Abstract
Helicobacter pylori causes numerous alterations in gastric epithelial cells through processes that are dependent on activity of the cag type IV secretion system (T4SS). Filamentous structures termed “pili” have been visualized at the interface between H. pylori and gastric epithelial cells, and previous studies suggested that pilus formation is dependent on the presence of the cag pathogenicity island (PAI). Thus far, there has been relatively little effort to identify specific genes that are required for pilus formation, and the role of pili in T4SS function is unclear. In this study, we selected 7 genes in the cag PAI that are known to be required for T4SS function and investigated whether these genes were required for pilus formation. cagT, cagX, cagV, cagM, and cag3 mutants were defective in both T4SS function and pilus formation; complemented mutants regained T4SS function and the capacity for pilus formation. cagY and cagC mutants were defective in T4SS function but retained the capacity for pilus formation. These results define a set of cag PAI genes that are required for both pilus biogenesis and T4SS function and reveal that these processes can be uncoupled in specific mutant strains.
INTRODUCTION
Helicobacter pylori is a curved, Gram-negative bacterium that persistently colonizes the gastric mucosa in about 50 percent of humans (1, 2). Most persons colonized with H. pylori remain asymptomatic, but the presence of H. pylori is associated with an increased risk of gastric adenocarcinoma, gastric lymphoma, and peptic ulcer disease (3). Gastric adenocarcinoma is the second leading cause of cancer-related death worldwide (4). H. pylori strains containing a 40-kb chromosomal region known as the cag pathogenicity island (PAI) are associated with an increased risk of gastric cancer or ulcer disease compared to strains that lack the cag PAI (5–7). The cag PAI encodes the effector protein CagA and multiple proteins that constitute a type IV secretion system (T4SS) (8–11).
CagA is the only known effector protein translocated by the H. pylori cag T4SS (12). Upon entry into gastric epithelial cells, CagA undergoes phosphorylation by host cell kinases at EPIYA motifs (13, 14). Phosphorylated and nonphosphorylated forms of CagA can interact with multiple cellular proteins, resulting in an array of phenotypic changes in the epithelial cells (12). These include remodeling of the actin cytoskeleton, alterations in cellular morphology (including an elongated cell shape known as the hummingbird phenotype) (14), increased cell motility (15), and a transition of polarized epithelial monolayers to an invasive phenotype (16).
The cag T4SS has an important role in activation of proinflammatory signal transduction pathways in gastric epithelial cells. Interaction between cag PAI-positive H. pylori and gastric epithelial cells results in upregulated expression of multiple cytokines, including the proinflammatory cytokine interleukin 8 (IL-8) (17–19). IL-8 induction is thought to be triggered by the entry of H. pylori peptidoglycan into host cells through a cag T4SS-dependent process (20), and CagA may also stimulate IL-8 production (21–23).
T4SSs are present in multiple Gram-negative species, including Agrobacterium tumefaciens, Legionella pneumophila, Bordetella pertussis, and Brucella suis (24, 25). These T4SSs can translocate protein, DNA, or both into eukaryotic cells. The A. tumefaciens VirB-VirD secretion system and related conjugation systems have been studied in the most detail, and these serve as models for understanding other T4SSs (26–28). The A. tumefaciens T4SS consists of 12 proteins encoded by the virB and virD operons. Several genes within the H. pylori cag PAI demonstrate sequence similarity to genes that encode components of T4SSs in other bacterial species (7–9), but the level of sequence relatedness is very weak in most cases.
When H. pylori is cocultured with gastric epithelial cells, filamentous structures can be detected at the bacterium-host cell interface (29–37). The assembly of these structures is dependent on the presence of the cag PAI (31, 32, 34–36), and CagA has been visualized at the tips of the structures (29, 31, 37). Based on these observations, it has been suggested that the structures are components of the cag T4SS utilized for translocation of CagA into host cells, and the structures have been termed “pili,” analogous to pilus-like structures in the A. tumefaciens T4SS (38). There is considerable uncertainty about the composition of H. pylori pili. The major pilin protein in H. pylori was suggested to be CagC, based on weak sequence similarities between CagC and the major pilus protein (VirB2) in T4SSs of other bacterial species (39, 40). CagC has been localized to the surface of H. pylori (39) but has not been localized to the pili that form at the interface between H. pylori and gastric epithelial cells.
Thus far, there has been relatively little effort to identify specific genes that are required for pilus formation in H. pylori. Several early reports commented that cagT, cagX, cagY, and cagα (also known as virB11 or HP0525) are required for pilus formation, but in each case, the data were not shown (31, 34, 36). More recently, one study reported that cagI and cagL are required for pilus production (35). Another recent study used a tetR-tetO system to regulate H. pylori gene expression and reported that the cagU-cagT operon is required for pilus production, but experiments were not done to determine which of these two genes is essential (32).
In summary, numerous studies have reported that H. pylori contact with gastric epithelial cells stimulates the formation of pili, and there is evidence that these pili are associated with the cag T4SS. Relatively little is known about the genetic requirements for pilus formation and the composition of pili, and the role of pili in cag T4SS function is unclear. The goal of this study was to analyze a set of genes in the cag PAI that are known to be required for T4SS function (19) and determine whether these genes are also required for pilus formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strains used in this study are listed in Table 1. Marked mutant strains (containing insertions of antibiotic cassettes into genes of interest), unmarked mutants, and complemented mutants were all derived from wild-type (WT) strain 26695. H. pylori strains were cultured on Trypticase soy agar plates supplemented with 5% sheep blood or Brucella agar plates supplemented with 5% fetal bovine serum at 37°C in room air containing 5% CO2. H. pylori mutant strains were selected based on resistance to chloramphenicol (5 μg/ml), kanamycin (10 μg/ml), metronidazole (7.5 μg/ml), or streptomycin (50 μg/ml). Escherichia coli strain DH5α, used for plasmid propagation, was grown on Luria-Bertani agar plates or in Luria-Bertani liquid medium supplemented with ampicillin (50 μg/ml), chloramphenicol (25 μg/ml), or kanamycin (25 μg/ml), as appropriate.
TABLE 1.
H. pylori cag mutant strains used in the current study
| Description of mutant | Gene mutated | Nucleotides deleted | Insertion position (nucleotide) | Complemented gene | Antibiotic resistance cassettea | Antibiotic resistanceb | Genotype of parental strain |
|---|---|---|---|---|---|---|---|
| Marked mutants | |||||||
| cagC::cat-rdx | cagC | 219 | cat-rdx | Chl | ΔrdxA | ||
| ΔcagM marked | cagM | 70–909 | cat-rdx | Chl | ΔrdxA | ||
| cagT::cat-rdx | cagT | 542 | cat-rdx | Chl | ΔrdxA | ||
| ΔcagV marked | cagV | 139–720 | cat-rpsL | Chl | ΔrpsL | ||
| ΔcagX marked | cagX | 139–1,470 | cat-rpsL | Chl | ΔrpsL | ||
| ΔcagY marked | cagY | 1–5,745 | cat-rpsL | Chl | ΔrpsL | ||
| Δcag3 marked | cag3 | 1–1,203 | cat-rpsL | Chl | ΔrpsL | ||
| Δcag PAI | cag PAI | All | cat-rdx | Chl | ΔrdxA | ||
| Unmarked mutants | |||||||
| ΔcagC unmarked | cagC | 78–341 | Met | ΔrdxA | |||
| ΔcagM unmarked | cagM | 70–909 | Met | ΔrdxA | |||
| ΔcagV unmarked | cagV | 139–720 | Strep | ΔrpsL | |||
| ΔcagX unmarked | cagX | 139–1,470 | Strep | ΔrpsL | |||
| Δcag3 unmarked | cag3 | 1–1,203 | Strep | ΔrpsL | |||
| Complemented mutants | |||||||
| ΔcagC unmarked complemented | cagC | cat | Chl, Met | ΔrdxA | |||
| ΔcagM unmarked complemented | cagM | cat | Chl, Met | ΔrdxA | |||
| cagT::cat-rdx complemented | cagT | cat | Kan, Chl | ΔrdxA | |||
| ΔcagV unmarked complemented | cagV | cat | Chl, Strep | ΔrpsL | |||
| ΔcagX unmarked complemented | cagX | cat | Chl, Strep | ΔrpsL | |||
| Δcag3 unmarked complemented | cag3 | cat | Chl, Strep | ΔrpsL |
Mutants generated using the cat-rdx cassette were derived from the ΔrdxA parental strain. Mutants generated using the cat-rpsL cassette were derived from the ΔrpsL parental strain.
