Abstract
Burkholderia pseudomallei, the etiologic agent of melioidosis, is an opportunistic pathogen that harbors a wide array of secretion systems, including a type II secretion system (T2SS), three type III secretion systems (T3SS), and six type VI secretion systems (T6SS). The proteins exported by these systems provide B. pseudomallei with a growth advantage in vitro and in vivo, but relatively little is known about the full repertoire of exoproducts associated with each system. In this study, we constructed deletion mutations in gspD and gspE, T2SS genes encoding an outer membrane secretin and a cytoplasmic ATPase, respectively. The secretion profiles of B. pseudomallei MSHR668 and its T2SS mutants were noticeably different when analyzed by SDS-PAGE. We utilized liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify proteins present in the supernatants of B. pseudomallei MSHR668 and B. pseudomallei ΔgspD grown in rich and minimal media. The MSHR668 supernatants contained 48 proteins that were either absent or substantially reduced in the supernatants of ΔgspD strains. Many of these proteins were putative hydrolytic enzymes, including 12 proteases, two phospholipases, and a chitinase. Biochemical assays validated the LC-MS/MS results and demonstrated that the export of protease, phospholipase C, and chitinase activities is T2SS dependent. Previous studies had failed to identify the mechanism of secretion of TssM, a deubiquitinase that plays an integral role in regulating the innate immune response. Here we present evidence that TssM harbors an atypical signal sequence and that its secretion is mediated by the T2SS. This study provides the first in-depth characterization of the B. pseudomallei T2SS secretome.
INTRODUCTION
Burkholderia pseudomallei is the etiologic agent of melioidosis in humans and animals and has been designated a tier 1 select agent in the United States because of its potential for misuse as a biological weapon (1–3). The Gram-negative organism is present in soil and water in tropical regions, particularly in Southeast Asia and northern Australia. Melioidosis often occurs in immunocompromised individuals and is acquired from an environmental source via cutaneous inoculation, inhalation, or ingestion. The resulting infection can be latent, chronic, or acute, and disease manifestation is largely dependent on the immune status of the host, the route of infection, and the infectious dose. Clinical presentations can vary widely and may include pneumonia, lymphadenitis, cellulitis, necrotizing fasciitis, prostatitis, osteomyelitis, encephalomyelitis, parotitis, and septicemia with disseminated abscess formation (3).
B. pseudomallei possesses a large genome (∼7 Mb) that encodes numerous virulence factors, including capsular polysaccharide, lipopolysaccharide, type III secretion system 1 (T3SS-1), T3SS-3, T6SS-1, TssM, type IV pili, MviN, PlcN3, type III O-PS, type IV O-PS, LfpA, ecotin, BPSL0918, SodC, BbfA, DsbA, RelA, SpoT, and BLF1 (4–12). Many of these virulence determinants are secretion systems or secreted proteins that promote survival in vivo by disarming elements of the host immune system. B. pseudomallei is a facultative intracellular pathogen that utilizes secreted proteins to survive and replicate inside host macrophages and epithelial cells (5, 13). TssM is a secreted deubiquitinase that is expressed inside host cells (14) and plays an important role in regulating the innate immune response in tissue culture and in mice (15). The B. pseudomallei tssM gene is physically linked to the T6SS-1 and T3SS-3 gene clusters, and its expression is coregulated with that of T6SS-1 (16). However, T6SS-1 and T3SS-3 are not responsible for TssM export, and the mechanism of its secretion is currently unknown (14, 15).
The type II secretion system (T2SS), also referred to as the main terminal branch of the general secretory pathway (Gsp), is widely distributed in Gram-negative bacteria and is encoded by 12 to 15 genes (17). Proteins exported by the T2SS typically harbor N-terminal Sec or Tat signal sequences that direct their translocation across the inner membrane into the periplasmic space (18). T2SS uses the energy derived from GspE-mediated ATP hydrolysis to transport folded periplasmic substrates across the GspD outer membrane channel to the extracellular milieu (17, 19, 20). The T2SS has been described as a “fiber assembly nanomachine” because the export of protein substrates is facilitated by the addition of pseudopili to a growing membrane-spanning periplasmic pseudopilus (19, 20). In a previous study, we genetically characterized the T2SS of B. pseudomallei 1026b and found that two distinct gsp loci were required for the export of protease, phospholipase C (PLC), and lipase activities (21). The 1026b genome was recently completed (22), and the gspD to -N and gspO loci correspond to BP1026B_I0008 to BP1026B_I0016, BP1026B_I0018, BP1026B_I0019, and BP1026B_I0298, respectively. The specific proteins secreted by T2SS in B. pseudomallei have not yet been determined, and the role of these exoproducts in the murine model of infection is unknown.
Here we present a proteomic analysis of the B. pseudomallei products that are exported in a T2SS-dependent manner. The genes encoding the T2SS outer membrane channel (GspD) and the cytoplasmic ATPase (GspE) were targeted for mutation, and the resulting mutants were assessed using SDS-PAGE, liquid chromatography-tandem mass spectrometry (LC-MS/MS), biochemical assays, and virulence testing in BALB/c mice. The results demonstrate that ∼50 hydrolytic enzymes and novel proteins are secreted by the T2SS in B. pseudomallei. In addition, we show that the virulence factor TssM is also exported by the T2SS. Taken together, the data suggest that the B. pseudomallei T2SS is important for survival and persistence in both the environment and the host.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli and B. pseudomallei were grown at 37°C on LB agar (Lennox formulation), LB broth (Lennox formulation), or M9 minimal salts (Difco) containing 2 mM MgSO4, 0.5 mM CaCl2, 0.5% Casamino Acids, and 0.4% glucose (M9CG). The LB broth used for the SDS-PAGE, LC-MS/MS, and biochemical assay experiments was processed with a 10,000-molecular-weight-cutoff (MWCO) Centricon Plus-20 (Millipore) centrifugal filter device to remove high-molecular-weight proteins prior to use. When appropriate, antibiotics were added at the following concentrations: 100 μg of ampicillin (Ap), 50 μg of carbenicillin (Cb), 25 μg of streptomycin (Sm), and 25 μg of kanamycin (Km) per ml for E. coli and 25 μg of polymyxin B (Pm) and 500 μg of Km per ml for B. pseudomallei. Skim milk agar plates were made by dissolving 30 g of skim milk powder (Oxoid) in 1 liter of distilled water, adding 15 g of agar (Sigma-Aldrich), and autoclaving at 121°C for 20 min. For induction studies, isopropyl-ß-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM. A 20-mg/ml stock solution of the chromogenic indicator 5-bromo-4-chloro-3-indolyl-ß-d-galactopyranoside (X-Gal) was prepared in N,N-dimethylformamide, and 40 μl was spread onto the surface of plate medium for blue/white screening in E. coli TOP10 or E. cloni 10G chemically competent cells. All manipulations with B. pseudomallei were carried out in a class II microbiological safety cabinet located in a designated biosafety level 3 (BSL-3) laboratory.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain or plasmid | Relevant characteristics or sequencea | Source, reference, or gene |
|---|---|---|
| E. coli TOP10 | General cloning and blue/white screening | Life Technologies |
| E. cloni 10G | General cloning and blue/white screening | Lucigen |
| E. coli S17–1 | Mobilizing strain with transfer genes of RP4 integrated on chromosome; Smr Tpr | 58 |
| B. pseudomallei | ||
| MSHR668 | Isolated in 1995 in Darwin, Australia, from the blood of a 53-year-old male patient with severe melioidosis encephalomyelitis (the patient survived); Pmr | 59 |
| 668 ΔgspD | MSHR668 derivative harboring a 1,476-bp gspD in-frame deletion mutation (ΔgspD) | This study |
| 668 ΔgspE | MSHR668 derivative harboring a 744-bp gspE deletion mutation (ΔgspE) | This study |
| Plasmids | ||
| pCR2.1-TOPO | 3,931-bp TA vector; pMB1 oriR; Kmr Apr | Life Technologies |
| pCR2.1-gspD | pCR2.1-TOPO containing 3,231-bp PCR product generated with gspD-FH and gspD-RXb | This study |
| pMo130 | Suicide vector for allelic exchange in Burkholderia; ColE1 ori RK2 oriT xylE sacB Kmr | 25 |
| pMogspD | pMo130 containing gspD amplified with gspD-FH and gspD-RXb and cloned into HindIII/XbaI sites | This study |
| pMoΔgspD | pMogspD digested with PstI and religated, resulting in a 1,476-bp in-frame deletion in gspD | This study |
| pMogspE-5′ | pMo130 containing 5′ gspE PCR product amplified with BpgspE-5FH and BpgspE-5RB and cloned into HindIII/BamHIb sites | This study |
| pMoΔgspE | pMogspE-5′ containing a 3′ gspE PCR product amplified with BpgspE-3FB and BpgspE-3RXb and cloned into BamHI/XbaI sites, resulting in a 744-bp deletion in gspE | This study |
| pMo168 | Broad-host-range plasmid; pBBR1 ori mob xylE Kmr | 25 |
| pMo168-gspD | pMo168 containing a HindIII/XbaI fragment from pCR2.1-gspD | This study |
| pBHR2 | Broad-host-range plasmid; Kmr | 60 |
| pBHR2-virAG | pBHR2 constitutively expressing virAG | 60 |
| pBHR2-tssM | pBHR2 constitutively expressing tssM | 14 |
| Primers (5′-3′)b | ||
| gspD-FH | CATGAAGCTTACGAGCGGATTCACCTTCTG | gspD |
| gspD-RXb | CATGTCTAGATCGAACGGCTCGATGTGGATGTC | gspD |
| BpgspE-5FH | CATGAAGCTTTCCTCGGCGACATTCCGTGGATCG | gspE |
| BpgspE-5RB | CATGGGATCCATCCACACCTCGAGCGTGTCC | gspE |
| BpgspE-3FB | TCGAGGATCCGATCGAATACGATCTGTCC | gspE |
| BpgspE-3RXb | CATGTCTAGAACGCGTCGATCGCTTCGAAACGGAAGG | gspE |
r, resistant; s, susceptible; Sm, streptomycin; Tp, trimethoprim; Pm, polymyxin B; Km, kanamycin; Ap, ampicillin.
