Abstract
Research into chronic infection by bacterial pathogens, such as Pseudomonas aeruginosa, uses various in vitro and live host models. While these have increased our understanding of pathogen growth, virulence, and evolution, each model has certain limitations. In vitro models cannot recapitulate the complex spatial structure of host organs, while experiments on live hosts are limited in terms of sample size and infection duration for ethical reasons; live mammal models also require specialized facilities which are costly to run. To address this, we have developed an ex vivo pig lung (EVPL) model for quantifying Pseudomonas aeruginosa growth, quorum sensing (QS), virulence factor production, and tissue damage in an environment that mimics a chronically infected cystic fibrosis (CF) lung. In a first test of our model, we show that lasR mutants, which do not respond to 3-oxo-C12-homoserine lactone (HSL)-mediated QS, exhibit reduced virulence factor production in EVPL. We also show that lasR mutants grow as well as or better than a corresponding wild-type strain in EVPL. lasR mutants frequently and repeatedly arise during chronic CF lung infection, but the evolutionary forces governing their appearance and spread are not clear. Our data are not consistent with the hypothesis that lasR mutants act as social “cheats” in the lung; rather, our results support the hypothesis that lasR mutants are more adapted to the lung environment. More generally, this model will facilitate improved studies of microbial disease, especially studies of how cells of the same and different species interact in polymicrobial infections in a spatially structured environment.
INTRODUCTION
Pseudomonas aeruginosa commonly causes nosocomial infections and is a particular danger for people with cystic fibrosis (CF), in whom it establishes chronic lung infections (1). These are virtually impossible to clear with current therapeutic regimes, due to ciliary malfunction, the build-up of adhesive mucus in the CF airways (2), antibiotic resistance, and P. aeruginosa's ability to produce protective polysaccharide capsules (3). People with CF experience decades of chronic infection with repeated episodes of acute pulmonary exacerbation (1). During this time, P. aeruginosa evolves and diversifies; mutants with altered production of virulence factors are commonly isolated from patients (4–11), as are mutants that are impaired in quorum sensing (QS) (9, 10, 12).
Our understanding of the evolutionary pressures on P. aeruginosa during chronic lung infection and how these may be mediated by population or wider microbial community structure in these spatially structured organs is currently limited (13–15). Yet the evolutionary ecology of lung infections is likely a key factor in morbidity and response to clinical interventions (13, 16).
To clarify the role played by different P. aeruginosa virulence factors and by intermicrobial interactions in chronic infection, we need model systems that closely mimic a lung environment but are tractable in the lab and amenable to high-throughput experiments. A variety of in vitro growth conditions and insect or rodent hosts have been used to study P. aeruginosa populations. The pros and cons of these systems are outlined in Table 1, along with those of an underused model host: ex vivo sections of porcine lung. This model is useful for several reasons. First, pigs are arguably better models for studying human disease than are rodents or invertebrates (17–19). Second, lungs can be obtained from butchers: since little or no lung tissue is used in food production, lungs are (i) cheap and (ii) a waste product whose use does not raise ethical questions about the slaughter of animals for research. Third, many small sections of tissue can be kept in culture for several weeks (20, 21). Finally, and crucially, the spatial structure of the tissue is retained, and microbes can be visualized within the tissue by conventional or confocal microscopy. Histopathological changes can also be examined.
TABLE 1.
Comparison of different model systems for studying pathogen social behavior and virulence
| Category or concern | Data for model system (reference) |
||||
|---|---|---|---|---|---|
| Petri dish/test tube | Live invertebrate | Live mammal | Human cell or tissue culture | Ex vivo pig lung culture | |
| Example studies | References 22–26 | Waxworm (27, 28), fruit fly (16), or nematode (29, 30) | Usually mouse (14, 31–36), occasionally other small mammals (37); also, a CF pig model exists (38) | References 39–41 | References 20, 21, 42 |
| Chemical environment | Can be controlled to mimic CF sputum (24, 25) | Not known | Mouse metabolome (17) and gene expression (19) very different from human | Controllable and can be made to mimic in vivo conditions | Metabolome more similar to human than a mouse is (17) |
| Spatial structure | Can be controlled, but artificial | Limited | Burn wounds, limited; lung infections, yes | Possible with scaffolding or organ sections | Very similar to human lung (18) |
| Immune system | None | Limited similarity to humans | Limited similarity to humans | Human | Very similar to human (18) but largely lost ex vivo |
| Infection time scale | Can study hundreds to thousands of generations | Acute: host dies very quickly | At best semichronic; rodent lung infections tend to be acute (days), though can sometimes last 1–4 weeks (43, 44); wound infections are usually limited to ca. 3 weeks (K. Rumbaugh, pers. commun.)a | Days to weeks | Not known |
| Large sample sizes (tens plus) possible? | Yes | Yes | No, due to cost and ethical considerations | Not usually | Yes |
| Cost | Low-medium | Low | High | Medium to set up, low to run | Low |
| Ease of method | Simple, requires only general microbiology techniques | Must learn how to inoculate but otherwise simple | Requires specialized expertise, an animal license and often a dedicated animal worker to carry out inoculation | Requires expertise and dedicated lab space/equipment to minimize risk of contaminating cell lines | Lungs are readily obtained from commercial butchers; we developed dissection, infection, and culture techniques in ca. 3 months |
| Ethical considerations | None | None | Yes—and limit sample size/infection duration | Minimal (donor informed consent must be obtained) | None if obtained from animals slaughtered for meat; little or no tissue is used for human consumption, so lungs are basically a waste product. |
| Review articles | References 45, 46 | References 37, 46, 47 | References 46, 48 | ||
Infection duration depends on local rules governing animal welfare; e.g., in the United Kingdom, animals must be euthanized when the symptoms of infection become too severe. pers. commun., personal communication.
