Abstract
Recently, iso-diabolic acid (13,16-dimethyl octacosanedioic acid) has been identified as a major membrane-spanning lipid of subdivisions 1 and 3 of the Acidobacteria, a highly diverse phylum within the Bacteria. This finding pointed to the Acidobacteria as a potential source for the bacterial glycerol dialkyl glycerol tetraethers that occur ubiquitously in peat, soil, lakes, and hot springs. Here, we examined the lipid composition of seven phylogenetically divergent strains of subdivision 4 of the Acidobacteria, a bacterial group that is commonly encountered in soil. Acid hydrolysis of total cell material released iso-diabolic acid derivatives in substantial quantities (11 to 48% of all fatty acids). In contrast to subdivisions 1 and 3 of the Acidobacteria, 6 out of the 7 species of subdivision 4 (excepting “Candidatus Chloracidobacterium thermophilum”) contained iso-diabolic acid ether bound to a glycerol in larger fractional abundance than iso-diabolic acid itself. This is in agreement with the analysis of intact polar lipids (IPLs) by high-performance liquid chromatography-mass spectrometry (HPLC-MS), which showed the dominance of mixed ether-ester glycerides. iso-Diabolic acid-containing IPLs were not identified, because these IPLs are not released with a Bligh-Dyer extraction, as observed before when studying lipid compositions of subdivisions 1 and 3 of the Acidobacteria. The presence of ether bonds in the membrane lipids does not seem to be an adaptation to temperature, because the five mesophilic isolates contained a larger amount of ether lipids than the thermophile “Ca. Chloracidobacterium thermophilum.” Furthermore, experiments with Pyrinomonas methylaliphatogenes did not reveal a major influence of growth temperature over the 50 to 69°C range.
INTRODUCTION
Isoprenoidal ether lipids ubiquitously occur in the membrane lipids of Archaea (1), but occasionally ether lipids also are detected in the bacterial domain, albeit with nonisoprenoidal chains (2, 3). Unusual glycerol dialkyl glycerol tetraethers (GDGTs) with n-alkyl chains containing 2-3 methyl groups instead of isoprenoidal chains (so-called branched GDGTs [brGDGTs]; e.g., structures 1 and 2 in Fig. 1) were identified for the first time in peat more than a decade ago (4) and subsequently turned out to occur ubiquitously in soil, peat, lake water and sediments, river water and sediments, and coastal marine sediments (5). brGDGTs also have been observed in thermophilic environments, such as terrestrial hot springs (6), where they are believed to be produced in situ by thermophilic bacteria (7, 8). Despite their widespread occurrence and potential applications in geochemistry and paleoclimatology (5), their microbial source still is unclear. The assessment of the stereochemistry of the glycerol units in brGDGTs revealed that it is the opposite of that of archaeal isoprenoidal GDGTs, suggesting that they must derive from Bacteria (9). A heterotrophic lifestyle of the source organism(s) of brGDGTs was suggested based on their natural stable carbon isotopic composition in peat (10) and soil (11) and natural labeling experiments (11, 12). The environmental abundance of Acidobacteria has led to the suggestion that these bacteria are the biological source of the brGDGTs (13). This hypothesis was recently supported by membrane lipid analysis of 13 species of subdivisions (SD) 1 and 3 of the Acidobacteria, which showed that the uncommon membrane-spanning lipid, 13,16-dimethyl octacosanedioic acid (iso-diabolic acid), is a major lipid in all species studied (14). This lipid can be considered a building block of the brGDGTs but occurs in predominantly ester- and not ether-bound form in SD 1 and 3 Acidobacteria. In 3 of the 13 analyzed strains, small amounts of ether-bound iso-diabolic acid, including brGDGT 1, were detected after hydrolysis of the cells. However, the brGDGT distribution in soils is much more complex, and the presence of additional (acido)bacteria might explain the presence of the full complement of brGDGTs in the environment.
FIG 1.
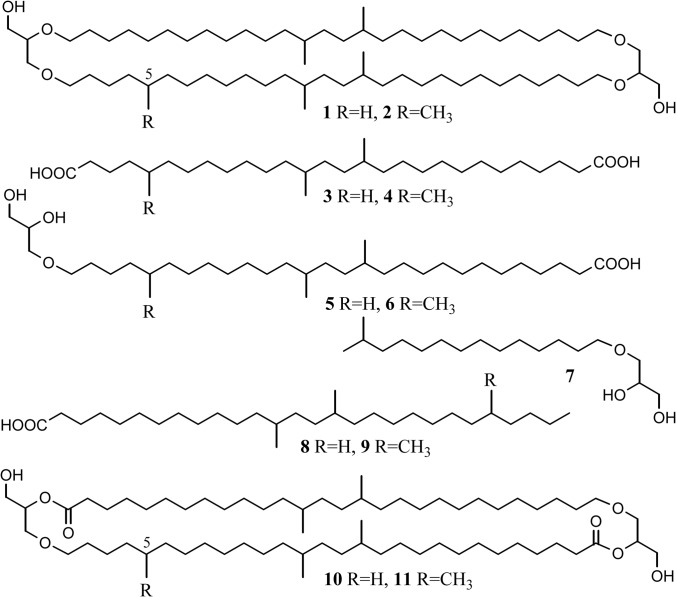
Structures of lipids mentioned in the text. Structures 1 and 2 are brGDGTs ubiquitously occurring in the environment. Structures 3 and 4 are iso-diabolic acids. Structures 5 and 6 are iso-diabolic acids ether bound to a glycerol moiety at the sn1 position. Structure 7 is a C15 iso fatty acid ether bound to a glycerol moiety at the sn1 position. Structures 8 and 9 are derivatives of iso-diabolic acids 3 and 4 where one of the carboxylic groups is reduced. Structures 10 and 11 represent hypothetical structures showing the core of the membrane-spanning lipids of the SD 4 Acidobacteria based on the results reported in this paper.
Acidobacteria are a highly abundant and diverse phylum of the domain Bacteria (15–20). For example, a recent study of bacterial abundance of peat layers of a Siberian wetland using pyrosequencing of 16S rRNA genes revealed that 35 to 40% of the reads were from Acidobacteria (21). Using similar methods, the abundance of Acidobacteria in organic matter-rich, low-pH soils was reported to be over 60% (22). Because known whole genomes of Acidobacteria contain only one copy of the 16S rRNA gene, in contrast to many other bacteria, their abundance may even be underestimated by these methods (23). The Acidobacteria have been divided into 26 SD, based mainly on environmental sequences (24), but only six of these contain taxonomically characterized representatives. For SD 1, eight genera have been defined, Acidobacterium (25), Acidicapsa (26), “Acidipila” (27), Bryocella (28), Edaphobacter (29), Granulicella (30, 31), Telmatobacter (32), and Terriglobus (33, 34), while only 1 to 3 genera have been characterized for SD 3 (Bryobacter [35]), 8 (Holophaga [36], Geothrix [37] and Acanthopleuribacter [38]), 10 (Thermotomaculum [39]), and 23 (Thermoanaerobaculum [40]). For SD 4, the number of known genera recently has been expanded. Four genera now have been defined. The thermophilic “Ca. Chloracidobacterium thermophilum” was enriched from a hot spring and represents the first phototrophic acidobacterium (41). Blastocatella fastidosa, an aerobic chemoorganoheterotroph (42), and two Aridibacter species (43) were isolated from semiarid savannah soils. The thermophile Pyrinomonas methylaliphatogenes was isolated from a geothermally heated soil and possesses a chemoheterotrophic and obligately aerobic metabolism (44). Molecular ecological studies based on 16S rRNA genes have indicated that, in wetlands, the most abundant Acidobacteria members fall in SD 1 and 3 (21), whereas in lakes SD 1, 6, and 7 thrive (45). In soils, SD 1 to 4 and 6 are the most dominant, with SD 4 contributing, on average, 20 to 30% of total Acidobacteria depending on the method used (i.e., clone libraries or pyrosequencing) (19). In contrast to most other SDs, the relative abundance of SD 4 increased with increasing soil pH, and at pHs above 7, 16S rRNA sequences derived from members of this SD typically represent more than half of all acidobacterial sequences (19). Thus, the lipids produced by Acidobacteria of SD 4 may form a major source of the unusual ether lipids in soil. Here, we describe in detail the lipid composition of five previously classified bacteria and two newly isolated strains, all belonging to the Acidobacteria SD 4, and discuss their distributions.
MATERIALS AND METHODS
Cultures.
The acidobacterial strains used in this study are listed in Table 1. Blastocatella fastidiosa A2_16T, Aridibacter famidurans A22_HD_4HT, Aridibacter kavangonensis Ac_23_E3T, and two other acidobacterial strains from semiarid soils from Namibia were grown at the DSMZ at 28°C by moderate shaking for 9 to 14 days, depending on the strain. All strains were grown in liquid SSE-HD (1:10) medium that was based on a soil solution equivalent (SSE) (46) with an increased iron content and supplemented with 0.25 g liter−1 yeast extract (Difco Laboratories Inc., Detroit, MI), 0.5 g liter−1 of peptone (Difco), 0.1 g liter−1 glucose (Sigma-Aldrich, Steinheim, Germany), 0.1 ml liter−1 10 vitamin solution (47), and 1 ml liter−1 trace element solution SL 10 (48). Ten mM 2-(4-morpholino)ethanesulfonic acid (MES; Sigma) or 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Sigma) was used to buffer the medium at pH 5.5 (B. fastidiosa strain Ac_28_D10T) or 6.5 (Aridibacter famidurans and A. kavangonensis strain Ac_23_E3T), respectively. Biomass was harvested by centrifugation (9,000 × g, 30 min; Avanti-J26 XPI; Beckman Coulter), frozen (−20°C overnight), and lyophilized (0.05 mbar at −30°C).