Restriction enzyme sites in the linker regions are underlined.
DNA manipulation.
Restriction enzymes (Roche Molecular Biochemicals), Antarctic phosphatase (New England BioLabs), and T4 DNA ligase (Roche Molecular Biochemicals) were used according to the manufacturer's instructions. When necessary, the End-It DNA end repair kit (Epicentre) was used to convert 5′ or 3′ protruding ends to blunt-ended DNA. DNA fragments used in cloning procedures were excised from agarose gels and purified with a Geneclean III kit (MP Biomedicals). Bacterial genomic DNA was prepared by using a previously described protocol (23). Plasmids were purified from overnight cultures by using a Wizard Plus SV miniprep DNA purification system (Promega).
PCR amplifications.
PCR primers are shown in Table 1. PCR products were sized and isolated using agarose gel electrophoresis, cloned using the pCR2.1-TOPO TA cloning kit (Life Technologies), and transformed into chemically competent E. coli TOP10 or E. cloni 10G. PCR amplifications were performed in a final reaction volume of 50 μl containing 1× FailSafe PCR PreMix D (Epicentre), 1.25 U FailSafe PCR enzyme mix (Epicentre), 1 μM PCR primers, and approximately 200 ng of genomic DNA. Colony PCR was utilized to screen for B. pseudomallei deletion mutants. Briefly, sucrose-resistant and Km-sensitive colonies were resuspended in 50 μl of water, and 5 μl was added to the PCR rather than purified genomic DNA. PCR cycling was performed using a PTC-150 minicycler with a Hot Bonnet accessory (MJ Research, Inc.) and heated to 97°C for 5 min. This was followed by 30 cycles of a three-temperature cycling protocol (97°C for 30 s, 55°C for 30 s, and 72°C for 1 min) and one cycle at 72°C for 10 min. For PCR products larger than 1 kb, an additional 1 min per kb was added to the extension time.
Construction of B. pseudomallei mutants.
Gene replacement experiments with B. pseudomallei were performed using the sacB-based vector pMo130, as previously described (24–26). Recombinant derivatives of pMo130 (Table 1) were electroporated into E. coli S17-1 (12.25 kV/cm) and conjugated with B. pseudomallei MSHR668 for 8 h. Pm was used to counterselect E. coli S17-1. Optimal conditions for resolution of the sacB constructs were found to be LB agar lacking NaCl and containing 10% (wt/vol) sucrose, with incubation at 25°C for 4 days. B. pseudomallei deletion mutants were identified by colony PCR using the primers flanking the deleted regions of gspD and gspE (Table 1). As expected, the PCR products generated from the mutant strains were smaller than those obtained from the wild-type strain. The mutants generated in this study did not display any noticeable growth phenotype in LB broth or on LB agar (see Fig. S1 in the supplemental material).
Protease plate assay and API 20 NE gelatinase activity.
Sterile toothpicks were used to transfer bacterial growth from LB agar plates to 3% skim milk agar plates and protease activity was detected as a zone of clearing around isolated colonies after incubation for 24 h at 37°C. API 20 NE (bioMérieux) strips were inoculated and read according to the manufacturer's recommendations. Gelatin hydrolysis in the GEL microtube was detected by the uniform diffusion of black pigment in the microtube.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
MSHR668, 668 ΔgspD, and 668 ΔgspE were grown for ∼22 h in 3 ml LB broth, and 668 ΔgspD(pMo168-gspD) was grown in 3 ml LB broth with 500 μg/ml Km. One ml of culture was pelleted in a microcentrifuge, and the supernatant was filtered through a low-protein-binding 0.45-μm Millex-HV PVDF filter apparatus (Millipore). Two hundred microliters of supernatant was precipitated with 10% trichloroacetic acid (TCA), and the resulting pellet was washed with acetone. The protein pellet was resuspended in 1× Tris-glycine SDS sample buffer (Life Technologies), boiled for 2 min, and loaded onto a 4 to 20% Precise protein gel (Thermo Scientific). Electrophoresis was performed with 1× Tris-HEPES SDS buffer (Thermo Scientific), and the gel was stained with a colloidal blue staining kit (Life Technologies). The EZ-Run protein marker (Fisher Scientific) was used as a size standard for unknown proteins in SDS-PAGE.
Denaturation and digestion of supernatant proteins for proteomic analysis.
B. pseudomallei MSHR668 and 668 ΔgspD were grown in LB broth or M9CG broth, and the supernatants were processed as described for SDS-PAGE above, except that TCA precipitation was not performed. The filter-sterilized supernatants were shipped to Bioproximity, LLC (Chantilly, VA), for proteomic analysis. Samples were prepared for digestion using the filter-assisted sample preparation (FASP) method (27). Briefly, the samples were suspended in 8 M urea, 50 mM Tris-HCl (pH 7.6), 3 mM dithiothreitol (DTT), sonicated briefly, and incubated in a Thermo-Mixer at 40°C and 1,000 rpm for 20 min. Samples were centrifuged, and the supernatant was transferred to a 30,000-MWCO Amicon device (Millipore) and centrifuged at 13,000 × g for 30 min. The remaining sample was buffer exchanged with 8 M urea, 100 mM Tris-HCl (pH 7.6) and then alkylated with 15 mM iodoacetamide. The urea concentration was reduced to 2 M. The entirety of the samples, each containing about 40 μg of total protein, was digested using 1 mg of trypsin, overnight, at 37°C on the Thermo-Mixer at 1,000 rpm. Digested peptides were collected by centrifugation.
Peptide desalting.
A portion of the digested peptides, about 20 μg, were desalted using C18 stop-and-go extraction (STAGE) tips (28). Briefly, for each sample a C18 STAGE tip was activated with methanol and then conditioned with 60% acetonitrile, 0.5% acetic acid followed by 5% acetonitrile, 0.5% acetic acid. Samples were loaded onto the tips and desalted with 0.5% acetic acid. Peptides were eluted with 60% acetonitrile, 0.5% acetic acid and lyophilized in a SpeedVac (Thermo Savant) to dryness, approximately 2 h.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Each digestion mixture was analyzed by ultra-high-performance liquid chromatography (UHPLC)-MS/MS. LC was performed on an Easy-nLC 1000 UHPLC system (Thermo). Mobile phase A was 97.5% MilliQ water, 2% acetonitrile, 0.5% acetic acid. Mobile phase B was 99.5% acetonitrile, 0.5% acetic acid. The 240-min LC gradient ran from 0% B to 35% B over 200 min and then to 80% B for the remaining time. The samples were then loaded directly onto the column. The column was 50 cm by 50 mm (inside diameter0, packed with 2 mm C18 medium (Thermo Fisher), and heated to 40°C throughout the analysis. The LC was interfaced to a quadrupole-Orbitrap mass spectrometer (Q-Exactive; Thermo Fisher) via nano-electrospray ionization. An electrospray voltage of 2.2 kV was applied. The mass spectrometer was programmed to acquire, by data-dependent acquisition, tandem mass spectra from the top 20 ions in the full scan from 350 to 1,600 m/z. Dynamic exclusion was set to 60 s, singly charged ions were excluded, the isolation width was set to 1.6 Da, the full MS resolution was set to 70,000, and the MS/MS resolution was set to 17,500. Normalized collision energy was set to 25, automatic gain control to 1e6, maximum fill MS to 20 ms, maximum fill MS/MS to 60 ms, and the underfill ratio to 0.1%.
Data processing and library searching.
Mass spectrometer RAW data files were converted to MGF format using msconvert (29). Detailed search parameters are printed in the search output XML files. Briefly, all searches required a 20-ppm precursor mass tolerance, a 0.05-Da fragment mass tolerance, strict tryptic cleavage, 0 or 1 missed cleavage, fixed modification of cysteine alkylation, variable modification of methionine oxidation, and expectation value scores of 0.01 or lower. MGF files were searched using sequence libraries listed below. MGF files were searched using X!!Tandem (30) using both the native (31) and k-score (32) scoring algorithms and by OMSSA (open mass spectrometry search algorithm) (33). All searches were performed on Amazon Web Services-based cluster compute instances using the Proteome Cluster interface. XML output files were parsed and nonredundant protein sets determined using MassSieve (34). Proteins were required to have 1 or more unique peptides across the analyzed samples with expectation value scores of 0.01 or less. The samples were searched against the B. pseudomallei (strain 668) sequence library from UniProt. The relative amount of each protein present in the MSHR668 and 668 ΔgspD samples was presented as a fold change value, and we established a ≥3-fold change cutoff to identify proteins that were differentially secreted by MSHR668 and 668 ΔgspD in rich and minimal media. LC-MS/MS was performed on the supernatants from MSHR668 and 668 ΔgspD grown in rich and minimal media on two separate occasions.
Quantitative enzyme assays.
MSHR668, 668 ΔgspD, 668 ΔgspE, and 668 ΔgspD(pMo168-gspD) were grown on three separate occasions as described in the SDS-PAGE section, and the supernatants were filtered and placed at −70°C until use. The three sets of supernatants were assayed for protease, phospholipase C (PLC), and chitinase activity. Protease activity in the samples was assessed with the protease assay kit (G-Biosciences), which uses a dye-labeled protein substrate. The proteases in the sample release dye-labeled peptides, and the absorbance is measured at 570 nm for the determination of protease activity. Mass spectrometry-grade trypsin, provided in the kit, was used as a protease standard to generate a protease activity standard curve. PLC activity in the samples was assessed by p-nitrophenylphosphorylcholine hydrolysis and was monitored spectrophotometrically by the measurement of p-nitrophenol at 410 nm (35). Chitinase activity in the sample was determined using a chitinase assay kit (Sigma-Aldrich). The kit allows the detection of exochitinase activity, endochitinase activity, and chitobiosidase activity using the substrates 4-nitrophenyl N-acetyl-β-d-glucosamide, 4-nitrophenyl β-d-N,N′,N″-triacetylchitotriose, and 4-nitrophenyl N,N′-diacetyl-β-d-chitobioside, respectively. One unit of chitinase activity will release 1 mmol of p-nitrophenol from the appropriate substrate per min at pH 4.8 at 37°C. For statistical analysis of the data, a linear mixed model was fitted for each substrate with a single fixed effect for the strains. A random effect was included to model the influence of the experiment using a variance components covariance structure. Least-squares means contrasts of the four levels of each strain were estimated for the comparison of each pair of strains. P values were adjusted by simulation to account for multiple comparisons, and P values of <0.05 were considered significant.