We developed this model for quantitative studies of P. aeruginosa growth and exoproduct production. We focused on the well-characterized PAO1 wild-type (WT) strain and two lasR mutant strains which do not respond to the QS signal N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) (49). QS controls the expression of various exoproducts (49) and facilitates the establishment of acute infection (31–36). However, mutants that have lost LasR function and so do not respond to 3-oxo-C12-HSL commonly arise in chronic CF infections (9, 10, 12) and ventilator-associated pneumonia (50). There is debate over whether lasR mutants are social “cheats” that benefit from the presence of WT cells (51, 52) or whether they are adapted to the chronic lung environment. Resolving this question is important because it will affect the likely clinical success of QS inhibitors, which have been suggested as novel antivirulence agents and antibiotic adjuvants (53, 54). We therefore compared the growth of lasR mutants with that of the WT in single-genotype and mixed infections in ex vivo pig lungs (EVPL). We also measured the production of 3-oxo-C12-HSL and of two groups of virulence factors whose expression is regulated by QS and that are linked with virulence in acute infection or with acute exacerbation and declining lung function in people with CF: tissue-degrading proteases (55) and redox-active phenazines (55–57). We also assayed for the siderophores pyoverdine and pyochelin, since these have been shown to be necessary for acute infection (27, 58) and their role in chronic infection has been much discussed (59–62).
We report three key results: (i) we can detect differential production of 3-oxo-C12-HSL, protease, and phenazine compounds by WT and QS mutant P. aeruginosa colonizing EVPL; (ii) consistent with this, lasR mutants cause less pathological change to the host tissue; and (iii) lasR mutants grow as well as or better than the WT in EVPL in single infections, and a marked lasR mutant had fitness equal to that of the WT in a mixed infection. Therefore, in this context, lasR mutants do not behave as social cheats: rather, they grow well in this chronic infection model.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The Nottingham PAO1 strain of P. aeruginosa was used as the wild type (WT). A PAO1 mutant, carrying a gentamicin resistance cassette inserted into the lasR gene, was used as a marked lasR-null mutant (lasR::Gm) (26). For comparison, an unmarked PAO1 clone with a clean deletion of lasR (ΔlasR) was also used. Preliminary work suggested that levels of phenazines produced in our infection model were too low to be assayed via spectrophotometry, so we used PAO1 WT and PAO1ΔlasR strains carrying a reporter construct for one of the main phenazine biosynthetic operons (phzA1-luxCDABE fusion; S. Higgins, S. Heeb, G. Rampioni, P. Williams, N. Krasnogor, and M. Cámara, unpublished data). Infected cubes of lung tissue were cultured in artificial sputum medium (ASM) (24) for 24 h at 37°C on an orbital shaker. ASM mimics the chemical composition of CF sputum but is not viscous. All media used were supplemented with 50 μg/ml ampicillin to minimize the growth of any resident bacteria present in the lung cubes.
Preliminary work and observations.
Lungs were purchased from a butcher (A Holmes and Son, Coalville, Leicestershire, United Kingdom). We conducted preliminary work on five lungs and determined that the tissue was healthy and not damaged by the process of dissecting and preparing tissue (see Fig. S1 in the supplemental material). We used the work of Nunes et al. (42) as a starting point to develop a protocol for dissecting out relatively regular cubes of alveolar tissue of approximately 5 mm3 (125 μl), inoculating with ca. 104 to 105 P. aeruginosa cells, and culturing the infected cubes in ASM for up to 7 days. Finally, we determined that we could visualize luminescent reporter bacteria in the cubes, homogenize infected tissue to recover live bacteria, and conduct quantitative assays with lung homogenate for the presence of 3-oxo-C12 HSL, total protease, pyocyanin, and light production by luminescent (lux) reporters. We also verified that resident lung bacteria were present at very low levels, being almost entirely outcompeted by P. aeruginosa in infected tissue.