TABLE 1.
Acidobacteria of SD 4 used in this study
| Species | Origin | Substrates used | Temp (°C) |
pH |
Reference | ||
|---|---|---|---|---|---|---|---|
| Range | Optimal | Range | Optimal | ||||
| Blastocatella fastidiosa A2_16T (DSM 25172T) | Pastureland soil, Erichsfelde, central Namibia | Complex protein substrates, protocatechuateb | 14–40 | 29–35 | 4.0–10.0 | 5.0–7.5 | 42 |
| Aridibacter famidurans A22_HD_4HT (DSM 26555T) | Pastureland soil, Erichsfelde, central Namibia | Complex protein substrates, protocatechuate, N-acetylgalactosamine, rhamnose, xyloseb | 15–44 | 24–36 | 4.0–9.0 | 5.5–9.0 | 43 |
| Aridibacter kavangonensis Ac_23_E3T (DSM 26558T) | Fallow soil, Mashare, northern Namibia | Complex protein substrates, protocatechuate, N-acetylgalactosamine, maltose, rhamnose, fumarate, isovalerate, laminarinb | 12–44 | 36–44 | 3.5–10.0 | 5.5–8.0 | 43 |
| Unclassified Acidobacteria bacterium Ac_11_E3a | Bushveld soil, Mashare, northern Namibia | Casamino Acids, casein hydrolysate, yeast, peptone | 11–53 | 35–45 | 4.7–8.1 | 5.4–7.0 | |
| Unclassified Acidobacteria bacterium Ac_28_D10a | Agricultural soil, Mashare, northern Namibia | Casamino Acids, yeast, proline, protocatechuate | 17–40 | 29–35 | 4.3–9.4 | 5.5–7.9 | |
| Pyrinomonas methylaliphatogenes K22T (DSM 25857T) | Geothermal soil, New Zealand | Simple mono- and oligosaccharides and a limited number of complex protein substrates | 50–69 | 65 | 4.1–7.8 | 6.5 | 44 |
| “Ca. Chloracidobacterium thermophilum” | Hot spring, Yellowstone, WY | Peptone, yeast extract, 2-oxoglutarate, bicarbonate, thioglycolate | 45–60 | 50–55 | ND | 8.5 | |
Pyrinomonas methylaliphatogenes K22T was isolated from a geothermally heated soil (68°C, pH 6.9) collected from Mt. Ngauruhoe, an active strato-volcano located in the Tongariro volcano complex on the North Island of New Zealand. Cells were grown at 60°C as described previously (44) using basal liquid FS1V medium with the addition of 0.1 g liter−1 Casamino Acids (Difco) and 0.5 g liter−1 glucose in an oxic headspace (1:1 ratio of headspace to medium) (49). Subsequently, this bacterium also was grown at three different temperatures (50, 60, and 69°C). The cells then were centrifuged at 5,000 rpm for 30 min and the supernatant decanted off. The subsequent pellet was lyophilized overnight.
“Ca. Chloracidobacterium thermophilum” was isolated from microbial mats in alkaline siliceous hot springs in Yellowstone National Park, WY, USA (41). The enrichment culture was grown at 53°C as described previously (50). However, carbon and nitrogen sources were changed to 50 mg liter−1 peptone and yeast extract of each 365 mg liter−1 2-oxoglutarate and 625 mg liter−1 bicarbonate. Thioglycolate (125 mg liter−1) was added as a reduced sulfur source. Cells of “Ca. Chloracidobacterium thermophilum” were separated from the other members of the enrichment (predominantly Anoxybacillus sp.; ca. 20%) by Percoll density centrifugation (50).
Tree calculation.
Almost-full-length 16S rRNA gene fragments of two strains (Ac_11_E3a and Ac_28_D10a) isolated at the DSMZ were amplified by colony PCR with primers 8f and 1492r (51). Sequences of purified PCR products (ExoSAP-IT; USB, Cleveland, OH) were determined by Sanger sequencing on an AB 3730 DNA analyzer (Applied Biosystems, Foster City, CA) using the AmpliTaq FS BigDye Terminator cycle sequencing kit (Applied Biosystems). The 16S rRNA gene sequences of strains Ac_11_E3 and Ac_28_D10, together with those published for the other strains, were added to the small-subunit rRNA nonredundant reference database SILVA, version 108 (www.arb-silva.de) (52), in the ARB software environment (53). After automated alignment with the Fast aligner tool, the alignment was manually refined based on secondary structure information. A phylogenetic tree was calculated using the neighbor-joining algorithm (termini filter; 41,484 valid positions between positions 60 and 1438 of the Escherichia coli 16S rRNA reference gene; 1,000 bootstrap resamplings).
Lipid analysis.
For all studied strains, lyophilized cells were hydrolyzed with 1 N HCl in methanol by refluxing for 3 h by following the procedure described previously (14). The extracts obtained were methylated with diazomethane to transform fatty acids into methyl esters, and an aliquot was silylated with N,O-bis(trimethylsilyl)-trifluoroacetamide (BSTFA) in pyridine at 60°C for 20 min and analyzed by gas chromatography (GC) and GC-mass spectrometry (GC-MS) using conditions previously described (14). Another aliquot of the methylated extract was separated over an activated Al2O3 column using dichloromethane (DCM) and DCM-methanol (1:1, vol/vol/) to give an apolar and polar fraction, respectively. The apolar fraction was used to determine the double-bond positions of the monounsaturated fatty acid methyl esters (FAMEs) using the mass spectra of their dimethyl disulfide derivatives as described by Nichols et al. (54). The polar fraction was dissolved in hexane-propanol (99:1, vol/vol), filtered over a 0.45-μm-pore-size polytetrafluoroethylene filter, and analyzed by high-performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry (HPLC–APCI-MS) for brGDGTs.
For all strains, intact polar lipids were extracted from the lyophilized cells using a modified Bligh-Dyer technique (55) as described by Pitcher et al. (56). An aliquot of the obtained extract was dissolved in hexane–2-propanol–water (72:27:1), filtered through a 0.45-μm-pore-size regenerated cellulose filter, and analyzed by HPLC–electrospray ionization-MSn using conditions previously described (14).
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of the acidobacterial strains Ac_11_E3 and Ac_28_D10 are KF840370 and KF840371, respectively.
RESULTS
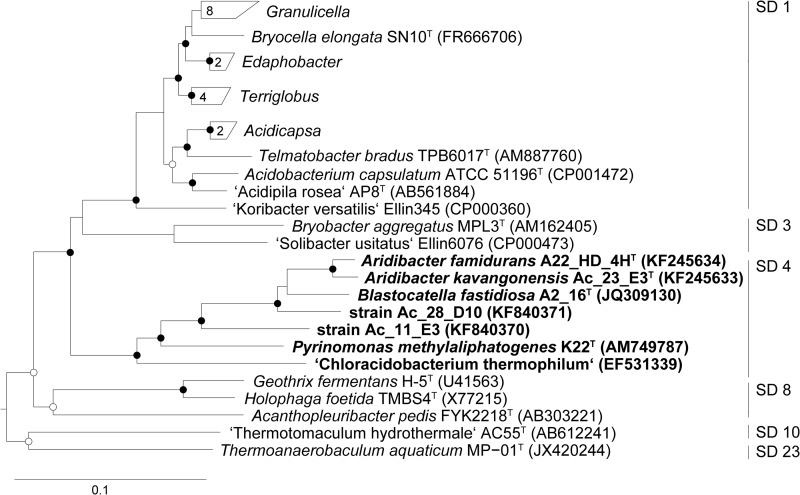
Seven strains of bacteria belonging to Acidobacteria SD 4 were analyzed for their lipid compositions; five are species that have previously been characterized (Blastocatella fastidiosa [42], Pyrinomonas methylaliphatogenes [44], “Ca. Chloracidobacterium thermophilum” [41], Aridibacter famidurans, and Aridibacter kavangonensis [43]), and two are novel strains isolated from soils in Namibia (Table 1). Figure 2 depicts their phylogenetic relationship based on the 16S rRNA gene and the position of SD 4 relative to other characterized phylogenetic branches within the phylum Acidobacteria. The maximum phylogenetic diversity within the cited SD 4 strains is quite large, with up to >20% sequence dissimilarity, which is substantially larger than that observed for SD 1 and 3 Acidobacteria (Fig. 2).
FIG 2.
Rooted neighbor-joining phylogenetic tree (Felsenstein correction) based on almost-full-length 16S rRNA gene sequences showing the investigated strains of Acidobacteria SD 4 (boldface) in relation to other described acidobacterial taxa. Open and closed circles indicate bootstrap values (expressed as percentages of 1,000 replicates) of >70% and >90%, respectively. The following sequences were used as the outgroup: Planctomyces brasiliensis DSM5305T (AJ231190), Planctomyces maris DSM8797T (AJ231184), and Planctomyces limnophilus DSM 3776T (CP001744). The bar indicates 10% nucleotide divergence.
Lipids released by acid hydrolysis.