Immunoblotting with murine anti-Hcp1 and anti-TssM sera.
Wild-type and mutant B. pseudomallei strains were grown for 8 h in 14-ml snap-cap tubes containing 3 ml of LB for immunoblotting experiments. One-milliliter volumes were transferred to microcentrifuge tubes, and supernatants were obtained after centrifugation at 14,000 rpm for 2 min at 25°C. Supernatants were filter sterilized through 0.45-μm Millex HV PVDF filters, and 100 μl was precipitated at −20°C for 30 min using 10% TCA. Protein pellets were collected by centrifugation (14,000 rpm, 10 min) and washed once with cold acetone. The protein pellets were resuspended in 1× Tris-glycine-SDS sample buffer and loaded onto a 4 to 20% Precise protein gel, and electrophoresis was performed as described above. Proteins were transferred to Invitrolon polyvinylidene difluoride by using an XCell SureLock apparatus (Life Technologies). The membranes were blocked with 3.5% skim milk powder (EMD Chemicals, Inc.)–0.1% Tween 20 (Sigma), incubated with a 1:15,000 dilution of anti-Hcp1 murine polyclonal sera antibody (16) or a 1:5,000 dilution of anti-TssM murine polyclonal sera antibody (14), and washed three times with blocking buffer. The membranes were then reacted with a 1:5,000 dilution of peroxidase-labeled goat anti-mouse IgG (γ; KPL), washed three times with blocking buffer, washed once with PBS, and then developed with 3,3′,5,5′-tetramethylbenzidine membrane peroxidase substrate (KPL). Images were acquired using GeneSnap software on a U:Genius3 Imaging System (Syngene).
SDS sensitivity of B. pseudomallei wild-type and T2SS mutants.
Outer membrane integrity was assessed by growing strains in the presence of SDS in liquid medium as described by Ize et al. (36) with minor modifications. Briefly, strains were grown overnight in LB broth, diluted to an optical density at 600 nm (OD600) of 0.05 in LB broth containing SDS, and grown aerobically for 4 h. The OD600 of cultures grown in the absence of SDS after 4 h was typically between 0.65 and 0.75, and there was no significant difference in optical density (in the absence of SDS) between any of the strains tested. One hundred percent survival was defined as the OD600 of each strain after 4 h growth in LB without SDS. The mean percent survival between the mutants 668 ΔgspD and 668 ΔgspE and the control strain MSHR668 for increasing levels of SDS (0.01, 0.02, 0.03, and 0.04%) was compared using a general linear model. Comparisons of the mutants with the control strain were made for each level of SDS (0.01, 0.02, 0.03, and 0.04%). P values were two sided, were adjusted by Dunnett's correction to account for multiple comparisons, and were considered significant at a P value of <0.05.
BALB/c mouse challenge studies.
Bacterial strains were grown overnight in LB broth and serially diluted in PBS, and aliquots were spread onto LB agar plates to determine the number of CFU present. Six- to 8-week-old female BALB/c mice, 10 per group, were challenged by the intraperitoneal (i.p.) route with 101 to 104 CFU of B. pseudomallei MSHR668, 668 ΔgspD, and 668 ΔgspE. The animals were observed at least once daily, and moribund animals were euthanized by CO2 exposure. On day 21, the surviving animals from each group were euthanized with CO2. A Bayesian probit analysis was performed for each strain to estimate the lethal dose response curve and the 50% lethal dose (LD50). Prior distributions for each parameter were assumed to be independent, weakly informative Cauchy distributions with a center of 0 and a scale of 10. By using samples from the posterior distributions of the slope and intercept parameters from the probit analysis, the median and 95% credible intervals of the range of dose responses were estimated.
Research was conducted under an IACUC-approved protocol in compliance with the Animal Welfare Act, PHS policy, and other federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted, USAMRIID, is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International, and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals (37).
RESULTS
Construction and initial characterization of B. pseudomallei MSHR668 T2SS mutants.
The overall goal of this study was to characterize the B. pseudomallei proteins that require the T2SS for export outside the cell (i.e., the T2SS secretome). In a previous study, we isolated and characterized Tn5-OT182 mutants of B. pseudomallei 1026b that could not secrete protease activity on 3% skim milk agar plates (21). All of the transposon insertions mapped to two distinct T2SS loci, gspD-N and gspO, and further characterization demonstrated that the secretion of protease and phospholipase C activities was dependent on the T2SS. No further studies have been conducted on the B. pseudomallei T2SS, and the full complement of proteins secreted by this system is unknown. Transposon mutations often exhibit polar effects on downstream genes, and strain 1026b is weakly virulent by the i.p. route of infection in mice. As a result, in this study we constructed in-frame, internal deletion mutations in gspD and gspE in strain MSHR668 (Fig. 1A). This strain is genetically tractable and is highly virulent by the aerosol and i.p. routes of infection in BALB/c mice (Christopher K. Cote and David M. Waag, personal communication). Figure 1B shows that MSHR668 secretes protease activity on 3% skim milk agar, while no protease activity is associated with 668 ΔgspD and 668 ΔgspE. Gelatin was also hydrolyzed by MSHR668 but not by 668 ΔgspD and 668 ΔgspE (Fig. 1B). The results demonstrate that MSHR668 secretes a protease(s) that hydrolyzes both casein and gelatin. In addition, the protease secretion phenotypes of 668 ΔgspD and 668 ΔgspE provide evidence that we have constructed bona fide T2SS mutants in the MSHR668 background (21).
FIG 1.
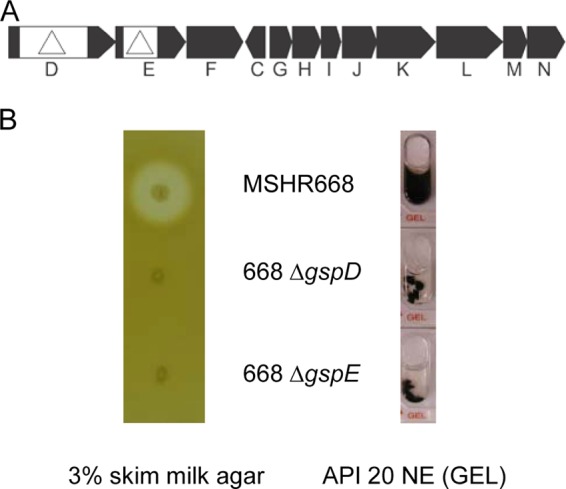
Genetic map of the gspD-N locus and screening of the extracellular proteolytic activities of ΔgspD and ΔgspE mutants. (A) Genetic map of the B. pseudomallei MSHR668 T2SS genes gspD to -N, corresponding to locus tags BURPS668_0007 to BURPS668_0010 and BURPS668_0012 to BURPS668_0019, respectively. The location and direction of transcription of the genes are represented by black arrows, and the gene names are shown below. The deletion mutations constructed in gspD and gspE in this study are shown as white boxes with triangles. (B) Secretion of proteolytic activity against casein and gelatin was assessed following growth of strains on a 3% skim milk agar plate and in an API 20 NE GEL microtube, respectively. Protease activity on the 3% skim milk agar plate was detected by a light zone of clearing around the isolated colonies and protease activity in the API 20 NE GEL microtube was detected by the uniform diffusion of black pigment in the microtube. The images are representative of three independent 3% skim milk agar plates and API 20 NE strips.
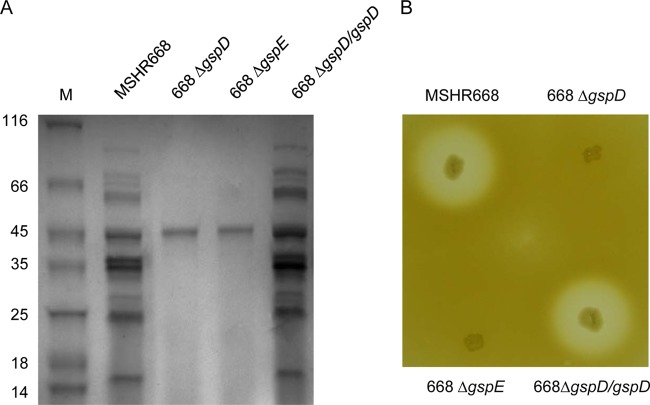
We next examined the proteins present in the supernatants of MSHR668, 668 ΔgspD, and 668 ΔgspE grown to stationary phase in LB broth. There were no obvious growth differences between these strains in this media (see Fig. S1 in the supplemental material). The bacterial supernatants were filtered, precipitated with TCA, and analyzed using SDS-PAGE (Fig. 2A). The MSHR668 supernatant contained >10 proteins, ranging in size from ∼15 to 90 kDa (Fig. 2A, first lane). By comparison, only one protein of ∼46 kDa was identifiable in the 668 ΔgspD and 668 ΔgspE supernatants (Fig. 2A, second and third lanes). We next attempted to complement the ΔgspD mutation by providing the wild-type gspD gene in trans on the broad-host-range plasmid pMo168 (Table 1). Figure 2 demonstrates that the wild-type gspD gene restored the ΔgspD secretion phenotype in rich broth (Fig. 2A) and protease activity on 3% skim milk agar (Fig. 2B). We conclude from this result that the ΔgspD mutation does not have a polar effect on downstream T2SS genes and that GspD is essential for the function of the T2SS in B. pseudomallei. Furthermore, the results indicate that the T2SS is responsible for the secretion of numerous proteins when B. pseudomallei is grown in rich broth.
