Abstract
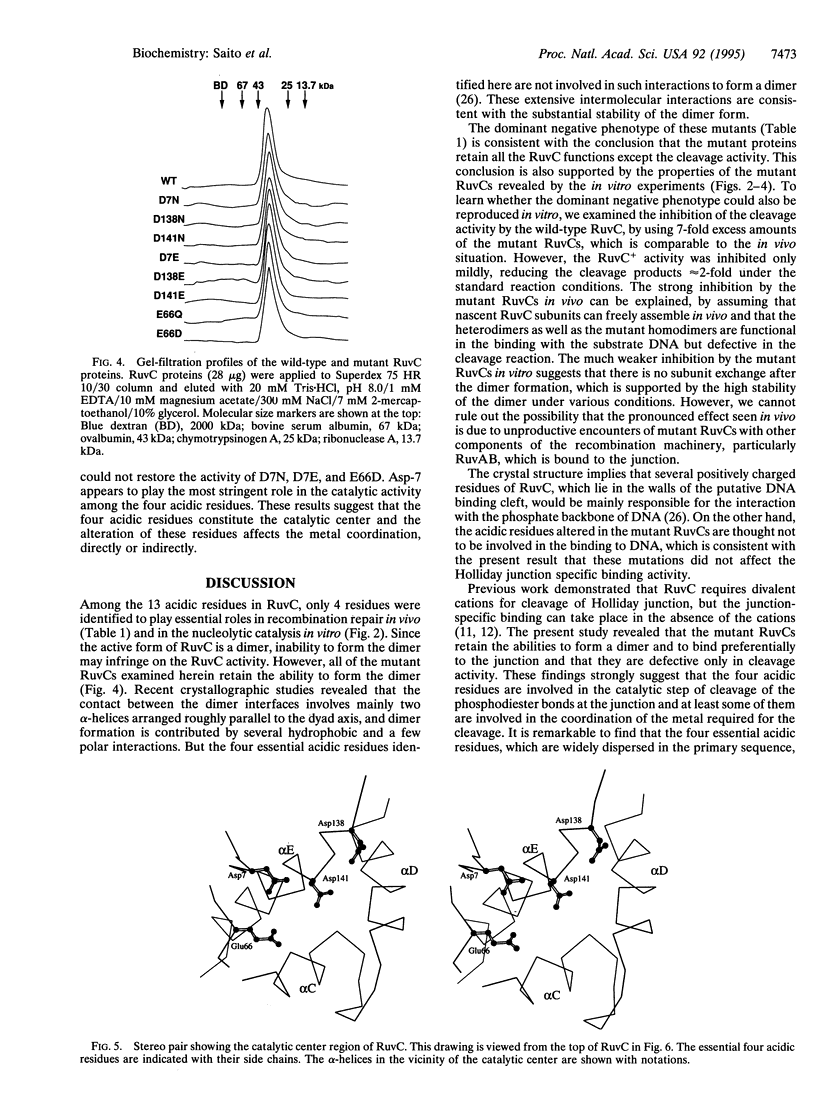
Escherichia coli RuvC protein is a specific endonuclease that resolves Holliday junctions during homologous recombination. Since the endonucleolytic activity of RuvC requires a divalent cation and since 3 or 4 acidic residues constitute the catalytic centers of several nucleases that require a divalent cation for the catalytic activity, we examined whether any of the acidic residues of RuvC were required for the nucleolytic activity. By site-directed mutagenesis, we constructed a series of ruvC mutant genes with similar amino acid replacements in 1 of the 13 acidic residues. Among them, the mutant genes with an alteration at Asp-7, Glu-66, Asp-138, or Asp-141 could not complement UV sensitivity of a ruvC deletion strain, and the multicopy mutant genes showed a dominant negative phenotype when introduced into a wild-type strain. The products of these mutant genes were purified and their biochemical properties were studied. All of them retained the ability to form a dimer and to bind specifically to a synthetic Holliday junction. However, they showed no, or extremely reduced, endonuclease activity specific for the junction. These 4 acidic residues, which are dispersed in the primary sequence, are located in close proximity at the bottom of the putative DNA binding cleft in the three-dimensional structure. From these results, we propose that these 4 acidic residues constitute the catalytic center for the Holliday junction resolvase and that some of them play a role in coordinating a divalent metal ion in the active center.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ariyoshi M., Vassylyev D. G., Iwasaki H., Nakamura H., Shinagawa H., Morikawa K. Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli. Cell. 1994 Sep 23;78(6):1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Luo L. Identification of residues in the Mu transposase essential for catalysis. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6654–6658. doi: 10.1073/pnas.91.14.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Dunderdale H. J., West S. C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993 Sep 24;74(6):1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- Benson F., Collier S., Lloyd R. G. Evidence of abortive recombination in ruv mutants of Escherichia coli K12. Mol Gen Genet. 1991 Feb;225(2):266–272. doi: 10.1007/BF00269858. [DOI] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Dunderdale H. J., Benson F. E., Parsons C. A., Sharples G. J., Lloyd R. G., West S. C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991 Dec 19;354(6354):506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- Dyda F., Hickman A. B., Jenkins T. M., Engelman A., Craigie R., Davies D. R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994 Dec 23;266(5193):1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Nakata A., Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992 Nov;6(11):2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991 Dec;10(13):4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., Steitz T. A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- Kanaya S., Kohara A., Miura Y., Sekiguchi A., Iwai S., Inoue H., Ohtsuka E., Ikehara M. Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J Biol Chem. 1990 Mar 15;265(8):4615–4621. [PubMed] [Google Scholar]
- Katayanagi K., Ishikawa M., Okumura M., Ariyoshi M., Kanaya S., Kawano Y., Suzuki M., Tanaka I., Morikawa K. Crystal structures of ribonuclease HI active site mutants from Escherichia coli. J Biol Chem. 1993 Oct 15;268(29):22092–22099. [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Nakamura H., Ikehara M., Matsuzaki T., Morikawa K. Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution. J Mol Biol. 1992 Feb 20;223(4):1029–1052. doi: 10.1016/0022-2836(92)90260-q. [DOI] [PubMed] [Google Scholar]
- Katayanagi K., Okumura M., Morikawa K. Crystal structure of Escherichia coli RNase HI in complex with Mg2+ at 2.8 A resolution: proof for a single Mg(2+)-binding site. Proteins. 1993 Dec;17(4):337–346. doi: 10.1002/prot.340170402. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994 Sep;58(3):401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Eggleston A. K. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993 Jan;12(1):17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi V., Usdin M. T., Harington A., Dudding L. R. Site-directed mutagenesis of the conserved Asp-443 and Asp-498 carboxy-terminal residues of HIV-1 reverse transcriptase. Nucleic Acids Res. 1990 Sep 25;18(18):5359–5363. doi: 10.1093/nar/18.18.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C. B., Thaler D. S., Dahlquist F. W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989 May;171(5):2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz O., Cromme F. V., Grüninger-Leitch F., Le Grice S. F. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989 Nov 6;257(2):311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Takahagi M., Iwasaki H., Shinagawa H. Structural requirements of substrate DNA for binding to and cleavage by RuvC, a Holliday junction resolvase. J Biol Chem. 1994 May 27;269(21):15132–15139. [PubMed] [Google Scholar]
- Tsaneva I. R., Müller B., West S. C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992 Jun 26;69(7):1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- Whitby M. C., Ryder L., Lloyd R. G. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993 Oct 22;75(2):341–350. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]