Ethics statement.
All lung material was purchased from a retail butcher and was sourced from animals already slaughtered for meat; ethical approval was therefore not required for this study.
Preparation of lung material.
The final protocol for the preparation, inoculation, and culture of EVPL is shown in Fig. 1. Cubes of approximately 5 mm3 were dissected from the ventral surface of the left caudal lobe of three sets of lungs using a sterile mounted razor blade. Large bronchioles and veins were avoided in order to keep the cubes as comparable as possible. Prior to dissection, the ventral surface of the pleura was briefly (<1 s) seared with a hot pallet knife to kill surface contaminants from the abbatoir or butcher's shop. This also rendered the pleura easier to cut. During dissection, the tissue to be used was washed three times with cell culture medium (1:1 mix of RPMI 1640 and Dulbecco's modified Eagle medium [DMEM]; Sigma-Aldrich). The cubes were then washed for a fourth time in ASM. Preliminary work confirmed that searing and washing did not cause any visible damage to the pleura (light microscopy of formalin-fixed tissue) and that these processes reduced the numbers of contaminating and/or resident bacterial cells present in the cubes. We aliquoted 400 μl ASM supplemented with 0.8% agarose to individual wells of a sterile 24-well plate (to provide a soft surface for the tissue to sit on) and placed cubes singly in wells on this surface. Cubes were covered with 500 μl liquid ASM. As a control experiment to explore the growth of the bacterial strains in the absence of lung tissue, cultures were set up exactly as described above, but in place of the lung cube, an extra 125 μl liquid ASM was added. Three experimental replicates of this experiment were performed; in each case, five populations each of WT, lasR::Gm, ΔlasR, and WT plus lasR::Gm bacteria in a 1:1 mix were inoculated.
FIG 1.
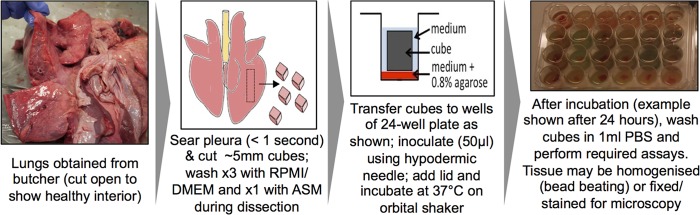
Schematic of the final protocol for preparation, infection, and culture of EVPL.
Inoculation of lung tissue.
Bacterial strains were grown overnight in lysogeny broth (LB), washed twice in phosphate-buffered saline (PBS), and resuspended in ASM. Cubes were inoculated with ca. 104 washed overnight-culture cells in 50 μl ASM—or as a mock-infection control, with 50 μl sterile ASM—using a 30-gauge needle attached to a disposable 1-ml syringe. Cubes were then incubated for 24 h at 37°C on an orbital shaker. Figure S2 in the supplemental material shows a schematic of the experiment.
Assays.
After incubation, cubes were rinsed in 1 ml PBS to remove loosely adhering cells. Growth of bacteria was assayed by homogenizing cubes individually in 500 μl phosphate-buffered saline with metal bead tubes (Cambio) using a Precellys24 homogenizer, serially diluting the homogenate and plating aliquots on LB plates to obtain single colonies. To score the relative frequencies of WT and lasR mutant cells in mixed infections, aliquots were replica plated onto LB plus 20 μg/ml gentamicin. In mixed infections, the relative fitness of the mutant was calculated as follows: v = [x2(1 − x1)]/[x1(1 − x2)], where x1 and x2 are the initial and final frequencies of the mutant in the population, respectively. When the two genotypes have equal fitness, x1 = x2 and v = 1. v values of <1 reflect the mutant being outcompeted by the WT, and values of >1 indicate that the mutant outcompetes the WT. To quantify QS signals, total protease, pyocyanin, and the siderophores pyoverdin and pyochelin, an aliquot of the homogenate was diluted 10-fold in PBS and filtered using a 0.2-μm syringe-driven filter unit to remove cells. This was stored at −20°C. The amount of the QS signal 3-oxo-C12-HSL in the diluted homogenates was quantified using the pSB1075 Escherichia coli biosensor (63); briefly, 100 μl of each homogenate was mixed with 100 μl of an overnight biosensor culture diluted to an optical density at 600 nm (OD600) of ∼0.1, and the luminescence/OD600 for each culture measured after 30 min of incubation at 37°C in a 96-well plate. The assay was calibrated using purified 3-oxo-C12-HSL. To measure total protease, 100 μl homogenate was mixed with 5 mg azocasein dissolved in 900 μl 100 mM Tris-HCl plus 1 mM CaCl2, and the mixture was incubated with shaking for 15 min at 37°C; the reaction was then stopped by adding 500 μl 10% trichloroacetic acid, and the absorbance of the supernatant was read at 400 nm. This assay was calibrated using known concentrations of purified proteinase K. Pyocyanin was quantified by measuring absorbance of homogenates at 695 nm, pyoverdine by exciting with light at 400 nm and measuring fluorescence at 460 nm (64) and pyochelin by exciting at 350 nm and measuring fluorescence at 430 nm (65). To assay activity of the phzA1 phenazine operon by reporter bacteria, aliquots of nonfiltered, undiluted homogenate were assayed for luminescence. Spectrophotometric assays were carried out using either a Tecan Infinite F200 Pro instrument (3-oxo-C12-HSL biosensor, protease, luminescence) or a Molecular Devices SpectraMax M2 instrument (pyoverdine, pyochelin, and pyocycanin). Finally, to assess tissue damage and bacterial growth, cubes were fixed in formalin, sectioned, and stained with hematoxylin and eosin (H&E) and Gram's stain and inspected under a light microscope (Nikon Eclipse 50i with Digital Sight DS-U3 camera).