Chl, chloramphenicol; Met, metronidazole; Strep, streptomycin; Kan, kanamycin.
Cell culture methods.
AGS human gastric epithelial cells were grown in RPMI medium containing 10% fetal bovine serum (FBS) and 10 mM HEPES buffer.
Mutagenesis of H. pylori cag genes.
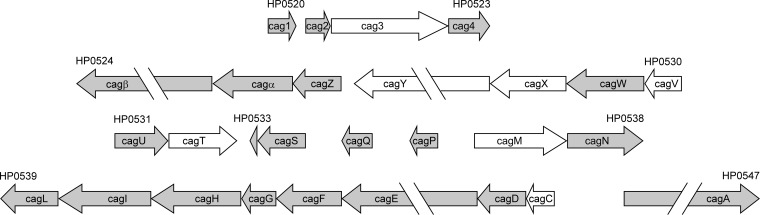
Seven genes within the cag PAI were mutated in the current study. As shown in Fig. 1, these seven genes are distributed at multiple sites with the cag PAI and are found within several different operons. To mutate cag genes, we utilized two contraselectable mutagenesis strategies. We generated mutants in cagC, cagM, and cagT through the use of a cat-rdxA cassette (35). In brief, we generated a metronidazole-resistant strain (ΔrdxA) by deleting the rdxA gene and then introduced a cat-rdx cassette into the relevant cag genes, as described further below. This cassette confers resistance to chloramphenicol via the chloramphenicol acetyltransferase (cat) gene from Campylobacter coli and susceptibility to metronidazole mediated by an intact rdxA gene (HP0954) from H. pylori 26695. We generated strains carrying mutations in cagV, cagX, and cag3 through the use of a cat-rpsL cassette (29, 41, 42). In brief, we generated a streptomycin-resistant strain of 26695 by introducing a mutation conferring streptomycin resistance (A-to-G substitution at nucleotide 128, resulting in Lys43Arg) into HP1197 (rps12 or rpsL). We then introduced a cat-rpsL cassette, conferring chloramphenicol resistance and containing the WT rpsL gene (which confers dominant streptomycin susceptibility), into the relevant cag genes, as described further below.
FIG 1.
Relative locations of mutated genes within the cag PAI. Individual cag genes and their orientations are shown. Gene designations (e.g., HP0520) indicate gene numbers in H. pylori strain 26695. Genes mutated in this study are shown in white. Gene length and spacing between operons are drawn to scale. Several genes (cagβ, cagY, cagE, and cagA) are only partially displayed due to their large size. The operon structure is based on the work of Sharma et al. and Ta et al. (53, 54).
To generate strains carrying deletion mutations in cagM, cagV, cagX, and cag3, we synthesized plasmids containing approximately 500 bp of sequence upstream and downstream from genes targeted for mutagenesis (GenScript USA Inc., Piscataway, NJ). Introduction of EcoRI and PstI sites flanking the cag gene sequences facilitated insertion into the pUC57 vector. In each case, we deleted a majority of the coding sequence from the target gene of interest and replaced it with a multiple cloning site containing SacI, BamHI, and XmaI sites. For each gene, the deleted nucleotides are listed in Table 1. To generate marked mutants, we cloned either the cat-rdx cassette or the cat-rpsL cassette into the multiple cloning site contained within each targeted gene, and the resulting plasmids (which fail to replicate in H. pylori) were used to transform either the H. pylori ΔrdxA strain or the rpsL mutant, respectively, followed by selection on chloramphenicol. To generate unmarked mutants, we transformed the marked mutants (containing cat-rdx or cat-rpsL cassettes in appropriate sites within the cag PAI) with the plasmids described above (lacking the cat-rdx or cat-rpsL cassette), and selected metronidazole- or streptomycin-resistant transformants (exhibiting a loss of chloramphenicol resistance). A cagY mutant strain was generated as described previously (29).
To construct a cagC marked mutant, we PCR amplified cagC along with approximately 0.5 kb of flanking DNA from H. pylori 26695 genomic DNA using AmpliTaq Gold (ABI) and cloned the PCR product into pGEM-T Easy (Promega), resulting in the plasmid pSFTC1. We inserted the cat-rdx cassette into an endogenous EcoRV site at nucleotide 219, generating plasmid pSFTC1–2. Transformation of the H. pylori ΔrdxA strain (35) and selection on chloramphenicol resulted in isolation of a cagC marked mutant. To generate the unmarked mutant, pSFTC1–2 was digested with XcmI and NheI restriction sites located near the 5′ and 3′ termini of the cagC gene, respectively. The ends were blunted with Klenow and ligated, resulting in excision of the cat-rdx cassette and cagC nucleotides 78 to 340. The resulting plasmid, pSFTC1–3, was used to transform the cagC-marked mutant, and metronidazole selection yielded a mutant in which cagC was deleted (cagC unmarked mutant).
To construct a cagT mutant strain, we PCR amplified cagT along with approximately 0.5 kb of flanking DNA from H. pylori 26695 genomic DNA using AmpliTaq Gold (ABI) and cloned the PCR product into pGEM-T Easy (Promega), resulting in plasmid pSFTC2. The cat-rdx cassette was cloned into the BclI site at nucleotide 542 to yield pSFTC2–1. The H. pylori ΔrdxA strain (35) was transformed with this plasmid. Selection on chloramphenicol resulted in isolation of a cagT marked mutant.
To complement the mutant strains, we introduced the relevant intact genes into the ureA chromosomal locus, which is located about 470 kb from the cag PAI in H. pylori strain 26695. Complementation was accomplished by using plasmids derived from pAD1 (35, 43). To complement the cagT marked mutant, we modified the pAD1 plasmid to contain a kanamycin resistance cassette, restriction sites to allow cloning of a gene of interest into a site downstream from the ureA promoter and a ribosomal binding site, and flanking sequences derived from the ureA and ureB loci; this plasmid is designated pADK. We also used an additional plasmid derived from pAD1 (35, 43), pADC. pADC is similar to pADK but contains a chloramphenicol resistance cassette. Plasmids derived from pADC were constructed to allow expression of cagC, cagM, cagV, cagX, and cag3. Site-directed mutagenesis was used to introduce an A279T nucleotide mutation into cagM and an A444T mutation into cagX; these silent mutations eliminated intergenic XbaI sites to facilitate cloning.
Generation of rabbit polyclonal antiserum.
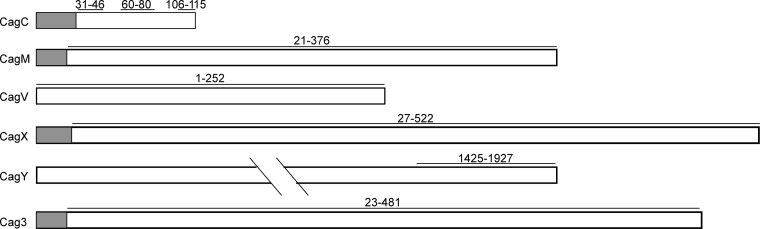
Rabbit anti-CagT antiserum has been described previously (32). To generate CagC antiserum, three peptides representing residues 31 to 46, 60 to 80, and 106 to 115 of CagC were synthesized (39, 40) (Covance, Princeton, NJ) (Fig. 2). These peptides are predicted to be surface exposed using various topology prediction programs. Rabbits were immunized with a mixture of the three peptides. Cag3, CagX, and CagM derived from H. pylori 26695 were expressed individually from the pET151/D-TOPO vector (Life Technologies, formerly Invitrogen) as His-tagged fusion proteins lacking the N-terminal signal sequence. Cag proteins were purified using nickel chromatography. Full-length CagV and the C-terminal 502 amino acids of CagY were expressed as glutathione S-transferase (GST) fusion proteins from the pGEX-6P-1 vector (GE Healthcare, formerly Amersham). GST fusion proteins were purified using glutathione beads (44). Rabbits were then immunized with purified His-tagged Cag proteins or GST-tagged Cag proteins, as approved by the Institutional Animal Care and Use Committee of Vanderbilt University School of Medicine.
FIG 2.
Peptides or recombinant proteins used for generating polyclonal antisera. Antisera against six Cag proteins were generated as described in Materials and Methods. Anti-CagC serum was generated by immunizing rabbits with three peptides corresponding to the regions illustrated by horizontal black lines. Antisera to the other five proteins were generated by immunizing rabbits with recombinant proteins corresponding to the regions indicated (horizontal black lines). Shaded regions indicate predicted signal sequences. Numbers indicate amino acid positions within the indicated proteins.