Figure 3 shows two examples of typical gas chromatograms of total lipid fractions obtained after acid hydrolysis of cells (i.e., for P. methylaliphatogenes and Aridibacter famidurans). All strains contained iso-C15 as a dominant regular fatty acid, with the unsaturated counterpart, iso-C15:1Δ9c, present in the mesophilic but not in the thermophilic strains (Table 2). The fatty acid distribution of P. methylaliphatogenes (Fig. 3a) and, to a lesser extent, of strain Ac_28_D10 deviates from the other investigated strains because it also contains relatively large amounts of longer iso fatty acids, i.e., iso-C17:0, iso-C19:0, and the uncommon iso-C21:0 fatty acid. The latter fatty acid also was encountered in low relative abundance (ca. 2%) in three other investigated strains (Table 2). In the mesophilic strain, n-C16:1Δ9 also was present as a relatively abundant fatty acid (Fig. 3b and Table 2). In addition to these regular fatty acids, the more unusual, later-eluting (Fig. 3a) lipid, 13,16-dimethyloctacosanedioic acid (or iso-diabolic acid 3), was detected in various amounts (1 to 47% of total lipids) (Table 2).
FIG 3.
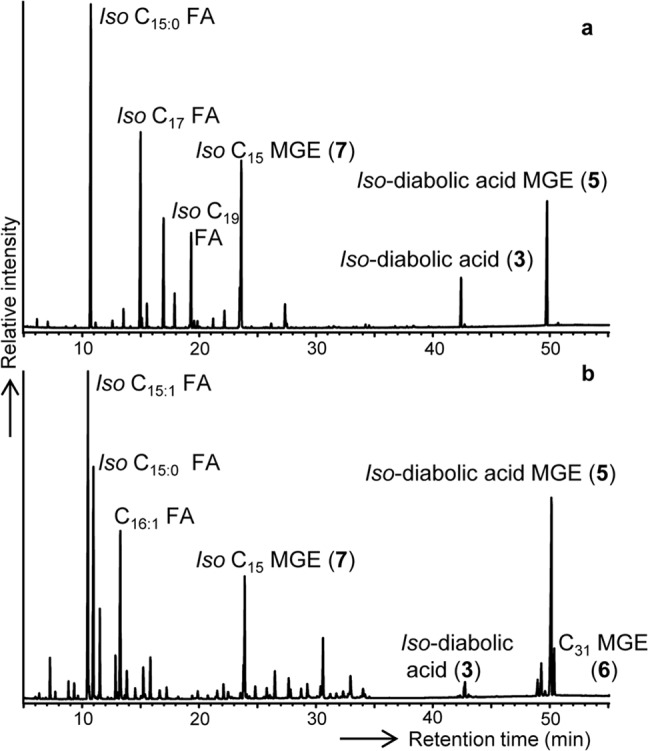
Gas chromatograms of lipids released after acid hydrolysis of whole-cell material of P. methylaliphatogenes K22T (a) and Aridibacter famidurans A22_HD_4HT (b). Carboxylic groups were derivatized to the corresponding methyl esters, and alcohol moieties were derivatized to trimethyl silyl ethers prior to gas chromatographic analysis. Numbers refer to structures shown in Fig. 1.
TABLE 2.
Relative abundance of fatty acids and ether lipids after acid hydrolysis of cell material and general characteristics of the membrane lipids in the studied SD 4 Acidobacteria
| Component | % of total lipidsa in strainb: |
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Fatty acids | |||||||
| iso-C13 | 1.7 | 1.6 | |||||
| C14:1Δ9 | 0.8 | ||||||
| C14:0 | 2.9 | ||||||
| iso-C15:1Δ9c | 9.6 | 6.6 | 19.0 | 8.7 | 16.8 | ||
| iso-C15:1Δ9tr | 0.4 | 0.7 | 0.3 | 0.4 | |||
| iso-C15:0 | 13.1 | 18.9 | 12.5 | 23.4 | 22.8 | 30.6 | 35.6 |
| anteiso-C15 | 1.2 | ||||||
| iso-C16 | 1.6 | 0.2 | 1.9 | 4.6 | |||
| C16:1Δ9 | 10.1 | 3.1 | 10.5 | 10.5 | 5.8 | 0.9 | |
| C16:0 | 1.0 | 1.2 | 1.8 | 1.3 | 4.7 | 1.1 | 4.1 |
| iso-C17:1Δ9 | 3.4 | 2.1 | 0.7 | 1.8 | 4.3 | ||
| iso-C17:0 | 2.4 | 0.6 | 1.1 | 5.4 | 16.1 | 2.5 | |
| anteiso-C17:0 | 1.4 | 1.1 | 0.6 | ||||
| C18:1Δ9 | 4.1 | ||||||
| C18:0 | 0.8 | 2.1 | |||||
| iso-C19:1Δ9 | 1.1 | ||||||
| iso-C19:0 | 6.8 | ||||||
| C20:1Δ9 | 1.2 | ||||||
| C20:0 | 0.9 | 0.5 | 1.1 | ||||
| iso-C21:1Δ9 | 0.8 | ||||||
| iso-C21:0 | 2.1 | 1.8 | 1.7 | 4.4 | 2.6 | ||
| iso-Diabolic acid (3)c | 1.8 | 1.8 | 1.6 | 1.8 | 1.0 | 3.8 | 46.5 |
| 5-Methyl iso-diabolic acid (4) | 1.2 | ||||||
| Ethers | |||||||
| iso-C15-MGE (7) | 21.6 | 20.7 | 15.9 | 15.7 | 19.5 | 14.9 | |
| iso-C16-MGE | 4.3 | 1.2 | 2.6 | 1.2 | |||
| C16-MGE | 2 | 3.3 | 4.6 | 3.6 | 2.1 | ||
| iso-C17-MGE | 7.3 | 0.2 | 2.2 | 3.3 | 2.8 | 1.9 | |
| anteiso-C17-MGE | 2.9 | 0.9 | 2.1 | 0.7 | |||
| iso-Diabolic acid-MGE (5) | 18.9 | 25.3 | 20.2 | 15.4 | 5.0 | 17.2 | |
| 5-Methyl iso-diabolic acid-MGE (6) | 4.6 | 3.4 | 1.8 | 3.8 | |||
| Monounsaturationd (%) | 27 | 21 | 36 | 24 | 31 | 0 | 1 |
| Membrane spanningd (%) | 21 | 31 | 24 | 18 | 9 | 20 | 48 |
| Ether moietiesd (%) | 40 | 34 | 30 | 29 | 23 | 21 | 0 |
Normalized to the sum of the components listed. Values for major components (i.e., ≥5%) are underlined.
Strains: 1, Blastocatella fastidiosa A2_16T (DSM 25172T); 2, unclassified Acidobacteria bacterium Ac_11_E3; 3, Aridibacter famidurans A22_HD_4HT; 4, Aridibacter kavangonensis Ac_23_E3T; 5, unclassified Acidobacteria bacterium Ac_28_D10; 6, Pyrinomonas methylaliphatogenes K22T (DSM 25857T); 7, “Ca. Chloracidobacterium thermophilum.”
Numbers in parentheses refer to structures shown in Fig. 1.
Calculated on a molar basis, where membrane-spanning lipids are counted as two molecules.
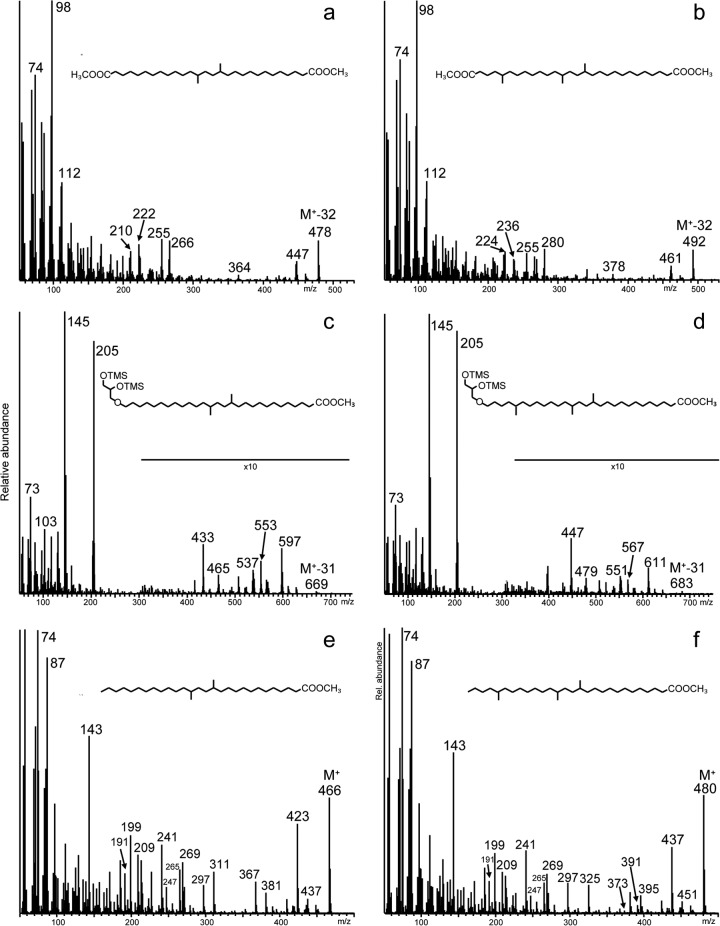
Strikingly, acid hydrolysis of cell material released not only fatty acids and iso-diabolic acid 3 but also substantial amounts of monoalkyl glycerol ethers (MGE), except for “Ca. Chloracidobacterium thermophilum,” in which no ether lipids were detected (Table 2). The ether lipids were MGE derivatives of the abundant saturated fatty acids, with iso-C15 MGE (7) and the MGE derivative (5) of iso-diabolic acid 3 as the most abundant representatives (Table 2 and Fig. 3). MGE 5 was previously (14) tentatively identified in two species of SD 1 Acidobacteria by its mass spectrum (Fig. 4c), which was virtually identical to that of 15,16-dimethyl-28-glyceryloxydodecanoic acid (57) but had a deviating retention time. In the two SD 1 species, MGE 5 represented only ca. 3% of the lipids (14), whereas in the SD 4 species investigated here, MGE 5 represents 5 to 26% of the lipids (Table 2). To confirm fully its structural resemblance with iso-diabolic acid 3, a fraction enriched in MGE 5 (as the methyl ester) was subjected to reduction with LiAlH4 to convert the methyl ester to an alcohol. This was followed by treatment with HI and H2-PtO2, which yielded the hydrocarbon 13,16-dimethyloctacosane, as confirmed by mass spectral analysis and relative retention time data (4).