Statistical analysis.
Analysis of variance (ANOVA) with type II sums of squares (car package [66] in R 2.14.0 [67]) was used to test for main effects of lung and inoculum (sterile ASM and/or the different bacterial strains) and for differential effect of inoculum in different lungs (lung-inoculum interaction when the sterile ASM control was included in the analysis, lung-strain interaction when the sterile ASM control was excluded) on dependent variables. Data on total number of CFU, relative fitness of the lasR::Gm mutant, 3-oxo-C12-HSL concentration, protease concentration, and phenazine reporter expression were transformed using the natural logarithm when they were used as dependent variables, in order to meet the assumptions of ANOVA. All P values are given for two-tailed tests.
RESULTS
P. aeruginosa causes visible tissue damage.
As shown in Figure S1 in the supplemental material, fresh uninfected lung cubes appeared healthy, with open alveoli surrounded by thin, well-defined epithelium. To explore the effects of lasR-mediated QS on growth and virulence, cubes were inoculated with ca. 104 washed overnight-culture cells of WT PAO1, two independent lasR mutants (the insertional mutant PAO1 lasR::Gm and the clean knockout PAO1 ΔlasR), a mix of PAO1 and PAO1 lasR::Gm, or a phenazine bioreporter strain constructed in a WT or ΔlasR background. As a mock infection control, cubes were inoculated with 50 μl sterile artificial sputum medium (ASM). A visual overview of the final dissection and infection protocol is given in Fig. 1, and a schematic of the experimental design is given in Fig. S2.
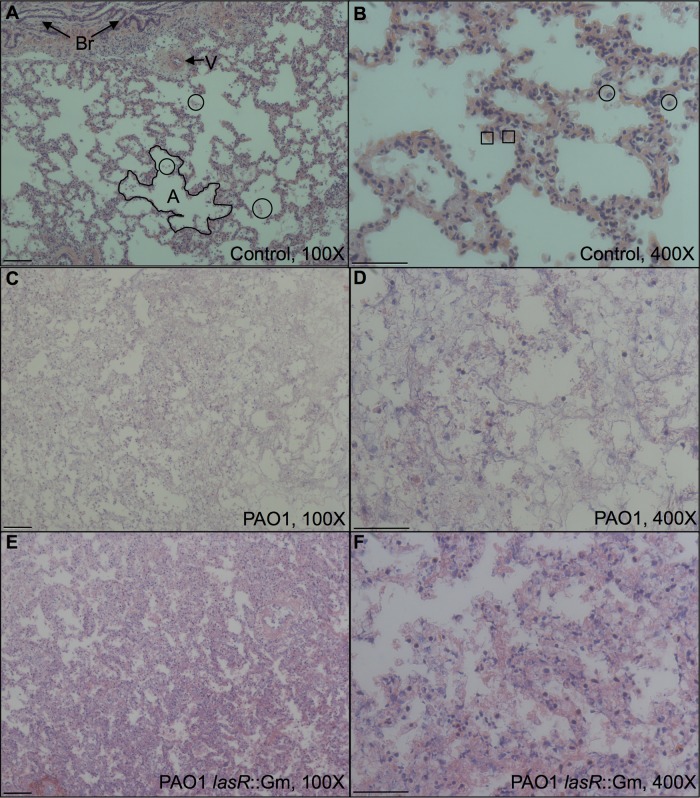
Mock-infected cubes retained their gross structural integrity over 24 h at 37°C, while infected cubes lost their shape and became soft, with visible green P. aeruginosa growth (see Fig. S3 in the supplemental material). Microscopy of fixed and stained tissue sections showed that mock-infected lungs showed minimal histopathological changes compared with lung tissue that was fixed and sectioned prior to infection, with only small amounts of apoptotic/necrotic debris in the alveoli (Fig. 2A and B); in most cases, Gram staining did not show the presence of resident bacteria. Some sections of mock-infected tissue exhibited areas of reduced alveolar volume reminiscent of areas of inflammation in living tissue, and the least histologically normal sample also contained large numbers of Gram-negative rods; it is highly unlikely that these were P. aeruginosa, since we never recovered P. aeruginosa when we plated out mock-infected lung homogenate. As exemplified in Fig. 2C and D, sections of tissue infected with PAO1 WT had no remaining alveolar structure, and far fewer cell nuclei were visible than in mock-infected tissue. Tissue preservation appeared slightly better in lung tissues infected with the lasR::Gm mutant; as exemplified in Fig. 2E and F, these were more reminiscent of highly inflamed tissue. In infected tissues, Gram staining revealed large numbers of Gram-negative rods, which we presume to be P. aeruginosa (see Fig. S4 in the supplemental material).