Immunoblot analysis.
To detect production of Cag proteins, individual samples were separated by SDS-PAGE (4 to 20% gradient), transferred to a nitrocellulose membrane, and subsequently immunoblotted using rabbit polyclonal antiserum raised against the indicated recombinant Cag protein. To confirm similar loading of samples, immunoblotting using a rabbit polyclonal antiserum to H. pylori HspB, a GroEL homolog, was utilized (45). Horseradish peroxidase-conjugated anti-rabbit IgG was used as the second antibody. Signals were generated by an enhanced chemiluminescence reaction and detection by exposure to X-ray film.
IL-8 secretion by gastric cells in contact with H. pylori.
H. pylori strains were cocultured with AGS cells at a multiplicity of infection of 100:1, and IL-8 secretion was analyzed using an anti-human IL-8 sandwich enzyme-linked immunosorbent assay (ELISA) (R&D). The levels of IL-8 secreted by AGS cells in response to isogenic cag mutant strains were compared to levels secreted by AGS cells in response to the respective parental rdxA mutant or rpsL mutant strains from which the cag mutant strains were derived. Results are shown below as means ± standard deviations (SD), based on data from 3 to 5 experiments.
Scanning electron microscopy of H. pylori in contact with gastric epithelial cells.
Samples were prepared as described previously (35). H. pylori and AGS human gastric epithelial cells were cocultured at a multiplicity of infection (MOI) of 100:1 on tissue culture-treated coverslips (BD Biosciences) for 4 h at 37°C in the presence of 5% CO2. Cells were fixed with 2.0% paraformaldehyde–2.5% glutaraldehyde in 0.05 M sodium cacodylate buffer for 1 h at 37°C. Coverslips were washed with sodium cacodylate buffer, and secondary fixation was performed with 1% osmium tetroxide at room temperature for 15 min. Coverslips were washed with sodium cacodylate buffer and dehydrated with sequential washes of increasing concentrations of ethanol. Samples were then dried at the critical point, mounted onto sample stubs, grounded with a thin strip of silver paint at the sample edge, and sputter coated with gold/palladium before viewing with an FEI Quanta 250 field emission gun scanning electron microscope. Image analysis was performed using the Image J software program. For quantitative analysis of the number of visible pili per bacterial cell, images of at least 20 adherent bacterial cells, derived from three separate experiments, were analyzed. When selecting images for quantitation, we chose images that allowed a clear view of a relatively large region of the bacterium-host interface, thereby allowing the maximum number of structures to be visualized. To evaluate mutants with no visible pili, at least 100 adherent bacteria were visualized. For quantitative analysis of the proportion of bacteria with visible pili, images of at least 10 fields, derived from three separate experiments, were analyzed.
RESULTS
Pilus formation by parental H. pylori strains.
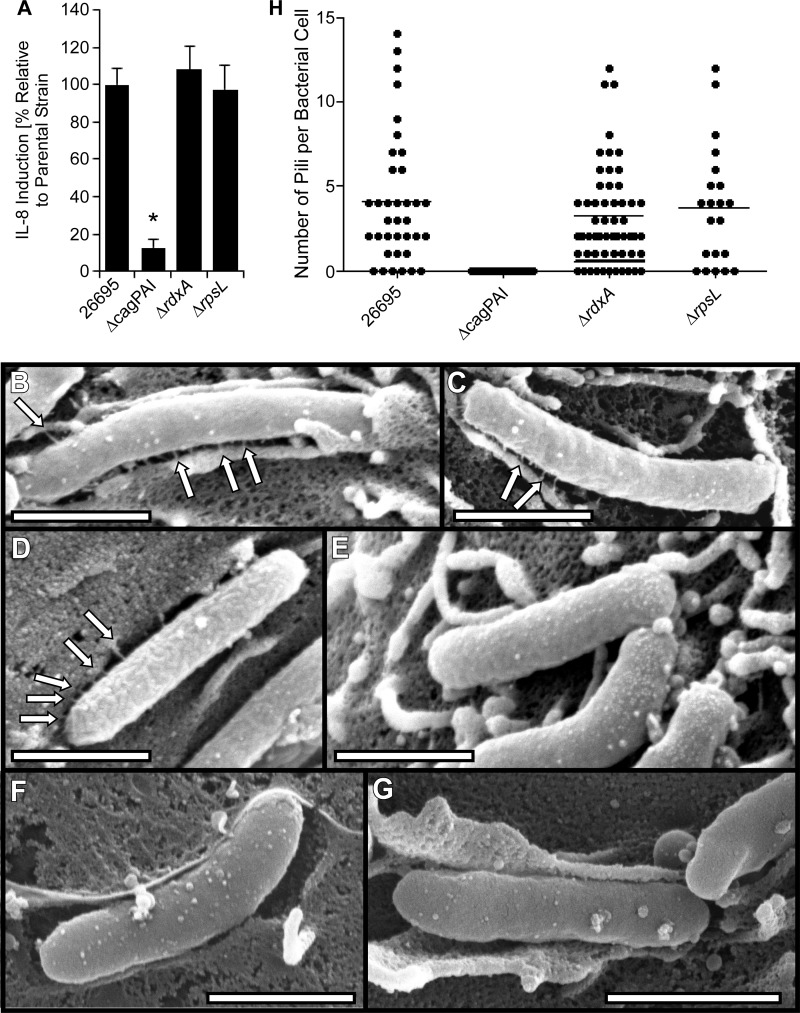
In the current study, we mutated multiple genes in the cag PAI by using procedures that were dependent on the generation of either metronidazole-resistant or streptomycin-resistant H. pylori strains as an initial step (29, 35, 41, 42). To ensure that introduction of mutations encoding metronidazole or streptomycin resistance did not alter the activity of the cag T4SS or pilus formation, we cocultured the metronidazole- or streptomycin-resistant strains with AGS cells and then analyzed IL-8 production (a T4SS-dependent phenotype) as well as pilus formation; the WT strain 26695 and a Δcag PAI mutant strain (35) were tested in parallel as controls. As expected, the WT strain, metronidazole-resistant strain (ΔrdxA), and streptomycin-resistant strain (rpsL mutant) each stimulated IL-8 secretion, whereas the Δcag PAI mutant strain did not (Fig. 3). To evaluate pilus formation, we cocultured the bacteria with AGS cells and then analyzed the adherent bacteria by scanning electron microscopy (SEM) as described in Materials and Methods. As expected, the WT strain produced pili, whereas the Δcag PAI mutant strain did not (Fig. 3 and Table 2). The rdxA and rpsL mutant strains each retained the capacity to assemble pili at the bacterium-host cell interface. Consistent with previous results (29, 35), the pili were approximately 13 nm in width and 75 nm in length, with an average of about 3 or 4 pili visualized per adherent bacterial cell (ranging from no visible pili to >10 visible pili per bacterium) (Table 2 and Fig. 3H). The number of pili visualized per bacterium is likely to be substantially less than the total number of pili that are actually present.
FIG 3.
Characterization of parental strains. (A) AGS cells were infected with the indicated strains of H. pylori for 4 h, and levels of secreted IL-8 were quantified by ELISA of cell culture supernatants. Levels of IL-8 secreted in response to rdxA or rpsL mutant strains were compared to the levels of IL-8 secreted in response to WT strain 26695. The rdxA and rpsL mutants induced levels of IL-8 production similar to that of the wild-type 26695 strain from which they were derived, and the Δcag PAI mutant induced significantly lower levels of IL-8 production. *, P < 0.01 in comparison to results for wild-type strain 26695 (Student's t test). (B to G) H. pylori strains were cocultured with AGS cells, and pilus formation at the bacterium-host cell interface was analyzed by SEM. Wild-type H. pylori 26695 (B), the rpsL mutant (C), the ΔrdxA mutant (D), or the Δcag PAI mutant (E, F, and G) is shown. (H) Number of pili visualized per bacterium. Bars, 1 μm. Arrows point to pili.
TABLE 2.