FIG 2.
The T2SS mutants exhibit protein secretion deficiencies, and 668 ΔgspD can be complemented in trans. (A) SDS-PAGE of filtered and concentrated LB broth supernatant proteins. Bacterial supernatant proteins were loaded on a 4 to 20% polyacrylamide gel, subjected to SDS-PAGE, and stained with colloidal Coomassie blue. The relative migrations of molecular mass protein standards (lane M; in kDa) are indicated on the left. (B) Detection of casein hydrolysis on 3% skim milk agar plate. The protein gel and the 3% skim milk plate are representative of at least 3 separate experiments.
Proteomic analysis of MSHR668 and 668 ΔgspD grown in rich and minimal media reveals 48 putative T2SS substrates.
We next utilized LC-MS/MS to identify the proteins present in the supernatants of B. pseudomallei MSHR668 and ΔgspD grown in rich medium (LB broth) and minimal medium (M9CG broth). When the strains were grown in rich medium, there were 39 proteins present at ≥3-fold-higher levels in the MSHR668 supernatant than in the 668 ΔgspD supernatant (Table 2). In fact, ∼70% of these proteins were found exclusively in the MSHR668 supernatant. Numerous hydrolytic enzymes were present in the rich-medium T2SS secretome, including 12 distinct peptidases from the M1, M4, M23A, M28, S10, S15, and S8/S53 families (Table 2). MprA, an immunogenic serine metalloprotease (38, 39), was one of the proteases exported by the T2SS in rich medium. Two nonhemolytic phospholipase C enzymes, PlcN1 and PlcN2 (40), were also secreted in a T2SS-dependent fashion in rich medium (Table 2). The T2SS secretome in rich medium also included the acid phosphatase AcpA (41), a pectinacetylesterase, a neuraminidase/sialidase, levansucrase (SacB), a cholesterol oxidase, a Ser/Thr protein phosphatase, a chitinase, several glycosyl hydrolases, and three proteins with no known function. In comparison, 15 proteins were present at ≥3-fold-higher levels in the MSHR668 supernatant than in the 668 ΔgspD supernatant when the strains were grown in minimal medium (Table 2). Six of these proteins were also present in the rich-medium T2SS secretome (Table 2, boldface). Interestingly, the most abundant protein in the T2SS secretome in both rich and minimal media was the peptidase M28 family protein BURPS668_A0845. The T2SS secretome contains multiple proteins that are present only in rich medium (33 proteins) or in minimal medium (9 proteins), which likely reflects gene expression differences of the exported products under these distinct growth conditions (42). Taken together, the results show that the MSHR668 T2SS secretome in rich and minimal media is composed of 48 distinct proteins with an array of putative enzymatic activities.
TABLE 2.
MSHR668 supernatant proteins absent, or appreciably reduced, in the MSHR668 ΔgspD supernatant following growth in rich (LB) and minimal (M9CG) broth
| Fold changea | Locus tag | Productb | Signal sequencec |
|---|---|---|---|
| Rich medium | |||
| 252 | BURPS668_A0845 | Peptidase M28E, aminopeptidase | Sec |
| 114 | BURPS668_2745 | Non-hemolytic phospholipase C (PlcN1) | Tat |
| 85 | BURPS668_A2859 | Peptidase S15, X-Pro dipeptidyl-peptidase | Tat |
| 65 | BURPS668_3144 | Peptidase S10, serine carboxypeptidase | Sec |
| 58 | BURPS668_A1560 | Bacterial extracellular solute-binding, family 1 | Sec |
| 49 | BURPS668_3129 | Putative uncharacterized protein | Sec |
| 40 | BURPS668_A2191 | Peptidase M4, thermolysin family | Sec |
| 37 | BURPS668_A0995 | Glycoside hydrolase, family 37 protein | NCS |
| 29 | BURPS668_A0507 | Purine nucleoside permease family protein | NCS |
| 27 | BURPS668_3846 | Putative lipoprotein, DUF3443 domain | Sec |
| 19 | BURPS668_3884 | Acid phosphatase (AcpA) | Tat |
| 18 | BURPS668_3613 | Peptidase M1, alanine aminopeptidase/leukotriene A4 hydrolase | Sec |
| 18 | BURPS668_1221 | Pectinacetylesterase family protein | NCS |
| 17 | BURPS668_3454 | Peptidase S10, serine carboxypeptidase | Sec |
| 17 | BURPS668_A0147 | PhoPQ-activated pathogenicity-related protein, PqaA type | NCS |
| 15 | BURPS668_A1338 | Six-hairpin glycosidase-like domain protein | NCS |
| 14 | BURPS668_0358 | Nonhemolytic phospholipase C (PlcN2) | Tat |
| 13 | BURPS668_2554 | Putative exported protein | Sec |
| 12 | BURPS668_A2736 | Neuraminidase/sialidase | Sec |
| 9 | BURPS668_1791 | Protease-associated domain, PA | Sec |
| 8 | BURPS668_A0822 | Levansucrase (SacB) | NCS |
| 8 | BURPS668_0540 | Putative β-N-acetylglucosaminidase; glycosyl hydrolase, family 20 | Sec |
| 8 | BURPS668_0751 | Cholesterol oxidase | Tat |
| 6 | BURPS668_1311 | Pectin lyase-like superfamily | Sec |
| 6 | BURPS668_A1638 | Peptidase S15, X-Pro dipeptidyl-peptidase | Sec |
| 6 | BURPS668_A2497 | Peptidase S8/S53 domain protein | NCS |
| 6 | BURPS668_A0869 | Peptidase M23A, B-lytic metalloendopeptidase | NCS |
| 5 | BURPS668_0744 | Ser/Thr protein phosphatase family protein | Tat |
| 5 | BURPS668_A1006 | Aldehyde oxidase/xanthine dehydrogenase family protein | Tat |
| 5 | BURPS668_A2658 | Bacterial Ig-like, group 1 domain protein | Sec |
| 4 | BURPS668_A2630 | Putative N-acetylmuramoyl-l-alanine amidase | Sec |
| 4 | BURPS668_2041 | Putative exported protein | Sec |
| 4 | BURPS668_A0106 | Putative uncharacterized protein | Sec |
| 3 | BURPS668_1946 | Exported chitinase; glycosyl hydrolase, family 18 | Sec |
| 3 | BURPS668_1435 | Putative nitrite/sulfite reductase | |
| 3 | BURPS668_2874 | Cysteine synthase K/M family protein | |
| 3 | BURPS668_3648 | Ubiquinol-cytochrome c reductase, cytochrome c1 | Sec |
| 3 | BURPS668_A2861 | Serine metalloprotease MprA (Peptidase S8, subtilisin-related) | Sec |
| 3 | BURPS668_A0846 | Metallopeptidase domain protein | Sec |
| Minimal medium | |||
| 20 | BURPS668_A0845 | Peptidase M28E, aminopeptidase | Sec |
| 16 | BURPS668_3129 | Putative uncharacterized protein | Sec |
| 7 | BURPS668_3846 | Putative lipoprotein, DUF3443 domain | Sec |
| 6 | BURPS668_A1560 | Bacterial extracellular solute-binding, family 1 | Sec |
| 6 | BURPS668_3918 | Cytochrome c5 | NCS |
| 5 | BURPS668_A0759 | Chitin binding domain protein | NCS |
| 4 | BURPS668_2270 | Putative lipoprotein, Ysc84 actin-binding domain | Sec |
| 3 | BURPS668_A2191 | Peptidase M4, thermolysin family | Sec |
| 3 | BURPS668_A1718 | Cytochrome cd1-nitrite reductase-like, haem d1 domain protein | NCS |
| 3 | BURPS668_A1062 | DJ-1/PfpI family protein | NCS |
| 3 | BURPS668_3884 | Acid phosphatase (AcpA) | Tat |
| 3 | BURPS668_1157 | GntR family transcriptional regulator/aminotransferase | Sec |
| 3 | BURPS668_1132 | Epoxide hydrolase-like family protein | |
| 3 | BURPS668_0776 | Glucose-methanol-choline oxidoreductase domain protein | |
| 3 | BURPS668_0766 | Glycosyl transferase, family 51 |
Values are MSHR668/668 ΔgspD ratios; a value of 1 was assigned to proteins that were not present or were present below the limit of detection, to prevent dividing by zero.
Putative functional analysis was predicted by InterPro (61), and products in bold were present in MSHR668 supernatants in both media.
Proteins secreted by the T2SS often harbor N-terminal Sec or Tat signal sequences for translocation across the cytoplasmic membrane (17, 18). Once the exoproteins cross the cytoplasmic membrane, the signal sequences are removed, and they fold into their native conformation in the periplasmic space while awaiting transport by the T2SS. We identified Sec signal sequences on 24 proteins and Tat signal sequences on 7 proteins exported by MSHR668 in rich and minimal media, suggesting that 31/48 proteins are legitimate members of the T2SS secretome (Table 2). Twelve proteins were predicted to be “nonclassically secreted” by the SecretomeP 2.0 server (43), and 5 proteins had no identifiable signal sequence. It is known that proteins without Sec or Tat sequences can be transported across the cytoplasmic membrane, but the mechanism by which nonclassical secretion occurs is unclear (44). The fact that we have identified 12/48 proteins in the T2SS secretome without classical signal sequences suggests that the T2SS may export nonclassically secreted proteins across the outer membrane in B. pseudomallei.
Quantitative enzymatic assays confirm that protease, PLC, and chitinase activities are exported in a T2SS-dependent manner.
