24:748–756. 10.1021/bp070345m [DOI] [PubMed] [Google Scholar]
- 20.Gross E, Kastner DB, Kaiser CA, Fass D. 2004. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell 117:601–610. 10.1016/S0092-8674(04)00418-0 [DOI] [PubMed] [Google Scholar]
- 21.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517. 10.1002/yea.320080703 [DOI] [PubMed] [Google Scholar]
- 22.Ruohonen L, Toikkanen J, Outola M, Soderlund H, Keranen S. 1997. Enhancement of protein secretion in Saccharomyces cerevisiae by overproduction of Sso protein, a late-acting component of the secretory machinery. Yeast 13:337–351 [DOI] [PubMed] [Google Scholar]
- 23.Väremo L, Nielsen J, Nookaew I. 2013. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 41:4378–4391. 10.1093/nar/gkt111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nijkamp JF, van den Broek M, Datema E, de Kok S, Bosman L, Luttik MA, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne WHM, Klaassen P, Paddon CJ, Platt D, Kötter P, van Ham RC, Reinders MJ, Pronk JT, de Ridder D, Daran J-M. 2012. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK 113–7D, a model for modern industrial biotechnology. Microb. Cell Fact. 11:36. 10.1186/1475-2859-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21:936–939. 10.1101/gr.111120.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y, Wan Z, Coarfa C, Drabek R, Chen L, Ostrowski EA, Liu Y, Weinstock GM, Wheeler DA, Gibbs RA. 2010. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 20:273–280. 10.1101/gr.096388.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, Milosavljevic A, Gibbs RA, Yu F. 2012. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics 13:8. 10.1186/1471-2105-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Tyo KE, Martínez JL, Petranovic D, Nielsen J. 2012. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol. Bioeng. 109:1259–1268. 10.1002/bit.24409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlsen M, Jochumsen KV, Emborg C, Nielsen J. 1997. Modeling the growth and proteinase A production in continuous cultures of recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 55:447–454 [DOI] [PubMed] [Google Scholar]
- 30.Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. 2011. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 7:e1002202. 10.1371/journal.pgen.1002202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patil KR, Nielsen J. 2005. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. U. S. A. 102:2685–2689. 10.1073/pnas.0406811102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, Martínez JL, Liu Z, Petranovic D, Nielsen J. 2014. Balanced globin protein expression and heme biosynthesis improve production of human hemoglobin in Saccharomyces cerevisiae. Metab. Eng. 21:9–16. 10.1016/j.ymben.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 33.Rigoulet M, Aguilaniu H, Avéret N, Bunoust O, Camougrand N, Grandier-Vazeille X, Larsson C, Pahlman I-L, Manon S, Gustafsson L. 2004. Organization and regulation of the cytosolic NADH metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biochem. 256:73–81. 10.1023/B:MCBI.0000009888.79484.fd [DOI] [PubMed] [Google Scholar]
- 34.Tyo KEJ, Liu Z, Petranovic D, Nielsen J. 2012. Imbalance of heterologous protein folding and disulfide bond formation rates yields runaway oxidative stress. BMC Biol. 10:16. 10.1186/1741-7007-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overkamp KM, Bakker BM, Kötter P, Luttik MAH, Van Dijken JP, Pronk JT. 2002. Metabolic engineering of glycerol production in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 68:2814–2821. 10.1128/AEM.68.6.2814-2821.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799–804. 10.1126/science.1075090 [DOI] [PubMed] [Google Scholar]
- 37.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne J-B, Reynolds DB, Yoo J. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104. 10.1038/nature02800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira AP, Patil KR, Nielsen J. 2008. Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst. Biol. 2:17. 10.1186/1752-0509-2-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reference deleted.
- 40. Reference deleted.