Quantitative analysis of H. pylori pilia
| Strain name or description | Mean (± SE) no. of pili per bacterial cellb | % (± SE) of bacteria with visible pilic |
|---|---|---|
| 26695 | 4.1 ± 0.6 | 81.9 ± 5.9 |
| Δcag PAI | Not detected* | Not detected* |
| ΔrdxA | 3.1 ± 0.4 | 82.3 ± 3.2 |
| ΔrpsL | 3.8 ± 0.8 | 72.5 ± 8.6 |
| cag3 marked mutant | 0.02 ± 0.02* | 0.0 ± 0.0* |
| cag3 unmarked mutant | Not detected* | Not detected* |
| cag3 complemented mutant | 5.0 ± 1.1 | 75.0 ± 5.8 |
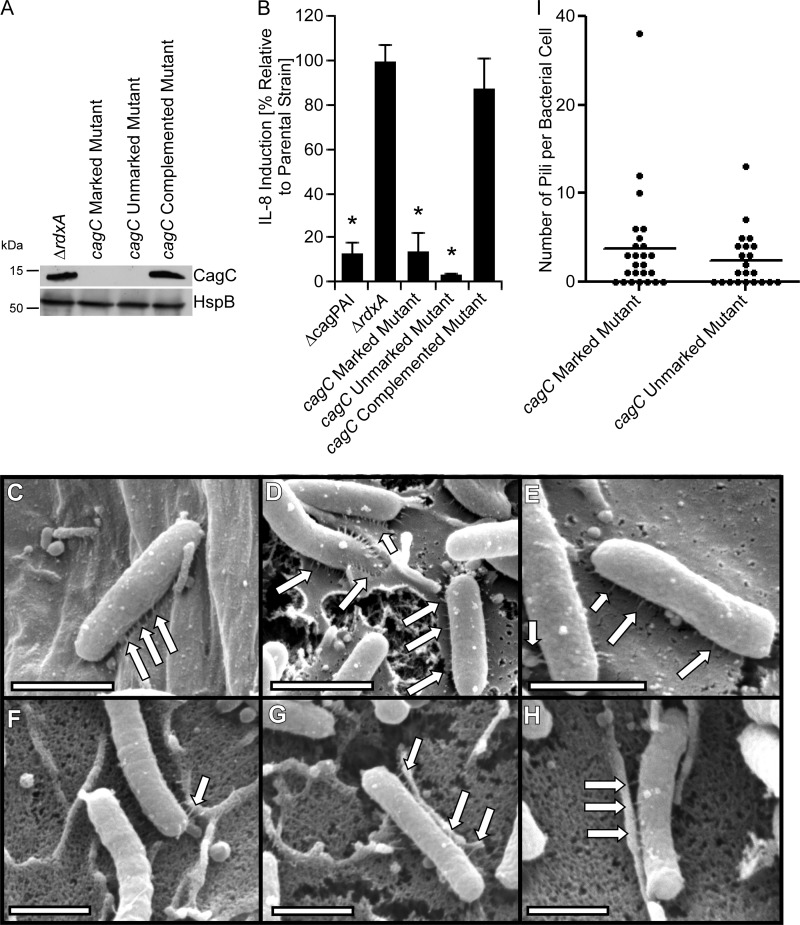
| cagC marked mutant | 3.8 ± 1.2 | 79.0 ± 6.1 |
| cagC unmarked mutant | 2.4 ± 0.7 | 70.8 ± 7.5 |
| cagM marked mutant | 0.1 ± 0.1* | 0.0 ± 0.0* |
| cagM unmarked mutant | Not detected* | Not detected* |
| cagM complemented mutant | 3.7 ± 0.9 | 85.8 ± 4.6 |
| cagT marked mutant | Not detected* | Not detected* |
| cagT complemented mutant | 5.7 ± 0.8* | 74.4 ± 5.4 |
| cagV marked mutant | Not detected* | Not detected* |
| cagV unmarked mutant | Not detected* | Not detected* |
| cagV complemented mutant | 4.8 ± 1.2 | 79.2 ± 8.6 |
| cagX marked mutant | Not detected* | Not detected* |
| cagX unmarked mutant | Not detected* | Not detected* |
| cagX complemented mutant | 4.4 ± 1.1 | 77.3 ± 8.4 |
| cagY marked mutant | 5.4 ± 1.1 | 81.7 ± 6.2 |
SE, standard error of the mean. *, P < 0.005 compared to results for the parental strain, based on two-tailed t test analysis.
Images of at least 20 adherent bacterial cells, derived from three separate experiments, were analyzed. Among the strains capable of pilus production, nonpiliated bacteria were included in the analysis.
Percent piliated bacteria were quantified based on images of at least 10 fields, derived from three separate experiments.
Analysis of CagC.
Previously it was noted that H. pylori CagC exhibits weak sequence relatedness to the A. tumefaciens major pilus protein VirB2, including the presence of two predicted transmembrane regions, a long predicted N-terminal signal sequence, and a C-terminal motif that exhibits weak sequence relatedness to the cyclization motif of other T4SS pilin proteins, and it was proposed that CagC corresponds to the major pilin in H. pylori (39, 40). Therefore, we hypothesized that a cagC mutant strain should be defective in pilus formation. To test this hypothesis, we generated marked and unmarked cagC mutant strains, as well as a complemented mutant strain. Immunoblotting confirmed that CagC was produced by the parental strain and complemented mutant strain but not by cagC mutant strains (Fig. 4). In contrast to the parental rdxA mutant strain, neither the marked nor the unmarked cagC mutants induced IL-8 production by AGS cells. Complementation of cagC restored the WT phenotype. Thus, consistent with previous results (19, 39), these data indicate that CagC is required for T4SS function (Fig. 4). We then analyzed the capacity of these strains to form pili when cultured with AGS cells (Fig. 4 and Table 2). The marked and unmarked cagC mutants produced pili that appeared similar to those produced by WT 26695 and the parental rdxA mutant strain. The dimensions of wild-type pili were 77.6 ± 3.3 nm (mean length ± standard error of the mean [SE]) and 13.6 ± 0.6 nm (mean width ± SE), and the dimensions of pili produced by the cagC mutant strain were 72.2 nm ± 3.2 nm and 15.0 ± 0.6 nm (P = 0.048 compared to the width of the wild-type pili). The biological significance of this small difference in width is uncertain. These findings indicate that CagC is not required for pilus formation.
FIG 4.
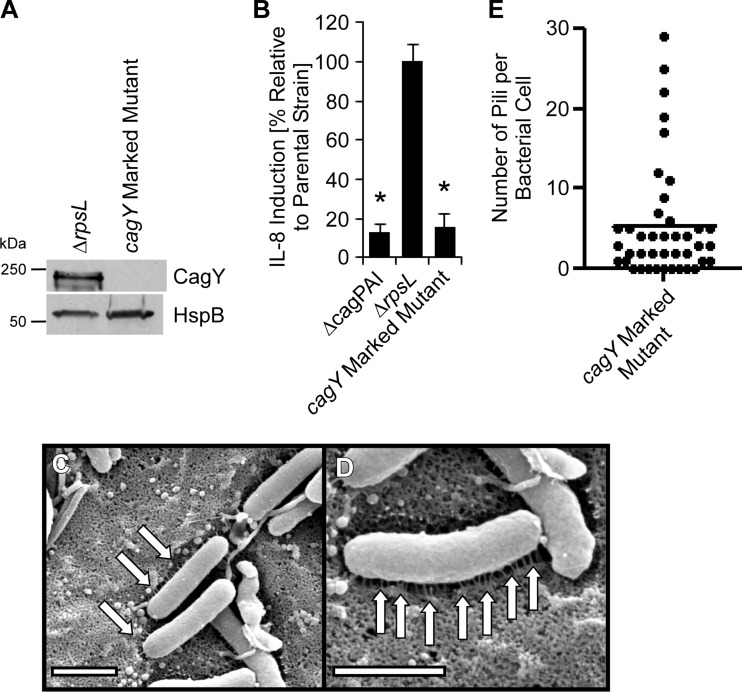
Analysis of cagC mutants. A marked cagC mutant strain (containing an antibiotic cassette inserted in cagC), unmarked cagC mutant, and complemented mutant were generated as described in Materials and Methods. (A) Production of CagC and a control protein (HspB) was assessed by immunoblot analysis. (B) AGS cells were infected with the indicated strains of H. pylori, and IL-8 secretion was quantified by ELISA of cell culture supernatants. *, P < 0.01 in comparison to findings for the parental strain (Student's t test). (C to H) H. pylori strains were cocultured with AGS cells, and pilus formation was then analyzed by SEM. Panels C to E show the cagC marked mutant. Panels F to H show the cagC unmarked mutant. (I) Number of pili visualized per bacterium. Bars, 1 μm. Arrows point to pili.
Analysis of CagY.