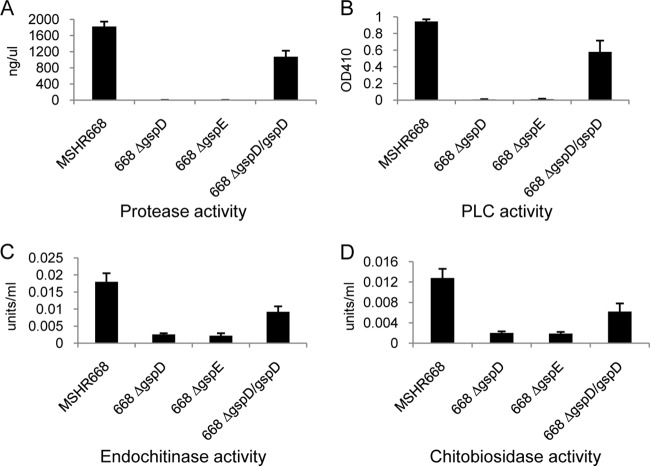
We next conducted quantitative enzymatic assays on B. pseudomallei strains grown in LB broth in an attempt to confirm that protease, PLC, and chitinase are in fact T2SS-dependent substrates. A dye-labeled protein substrate was used to assay for proteolytic activity, and we found that the MSHR668 supernatant contained 1,819 ng/μl of protease activity while the 668 ΔgspD and 668 ΔgspE supernatants contained no detectable protease activity (Fig. 3A). The T2SS mutants also exhibited no protease activity when grown on 3% skim milk agar or in the API 20 NE GEL cupule (Fig. 1B). We were able to complement the protease secretion phenotype of 668 ΔgspD by supplying gspD in trans (Fig. 3A, 668 ΔgspD/gspD), which is consistent with the result shown in Fig. 2B. Twelve distinct peptidases were exported by the T2SS in rich medium (Table 2), but it is not clear which of these is responsible for the proteolytic activity shown in Fig. 1B and Fig. 3A. The peptidases are predicted to be aminopeptidases, carboxypeptidases, and endopeptidases, and it is possible that they play a synergistic role in the hydrolysis of the protein substrates examined in this study. Further studies involving the construction of mutations in each protease gene will be necessary to understand the relative role each protease plays in substrate utilization.
FIG 3.
Quantitative enzymatic assay of secreted protease, PLC, endochitinase, and chitobiosidase activities. Bacterial strains were grown in LB broth for 22 h at 37°C with shaking (250 rpm), and the supernatants were filtered and assayed for enzymatic activity as described in Materials and Methods. (A) Protease assay. Values are presented as ng of protease activity per μl of culture supernatant. (B) PLC assay. p-Nitrophenylphosphorylcholine hydrolysis by PLC was monitored spectrophotometrically by the measurement of p-nitrophenol at 410 nm. (C) Endochitinase assay. One unit of activity releases 1 μmol of p-nitrophenol from 4-nitrophenyl β-d-N,N′,N″-triacetylchitotriose per min at pH 4.8 at 37°C. (D) Chitobiosidase activity. One unit of activity releases 1 μmol of p-nitrophenol from 4-nitrophenyl N,N′-diacetyl-β-d-chitobioside per min at pH 4.8 at 37°C. All numerical values are the means of three separate experiments performed in triplicate plus standard deviations (error bars). In all assays, MSHR668 was significantly different than 668 ΔgspD and 668 ΔgspE (P < 0.0001) and 668 ΔgspD/gspD was significantly different than 668 ΔgspD and 668 ΔgspE (P < 0.005).
Figure 3B shows that MSHR668 exports PLC activity against p-nitrophenylphosphorylcholine, but no PLC activity was detected in the supernatants of 668 ΔgspD and 668 ΔgspE. As expected, supplying gspD in trans resulted in complementation of the PLC secretion phenotype in 668 ΔgspD (Fig. 3B). Korbsrisate et al. demonstrated that B. pseudomallei PlcN1 and PlcN2 exhibit extracellular PLC activity (40). We found that PlcN1 and PlcN2 were secreted in a T2SS-dependent manner (Table 2), and it is likely that they both contribute to the PLC activity present in the supernatants of MSHR668 and 668 ΔgspD/gspD (Fig. 3B). Interestingly, PlcN3 (BURPS668_A0114) was not identified in the MSHR668 rich- or minimal-medium supernatants, even though it has a clearly defined Tat signal sequence.
Chitin is a polymer of β-(1-4)-N-acetyl-d-glucosamine units and is the second most abundant carbohydrate polymer in nature after cellulose (45, 46). It is a structural component of fungal cell walls, arthropod exoskeletons, and crustacean shells. In our LC-MS/MS studies, we identified two potential chitinolytic enzymes, BURPS668_0540 and BURPS668_1946, that were dependent on the T2SS for export in rich medium (Table 2). Endochitinase and chitobiosidase activities were >6-fold higher in the MSHR668 supernatant than in the 668 ΔgspD and 668 ΔgspE supernatants (Fig. 3C and D). In addition, these activities were >3-fold higher in the 668 ΔgspD/gspD supernatant than in the 668 ΔgspD supernatant. These results show that the T2SS is important for exporting endochitinase and chitobiosidase activities into the supernatant when B. pseudomallei is grown in rich medium (Fig. 3C and D). We predict that BURPS668_1946, a putative endo-β-N-acetylglucosaminidase, is responsible for the endochitinase activity presented in Fig. 3C. BURPS668_0540, a glycosyl hydrolase family 20 member, is a putative β-N-acetylglucosaminidase that may be responsible for the chitobiosidase activity shown in Fig. 3D. Interestingly, there was no significance difference in the secreted exochitinase activity of the strains used in this study (see Fig. S2 in the supplemental material), suggesting that only endochitinase and chitobiosidase activities are T2SS dependent.
Taken together, the quantitative enzyme studies support the LC-MS/MS results and demonstrate that protease, PLC, and chitinase activities are secreted by the T2SS. The data also support the notion that the B. pseudomallei T2SS exports numerous hydrolytic enzymes that break down a variety of macromolecular substrates.
Two collagenases, Burkholderia lethal factor (BLF1), and an iron-containing superoxide dismutase (SodB), are exported in a T2SS-independent manner in rich medium.
In 2004, Rainbow et al. (47) performed N-terminal sequencing on four proteins that were present in the supernatant of pathogenic B. pseudomallei but absent in the supernatant of nonpathogenic B. thailandensis. Three of these proteins, BURPS668_2041, BURPS668_A0759, and BURPS668_1946, were identified in this study as members of the T2SS secretome (Table 2). The last protein, BURPS668_A1216, was a protein with homology to known bacterial collagenases (47). We found two putative collagenases, including BURPS668_A1216, that were present in roughly equal amounts in the rich-medium supernatants of MSHR668 and 668 ΔgspD, suggesting that they are secreted in a T2SS-independent manner (Table 3). BLF1 is a B. pseudomallei cytotoxin that deamidates a specific glutamine residue in the eukaryotic translation initiation factor eIF4A, which abolishes helicase activity and inhibits translation (4). BLF1 does not harbor a signal sequence, and its mechanism of secretion is currently unknown. Here we demonstrate that BLF1 is not secreted by the T2SS, as it was readily detected in both the wild-type and T2SS mutant supernatants (Table 3). Furthermore, SodB and two hypothetical proteins were also secreted by MSHR668 and 668 ΔgspD (Table 3). Our proteomic results suggest that the virulence factor candidates shown in Table 3 are exported in a T2SS-independent manner.
TABLE 3.
Proteins with a ≤2-fold difference in the supernatants of MSHR668 and 668 ΔgspD grown in rich medium
| Locus tag | Producta | No. of hitsb |
|
|---|---|---|---|
| MSHR668 | 668 ΔgspD | ||
| BURPS668_A0988 | Collagenase | 10 | 17 |
| BURPS668_A1216 | Collagenase | 40 | 40 |
| BURPS668_2130 | Burkholderia lethal factor 1 (BLF1) | 45 | 24 |
| BURPS668_0930 | Superoxide dismutase (SodB) | 7 | 8 |
| BURPS668_0137 | Putative uncharacterized protein | 15 | 19 |
| BURPS668_A1199 | Putative uncharacterized protein | 8 | 4 |
Putative functional analysis predicted by InterPro (61).
Peptide spectral matches (PSMs) or the number of identification events specific for each protein.
TssM is secreted by the T2SS and contains an atypical signal sequence.
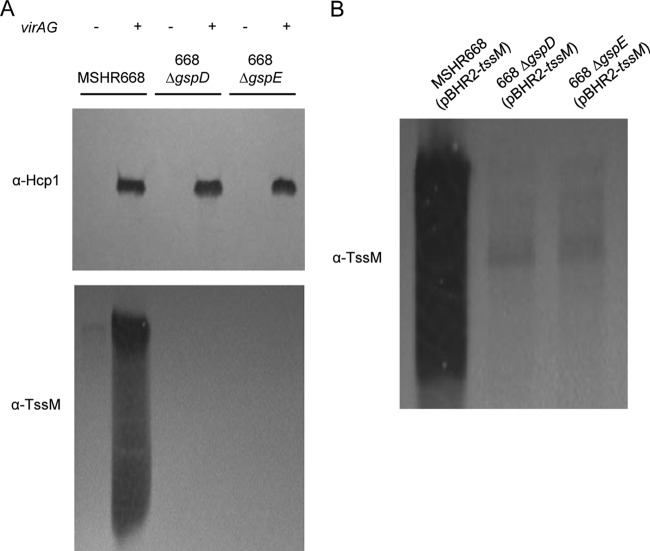
TssM is a secreted deubiquitinase that downregulates the innate immune response by interfering with the ubiquitination of signaling intermediates involved in TLR-mediated NF-κB activation (15). The B. pseudomallei tssM gene is located on chromosome 2 in the 4.1-kb region that separates the T6SS-1 and T3SS-3 gene clusters, but TssM is not dependent on either system for its secretion (14, 15). In addition, the tssM gene is coregulated with the T6SS-1 gene cluster and can be activated in vitro by overexpression of the VirAG two-component regulatory system (14, 16). We transformed MSHR668 and its T2SS mutants with pBHR2-virAG (Table 1) in order to examine the role of T2SS in TssM secretion. Hcp1, a protein exported by the T6SS-1 when pBHR2-virAG is provided in trans (16), was used a control for these experiments. As expected, Hcp1 was expressed and secreted by MSHR668, 668 ΔgspD, and 668 ΔgspE in a VirAG-dependent fashion (Fig. 4A). This result demonstrates that virAG overexpression activates the T6SS-1 gene cluster in MSHR668 and shows that the T2SS is not required for Hcp1 export. TssM was also expressed and secreted by MSHR668, but only when pBHR2-virAG was supplied in trans (Fig. 4A). The presence of multiple bands (or a smearing pattern) on TssM immunoblots has been previously described and likely represents processed derivatives of the secreted TssM (14, 15). Surprisingly, TssM was not secreted by 668 ΔgspD and 668 ΔgspE, suggesting that TssM is exported by the T2SS (Fig. 4A). In a previous study, we demonstrated that TssM can also be secreted in a virAG-independent manner by providing pBHR2-tssM (Table 1) in trans (14). We transformed MSHR668 and its T2SS mutants with pBHR2-tssM and found that TssM was secreted by MSHR668 but not by 668 ΔgspD and 668 ΔgspE (Fig. 4B). The results unequivocally show that secretion of TssM in B. pseudomallei is dependent on the T2SS.