- 41.Nemecek S, Marisch K, Juric R, Bayer K. 2008. Design of transcriptional fusions of stress sensitive promoters and GFP to monitor the overburden of Escherichia coli hosts during recombinant protein production. Bioprocess Biosyst. Eng. 31:47–53. 10.1007/s00449-007-0143-y [DOI] [PubMed] [Google Scholar]
- 42.Dürrschmid K, Reischer H, Schmidt-Heck W, Hrebicek T, Guthke R, Rizzi A, Bayer K. 2008. Monitoring of transcriptome and proteome profiles to investigate the cellular response of E. coli towards recombinant protein expression under defined chemostat conditions. J. Biotechnol. 135:34–44. 10.1016/j.jbiotec.2008.02.013 [DOI] [PubMed] [Google Scholar]
- 43.Mattanovich D, Gasser B, Hohenblum H, Sauer M. 2004. Stress in recombinant protein producing yeasts. J. Biotechnol. 113:121–135. 10.1016/j.jbiotec.2004.04.035 [DOI] [PubMed] [Google Scholar]
- 44.Toikkanen JH, Sundqvist L, Keränen S. 2004. Kluyveromyces lactis SSO1 and SEB1 genes are functional in Saccharomyces cerevisiae and enhance production of secreted proteins when overexpressed. Yeast 21:1045–1055. 10.1002/yea.1151 [DOI] [PubMed] [Google Scholar]
- 45.Xiao J, Xia H, Zhou J, Azmi IF, Davies BA, Katzmann DJ, Xu Z. 2008. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev. Cell 14:37–49. 10.1016/j.devcel.2007.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang D, Hurley JH. 2010. Structural role of the Vps4-Vta1 interface in ESCRT-III recycling. Structure 18:976–984. 10.1016/j.str.2010.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Vild C, Ju J, Zhang X, Liu J, Shen J, Zhao B, Lan W, Gong F, Liu M. 2012. Structural basis of molecular recognition between ESCRT-III-like protein Vps60 and AAA-ATPase regulator Vta1 in the multivesicular body pathway. J. Biol. Chem. 287:43899–43908. 10.1074/jbc.M112.390724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norgan AP, Davies BA, Azmi IF, Schroeder AS, Payne JA, Lynch GM, Xu Z, Katzmann DJ. 2013. Relief of autoinhibition enhances Vta1 activation of Vps4 via the Vps4 stimulatory element. J. Biol. Chem. 288:26147–26156. 10.1074/jbc.M113.494112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou J, Osterlund T, Liu Z, Petranovic D, Nielsen J. 2012. Heat shock response improves heterologous protein secretion in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 10.1007/s00253-012-4596-9 [DOI] [PubMed] [Google Scholar]
- 50.Ozcan S, Vallier LG, Flick JS, Carlson M, Johnston M. 1997. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast 13:127–137 [DOI] [PubMed] [Google Scholar]
- 51.Makarow M. 1985. Endocytosis in Saccharomyces cerevisiae: internalization of alpha-amylase and fluorescent dextran into cells. EMBO J. 4:1861–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tyo K, Liu Z, Magnusson Y, Petranovic D, Nielsen J. 2014. Protein uptake and degradation: impact on recombinant secretion in yeast. Appl. Microbiol. Biotechnol. 10.1007/s00253-014-5783-7 [DOI] [PubMed] [Google Scholar]
- 53.Segev N. 2009. Trafficking inside cells: pathways, mechanisms, and regulation. Springer, New York, NY [Google Scholar]
- 54.Feizi A, Österlund T, Petranovic D, Bordel S, Nielsen J. 2013. Genome-scale modeling of the protein secretory machinery in yeast. PLoS One 8:e63284. 10.1371/journal.pone.0063284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J-H, Lee D-E, Lee B-U, Kim H-S. 2003. Global analyses of transcriptomes and proteomes of a parent strain and an L-threonine-overproducing mutant strain. J. Bacteriol. 185:5442–5451. 10.1128/JB.185.18.5442-5451.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi S, Chen T, Zhang Z, Chen X, Zhao X. 2009. Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production. Metab. Eng. 11:243–252. 10.1016/j.ymben.2009.05.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.