FIG 2.
Micrographs of tissue after 24 h in ASM, fixed and stained with H&E, which colors nuclei dark blue and other structures (cytoplasm, collagen, etc.) pink. (A and B) Mock-infected control; (C and D) infected with WT P. aeruginosa; (E and F) infected with the lasR::Gm mutant. Panels A, C, and E show tissue at magnification ×100 with a 100 μM scale bar; panels B, D, and F show tissue at magnification ×400 with a 50 μM scale bar. In panel A, note two bronchioles (Br) with diagnostic folded epithelium of brush border, example of a blood vessel (V), and lace-like pattern of alveoli defined by thin epithelium (example outlined; A). Small patches of cellular debris are visible in the alveoli (three examples are circled). In panel B, occasional cells with horseshoe-shaped nuclei (circled) are visible, which may represent neutrophils, along with enucleate red blood cells (two examples are boxed). Note in panels C and D the loss of clear epithelium, lower number of nuclei, and a decreased volume of airspace. In panels E and F, this change is less extreme, with thickened outlines of epithelium still discernible.
lasR mutants do not show a growth disadvantage in EVPL.
We compared the fitness of WT and lasR mutant genotypes in single and mixed infections of EVPL. P. aeruginosa grew in the lungs (Fig. 3), reaching final densities of 6 × 105 to 4 × 109 CFU per cube (5 × 103 to 3 × 107 CFU per mm3 of tissue). The final density differed between strains (F3,36 = 4.48; P = 0.009) and between lungs (F2,36 = 19.2; P < 0.001); crucially, the different strains showed consistent differences in growth across the different lungs (interaction F6,36 = 0.830; P = 0.555). Post hoc Tukey honestly significant difference (HSD) comparisons showed that QS mutants grew as well (for the ΔlasR mutant, P = 0.297) or better (for the lasR::Gm mutant, P = 0.049) than the WT in single infections. The mixed infections were initiated with a mixture comprising ca. 60% WT/40% lasR::Gm mutant, and these frequencies did not change over the incubation period: the relative fitness of the mutant did not vary between lungs (ANOVA for effect of lung; F2,7 = 0.04; P = 0.97) and was not significantly different from 1 (post hoc t test, t = 0.62; P = 0.56).
FIG 3.
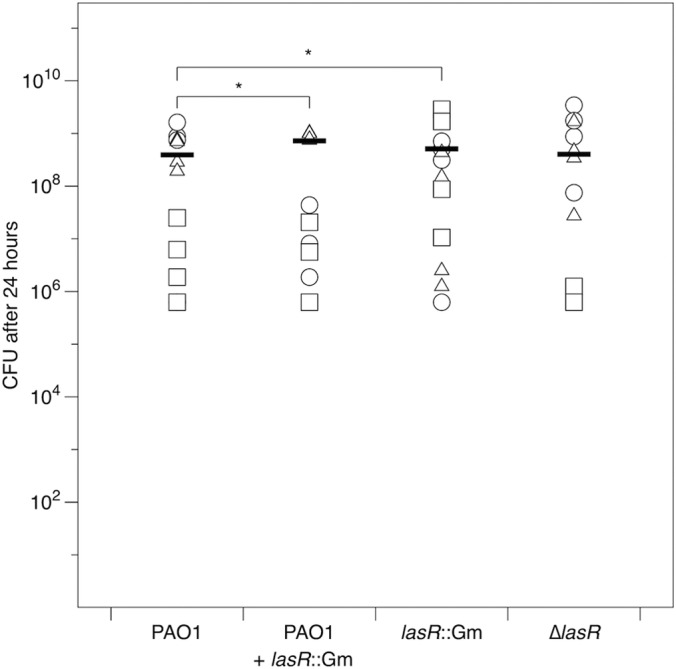
Number of CFU of P. aeruginosa recovered from ex vivo pig lung cubes after 24 h of incubation in artificial sputum medium. Different symbols show cubes from independent lungs, and bars denote overall means. Where pairwise differences between strains were found to be significant (P < 0.05) using Tukey HSD tests, this is indicated with an asterisk.