A recent study reported that CagY (a VirB10 homolog) in H. pylori strain J166 was required for T4SS function but was not required for pilus formation (29). To further investigate the role of CagY in pilus formation, we generated a cagY mutant derived from strain 26695. As expected, the cagY mutant was defective in the ability to stimulate IL-8 production by AGS cells (Fig. 5). Similar to results observed in the J166 strain background (29), the cagY mutant derived from strain 26695 retained the capacity to form pili (Fig. 5 and Table 2). These pili appeared similar to those produced by WT strain 26695. The dimensions of wild-type pili were 77.6 ± 3.3 nm (mean length ± SE) and 13.6 ± 0.6 nm (mean width ± SE), and the dimensions of pili produced by the cagY mutant strain were 73.0 nm ± 2.6 nm and 12.1 ± 0.5 nm (P = 0.02 compared to width of wild-type pili). The biological significance of this small difference in width is uncertain. These findings indicate that CagY is not required for pilus formation.
FIG 5.
Analysis of a cagY mutant. A marked cagY mutant strain (containing an antibiotic resistance cassette inserted into the cagY locus) was generated as described in Materials and Methods. (A) Production of CagY and a control protein (HspB) was assessed by immunoblot analysis. (B) AGS cells were infected with the indicated strains of H. pylori for 4 h, and IL-8 secretion was quantified by ELISA. *, P < 0.01 in comparison to results for the parental strain (Student's t test). (C and D) The marked cagY mutant was cocultured with AGS cells, and pilus formation was then analyzed by SEM. (E) Number of pili visualized per bacterium. Bars, 1 μm. Arrows point to pili.
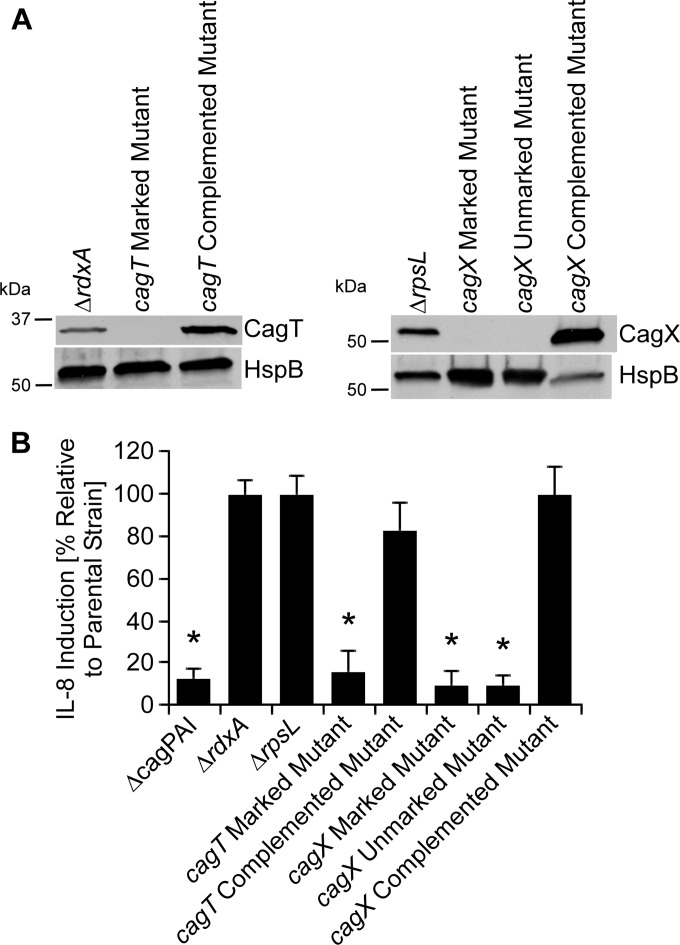
Analysis of CagX and CagT.
Based on the findings that CagC and CagY were each required for T4SS function but not pilus formation, we undertook further experiments to investigate the relationship between genes required for T4SS function and genes required for pilus formation. We first investigated the roles of CagX and CagT. These are considered to be homologs of the T4SS core complex proteins VirB9 and VirB7, respectively, but the level of sequence relatedness between CagT and VirB7 is very low (9, 11). We generated marked cagX and cagT mutant strains, as described in Materials and Methods. Unlike cagT, which is the terminal gene in the cagU-cagT operon, cagX is located in the middle of an operon (9, 11). Therefore, to minimize the possibility of polar effects that might be associated with the presence of an antibiotic cassette, we also generated an unmarked cagX mutant. Immunoblotting indicated that CagX and CagT were present in the rdxA and rpsL mutant parental strains but absent in the relevant cagX and cagT mutant strains; production of CagX and CagT was restored in the complemented mutant strains (Fig. 6). Consistent with previous reports (19), cagX and cagT mutants did not induce IL-8 production by AGS cells, and complemented mutants stimulated IL-8 induction in a manner similar to that observed in the parental strain.
FIG 6.
Immunoblot analysis of cagT and cagX mutants and requirement of these proteins for T4SS-dependent induction of IL-8 secretion. Marked cagT and cagX mutant strains (containing antibiotic cassettes inserted into cagT or cagX), an unmarked cagX mutant, and complemented mutant strains were generated as described in Materials and Methods. (A) Production of CagT and CagX and a control protein (HspB) was assessed by immunoblot analysis. (B) AGS cells were infected with the indicated strains of H. pylori for 4 h. Levels of secreted IL-8 were quantified by ELISA of cell culture supernatants. *, P < 0.01 in comparison to results for the parental strain (Student's t test).
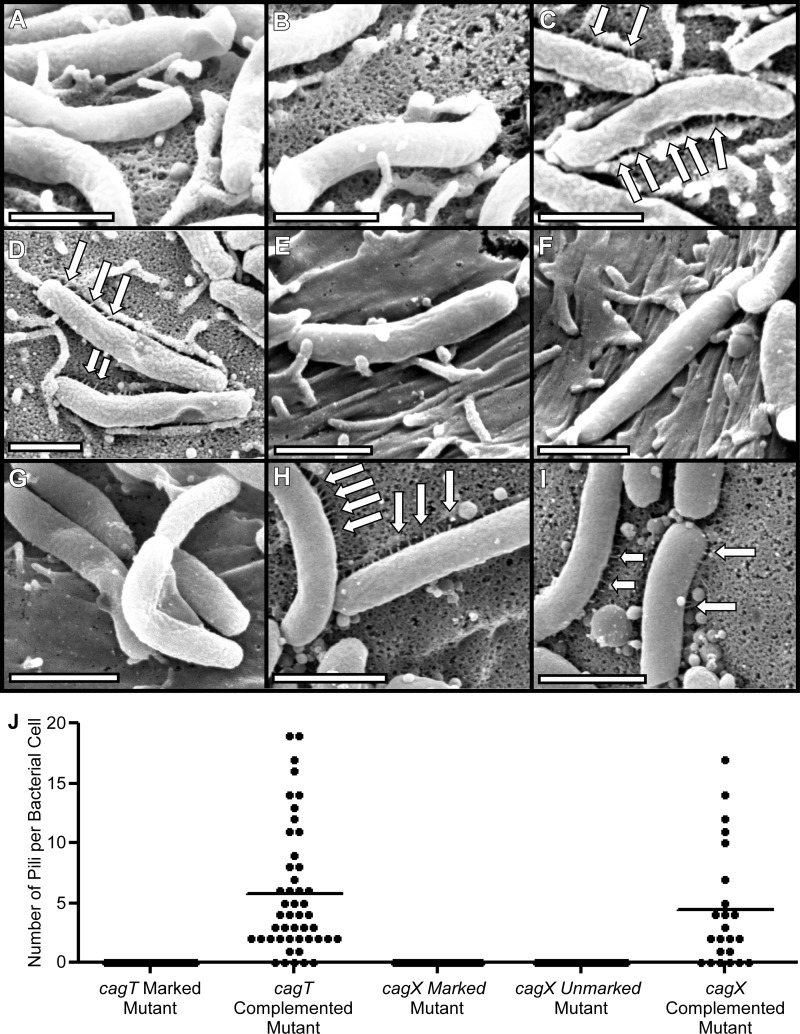
In contrast to the parental strains, the cagT and cagX mutants did not produce pili (Fig. 7 and Table 2). Complementation of cagT and cagX mutants restored the ability of the strains to produce pili when in the presence of gastric epithelial cells. These data indicate that CagX and CagT are required for both T4SS function and pilus formation.