FIG 4.
Immunoblot analysis of Hcp1 and TssM in B. pseudomallei supernatants. Strains were grown in LB for 8 h and supernatant proteins were separated by SDS-PAGE. The immunoblots were initially reacted with murine polyclonal anti-Hcp1 or murine polyclonal anti-TssM antisera and then with HRP-labeled goat anti-mouse IgG (γ). (A) Supernatant proteins from MSHR668, 668 ΔgspD, and 668 ΔgspE harboring pBHR2 (−) or pBHR2-virAG (+). (B) Supernatant proteins from MSHR668, 668 ΔgspD, and 668 ΔgspE harboring pBHR2-tssM. The images shown are representative of at least three separate immunoblotting experiments.
The proteins encoded by sequenced genomes are annotated bioinformatically prior to being deposited into sequence databases, which involves the prediction of signal peptides using computational methods such as SignalP or TatP (48). The TssM protein was not previously predicted to be a T2SS substrate because it was not annotated as possessing a classical Sec or a Tat signal sequence in the genome of B. pseudomallei MSHR668, K96243, 1710b, 1026b, 1106a, or MSHR305. Interestingly, the Phobius server (49) predicted a putative signal peptide on TssM (see Fig. S3 in the supplemental material). The N-terminal sequence of TssM, MNARRPAFGLIASHASRRRAVE, contained an N region (bold), an H region (italic), and a C region (underlined) (see Fig. S3 in the supplemental material). Phobius predicted the TssM cleavage site to be between amino acids 20 and 21 (RRA-VE). Thus, the Phobius server predicted a previously unidentified signal sequence on TssM that was not detected by the SignalP or TatP servers.
B. pseudomallei T2SS mutants exhibit an altered extracellular protein profile and compromised outer membrane integrity.
Sikora et al. (50) found that Vibrio cholerae T2SS mutants displayed extracellular protein profiles that were vastly different from those of wild-type strains. Their experiments revealed that V. cholerae T2SS mutants possessed leaky outer membranes that promoted leakage of periplasmic proteins and sensitivity to membrane-perturbing agents. We performed a LC-MS/MS analysis on proteins that were exclusively present, or considerably more abundant, in the rich-medium supernatant of 668 ΔgspD relative to MSHR668. While the SDS-PAGE result suggests that few such proteins exist (Fig. 2A, lanes 1 and 2), LC-MS/MS can identify proteins present at relatively low levels that may not be apparent by SDS-PAGE. In fact, our LC-MS/MS results identified 209 proteins that were present at ≥3-fold-higher levels in the 668 ΔgspD supernatant compared to the MSHR668 supernatant (see Table S1 in the supplemental material). The most abundant protein in the 668 ΔgspD supernatant was FliC, which is consistent with the molecular weight of the major protein identified by SDS-PAGE (Fig. 2A, second lane) (51). Thirteen additional flagellin-related proteins were present at elevated levels in the 668 ΔgspD supernatant, including FlgK, FliD, FlgG, FlgL, FlgF, FlgD, FlaE, FlgB, FlgC, FliK, and FlgJ. There was no difference in the motility of MSHR668 and 668 ΔgspD in LB medium with 0.3% agar, however (data not shown). There was also a 32-fold increase in the surface-associated type VI pilin protein, PilA (52), in the 668 ΔgspD supernatant relative to the MSHR668 supernatant (see Table S1 in the supplemental material). The most abundant class of proteins overrepresented in the 668 ΔgspD supernatant was ABC transporter periplasmic binding proteins (22 proteins). ABC transporters mediate the uptake of nutrients, growth factors, and trace elements and are dependent on periplasmic high-affinity substrate-binding proteins (53). The ABC periplasmic binding proteins in the 668 ΔgspD supernatant were predicted to be involved in the transport of amino acids, carbohydrates, oligopeptides, putrescine, glycerol-3-phosphate, ferric iron, sulfate, and molybdate (see Table S1 in the supplemental material). The LC-MS/MS extracellular protein profile of 668 ΔgspD suggests that B. pseudomallei T2SS mutants have a compromised outer membrane that promotes the leakage of periplasmic proteins and surface-associated proteins into the growth media.
Others have demonstrated that E. coli strains with outer membrane lesions are more sensitive to hydrophobic drugs and the detergent SDS than strains with an intact outer membrane (36, 54). We compared the survival of MSHR668, 668 ΔgspD, and 668 ΔgspE in LB broth containing 0, 0.01%, 0.02%, 0.03%, and 0.04% SDS in an attempt to assess the relative membrane integrity of the T2SS mutants. There was no difference in the survival of wild-type and the T2SS mutants in 0.01% SDS, but there was a significant difference in the survival of MSHR668 and the T2SS mutants in the presence of 0.02%, 0.03%, and 0.04% SDS (Fig. 5). MSHR668 was more resistant than 668 ΔgspD and 668 ΔgspE to 0.02 to 0.04% SDS, which suggests that the B. pseudomallei T2SS mutants have compromised outer membrane integrity.
FIG 5.
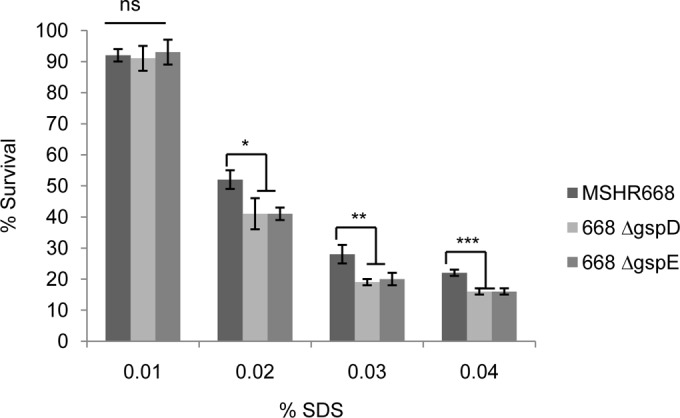
SDS sensitivity of B. pseudomallei wild type and T2SS mutants. Strains were grown overnight in LB broth, diluted to an OD600 of 0.05 in LB broth containing SDS, and grown aerobically for 4 h. One hundred percent survival is defined as the OD600 of each strain after 4 h growth in LB without SDS. Mean values ± standard deviations are plotted for 3 separate experiments. *, **, and ***, P < 0.05, P < 0.01, and P < 0.001, respectively. ns, not significant.
T2SS mutants are virulent in BALB/c mice.
The relative virulence of the B. pseudomallei T2SS mutants was assessed using BALB/c mice. Groups of 10 mice were inoculated by the i.p. route with 101 to 104 CFU of MSHR668, 668 ΔgspD, and 668 ΔgspE, and the animals were monitored for 21 days. Figure 6 shows the time to death with infectious doses of 102 CFU (∼1 LD50) and 104 CFU (∼100 LD50s). In general, the times to death of mice infected with wild-type and T2SS mutant strains were similar, and the LD50s of MSHR668, 668 ΔgspD, and 668 ΔgspE were 435 CFU, 276 CFU, and 381 CFU, respectively. Taken together, the data suggest that the B. pseudomallei T2SS is not a major virulence factor in BALB/c mice infected by the i.p. route.
FIG 6.
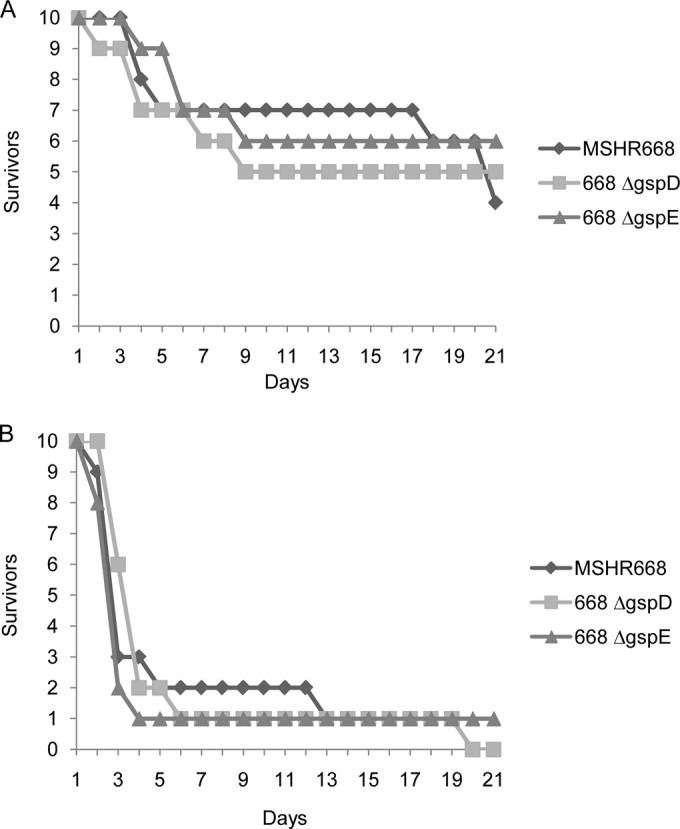
T2SS mutants are virulent in BALB/c mice. Groups of 10 BALB/c mice were infected i.p. with 102 CFU (A) or 104 CFU (B) of B. pseudomallei MSHR668, 668 ΔgspD, and 668 ΔgspE and observed for 21 days.