To determine whether the relative fitness of the lasR mutant was due to growth in lung tissue and not simply to growth in ASM, we performed a control experiment in which the cube of EVPL was replaced with a corresponding volume of ASM. Pure cultures of the lasR::Gm and ΔlasR mutants grew to approximately half the density of the WT (P < 0.001 and P = 0.013, respectively); the final density of the mixed WT plus lasR::Gm population did not differ significantly from that of the WT (P = 0.060). These results are shown in Figure S5 in the supplemental material; P values are from post hoc tests after a fully factorial ANOVA testing for the effects of strain and experimental replicate (strain F3,47 = 7.69; P < 0.001). The lasR::Gm mutant was not able to take advantage of the WT in mixed culture; its relative fitness did not differ in pure and mixed culture (fully factorial ANOVA including experimental replicate: strain F1,23 = 3.40; P = 0.078) and was significantly <1 in both cases (post hoc t test, t = 2.25; P = 0.034). This appeared to be due to the WT growing better in the absence of EVPL than in its presence, but further work is needed to explore this.
P. aeruginosa virulence factor expression in EVPL is QS dependent.
We used an E. coli bioreporter (63) to measure 3-oxo-C12-HSL in cell-free homogenates of mock-infected and infected lung. As shown in Fig. 4, the amount of signal produced differed between inocula (F2,36 = 28.6; P < 0.001) but not between lungs (main effect, F3, 36 = 0.50; P = 0.611; interaction, F6,36 = 1.39; P = 0.24). This was due to the WT infection producing more signal than the control or lasR mutant infections (Tukey HSD tests, P < 0.001; mutant infections did not differ from the control [P > 0.7]). The WT produced, on average, 16 nM 3-oxo-C12 HSL (range, 6 to 47 nM). We reran these analyses excluding the mock-infected control and included total CFU in the cube as a covariate to eliminate the possibility that any differences between strains were due to variability in population density; the results were unchanged. Many secreted molecules that have been linked with virulence in acute infection models or with acute exacerbations or more rapid decline of lung function in CF are under QS control. These include tissue-degrading proteases (55) and redox-active phenazine compounds (55–57). We then sought to determine whether mutations in lasR and concomitant loss of 3-oxo-C12-HSL led to decreased production of protease, phenazines, and siderophores in EVPL.
FIG 4.
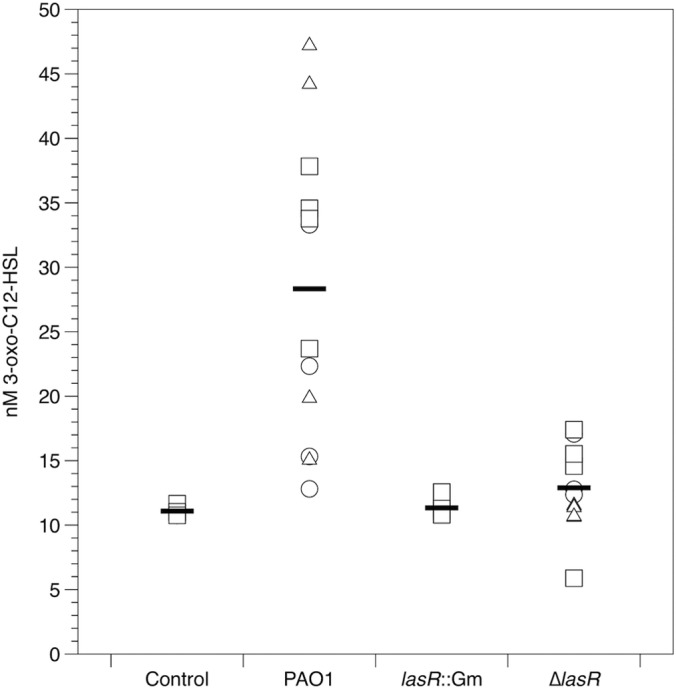
3-oxo-C12-HSL signal in mock-inoculated and P. aeruginosa-infected lung cubes after 24 h incubation. Different symbols show cubes from independent lungs, and bars denote overall means. The amount of signal in the WT-infected cubes was significantly greater than that in cubes infected with the other three strains (Tukey HSD tests, P < 0.001).