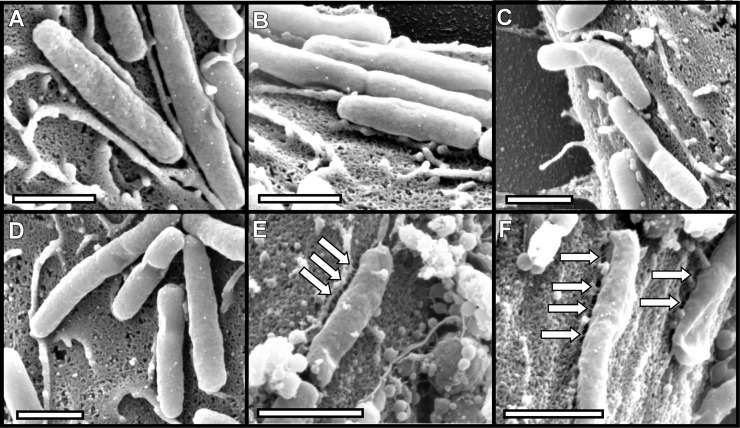
FIG 7.
Requirement of CagT and CagX for pilus formation. H. pylori strains were cocultured with AGS cells, and pilus formation was analyzed by SEM. (A and B) cagT marked mutant; (C and D) cagT complemented mutant; (E and F) cagX marked mutant; (G) cagX unmarked mutant; (H and I) cagX complemented mutant. Only the cagT and cagX complemented mutant strains produced pili. Arrows point to pili. Bars, 1 μm. (J) Number of pili visualized per bacterium.
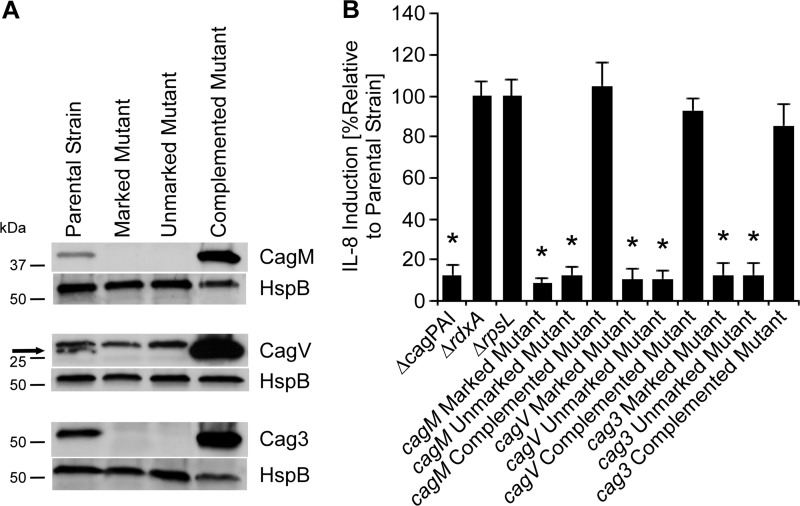
Analysis of CagV, CagM, and Cag3.
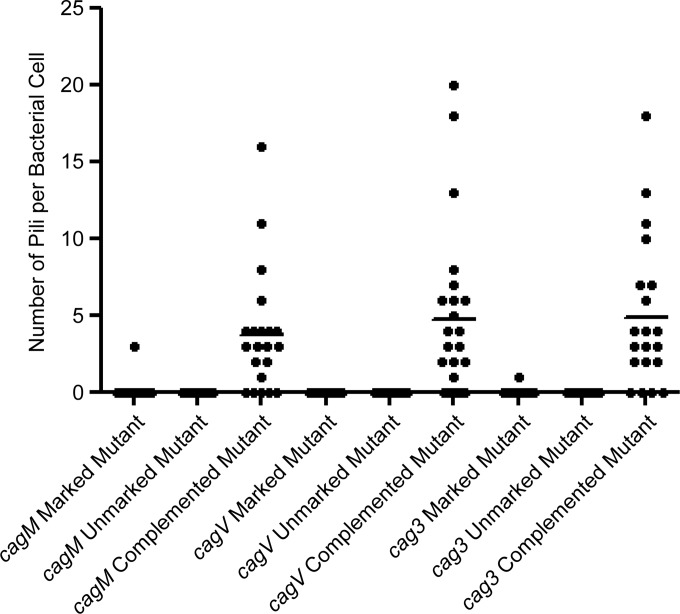
We next investigated the roles of three additional Cag proteins (CagV, CagM, and Cag3) that are reported to be required for T4SS function (19). CagV exhibits weak sequence relatedness to VirB8 (46), whereas CagM and Cag3 do not exhibit relatedness to components of T4SSs in other bacterial species. Immunoblotting indicated that cagV, cagM, and cag3 mutant strains did not express the corresponding Cag proteins and did not induce IL-8 production, whereas Cag protein production and IL-8 induction were restored in the complemented mutant strains (Fig. 8). Upon coculture with AGS cells, the cagV, cagM, and cag3 mutants did not produce pili (Fig. 9 to 12 and Table 2). In contrast, the complemented mutant strains regained the capacity for pilus production (Fig. 9 to 12 and Table 2). These data indicate that CagV, CagM, and Cag3 are required for both T4SS function and pilus formation.
FIG 8.
Immunoblot analysis of cagM, cagV, and cag3 mutants and requirement of these proteins for T4SS-dependent induction of IL-8 secretion. Marked cagM, cagV, and cag3 mutant strains (containing antibiotic cassettes inserted into these genes), unmarked mutants, and complemented mutants were generated as described in Materials and Methods. (A) Production of CagM, CagV, Cag3, and a control protein (HspB) was assessed by immunoblot analysis. Arrow designates CagV. (B) AGS cells were infected with the indicated strains, and IL-8 secretion was quantified by ELISA of cell culture supernatants. *, P < 0.01 in comparison to results for the parental strain (Student's t test).
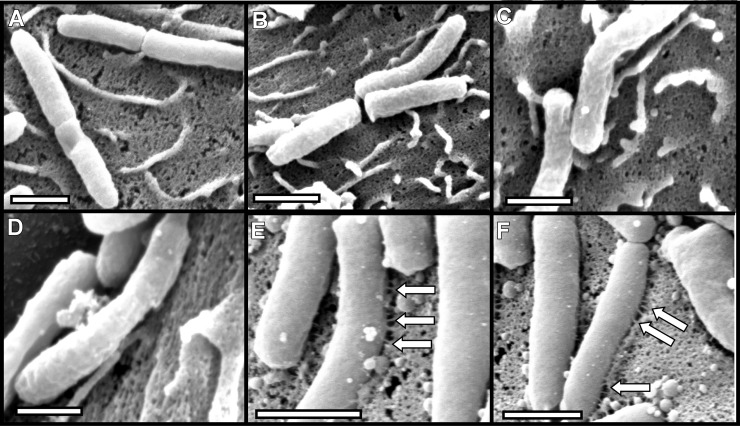
FIG 9.
Requirement of CagM for pilus formation. H. pylori strains were cocultured with AGS cells, and pilus formation was analyzed by SEM. (A and B) Marked cagM mutants; (C and D) unmarked cagM mutants; (E and F) complemented mutants. Bars, 1 μm. Arrows point to pili.
FIG 12.
Number of pili visualized per bacterium in studies of cagM, cagV, and cag3 mutant strains. Representative SEM images were used to enumerate the number of visible pili per bacterium.
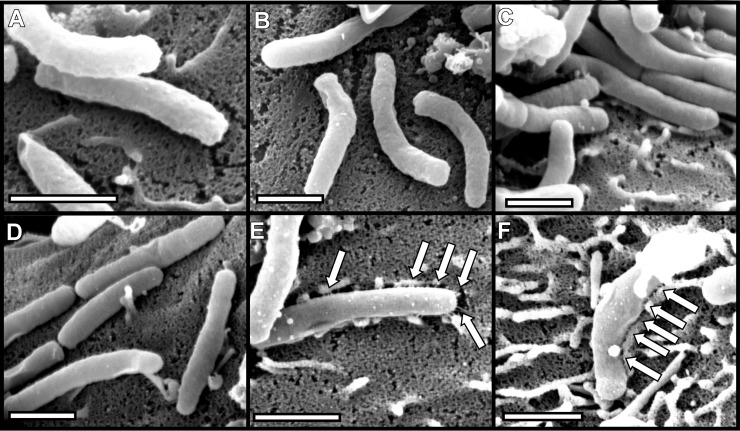
FIG 10.
Requirement of CagV for pilus formation. H. pylori strains were cocultured with AGS cells, and pilus formation was analyzed by SEM. (A and B) Marked cagV mutants; (C and D) unmarked cagV mutants; (E and F) complemented mutants. Bars, 1 μm. Arrows point to pili.
FIG 11.