FIG 4.
Mass spectra (corrected for background) of the methyl ester and TMS derivatives (where appropriate) of iso-diabolic acid (3) (a), 5-methyl iso-diabolic acid (4) (b), iso-diabolic acid MGE (5) (c), 5-methyl iso-diabolic acid MGE (6) (d), 13,16-dimethyl octacosanoic acid (e), and 13,16,24-trimethyl octacosanoic acid (f). The latter two components were formed by HI-LiAlH4 treatment of iso-diabolic acid MGE (5) and 5-methyl iso-diabolic acid MGE (6).
In addition to iso-diabolic acid 3 and its MGE derivative, we also detected two related components containing one additional methyl group (i.e., 4 and 6). This was apparent from their mass spectra (Fig. 4b and d), which revealed a shift of several fragment ions in the high-m/z region by 14 Th. To elucidate the position of the methyl group, a fraction containing MGE 6 was subjected to LiAlH4 followed by HI treatment and hydrogenation (described above). This yielded 5,13,16-trimethyloctacosane, as confirmed by mass spectral analysis and relative retention time data (4). This experiment revealed the position of the methyl group to be at C-5 but still did not elucidate the position of the additional methyl in the MGE derivative to be at C-5 or C-ω5. This was determined by direct HI treatment followed by hydrogenation, which generated the C31 monocarboxylic acid 9. Its mass spectrum, compared to that of the monocarboxylic acid 8 formed from MGE derivative 5, revealed that the additional methyl group is in the vicinity of the ether bond, resulting in structure 6. The mass spectral fragmentation pattern of a methylated iso-diabolic acid detected in “Ca. Chloracidobacterium thermophilum” (Table 2) also was consistent with a methyl group at position C-5.
The 5-methyl iso-diabolic acid MGE 6 was detected in 4 out of 5 mesophilic species, with strain Ac_11_E3 containing the highest relative amount of the methylated derivative. Because methylation at C-5 was detected for iso-diabolic acid from “Ca. Chloracidobacterium thermophilum,” B. fastidiosa and P. methylaliphatogenes were the only two species out of the seven investigated strains that did not contain 5-methyl lipids (Table 2).
Distribution of IPLs.
To characterize the intact polar lipids (IPLs) of all species of Acidobacteria investigated, the Bligh-Dyer solvent extracts were analyzed by HPLC/ESI-MSn. The IPLs were dominated by mixed ether-ester monoglycerides (Table 3). IPLs with phosphocholine (PC) head groups dominated, except for “Ca. Chloracidobacterium thermophilum,” for which the dominant IPLs were diacylglycerylhydroxy-methyl-(N,N,N)-trimethylalanine (DGTA) lipids. The overall number of carbon atoms in the acyl/alkyl groups of these IPLs is consistent with the dominant fatty acids and MGEs detected after acid hydrolysis (Table 2). However, no membrane-spanning IPLs (i.e., IPLs containing ester-bound iso-diabolic acid 3 or 4 or MGE 5 or 6) were detected in any of these Bligh-Dyer extracts.
TABLE 3.
Relative abundancesa and acyl/alkyl composition of IPLs in the seven species of SD 4 Acidobacteria
| IPLc | Speciesb |
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| DGTA | + (30:0) | ++ (30:0) | +++ (30:0, 32:0)d | ||||
| PE | + (32:0, 30:0, 33:1, 31:1, 30:1) | + (30:0, 30:1, 34:1) | + (30:1, 32:1, 30:0) | + (30:0, 32:0, 33:1) | + (30:0, 30:1) | + (30:0, 34:0) | |
| MMPE | + (30:0)d | ||||||
| DMPE | + (32:1, 30:0, 31:1, 30:1,32:0) | + (30:0, 30:1) | + (30:1, 32:1, 31:1, 30:0) | + (30:0, 32:0) | + (30:0, 30:1) | ||
| PC | +++ (30:1, 32:0, 32:1, 31:1, 30:0) | +++ (30:1, 30:0) | +++ (30:1, 31:1, 32:1) | +++ (30:0, 32:0) | +++ (30:1, 30:0, 32:0) | +++ (30:0, 32:0, 34:0) + (34:0, 32:0)d | |
| Unknown | +e | ++f | |||||
Abundance relative to the major peak in the LC-MS base peak chromatogram (+++, base peak; ++, 50 to 100% of base peak; +, 10 to 50% of base peak). Note that the mass spectral response factors for different IPL groups can be quite different. The predominant fatty acid composition, in order of relative abundance, is reported in parentheses as the total number of carbon atoms of the acyl/alkyl moieties and the number of double-bond equivalents.
Strains: 1, Blastocatella fastidiosa A2_16T (DSM 25172T); 2, Acidobacteria bacterium Ac_11_E3; 3, Aridibacter famidurans A22_HD_4HT; 4, Aridibacter kavangonensis Ac_23_E3T; 5, Acidobacteria bacterium Ac_28_D10; 6, Pyrinomonas methylaliphatogenes K22T (DSM 25857T); 7, “Ca. Chloracidobacterium thermophilum.”
IPLs were sn1-alkyl-sn2-acyl-glycerols, unless mentioned otherwise. IPLs are listed in order of elution. DGTA, dialylglycerylhydroxy-methyl-(N,N,N)-trimethylalanine; PE, phosphoethanolamine; MMPE, monomethylated PE; DMPE, dimethylated PE; PC, phosphocholine.
Diacyl IPL.
Characterized by m/z 1,283.
Characterized by m/z 1,366.
Branched GDGTs.
The acid-hydrolyzed biomass of some of the acidobacterial cultures was also analyzed for the presence of GDGTs by HPLC/APCI-MS using selected ion monitoring. However, we were unable to identify any of the brGDGTs 1 and 2 or any other brGDGT in the species investigated.
DISCUSSION
Chemotaxonomic relationships.
The fatty acid distributions of all studied Acidobacteria belonging to SD 4 show a quite consistent pattern: they all contain iso-C15:0 as an abundant fatty acid (13 to 36% of the total lipids) (Table 2). Five of them also contain iso-C15:1Δ9c as an abundant fatty acid (7 to 19%), while four of them contain C16:1Δ9 in substantial amounts (6 to 11%) (Table 2). iso-Diabolic acid 3 was detected in all examined species of SD 4 Acidobacteria in various amounts (1 to 47% of total lipids) (Table 2). This lipid was identified previously as an abundant lipid in Acidobacteria SD 1 and 3 (14) and in thermophilic Thermoanaerobacter species (58–60), in which they fulfill a role as membrane-spanning lipids. In these studies, iso-diabolic acid was detected only after hydrolysis of the cell material. In agreement with this mode of occurrence, a previous report on the lipids of “Ca. Chloracidobacterium thermophilum” likewise did not report iso-diabolic acid in the Bligh-Dyer extract (50), whereas after acid hydrolysis of cell material, as performed in this study, it comprises the most abundant lipid (Table 2). In contrast to “Ca. Chloracidobacterium thermophilum” and Acidobacteria SD 1 and 3 (14), the relative abundance of iso-diabolic acid is relatively low (1 to 4%) (Table 2) in the other investigated SD 4 species. However, in these other species iso-diabolic acid occurs relatively abundantly (5 to 25% of total lipids) (Table 2) in an ether-bound form as MGE derivative 5. This component was previously identified as a minor constituent in Acidobacteria SD 1 and 3 (14). In general, this observation seems to be characteristic for SD 4 Acidobacteria; all species, except “Ca. Chloracidobacterium thermophilum,” contain substantial amounts (21 to 40%) of ether lipids (Table 2). This is consistent with the analysis of IPLs in the Bligh-Dyer extract, which shows that the most dominant IPLs are mixed ether/ester lipids (Table 3).
These chemotaxonomic relationships are confirmed when cluster analysis is performed on the lipid distributions, including those of previously reported SD 1 and 3 Acidobacteria (14, 26) (Fig. 5). The lipid distributions of all SD 4 Acidobacteria form a clearly distinct cluster. The only exception is “Ca. Chloracidobacterium thermophilum”; its lipid distribution is more similar to that of various members of Acidobacteria SD 1. In the phylogenetic tree based on the 16S rRNA gene (Fig. 2), “Ca. Chloracidobacterium thermophilum” is also clearly separated from the other SD 4 Acidobacteria members (Fig. 2), although it is also distinct from SD 1 and 3 species. The distinct taxonomic position of “Ca. Chloracidobacterium thermophilum” is consistent with its physiological capabilities; it is the only known phototrophic member of the Acidobacteria (41), while all other species are heterotrophs.