Consistent with lower levels of tissue damage (Fig. 2), the lasR mutants produced less protease per capita than the WT (strain, F3,36 = 8.77; P < 0.001; lung, F2,36 = 12.5; P < 0.001; interaction, F6,36 = 1.28; P = 0.29), and this translated into much lower total protease in lung cubes (Fig. 5). Analysis including mock-infected cubes: inoculum, F4,45 = 148; P < 0.001; lung, F2,45 = 14.0; P < 0.001; interaction, F8,45 = 7.4; P < 0.001). On average, the total protease activities in cubes infected with the ΔlasR mutant and the lasR::Gm mutant were 16% and 12%, respectively, of that measured in WT-infected cubes. Mock-infected cubes contained no measurable protease (t = 0.581; P = 0.56), underlining the loss of immune activity in this model. We reran the per capita and total protease analyses excluding the mock-infected control and included total CFU in the cube as a covariate to eliminate the possibility that our results were due to variability in population density; the results for the main effects of lung and strain were unchanged, but the lung-strain interaction became nonsignificant in both cases (P > 0.1).
FIG 5.
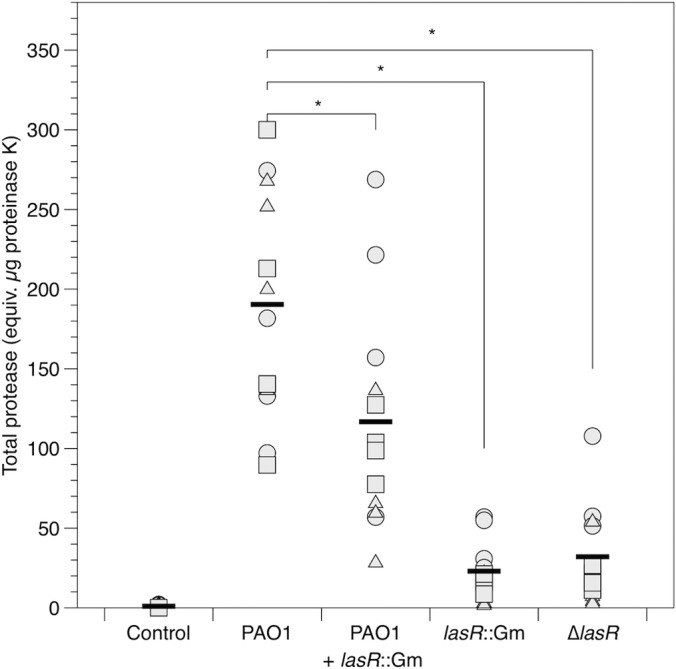
Total protease in mock-inoculated and P. aeruginosa-infected lung cubes after 24 h of incubation. Different symbols show cubes from independent lungs, and bars denote overall means. Where pairwise differences between strains were found to be significant (P < 0.005) using Tukey HSD tests, this is indicated with an asterisk.
Similar results were obtained for the phenazine compound pyocyanin (Fig. 6. Analysis including mock-infected cubes: inoculum, F4,45 = 12.7; P < 0.001; lung, F2,45 = 2.47; P = 0.096; interaction, F8,36 = 2.18; P = 0.047; dropping two outliers from the control group did not affect these results). Visual inspection of cubes infected with a luminescent reporter for the phenazine biosynthetic operon phzA1 using a photon-counting camera confirmed that this operon was expressed in infected cubes (Fig. 7A), and plating confirmed that phenazine reporter constructs grew to similar densities regardless of whether they were in a WT or ΔlasR genetic background. A quantitative assay showed that per-CFU expression of luminescence by the phzA1 reporter construct was lower in the ΔlasR background than in the WT (Fig. 7B, F1,12 = 37.9; P < 0.001); on average, expression in the ΔlasR background was 45% of that in the WT background, but the magnitude of this difference differed between lungs (main effect, F2,12 = 1.22; P = 0.33; interaction, F2,12 = 16.9; P < 0.001). Again, including total CFU in the cubes as a covariate did not affect the results for pyocyanin and phzA1 reporter expression. We could not detect the primary and secondary siderophores pyoverdine and pyochelin in lung homogenates using excitation/emission assays, which have been shown to be sensitive to ≥10 μM pyoverdine (F. Harrison, unpublished data) and ≥2 μM pyochelin (65).
FIG 6.
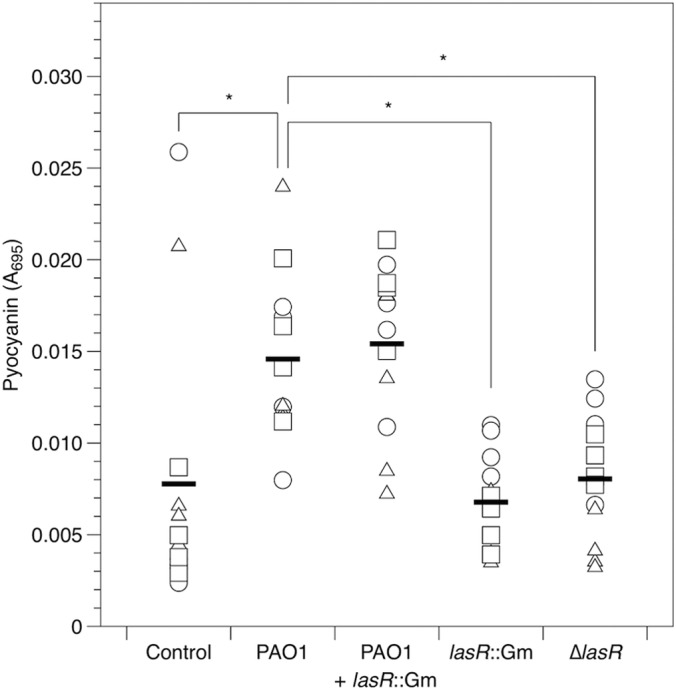
Pyocyanin (A695) in mock-inoculated and P. aeruginosa-infected lung cubes after 24 h of incubation. Different symbols show cubes from independent lungs, and bars denote overall means. Where pairwise differences between strains were found to be significant (P ≤ 0.006) using Tukey HSD tests, this is indicated with an asterisk.