DISCUSSION
Our proteomic analysis of the B. pseudomallei MSHR668 T2SS secretome revealed the presence of ∼50 proteins dependent on the T2SS for export. Numerous hydrolytic enzymes, including 12 putative peptidases, were members of the T2SS secretome. Previous studies suggest that MprA, a serine metalloprotease, is responsible for the majority of protease activity in B. pseudomallei supernatants (39, 55, 56). However, the results presented here indicate that MprA is not the only protease exported by the B. pseudomallei T2SS (Table 2). Valade et al. constructed a mprA mutant and showed that a small amount of protease activity was still present in the supernatant when it was grown in rich medium, which is consistent with our results (56). Although MprA is likely responsible for most of the protease activity present in B. pseudomallei supernatants, we speculate that one or more of the T2SS-dependent peptidases shown in Table 2 also play a minor role in the hydrolysis of protein substrates. It is also worth mentioning here that the BURPS668_A2859 gene, encoding a serine S15 family X-Pro dipeptidyl-peptidase, is located immediately downstream of mprA and is also part of the T2SS secretome (Table 2). It is not uncommon for proteins encoded by genes in the same operon to interact or have a common function. In any case, determining the relative contribution of the 12 peptidases in the overall protease activity will require further studies.
It is unlikely that our proteomic analysis has identified all of the proteins secreted by the T2SS, as some of the genes encoding potential T2SS substrates are probably expressed poorly under the growth conditions employed in this study. Ooi et al. (42) recently reported that a large proportion of the B. pseudomallei transcriptome exhibits condition-dependent expression. Thus, it is possible that additional T2SS-dependent B. pseudomallei exoproducts will be identified by employing alternative growth conditions. Here we found 33 proteins that were exported exclusively in rich medium (Table 2), which is presumably due to medium-dependent expression differences of the genes encoding secreted products. In fact, many of these same genes exhibited decreased expression in minimal medium compared to rich medium in B. pseudomallei K96243 (42). While additional growth conditions will be required to elucidate the complete repertoire of T2SS exoproducts, we have provided a baseline T2SS secretome in rich and minimal media that can be built upon (or modified) in future studies.
One of the surprising findings presented here is that TssM is secreted by the T2SS (Fig. 4). TssM is a deubiquitinase that is expressed and secreted inside host cells (14), where it plays an important role in regulating the innate immune response in tissue culture and in mice (15). The tssM gene is physically linked to both the T6SS-1 and T3SS-3 gene clusters in B. pseudomallei, and its expression is coregulated with that of T6SS-1 (14, 16). Interestingly, neither secretion system is involved in TssM export (14, 15). Determining the mechanism of TssM secretion has been problematic, because tssM is poorly expressed in vitro, but it can be artificially “turned on” by overexpressing the two-component regulatory system virAG or tssM in trans from pBHR2 (14, 16). We demonstrate here that TssM is secreted in rich medium in a T2SS-dependent manner, but only when virAG or tssM is overexpressed in trans (Fig. 4). The mechanism of TssM secretion has also been an enigma due to the lack of an obvious Sec or Tat signal sequence at its N terminus (18). Here we used the Phobius web server (49), which uses transmembrane topology and signal peptide predictors, to identify a putative signal sequence on TssM. The results presented here demonstrate for the first time that TssM harbors an N-terminal signal sequence and that it is secreted in a T2SS-dependent manner.
TssM was first described in Burkholderia mallei as a secreted enzyme with the ability to hydrolyze multiple ubiquitinated substrates (14). The mechanism of TssM secretion in B. mallei is currently unknown, but the results presented here strongly suggest that it also uses the T2SS for export. Unlike B. pseudomallei, the T2SS of B. mallei reportedly contains mutations that prevent it from functioning properly (57). B. mallei is a host-adapted clone of B. pseudomallei and is a relatively poor secretor of protease and PLC activities (57). In 2004, the genome of B. mallei ATCC 23344 was annotated as containing the entire gsp cluster found in B. pseudomallei. However, comparison of the B. mallei gspD-N genes to those of B. pseudomallei K96243 (6) and 1026b (21) revealed that whereas most of the genes were nearly identical, the B. mallei gspJ gene contained a frameshift in the 3′ region of the gene, and the gspL gene contained a 54-base insertion for 18 amino acids (57). It was originally thought that these differences might account for B. mallei's protease and PLC secretion differences, but upon reanalysis of the B. mallei ATCC 23344 genomic data, we found that gspJ and gspL are no longer annotated as having these mutations (48). We suspect that genes with insertions or frameshifts, such as gspJ and gspL, were flagged for a more comprehensive closure effort to confirm the true status of such genes and that the B. mallei ATCC 23344 genome manuscript was written before the closure review. Thus, it appears that the B. mallei T2SS is functional and that it exports TssM, but this will have to be confirmed in future studies. Finally, the B. mallei genome has undergone numerous deletion and rearrangement events during the host adaptation process, and two of the genes that were “lost” are mprA and plcN2 (39, 40, 57). We propose that the loss of these genes might be responsible for the protease and PLC secretion deficiencies of B. mallei rather than the putative mutations in the B. mallei T2SS gene cluster (57).
Human melioidosis patient serum samples recognize both TssM and MprA, indicating that they are produced in vivo and are immunogenic (15, 38). This indicates that T2SS is active in vivo and that it might play a role in virulence. Previously we showed that the T2SS plays a minor role in the acute hamster model of melioidosis (21), but no studies have examined the role of T2SS in the BALB/c model of infection. Here we demonstrate that B. pseudomallei T2SS mutants are actually slightly more virulent than the wild-type strain in BALB/c mice by the i.p. route of infection. The LD50s for 668 ΔgspD and 668 ΔgspE were 276 and 381 CFU, respectively. By comparison, the LD50 of MSHR668 was 435 CFU. Interestingly, BALB/c mice infected intranasally with a B. pseudomallei tssM mutant exhibited hyperinflammation and died faster than mice infected with the wild-type strain (15). These results are consistent with TssM's role as a suppressor of the inherent innate immune response to B. pseudomallei infection. Tan et al. suggested that TssM should be viewed as a virulence factor because it prevents premature host death brought on by overt inflammation (15). Thus, the lack of TssM export by our T2SS mutants might mask the role of other T2SS-exported proteins as virulence factors in the murine model of infection.
Taken together, the results of our proteomic analysis of the B. pseudomallei T2SS secretome suggest that it plays an important role both in vivo and in vitro. The T2SS-dependent hydrolytic enzymes identified in this study are likely to be important for scavenging nutrients in soil and water and probably play an important role in the environmental persistence of this opportunistic pathogen.
Supplementary Material
ACKNOWLEDGMENTS
This project received support from DTRA/JSTO-CBD proposal number CBCALL12-LS1-2-0070 (to D.D.). This project was also supported in part by awards AI065359 (to M.N.B) and AI091783 (to P.J.B.) from the National Institute of Allergy and Infectious Diseases.
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
Footnotes
Published ahead of print 27 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01739-14.