FIG 5.
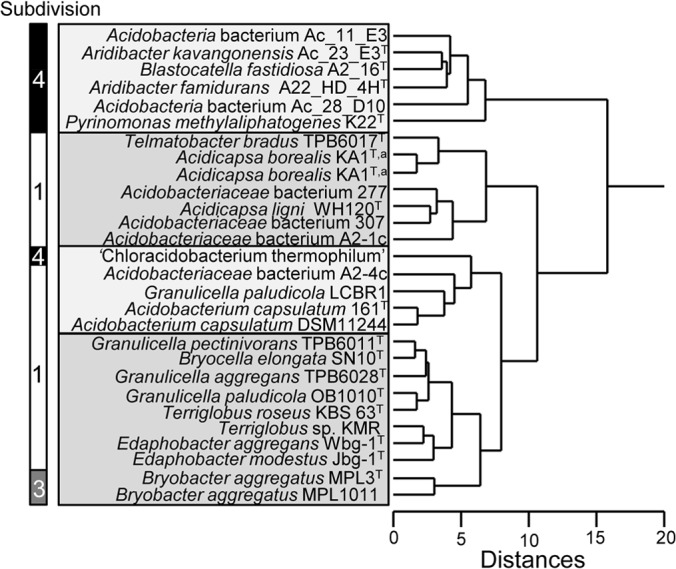
Cluster analysis of the distribution of the lipids released by acid hydrolysis of cell material of the Acidobacteria of SD 4 compared to results of Acidobacteria SD 1 and 3 reported previously (14, 26) using an identical method of lipid analysis. The input of the cluster analysis was the Bray-Curtis similarity matrix of lipid profiles (percentage of total lipids, as in Table 2). A hierarchical clustering was performed in SYSTAT 13 using Euclidian distance and the average linking method. A superscript letter “a” indicates that two different batches of cultures were studied.
The IPL compositions of the SD 4 Acidobacteria are also in line with the cluster analysis of the lipid distribution; “Ca. Chloracidobacterium thermophilum” is the only species that contains predominantly diacyl lipids, whereas the other examined species contain mixed ether/ester lipids. Furthermore, “Ca. Chloracidobacterium thermophilum” contains predominantly diacylglycerylhydroxymethyl-N,N,N-trimethyl-β-alanine (DGTA) lipids, whereas all other species show a dominance of phosphocholine IPLs (Table 3). However, it should be noted that the reported IPL distribution probably represents a biased view of the membrane lipid composition, because IPLs containing membrane-spanning lipids were not detected, whereas direct acid hydrolysis of cells generated substantial amounts of these lipids (9 to 48%) (Table 2). As discussed previously for SD 1 and 3 Acidobacteria species (14), this may be caused by relatively large and polar head groups, which may render the IPLs containing membrane-spanning lipids nonextractable using the Bligh-Dyer protocol. Despite this bias, there is generally a good overlap between the reported acyl/alkyl composition of the IPLs (Table 3) and the lipid composition (Table 2); the IPLs seem to contain mainly C15 and, to a lesser extent, C17 acyl/alkyl chains, as can be tentatively concluded from the total number of acyl/alkyl carbons of C30 and C32.
Variation in lipid composition: influence of environmental variables.
The membrane lipids of SD 4 Acidobacteria are quite distinct from the diacyl glycerol membrane lipids that characterize most bacteria. First, they contain a substantial amount of membrane-spanning lipids (9 to 48%) (Table 2). Second, they contain a high percentage of ether linkages (up to 40%) (Table 2). In contrast to the Archaea, membrane-spanning lipids are uncommon in the bacterial domain, but diabolic or iso-diabolic acid, acids connecting two glycerol moieties, do occur in Butyrivibrio species (61), Sarcina ventriculi (62), members of the Thermotogales (2, 57, 63–65), Thermoanaerobacter species (58, 59, 62), Acidobacteria SD 1 and 3 (14), and Acidobacteria SD 4 (this work). Ether membrane lipids are the hallmark of the Archaea (1, 5), but an increasing number of bacterial species has been shown to contain diether, tetraether, or mixed ether/ester lipids. These include (but are not restricted to) Ammonifex degensii (66), Aquifex pyrophilus (67), Thermotoga species (2, 57), several sulfate-reducing bacteria (68–70), Mycoplasma fermentans (71), anammox bacteria (3), Acidobacteria SD 1 and 3 (14), and Acidobacteria SD 4 (this work).
Classically, the presence of membrane-spanning and ether-bound lipids is seen as an adaptation to high temperatures or other extreme conditions, as is the case for isoprenoidal tetraether lipids of Archaea (72). Consistent with this idea, most bacterial species that contain membrane-spanning lipids are moderate or extreme thermophiles, although Butyrivibrio species and most cultured Acidobacteria are mesophilic. In a study of different species of the order Thermotogales (57), it was shown that in Thermotoga spp., the core membrane lipids were characterized by the presence of both ester and ether bonds, whereas no ether bonds occurred in the phylogenetically related Thermosipho and Fervidobacterium spp. Therefore, both the occurrence of membrane-spanning lipids and the presence of ether bonds in bacteria do not seem to be an adaptation to temperature alone.
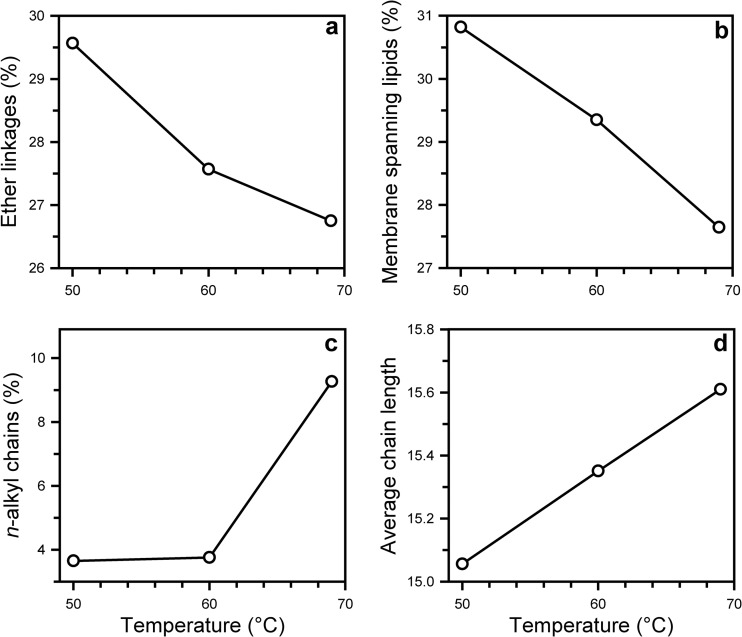
In this study, we examined two thermophilic species of the SD 4 Acidobacteria. “Ca. Chloracidobacterium thermophilum,” grown at 53°C, has the highest percentage of membrane-spanning lipids (48%) (Table 2), but its membrane lipids do not contain ether bonds. Compared to the mesophilic species, P. methylaliphatogenes, grown at 60°C, has a moderate percentage of membrane-spanning lipids (20%) (Table 2) but a lower total number of ether bonds (21%) (Table 2). The most distinct difference in the composition of the thermophilic species compared to the mesophiles is that they contain very few unsaturated lipids (Table 2). To examine the influence of growth temperature on the membrane lipid composition further, P. methylaliphatogenes was grown at three temperatures in the 50 to 69°C range. Subtle changes in the membrane lipid composition were detected, but in contrast with classical ideas on membrane adaptation, a decreasing rather than an increasing trend in the percentage of membrane-spanning lipids and ether bonds with increasing temperature was observed (Fig. 6a and b). Only a small increase in the number of n-alkyl chains (Fig. 6c) and a slight increase in the average chain length (Fig. 6d), determining the thickness of the membrane, were apparent with increasing temperature. Thus, the lipid data of the SD 4 Acidobacteria not only indicate that the occurrence of membrane-spanning lipids and the presence of ether bonds in bacteria are adaptations to temperature but suggest that other (including genetic) factors probably also play a role.
FIG 6.
Membrane lipid characteristics of P. methylaliphatogenes K22T as a function of growth temperature. (a) Fraction of ether linkages; (b) fraction of membrane-spanning lipids; (c) fraction of n-alkyl chains; (d) average chain length. The average chain length (number of carbon atoms) was calculated by dividing the chain length of the membrane lipids by a factor of two and by ignoring methyl substituents.