Requirement of Cag3 for pilus formation. H. pylori strains were cocultured with AGS cells, and pilus formation was analyzed by SEM. (A and B) Marked cag3 mutants; (C and D) unmarked cag3 mutants; (E and F) complemented mutants. Bars, 1 μm. Arrows point to pili.
DISCUSSION
When cocultured in vitro with gastric epithelial cells, H. pylori forms structures termed “pili” at the bacterium-host cell interface (29–37). Previous studies reported that a cag PAI mutant strain does not produce pili (29, 31–36), but thus far, there has been very little effort to identify specific genes that are required for pilus production. In the current study, we analyzed 7 genes in the cag PAI that are known to be required for T4SS function (19) and tested the hypothesis that each of these genes would also be required for pilus production. Several of the gene products selected for analysis exhibit sequence relatedness to components of T4SSs in non-H. pylori bacterial species, whereas others (such as Cag3 and CagM) are unrelated to T4SS components in non-H. pylori species (9, 11). We report that five of the mutant strains analyzed (containing deletions in cagM, cagT, cagV, cagX, and cag3) failed to produce pili, and pilus production was restored by complementation of the 5 mutant strains. Because the complementation assays were performed by inserting an intact copy of the wild-type allele into an exogenous locus on the chromosome (ureA), these experiments indicate that the mutated genes are required for T4SS function and pilus formation and that the observed effects were not attributable to polar effects of the mutations. A previous study reported that two additional genes in the cag PAI (cagL and cagI) are also required for pilus production (35). The genes required for pilus production are scattered throughout the cag PAI and are located in multiple different operons (Fig. 1). Collectively, these data reveal a set of genes within the cag PAI that are required for pilus production.
The current results, when taken together with evidence from previous studies, provide a body of evidence supporting the view that the pili are part of the cag T4SS encoded by the cag PAI. First, mutant strains that fail to produce pili are consistently defective in a cag T4SS-dependent phenotype (IL-8 induction in gastric epithelial cells). Second, several previous studies have shown that CagA is localized at or near the tips of the pili (29, 31, 37). Third, changes in environmental conditions (such as a reduced iron concentration) lead to an increase in pilus production, and this is associated with an increase in cag T4SS activity (33). Finally, the width of the structures visualized in the current study (about 13 to 14 nm in diameter) is similar to the reported width of pilus structures associated with type IV secretion systems in Agrobacterium tumefaciens and other bacteria (8 to 12 nm) (38).
The pilus structures visualized in the current study are slightly variable in size. This may be due to uneven metal coating, differences in the depth of field, or true biologic diversity. One of the important findings in the current study is that we do not visualize any structures at the bacterium-host interface when imaging a cagX, cagT, cagM, cagV, or cag3 mutant strain. If multiple different types of structures were produced by H. pylori under the conditions of these experiments, we would expect to see some structures still produced by these mutant strains. Thus, we do not have evidence pointing to the production of multiple different types of structures at the H. pylori-host cell interface under these conditions. If multiple structures are produced, we are visualizing the most common type of structure, and multiple genes in the cag PAI are required for production of these structures.
The H. pylori pili visualized in the current study appear similar to those that have been visualized in previous studies, and multiple different groups have reported that CagA is localized at or near the tips of the pili (29, 31, 37). However, there are several issues that deserve comment. First, there are differences when comparing the imaging methodology used in various studies. The images in the current study and several other studies were generated by coating with a thin layer of gold and palladium (29, 32, 33, 35), whereas the images in several other papers were generated by coating with carbon (31, 34). This difference may account at least in part for the increased width of structures that were described in a previous study compared to the width of structures visualized when using gold and palladium coating (29, 30, 32, 35). The current study shows exclusively images of structures that are produced when H. pylori is in contact with gastric epithelial cells. In contrast, some previous publications have also shown images of structures produced by H. pylori grown in pure culture (34, 36). Thus far, we have not visualized pili when H. pylori is grown in pure culture. It seems possible that the structures produced by bacteria grown in pure culture may differ from structures produced by bacteria cocultured with gastric epithelial cells.
The exact composition of H. pylori pili remains very poorly understood. Prior to the current study, it was suggested that CagC might be the major pilin subunit (39), but this seems unlikely since cagC mutant strains still produce pili. A major challenge in defining the composition of pili is the limitation in available methods for investigating this topic. Thus far, the main approach has involved labeling the pili with antibodies directed against specific Cag proteins that are involved in T4SS-dependent phenotypes. This approach is only feasible if specific antisera suitable for use in immunogold labeling studies are available, and this approach would not be successful in identifying pilus components if they were encoded by genes outside the cag PAI. In previous studies, four proteins (CagL, CagT, CagX, and CagY) were reported to be localized to pili, based on immunologic detection using either electron microscopic or confocal microscopic imaging methods (31, 34, 36). In several cases, these proteins were localized to surface structures of H. pylori that were cultured in the absence of gastric epithelial cells (34, 36), and there is uncertainty about whether or not such structures are identical to the more numerous pili that form when H. pylori is in contact with gastric epithelial cells (30).
As an alternate to immunogold labeling, it is possible that pilus components might be identified if overexpression of a component led to changes in the number of pili per bacterium or changes in pilus dimensions. Several of the complemented mutant strains generated in the current study appeared to produce increased levels of individual Cag proteins, based on Western blot analysis. However, these complemented mutants did not exhibit any substantial alterations in pilus number or pilus dimensions compared to the wild-type or parental strains. We detected a statistically significant increase in the number of pili produced by a cagT complemented mutant (and a small decrease in pilus length) in comparison to the WT strain (Table 2), but the small magnitude of these changes is unlikely to be biologically significant. One current hypothesis is that the pili may be comprised of CagL and CagI (35, 47). In support of this hypothesis, one previous study reported detection of CagL as a minor component of pili (31), CagL interacts with CagI (35, 48), and both of these proteins are capable of binding to integrins on host cells (31, 35, 37).
Surprisingly, we found that cagY and cagC are required for T4SS function but dispensable for pilus formation. Similarly, a previous study reported that cagH was required for T4SS function but not pilus formation (35). These data indicate that T4SS function and pilus production can be uncoupled in specific mutant strains. Mutant strains in which T4SS function and pilus production are uncoupled are potentially valuable tools for dissecting the assembly and function of the T4SS. The C-terminal region of CagY exhibits sequence relatedness to VirB10 (5, 9), which is a major component of the core complex in other T4SSs (49). In the A. tumefaciens VirB/D T4SS, VirB10 acts both as a structural component and as a gating element controlling substrate transfer (50), and specific mutations in VirB10 block pilus formation (50–52). The current results, as well as findings of recent studies of cagY in a different H. pylori strain background (29), indicate that cagY is required for T4SS function but is not required for pilus production. This suggests that there may be important differences in the mechanisms of T4SS pilus synthesis in H. pylori from those of synthesis of pili in T4SSs involved in conjugation. Moreover, H. pylori CagY is substantially larger in size than VirB10 proteins in other bacterial species, which suggests that CagY may have specialized functions different from those of VirB10 in other species. CagC exhibits weak sequence relatedness to VirB2, the major component of pili in the A. tumefaciens VirB/D T4SS (9, 39, 40). The current data confirm that CagC has an important function required for T4SS activity but suggest that CagC is not the major component of the H. pylori pili visualized in this study. It will be important in future studies to analyze the cagY and cagC mutants further in an effort to identify the defects that lead to a loss of T4SS function.
In summary, the current data provide new insights into the genetic requirements for pilus formation in H. pylori and provide additional evidence that these pili are components of the cag T4SS. These data also highlight numerous differences between the H. pylori cag T4SS and T4SSs in other bacterial species. In future work, it will be important to more clearly define the composition of cag T4SS-associated pili and to better define the role of these structures in T4SS function.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI068009, AI039657, and CA116087, the Department of Veterans Affairs, and the Medical Center for Postgraduation Education, Poland (CMKP grant 501-2-1-08-39/08). Antibody production was supported by the National Center for Research Resources, grant UL1RR024975-01, and is now at the National Center for Advancing Translational Sciences, grant 2UL1TR000445-06. Experiments in the Vanderbilt Cell Imaging Shared Resource were supported by Vanderbilt University Digestive Disease Research Center (NIH grant P30DK058404) and the Vanderbilt University Ingram Cancer Center (NIH grant P30 CA068485).