REFERENCES
- 1.CDC. 2012. Possession, use, and transfer of select agents and toxins; biennial review. Final rule. Fed. Regist. 77:61083–61115 http://www.gpo.gov/fdsys/pkg/FR-2012-10-05/pdf/2012-24389.pdf. [PubMed] [Google Scholar]
- 2.Sprague LD, Neubauer H. 2004. Melioidosis in animals: a review on epizootiology, diagnosis and clinical presentation. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:305–320. 10.1111/j.1439-0450.2004.00797.x. [DOI] [PubMed] [Google Scholar]
- 3.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N. Engl. J. Med. 367:1035–1044. 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 4.Cruz-Migoni A, Hautbergue GM, Artymiuk PJ, Baker PJ, Bokori-Brown M, Chang CT, Dickman MJ, Essex-Lopresti A, Harding SV, Mahadi NM, Marshall LE, Mobbs GW, Mohamed R, Nathan S, Ngugi SA, Ong C, Ooi WF, Partridge LJ, Phillips HL, Raih MF, Ruzheinikov S, Sarkar-Tyson M, Sedelnikova SE, Smither SJ, Tan P, Titball RW, Wilson SA, Rice DW. 2011. A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A. Science 334:821–824. 10.1126/science.1211915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galyov EE, Brett PJ, DeShazer D. 2010. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 64:495–517. 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- 6.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U. S. A. 101:14240–14245. 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ireland PM, Marshall L, Norville I, Sarkar-Tyson M. 23 January 2014. The serine protease inhibitor Ecotin is required for full virulence of Burkholderia pseudomallei. Microb. Pathog. 10.1016/j.micpath.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Ireland PM, McMahon RM, Marshall LE, Halili M, Furlong E, Tay S, Martin JL, Sarkar-Tyson M. 2014. Disarming Burkholderia pseudomallei: structural and functional characterization of a disulfide oxidoreductase (DsbA) required for virulence in vivo. Antioxid. Redox Signal. 20:606–617. 10.1089/ars.2013.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazar Adler NR, Dean RE, Saint RJ, Stevens MP, Prior JL, Atkins TP, Galyov EE. 2013. Identification of a predicted trimeric autotransporter adhesin required for biofilm formation of Burkholderia pseudomallei. PLoS One 8:e79461. 10.1371/journal.pone.0079461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller CM, Conejero L, Spink N, Wand ME, Bancroft GJ, Titball RW. 2012. Role of RelA and SpoT in Burkholderia pseudomallei virulence and immunity. Infect. Immun. 80:3247–3255. 10.1128/IAI.00178-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norville IH, Breitbach K, Eske-Pogodda K, Harmer NJ, Sarkar-Tyson M, Titball RW, Steinmetz I. 2011. A novel FK-506-binding-like protein that lacks peptidyl-prolyl isomerase activity is involved in intracellular infection and in vivo virulence of Burkholderia pseudomallei. Microbiology 157:2629–2638. 10.1099/mic.0.049163-0. [DOI] [PubMed] [Google Scholar]
- 12.D'Cruze T, Gong L, Treerat P, Ramm G, Boyce JD, Prescott M, Adler B, Devenish RJ. 2011. Role for the Burkholderia pseudomallei type three secretion system cluster 1 bpscN gene in virulence. Infect. Immun. 79:3659–3664. 10.1128/IAI.01351-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allwood EM, Devenish RJ, Prescott M, Adler B, Boyce JD. 2011. Strategies for Intracellular Survival of Burkholderia pseudomallei. Front. Microbiol. 2:170. 10.3389/fmicb.2011.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shanks J, Burtnick MN, Brett PJ, Waag DM, Spurgers KB, Ribot WJ, Schell MA, Panchal RG, Gherardini FC, Wilkinson KD, DeShazer D. 2009. Burkholderia mallei tssM encodes a putative deubiquitinase that is secreted and expressed inside infected RAW 264.7 murine macrophages. Infect. Immun. 77:1636–1648. 10.1128/IAI.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan KS, Chen Y, Lim YC, Tan GY, Liu Y, Lim YT, Macary P, Gan YH. 2010. Suppression of host innate immune response by Burkholderia pseudomallei through the virulence factor TssM. J. Immunol. 184:5160–5171. 10.4049/jimmunol.0902663. [DOI] [PubMed] [Google Scholar]
- 16.Burtnick MN, Brett PJ, Harding SV, Ngugi SA, Ribot WJ, Chantratita N, Scorpio A, Milne TS, Dean RE, Fritz DL, Peacock SJ, Prior JL, Atkins TP, DeShazer D. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79:1512–1525. 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korotkov KV, Sandkvist M, Hol WG. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10:336–351. 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudva R, Denks K, Kuhn P, Vogt A, Müller M, Koch HG. 2013. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164:505–534. 10.1016/j.resmic.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Campos M, Cisneros DA, Nivaskumar M, Francetic O. 2013. The type II secretion system—a dynamic fiber assembly nanomachine. Res. Microbiol. 164:545–555. 10.1016/j.resmic.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Nivaskumar M, Francetic O. 3 January 2014. Type II secretion system: a magic beanstalk or a protein escalator. Biochim. Biophys. Acta S0167–4889(13)00447–3. 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 21.DeShazer D, Brett PJ, Burtnick MN, Woods DE. 1999. Molecular characterization of genetic loci required for secretion of exoproducts in Burkholderia pseudomallei. J. Bacteriol. 181:4661–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, Radey M, Rohmer L, Haugen E, Gillett W, Wuthiekanun V, Peacock SJ, Kaul R, Miller SI, Manoil C, Jacobs MA. 2012. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One 7:e36507. 10.1371/journal.pone.0036507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson K. 1987. Preparation of genomic DNA from bacteria, p 2.4.1–2.4.5 In Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (ed), Current protocols in molecular biology. John Wiley & Sons, New York, NY. [DOI] [PubMed] [Google Scholar]
- 24.Logue C-A, Peak IR, Beacham IR. 2009. Facile construction of unmarked deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates. J. Microbiol. Methods 76:320–323. 10.1016/j.mimet.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Hamad MA, Zajdowicz SL, Holmes RK, Voskuil MI. 2009. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 430:123–131. 10.1016/j.gene.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeShazer D, Brett PJ, Carlyon R, Woods DE. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wisniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat. Methods 6:359–362. 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 28.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 29.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, et al. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30:918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjornson RD, Carriero Nj, Colangelo C, Shifman M, Cheung K-H, Miller PL, Williams K. 2008. X!!Tandem, an improved method for running X!Tandem in parallel on collections of commodity computers. J. Proteome Res. 7:293–299. 10.1021/pr0701198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig R, Beavis RC. 2003. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20:1466–1467. 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 32.MacLean B, Eng JK, Beavis RC, McIntosh M. 2006. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 22:2830–2832. 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 33.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. 2004. Open mass spectrometry search algorithm. J. Proteome Res. 3:958–964. 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 34.Slotta DJ, McFarland MA, Markey SP. 2010. MassSieve: panning MS/MS peptide data for proteins. Proteomics 10:3035–3039. 10.1002/pmic.200900370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurioka S, Matsuda M. 1976. Phospholipase C assay using p-nitrophenylphosphorylcholine together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal. Biochem. 75:281–289. 10.1016/0003-2697(76)90078-6. [DOI] [PubMed] [Google Scholar]
- 36.Ize B, Stanley NR, Buchanan G, Palmer T. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183–1193. 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 37.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 38.Chin CY, Tan SC, Nathan S. 2012. Immunogenic recombinant Burkholderia pseudomallei MprA serine protease elicits protective immunity in mice. Front. Cell. Infect. Microbiol. 2:85. 10.3389/fcimb.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MA, Liu Y. 2000. Sequencing and characterization of a novel serine metalloprotease from Burkholderia pseudomallei. FEMS Microbiol. Lett. 192:67–72. 10.1111/j.1574-6968.2000.tb09360.x. [DOI] [PubMed] [Google Scholar]
- 40.Korbsrisate S, Tomaras AP, Damnin S, Ckumdee J, Srinon V, Lengwehasatit I, Vasil ML, Suparak S. 2007. Characterization of two distinct phospholipase C enzymes from Burkholderia pseudomallei. Microbiology 153:1907–1915. 10.1099/mic.0.2006/003004-0. [DOI] [PubMed] [Google Scholar]
- 41.Burtnick MN, Bolton AJ, Brett PJ, Watanabe D, Woods DE. 2001. Identification of the acid phosphatase (acpA) gene homologues in pathogenic and non-pathogenic Burkholderia spp. facilitates TnphoA mutagenesis. Microbiology 147:111–120. [DOI] [PubMed] [Google Scholar]
- 42.Ooi WF, Ong C, Nandi T, Kreisberg JF, Chua HH, Sun G, Chen Y, Mueller C, Conejero L, Eshaghi M, Ang RM, Liu J, Sobral BW, Korbsrisate S, Gan YH, Titball RW, Bancroft GJ, Valade E, Tan P. 2013. The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 9:e1003795. 10.1371/journal.pgen.1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 5:58. 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Chen H, Xia Y, Cui J, Gu Z, Song Y, Chen YQ, Zhang H, Chen W. 2013. How are the non-classically secreted bacterial proteins released into the extracellular milieu? Curr. Microbiol. 67:688–695. 10.1007/s00284-013-0422-6. [DOI] [PubMed] [Google Scholar]
- 45.Adrangi S, Faramarzi MA. 2013. From bacteria to human: a journey into the world of chitinases. Biotechnol. Adv. 31:1786–1795. 10.1016/j.biotechadv.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Frederiksen RF, Paspaliari DK, Larsen T, Storgaard BG, Larsen MH, Ingmer H, Palcic MM, Leisner JJ. 2013. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology 159:833–847. 10.1099/mic.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 47.Rainbow L, Wilkinson MC, Sargent PJ, Hart CA, Winstanley C. 2004. Identification and expression of a Burkholderia pseudomallei collagenase in Escherichia coli. Curr. Microbiol. 48:300–304. 10.1007/s00284-003-4192-4. [DOI] [PubMed] [Google Scholar]
- 48.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122. 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Käll L, Krogh A, Sonnhammer EL. 2007. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 35:W429–W432. 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sikora AE, Lybarger SR, Sandkvist M. 2007. Compromised outer membrane integrity in Vibrio cholerae type II secretion mutants. J. Bacteriol. 189:8484–8495. 10.1128/JB.00583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brett PJ, Mah DC, Woods DE. 1994. Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect. Immun. 62:1914–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Essex-Lopresti AE, Boddey JA, Thomas R, Smith MP, Hartley MG, Atkins T, Brown NF, Tsang CH, Peak IR, Hill J, Beacham IR, Titball RW. 2005. A type IV pilin, PilA, contributes to adherence of Burkholderia pseudomallei and virulence in vivo. Infect. Immun. 73:1260–1264. 10.1128/IAI.73.2.1260-1264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eitinger T, Rodionov DA, Grote M, Schneider E. 2011. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol. Rev. 35:3–67. 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 54.Stanley NR, Findlay K, Berks BC, Palmer T. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183:139–144. 10.1128/JB.183.1.139-144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sexton MM, Jones AL, Chaowagul W, Woods DE. 1994. Purification and characterization of a protease from Pseudomonas pseudomallei. Can. J. Microbiol. 40:903–910. 10.1139/m94-145. [DOI] [PubMed] [Google Scholar]
- 56.Valade E, Thibault FM, Gauthier YP, Palencia M, Popoff MY, Vidal DR. 2004. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J. Bacteriol. 186:2288–2294. 10.1128/JB.186.8.2288-2294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, Davidsen TD, Deboy RT, Dimitrov G, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Khouri H, Kolonay JF, Madupu R, Mohammoud Y, Nelson WC, Radune D, Romero CM, Sarria S, Selengut J, Shamblin C, Sullivan SA, White O, Yu Y, Zafar N, Zhou L, Fraser CM. 2004. Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. U. S. A. 101:14246–14251. 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon R, Quandt J, Klipp W. 1989. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene 80:161–169. 10.1016/0378-1119(89)90262-X. [DOI] [PubMed] [Google Scholar]
- 59.Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, Currie BJ, Wagner DM, Keim P. 2008. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics 9:566. 10.1186/1471-2164-9-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrázek J, Nierman WC, DeShazer D. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64:1466–1485. 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 61.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJA, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong S-Y. 2012. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40:D306–D312. 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 63.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.