Acidobacteria as a potential source for branched GDGTs.
brGDGTs (e.g., 1 and 2) occur ubiquitously in soil, peat bogs, and lakes (5). Their distribution is used to reconstruct past pH and temperature based on a set of empirical relationships (73–75), which are thought to reflect the ability of bacteria in soil and lake water to adjust their membrane composition in response to temperature and pH. Acidobacteria have been proposed as candidates for the production of brGDGTs (13), and this has been supported by the recent identification of its “building block” iso-diabolic acid 3 in SD 1 and 3 Acidobacteria (14). Although small amounts of brGDGT 1 were detected in a few species, iso-diabolic acid 3 occurred predominantly in an ester-bound form and not in an ether-bound form, indicating that other Acidobacteria members are probably the origin of the brGDGTs. This was one of the reasons to perform this study. It showed that SD 4 Acidobacteria do not produce brGDGTs, at least not the seven species that we investigated. However, six of the seven investigated species produce lipids in which iso-diabolic acid 3 or its methylated counterpart 4 occur ether bound to a glycerol moiety (i.e., MGEs 5 and 6) in relatively large amounts (i.e., 9 to 30%) (Table 2). Such moieties reflect important structural units of the brGDGTs 1 and 2. Strikingly, the ether-bound iso-diabolic acid moiety occurs only at the sn1 but not at the sn2 position of glycerol. Apparently, while most of the SD 4 Acidobacteria are able to produce the ether bond at the sn1 position enzymatically, they lack the enzyme(s) able to produce ether bonds at the sn2 position. Consequently, the diester/diether lipids 10 and 11, composed of two esterified MGE 5 and 6 units, which are presumed to be important constituents of the membrane lipids of SD 4 Acidobacteria, have the closest structural resemblance to brGDGTs 1 and 2.
Another apparent mismatch with the GDGTs occurring in SD 1 Acidobacteria and brGDGTs occurring in the environment is that only GDGT 1 was detected in the Acidobacteria (14), whereas brGDGTs with additional methyl substituents (such as 2) occur widely in the environment (73, 76). This additional methylation occurs at one (i.e., 2) or both alkyl chains at C-5, although recently brGDGTs with the methylation at C-6 also have been reported (77). The detection of the 5-methyl iso-diabolic acid (i.e., 4) and MGE 6 in five out of seven species of SD 4 Acidobacteria now, for the first time, reveals that an additionally methylated iso-diabolic acid or its ether derivative is biosynthesized by Acidobacteria. Interestingly, the two thermophilic species produce no (i.e., P. methylaliphatogenes) or only small amounts (i.e., “Ca. Chloracidobacterium thermophilum”) of additionally methylated iso-diabolic acid or its derivative (i.e., 4 and 6) (Table 2). Four of the five mesophilic SD 4 Acidobacteria produce these components, with strain Ac_11_E3 containing them at the highest relative abundance (Table 2). This is in agreement with the distributions of brGDGTs in the environment, which generally reveals an increase in the degree of additional branching with decreasing temperature (73–75). The mesophilic species B. fastidiosa is, however, an exception in this respect, since it does not contain structure 4 or 6 (Table 2). This suggests that although there apparently is strong environmental control of brGDGT composition (73–75), there also are genetic factors involved. In the species investigated, we did not identify any additionally methylated iso-C15 fatty acid or iso-C15 MGE. This suggests that, in the biosynthesis of the membrane lipids, the methylation of C-5 occurs after the head-to-head condensation of two iso-C15 fatty acids to iso-diabolic acid 3, i.e., after the membrane-spanning lipid has been synthesized.
Our finding of ether-bound iso-diabolic acid and its 5-methyl derivative as important membrane lipids of SD 4 Acidobacteria further closes the gap between the presumed origin of brGDGTs in the environment and the occurrence of related lipids in bacteria. Presently, we still lack known Acidobacteria members that are able to produce glycerol membrane lipids that are ether linked at the sn2 position (although some SD 1 species are able to produce small amounts of GDGT 1) and Acidobacteria that produce membrane-spanning lipids containing cyclopentane moieties formed by internal cyclization (9). Further studies of the lipids of newly cultivated Acidobacteria may lead to identification of the bacterial sources of the ubiquitous brGDGTs in the environment. This will allow a more fundamental study of the environmental and genetic controls on the distribution of these lipids that are currently widely applied in paleoenvironment and paleoclimate studies (5).
ACKNOWLEDGMENTS
Soil samples from Namibia were taken under collection permits 1358/2009 and 1569/2011 and exported under permits ES 24478 (6 April 2009) and ES 25691 (12 April 2011). Characterization of strains A2_16, A22_HD_4H, Ac_11_E3, Ac_23_E3, and Ac_28_D10 was performed under the Material Transfer Agreements of the NBRI (National Botanical Research Institute, Namibia) of 5 April 2012.
We thank Alicia Geppert (DSMZ) for help with cultivating Acidobacteria strains for this study. We greatly appreciate the assistance of National Park Service personnel.
The research leading to these results received funding from the European Research Council (ERC) under the European Union's Seventh Framework Program (FP7/2007-2013), ERC grant agreement 226600. J.O. received support from the BMBF programs Biolog/BIOTA (01LC0621C) and TFO (01LL0912M). Studies of “Ca. Chloracidobacterium thermophilum” were supported by grant DE-FG02-94ER20137 from the U.S. Department of Energy to D.A.B. and were conducted under Yellowstone National Park research permits YELL-0129 and YELL-5494 (to David M. Ward, Montana State University). GNS staff acknowledge support from the DCF Geothermal Resources of New Zealand (GRN) work program.
Footnotes
Published ahead of print 13 June 2014
REFERENCES
- 1.Koga Y, Morii H. 2005. Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects. Biosci. Biotechnol. Biochem. 69:2019–2034. 10.1271/bbb.69.2019 [DOI] [PubMed] [Google Scholar]
- 2.DeRosa M, Gambacorta A, Huber R, Lanzotti V, Nicolaus B, Stetter KO, Trincone A. 1988. A new 15,16-dimethyl-30-glyceryloxytriacontanoic acid from lipids of Thermotoga maritima. J. Chem. Soc. Chem. Commun. 1988:1300–1301 [Google Scholar]
- 3.Sinninghe Damsté JS, Rijpstra WIC, Geenevasen JAJ, Strous M, Jetten MSM. 2005. Structural identification of ladderane and other membrane lipids of planctomycetes capable of anaerobic ammonium oxidation (anammox). FEBS J. 272:4270–4283. 10.1111/j.1742-4658.2005.04842.x [DOI] [PubMed] [Google Scholar]
- 4.Sinninghe Damsté JS, Hopmans EC, Pancost RD, Schouten S, Geenevasen JAJ. 2000. Newly discovered non-isoprenoid glycerol dialkyl glycerol tetraether lipids in sediments. Chem. Commun. 2000:1683–1684. 10.1039/B004517I [DOI] [Google Scholar]
- 5.Schouten S, Hopmans EC, Sinninghe Damsté JS. 2013. The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: a review. Org. Geochem. 54:19–61. 10.1016/j.orggeochem.2012.09.006 [DOI] [Google Scholar]
- 6.Schouten S, van der Meer MT, Hopmans EC, Rijpstra WI, Reysenbach AL, Ward DM, Sinninghe Damsté JS. 2007. Archaeal and bacterial glycerol dialkyl glycerol tetraether lipids in hot springs of Yellowstone National Park. Appl. Environ. Microbiol. 73:6181–6191. 10.1128/AEM.00630-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang CL, Wang J, Dodsworth JA, Williams AJ, Zhu C, Hinrichs KU, Zheng F, Hedlund BP. 2013. In situ production of branched glycerol dialkyl glycerol tetraethers in a great basin hot spring (U.S.A.). Front. Microbiol. 4:181. 10.3389/fmicb.2013.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedlund BP, Paraiso JJ, Williams AJ, Huang Q, Wei Y, Dijkstra P, Hungate BA, Dong H, Zhang CL. 2013. Wide distribution of autochthonous branched glycerol dialkyl glycerol tetraethers (bGDGTs) in US Great Basin hot springs. Front. Microbiol. 4:222. 10.3389/fmicb.2013.00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weijers JWH, Schouten S, Hopmans EC, Geenevasen JAJ, David ORP, Coleman JM, Pancost RD, Sinninghe Damsté JS. 2006. Membrane lipids of mesophilic anaerobic bacteria thriving in peats have typical archaeal traits. Environ. Microbiol. 8:648–657. 10.1111/j.1462-2920.2005.00941.x [DOI] [PubMed] [Google Scholar]
- 10.Pancost RD, Sinninghe Damsté JS. 2003. Carbon isotopic compositions of prokaryotic lipids as tracers of carbon cycling in diverse settings. Chem. Geol. 195:29–58. 10.1016/S0009-2541(02)00387-X [DOI] [Google Scholar]