We thank Douglas Berg and Jay Solnick for providing plasmid DNA.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 2 June 2014
REFERENCES
- 1.Atherton JC, Blaser MJ. 2009. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J. Clin. Invest. 119:2475–2487. 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–1873. 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peek RM, Jr, Blaser MJ. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28–37. 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs CS, Mayer RJ. 1995. Gastric carcinoma. N. Engl. J. Med. 333:32–41. 10.1056/NEJM199507063330107. [DOI] [PubMed] [Google Scholar]
- 5.Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, Bukanov NO, Drazek ES, Roe BA, Berg DE. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37–53. [DOI] [PubMed] [Google Scholar]
- 6.Blaser MJ. 2005. The biology of cag in the Helicobacter pylori-human interaction. Gastroenterology 128:1512–1515. 10.1053/j.gastro.2005.03.053. [DOI] [PubMed] [Google Scholar]
- 7.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. U. S. A. 93:14648–14653. 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourzac KM, Guillemin K. 2005. Helicobacter pylori-host cell interactions mediated by type IV secretion. Cell Microbiol. 7:911–919. 10.1111/j.1462-5822.2005.00541.x. [DOI] [PubMed] [Google Scholar]
- 9.Fischer W. 2011. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J. 278:1203–1212. 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 10.Tegtmeyer N, Wessler S, Backert S. 2011. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 278:1190–1202. 10.1111/j.1742-4658.2011.08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terradot L, Waksman G. 2011. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J. 278:1213–1222. 10.1111/j.1742-4658.2011.08037.x. [DOI] [PubMed] [Google Scholar]
- 12.Hatakeyama M. 2004. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 4:688–694. 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 13.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497–1500. 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 14.Segal ED, Cha J, Lo J, Falkow S, Tompkins LS. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 96:14559–14564. 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Churin Y, Al-Ghoul L, Kepp O, Meyer TF, Birchmeier W, Naumann M. 2003. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J. Cell Biol. 161:249–255. 10.1083/jcb.200208039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagnoli F, Buti L, Tompkins L, Covacci A, Amieva MR. 2005. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc. Natl. Acad. Sci. U. S. A. 102:16339–16344. 10.1073/pnas.0502598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillemin K, Salama NR, Tompkins LS, Falkow S. 2002. Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc. Natl. Acad. Sci. U. S. A. 99:15136–15141. 10.1073/pnas.182558799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tummuru MK, Sharma SA, Blaser MJ. 1995. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol. Microbiol. 18:867–876. 10.1111/j.1365-2958.1995.18050867.x. [DOI] [PubMed] [Google Scholar]
- 19.Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337–1348. 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 20.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Memet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5:1166–1174. 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 21.Brandt S, Kwok T, Hartig R, Konig W, Backert S. 2005. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc. Natl. Acad. Sci. U. S. A. 102:9300–9305. 10.1073/pnas.0409873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SY, Lee YC, Kim HK, Blaser MJ. 2006. Helicobacter pylori CagA transfection of gastric epithelial cells induces interleukin-8. Cell Microbiol. 8:97–106. 10.1111/j.1462-5822.2005.00603.x. [DOI] [PubMed] [Google Scholar]
- 23.Lamb A, Yang XD, Tsang YH, Li JD, Higashi H, Hatakeyama M, Peek RM, Blanke SR, Chen LF. 2009. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 10:1242–1249. 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149. 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo HJ, Waksman G. 2004. Unveiling molecular scaffolds of the type IV secretion system. J. Bacteriol. 186:1919–1926. 10.1128/JB.186.7.1919-1926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451–485. 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015. 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–268. 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrozo RM, Cooke CL, Hansen LM, Lam AM, Gaddy JA, Johnson EM, Cariaga TA, Suarez G, Peek RM, Jr, Cover TL, Solnick JV. 2013. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog. 9:e1003189. 10.1371/journal.ppat.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson EM, Gaddy JA, Cover TL. 2012. Alterations in Helicobacter pylori triggered by contact with gastric epithelial cells. Front. Cell Infect. Microbiol. 2:17. 10.3389/fcimb.2012.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, Konig W, Backert S. 2007. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449:862–866. 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 32.McClain MS, Duncan SS, Gaddy JA, Cover TL. 2013. Control of gene expression in Helicobacter pylori using the Tet repressor. J. Microbiol. Methods 95:336–341. 10.1016/j.mimet.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noto JM, Gaddy JA, Lee JY, Piazuelo MB, Friedman DB, Colvin DC, Romero-Gallo J, Suarez G, Loh J, Slaughter JC, Tan S, Morgan DR, Wilson KT, Bravo LE, Correa P, Cover TL, Amieva MR, Peek RM., Jr 2013. Iron deficiency accelerates Helicobacter pylori-induced carcinogenesis in rodents and humans. J. Clin. Invest. 123:479–492. 10.1172/JCI64373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohde M, Puls J, Buhrdorf R, Fischer W, Haas R. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49:219–234. 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 35.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 7:e1002237. 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka J, Suzuki T, Mimuro H, Sasakawa C. 2003. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 5:395–404. 10.1046/j.1462-5822.2003.00286.x. [DOI] [PubMed] [Google Scholar]
- 37.Jimenez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, Terradot L, Fischer W, Haas R. 2009. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 5:e1000684. 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703–714. 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrzejewska J, Lee SK, Olbermann P, Lotzing N, Katzowitsch E, Linz B, Achtman M, Kado CI, Suerbaum S, Josenhans C. 2006. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J. Bacteriol. 188:5865–5877. 10.1128/JB.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. 2008. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J. Bacteriol. 190:2161–2171. 10.1128/JB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dailidiene D, Dailide G, Kersulyte D, Berg DE. 2006. Contraselectable streptomycin susceptibility determinant for genetic manipulation and analysis of Helicobacter pylori. Appl. Environ. Microbiol. 72:5908–5914. 10.1128/AEM.01135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Styer CM, Hansen LM, Cooke CL, Gundersen AM, Choi SS, Berg DE, Benghezal M, Marshall BJ, Peek RM, Jr, Boren T, Solnick JV. 2010. Expression of the BabA adhesin during experimental infection with Helicobacter pylori. Infect. Immun. 78:1593–1600. 10.1128/IAI.01297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loh JT, Forsyth MH, Cover TL. 2004. Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect. Immun. 72:5506–5510. 10.1128/IAI.72.9.5506-5510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busler VJ, Torres VJ, McClain MS, Tirado O, Friedman DB, Cover TL. 2006. Protein-protein interactions among Helicobacter pylori cag proteins. J. Bacteriol. 188:4787–4800. 10.1128/JB.00066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivie SE, McClain MS, Algood HM, Lacy DB, Cover TL. 2010. Analysis of a beta-helical region in the p55 domain of Helicobacter pylori vacuolating toxin. BMC Microbiol. 10:60. 10.1186/1471-2180-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buhrdorf R, Forster C, Haas R, Fischer W. 2003. Topological analysis of a putative virB8 homologue essential for the cag type IV secretion system in Helicobacter pylori. Int. J. Med. Microbiol. 293:213–217. 10.1078/1438-4221-00260. [DOI] [PubMed] [Google Scholar]
- 47.Barden S, Schomburg B, Conradi J, Backert S, Sewald N, Niemann HH. 2014. Structure of a three-dimensional domain-swapped dimer of the Helicobacter pylori type IV secretion system pilus protein CagL. Acta Crystallogr. D Biol. Crystallogr. 70:1391–1400. 10.1107/S1399004714003150. [DOI] [PubMed] [Google Scholar]
- 48.Pham KT, Weiss E, Jimenez Soto LF, Breithaupt U, Haas R, Fischer W. 2012. CagI is an essential component of the Helicobacter pylori Cag type IV secretion system and forms a complex with CagL. PLoS One 7:e35341. 10.1371/journal.pone.0035341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808. 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banta LM, Kerr JE, Cascales E, Giuliano ME, Bailey ME, McKay C, Chandran V, Waksman G, Christie PJ. 2011. An Agrobacterium VirB10 mutation conferring a type IV secretion system gating defect. J. Bacteriol. 193:2566–2574. 10.1128/JB.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garza I, Christie PJ. 2013. A putative transmembrane leucine zipper of agrobacterium VirB10 is essential for t-pilus biogenesis but not type IV secretion. J. Bacteriol. 195:3022–3034. 10.1128/JB.00287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie PJ. 2009. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 71:779–794. 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255. 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 54.Ta LH, Hansen LM, Sause WE, Shiva O, Millstein A, Ottemann KM, Castillo AR, Solnick JV. 2012. Conserved transcriptional unit organization of the cag pathogenicity island among Helicobacter pylori strains. Front. Cell Infect. Microbiol. 2:46. 10.3389/fcimb.2012.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]