- 11.Weijers JWH, Wiesenberg GLB, Bol R, Hopmans EC, Pancost RD. 2010. Carbon isotopic composition of branched tetraether membrane lipids in soils suggest a rapid turnover and a heterotrophic life style of their source organism(s). Biogeosciences 7:2959–2973. 10.5194/bg-7-2959-2010 [DOI] [Google Scholar]
- 12.Oppermann BI, Michaelis W, Blumenberg M, Frerichs J, Schulz HM, Schippers A, Beaubien SE, Kruger M. 2010. Soil microbial community changes as a result of long-term exposure to a natural CO2 vent. Geochim. Cosmochim. Acta 74:2697–2716. 10.1016/j.gca.2010.02.006 [DOI] [Google Scholar]
- 13.Weijers JWH, Panoto E, van Bleijswijk J, Schouten S, Rijpstra WIC, Balk M, Stams AJM, Sinninghe Damsté JS. 2009. Constraints on the biological source(s) of the orphan branched tetraether membrane lipids. Geomicrobiol. J. 26:402–414. 10.1080/01490450902937293 [DOI] [Google Scholar]
- 14.Sinninghe Damsté JS, Rijpstra WIC, Hopmans EC, Weijers JWH, Foesel BU, Overmann J, Dedysh SN. 2011. 13,16-Dimethyl octacosanedioic acid (iso-diabolic acid), a common membrane-spanning lipid of Acidobacteria subdivisions 1 and 3. Appl. Environ. Microbiol. 77:4147–4154. 10.1128/AEM.00466-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morales SE, Mouser PJ, Ward N, Hudman SP, Gotelli NJ, Ross DS, Lewis TA. 2006. Comparison of bacterial communities in New England Sphagnum bogs using terminal restriction fragment length polymorphism (T-RFLP). Microb. Ecol. 52:34–44. 10.1007/s00248-005-0264-2 [DOI] [PubMed] [Google Scholar]
- 16.Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. 2006. Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72:2110–2117. 10.1128/AEM.72.3.2110-2117.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraigher B, Stres B, Hacin J, Ausec L, Mahne I, van Elsas JD, Mandic-Mulec I. 2006. Microbial activity and community structure in two drained fen soils in the Ljubljana Marsh. Soil Biol. Biochem. 38:2762–2771. 10.1016/j.soilbio.2006.04.031 [DOI] [Google Scholar]
- 18.Pankratov TA, Belova SE, Dedysh SN. 2005. Evaluation of the phylogenetic diversity of prokaryotic microorganisms in Sphagnum peat bogs by means of fluorescence in situ hybridization (FISH). Microbiology 74:722–728. 10.1007/s11021-005-0130-8 [DOI] [PubMed] [Google Scholar]
- 19.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3:442–453. 10.1038/ismej.2008.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssen PH. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719–1728. 10.1128/AEM.72.3.1719-1728.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serkebaeva YM, Kim Y, Liesack W, Dedysh SN. 2013. Pyrosequencing-based assessment of the bacteria diversity in surface and subsurface peat layers of a Northern wetland, with focus on poorly studied phyla and candidate divisions. PLoS One 8:e63994. 10.1371/journal.pone.0063994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120. 10.1128/AEM.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vetrovsky T, Baldrian P. 2013. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e57923. 10.1371/journal.pone.0057923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barns SM, Cain EC, Sommerville L, Kuske CR. 2007. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 73:3113–3116. 10.1128/AEM.02012-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishimoto N, Kosako Y, Tano T. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22:1–7. 10.1007/BF02106205 [DOI] [PubMed] [Google Scholar]
- 26.Kulichevskaya IS, Kostina LA, Valaskova V, Rijpstra WIC, Sinninghe Damsté JS, de Boer W, Dedysh SN. 2012. Acidicapsa borealis gen. nov., sp. nov., and Acidicapsa ligni sp. nov., subdivision 1 Acidobacteria from Sphagnum peat and decaying wood. Int. J. Syst. Evol. Microbiol. 62:1512–1520. 10.1099/ijs.0.034819-0 [DOI] [PubMed] [Google Scholar]
- 27.Okamura K, Kawai A, Yamada T, Hiraishi A. 2011. Acidipila rosea gen. nov., sp nov., an acidophilic chemoorganotrophic bacterium belonging to the phylum Acidobacteria. FEMS Microbiol. Lett. 317:138–142. 10.1111/j.1574-6968.2011.02224.x [DOI] [PubMed] [Google Scholar]
- 28.Dedysh SN, Kulichevskaya IS, Mityaeva MA, Serkebaeva YM, Sorokin VV, Suzina NE. 2012. Bryocella elongata gen. nov., sp. nov., a novel member of subdivision 1 of the Acidobacteria isolated from a methanotrophic enrichment culture. Int. J. Syst. Evol. Microbiol. 62:654–664. 10.1099/ijs.0.031898-0 [DOI] [PubMed] [Google Scholar]
- 29.Koch IH, Gich F, Dunfield PF, Overmann J. 2008. Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., acidobacteria isolated from alpine and forest soils. Int. J. Syst. Evol. Microbiol. 58:1114–1122. 10.1099/ijs.0.65303-0 [DOI] [PubMed] [Google Scholar]
- 30.Pankratov TA, Dedysh SN. 2010. Granulicella paudicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymers-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 60:2951–2959. 10.1099/ijs.0.021824-0 [DOI] [PubMed] [Google Scholar]
- 31.Männistö MK, Rawat S, Starovoytov V, Häggblom MM. 2012. Granulicella arctica sp. nov., Granulicella mallensis sp. nov., Granulicella tundricola sp. nov. and Granulicella sapmiensis sp. nov., novel acidobacteria from tundra soil. Int. J. Syst. Evol. Microbiol. 62:2097–2106. 10.1099/ijs.0.031864-0 [DOI] [PubMed] [Google Scholar]
- 32.Pankratov TA, Kirsanova LA, Kaparullina EN, Kevbrin VV, Dedysh SN. 2012. Telmatobacter bradus gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria, and emended description of Acidobacterium capsulatum Kishimoto et al. Int. J. Syst. Evol. Microbiol. 62:430–437. 10.1099/ijs.0.029629-0 [DOI] [PubMed] [Google Scholar]
- 33.Eichorst SA, Breznak JA, Schmidt TM. 2007. Isolation and characterization of soil bacteria that define Terriglobus gen. nov. in the phylum Acidobacteria. Appl. Environ. Microbiol. 73:2708–2717. 10.1128/AEM.02140-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Männistö MK, Rawat S, Starovoytov V, Häggblom MM. 2011. Terriglobus saanensis sp. nov., an acidobacterium isolated from tundra soil. Int. J. Syst. Evol. Microbiol. 61:1823–1828. 10.1099/ijs.0.026005-0 [DOI] [PubMed] [Google Scholar]
- 35.Kulichevskaya IS, Suzina NE, Liesack W, Dedysh SN. 2010. Bryobacter aggregatus gen. nov., sp. nov., a peat-inhabiting, aerobic chemo-organotroph from subdivision 3 of the Acidobacteria. Int. J. Syst. Evol. Microbiol. 60:301–306. 10.1099/ijs.0.013250-0 [DOI] [PubMed] [Google Scholar]
- 36.Liesack W, Bak F, Kreft J-U, Stackebrandt E. 1994. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85–90 [DOI] [PubMed] [Google Scholar]
- 37.Coates JD, Ellis DJ, Gaw CV, Lovley DR. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49:1615–1622. 10.1099/00207713-49-4-1615 [DOI] [PubMed] [Google Scholar]
- 38.Fukunaga Y, Kurahashi M, Yanagi K, Yokota A, Harayama S. 2008. Acanthopleuribacter pedis gen. nov., sp. nov., a marine bacterium isolated from a chiton, and description of Acanthopleuribacteraceae fam. nov., Acanthopleuribacterales ord. nov., Holophagaceae fam. nov., Holophagales ord. nov. and Holophagae classis nov. in the phylum “Acidobacteria.”. Int. J. Syst. Evol. Microbiol. 58:2597–2601. 10.1099/ijs.0.65589-0 [DOI] [PubMed] [Google Scholar]
- 39.Izumi H, Nunoura T, Miyazaki M, Mino S, Toki T, Takai K, Sako Y, Sawabe T, Nakagawa S. 2012. Thermotomaculum hydrothermale gen. nov., sp. nov., a novel heterotrophic thermophile within the phylum Acidobacteria from a deep-sea hydrothermal vent chimney in the Southern Okinawa Trough. Extremophiles 16:245–253. 10.1007/s00792-011-0425-9 [DOI] [PubMed] [Google Scholar]
- 40.Losey NA, Stevenson BS, Busse HJ, Sinninghe Damsté JS, Rijpstra WI, Rudd S, Lawson PA. 2013. Thermoanaerobaculum aquaticum gen. nov., sp. nov., the first cultivated member of Acidobacteria subdivision 23, isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 63:4149–4157. 10.1099/ijs.0.051425-0 [DOI] [PubMed] [Google Scholar]
- 41.Bryant DA, Costas AMG, Maresca JA, Chew AGM, Klatt CG, Bateson MM, Tallon LJ, Hostetler J, Nelson WC, Heidelberg JF, Ward DM. 2007. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523–526. 10.1126/science.1143236 [DOI] [PubMed] [Google Scholar]
- 42.Foesel BU, Rohde M, Overmann J. 2013. Blastocatella fastidiosa gen. nov., sp. nov., isolated from semiarid savanna soil—the first described species of Acidobacteria subdivision 4. Syst. Appl. Microbiol. 36:82–89. 10.1016/j.syapm.2012.11.002 [DOI] [PubMed] [Google Scholar]
- 43.Huber KJ, Wust PK, Rohde M, Overmann J, Foesel BU. 2014. Aridibacter famidurans and Aridibacter kavangonensis, two novel species of Acidobacteria subdivision 4 isolated from semiarid savanna soil. Int. J. Syst. Evol. Microbiol. 10.1099/ijs.0.060236-0 [DOI] [PubMed] [Google Scholar]
- 44.Crowe MA, Power JF, Morgan XC, Dunfield PF, Lagutin K, Rijpstra WIC, Vyssotski M, Sinninghe Damsté JS, Houghton KM, Ryan JLJ, Stott MB. 2014. Pyrinomonas alimethylogenes gen. nov., sp. nov., a novel group 4 thermophilic member of the Acidobacteria from geothermal soils. Int. J. Syst. Evol. Microbiol. 64:220–227. 10.1099/ijs.0.055079-0 [DOI] [PubMed] [Google Scholar]
- 45.Zimmermann J, Portillo MC, Serrano L, Ludwig W, Gonzalez JM. 2012. Acidobacteria in freshwater ponds at Donana National Park, Spain. Microb. Ecol. 63:844–855. 10.1007/s00248-011-9988-3 [DOI] [PubMed] [Google Scholar]
- 46.Angle JS, Mcgrath SP, Chaney RL. 1991. New culture-medium containing ionic concentrations of nutrients similar to concentrations found in the soil solution. Appl. Environ. Microbiol. 57:3674–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tschech A, Pfennig N. 1984. Growth-yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137:163–167. 10.1007/BF00414460 [DOI] [Google Scholar]
- 49.Stott MB, Crowe MA, Mountain BW, Smirnova AV, Hou SB, Alam M, Dunfield PF. 2008. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ. Microbiol. 10:2030–2041. 10.1111/j.1462-2920.2008.01621.x [DOI] [PubMed] [Google Scholar]
- 50.Costas AMG, Tsukatani Y, Rijpstra WIC, Schouten S, Welander PV, Summons RE, Bryant DA. 2012. Identification of the bacteriochlorophylls, carotenoids, quinones, lipids, and hopanoids of “Candidatus Chloracidobacterium thermophilum.” J. Bacteriol. 194:1158–1168. 10.1128/JB.06421-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner S, Pryer KM, Miao VPW, Palmer JD. 1999. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small submit rRNA sequence analysis. J. Eukaryot. Microbiol. 46:327–338. 10.1111/j.1550-7408.1999.tb04612.x [DOI] [PubMed] [Google Scholar]
- 52.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Gloeckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371. 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nichols PD, Guckert JB, White DC. 1986. Determination of monounsaturated fatty acid double-bond position and geometry for microbial monocultures and complex consortia by capillary GC-MS of their dimethyl disulphide adducts. J. Microbiol. Methods 5:49–55. 10.1016/0167-7012(86)90023-0 [DOI] [Google Scholar]
- 55.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 56.Pitcher A, Hopmans EC, Schouten S, Sinninghe Damsté JS. 2009. Separation of core and intact polar archaeal tetraether lipids using silica columns: insights into living and fossil biomass contributions. Org. Geochem. 40:12–19. 10.1016/j.orggeochem.2008.09.008 [DOI] [Google Scholar]
- 57.Sinninghe Damsté JS, Rijpstra WIC, Hopmans EC, Schouten S, Balk M, Stams AJM. 2007. Structural characterization of diabolic acid-based tetraester, tetraether and mixed ether/ester, membrane-spanning lipids of bacteria from the order Thermotogales. Arch. Microbiol. 188:629–641. 10.1007/s00203-007-0284-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee S, Kang S, Kim JN, Jung S. 2002. Structural analyses of the novel phosphoglycolipids containing the unusual very long bifunctional acyl chain, α,ω-13,16-dimethyloctacosanedioate in Thermoanaerobacter ethanolicus. Bull. Korean Chem. Soc. 23:1778–1784. 10.5012/bkcs.2002.23.12.1778 [DOI] [Google Scholar]
- 59.Balk M, Heilig HGHJ, van Eekert MHA, Stams AJM, Rijpstra IC, Sinninghe Damsté JS, de Vos WM, Kengen SWM. 2009. Isolation and characterization of a new CO-utilizing strain, Thermoanaerobacter thermohydrosulfuricus subsp. carboxydovorans, isolated from a geothermal spring in Turkey. Extremophiles 13:885–894. 10.1007/s00792-009-0276-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jung S, Zeikus JG, Hollingsworth RI. 1994. A new family of very long chain α,ω-dicarboxylic acids is a major structural fatty acyl component of the membrane lipids of Thermoanaerobacter ethanolicus 39E. J. Lipid Res. 35:1057–1065 [PubMed] [Google Scholar]
- 61.Clarke NG, Hazlewood GP, Dawson RMC. 1980. Structure of diabolic acid-containing phospholipids isolated from Butyrivibrio sp. Biochem. J. 191:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung S, Hollingsworth RI. 1994. Structures and stereochemistry of the very long α,ω-bifunctional alkyl species in the membrane of Sarcina ventriculi indicate that they are formed by tail-to-tail coupling of normal fatty acids. J. Lipid Res. 35:1932–1945 [PubMed] [Google Scholar]
- 63.Huber R, Langworthy TA, König H, Thomm M, Woese CR, Sleytr UB, Stetter KO. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324–333. 10.1007/BF00409880 [DOI] [Google Scholar]
- 64.Windberger E, Huber R, Trincone A, Fricke H, Stetter KO. 1989. Thermotoga thermarum sp. nov. and Thermotoga neapolitana occurring in African continental solfataric springs. Arch. Microbiol. 151:506–512. 10.1007/BF00454866 [DOI] [Google Scholar]
- 65.Jeanthon C, Reysenbach A-L, L'Haridon S, Gambacorta A, Pace NR, Glénat P, Prieur D. 1995. Thermotoga subterranea sp. nov., a new thermophilic bacterium isolated from a continental oil reservoir. Arch. Microbiol. 164:91–97. 10.1007/BF02525313 [DOI] [PubMed] [Google Scholar]
- 66.Huber R, Rossnagel P, Woese CR, Rachel R, Langworthy TA, Stetter KO. 1996. Formation of ammonium from nitrate during chemolithoautotrophic growth of the extremely thermophilic bacterium Ammonifex degensii gen. nov. sp. nov. Syst. Appl. Microbiol. 19:40–49. 10.1016/S0723-2020(96)80007-5 [DOI] [PubMed] [Google Scholar]
- 67.Huber R, Wilharm T, Huber D, Trincone A, Burggraf S, Konig H, Rachel R, Rockinger I, Fricke H, Stetter KO. 1992. Aquifex pyrophilus gen. nov. sp. nov. represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst. Appl. Microbiol. 15:340–351. 10.1016/S0723-2020(11)80206-7 [DOI] [Google Scholar]
- 68.Langworthy TA, Holzer G, Zeikus JG, Tornabene TG. 1983. Iso-branched and anteiso-branched glycerol diethers of the thermophilic anaerobe Thermodesulfotobacterium commune. Syst. Appl. Microbiol. 4:1–17. 10.1016/S0723-2020(83)80029-0 [DOI] [PubMed] [Google Scholar]
- 69.Rutters H, Sass H, Cypionka H, Rullkotter J. 2001. Monoalkylether phospholipids in the sulfate-reducing bacteria Desulfosarcina variabilis and Desulforhabdus amnigenus. Arch. Microbiol. 176:435–442. 10.1007/s002030100343 [DOI] [PubMed] [Google Scholar]
- 70.Hamilton-Brehm SD, Gibson RA, Green SJ, Hopmans EC, Schouten S, Van der Meer MTJ, Shields JP, Sinninghe Damsté JS, Elkins JG. 2013. Thermodesulfobacterium geofontis sp. nov., a hyperthermophilic, sulfate-reducing bacterium isolated from Obsidian Pool, Yellowstone National Park. Extremophiles 17:251–263. 10.1007/s00792-013-0512-1 [DOI] [PubMed] [Google Scholar]
- 71.Wagner F, Rottem S, Held HD, Uhlig S, Zahringer U. 2000. Ether lipids in the cell membrane of Mycoplasma fermentans. Eur. J. Biochem. 267:6276–6286. 10.1046/j.1432-1327.2000.01709.x [DOI] [PubMed] [Google Scholar]
- 72.van de Vossenberg JLCM, Driessen AJM, Konings WN. 1998. The essence of being extremophilic: the role of the unique archaeal membrane lipids. Extremophiles 2:163–170. 10.1007/s007920050056 [DOI] [PubMed] [Google Scholar]
- 73.Weijers JWH, Schouten S, van den Donker JC, Hopmans EC, Sinninghe Damsté JS. 2007. Environmental controls on bacterial tetraether membrane lipid distribution in soils. Geochim. Cosmochim. Acta 71:703–713. 10.1016/j.gca.2006.10.003 [DOI] [Google Scholar]
- 74.Peterse F, van der Meer J, Schouten S, Weijers JW, Fierer N, Jackson RB, Kim JH, Sinninghe Damsté JS. 2012. Revised calibration of the MBT-CBT paleotemperature proxy based on branched tetraether membrane lipids in surface soils. Geochim. Cosmochim. Acta 96:215–229. 10.1016/j.gca.2012.08.011 [DOI] [Google Scholar]
- 75.Tierney J, Russell J, Eggermont H, Hopmans E, Verschuren D, Sinninghe Damsté JS. 2010. Environmental controls on branched tetraether lipid distributions in tropical east African lake sediments. Geochim. Cosmochim. Acta 74:4902–4918. 10.1016/j.gca.2010.06.002 [DOI] [Google Scholar]
- 76.Weijers JWH, Schouten S, Spaargaren OC, Sinninghe Damsté JS. 2006. Occurrence and distribution of tetraether membrane lipids in soils: implications for the use of the TEX86 proxy and the BIT index. Org. Geochem. 37:1680–1693. 10.1016/j.orggeochem.2006.07.018 [DOI] [Google Scholar]
- 77.De Jonge C, Hopmans EC, Stadnitskaia A, Rijpstra WI, Hofland R, Tegelaar E, Sinninghe Damsté JS. 2013. Identification of novel penta- and hexamethylated branched glycerol dialkyl glycerol tetraethers in peat using HPLC-MS2, GC-MS and GC-SMB-MS. Org. Geochem. 54:78–82. 10.1016/j.orggeochem.2012.10.004 [DOI] [Google Scholar]