FIG 7.
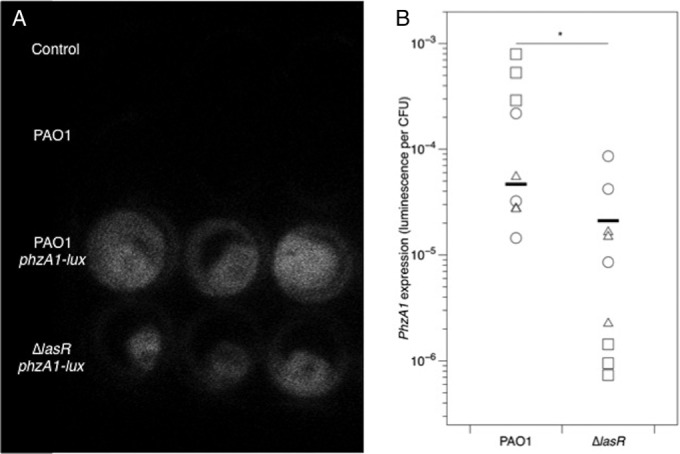
(A) Photon-counting image of cubes taken from one lung after 24 h of incubation. Mock-inoculated cubes and cubes infected with unlabeled NPAO1 show no luminescence, and cubes infected with phzA1-luxCDABE reporters show luminescence. (B) Per-CFU expression of phzA1 by P. aeruginosa in lung cubes (arbitrary luminescence units divided by CFU and blanked on samples from cubes infected with the unlabeled NPAO1). Different symbols show cubes from independent lungs, and bars denote overall means. The asterisk denotes a significant difference between strains in ANOVA (P < 0.001).
DISCUSSION
Tractability and potential of EVPL as an infection model.
Ex vivo sections of pig lung are a tractable model for studying P. aeruginosa growth and virulence. Mock-infected tissue was relatively histologically normal after 24 h of incubation in ASM at 37°C, and preliminary observations suggest that little further histological change occurs in mock-infected tissue after a further 6 days of incubation. P. aeruginosa cells could be visualized in EVPL using a light microscope and readily recovered from tissue. Cell-free suspensions of homogenized tissue could be assayed for a range of bacterial virulence factors. The relative growth of WT and lasR mutant bacteria in EVPL contrasted with the situation in ASM alone: in this setting, the WT outgrew the mutants by a factor of approximately 2:1, whereas in EVPL the mutants grew as well as or slightly better than the WT.
A key advantage of EVPL is the chance to study bacterial virulence factor production, growth, and cell-cell interactions in a spatially structured environment. A diverse literature has explored the potential effects on bacterial gene expression, growth, and virulence of interactions between cells of the same or different species (4, 16, 27, 68–73) and how population structure can affect these interactions (22, 23, 35, 74). However, while it is clear that the CF infection community is spatially structured on a gross anatomical level (15, 75, 76), we do not know whether this community is spatially ordered at a scale relevant to bacterial cell-cell interactions. This means that it is hard to assess the likely efficacy of proposed clinical interventions that rely on disrupting cell-cell interactions, such as QS inhibitors (54). Further, spatial structuring of bacterial populations will affect other processes relevant to the development of chronic infection, such as the dynamics of bacteriocin-producing and -sensitive strains (77), plasmid transfer (78), and the evolution of antibiotic resistance (79, 80). The potential to manipulate the infection community inoculated into EVPL, to study its evolution using conventional and confocal microscopy of sections taken at various times postinoculation, and to correlate aspects of community diversity and structure with histopathology and levels of virulence factors represents a significant opportunity to study the extent and consequences of cell-cell interactions in lung tissue.
Clearly, EVPL also has limitations, and future work must identify these, circumvent them where possible, and clarify the extent to which EVPL represents a chronically infected human lung. First, we must acknowledge the high variance in the data presented in this article. Since we had no a priori expectations of the likely level of variability or ease of replication, this should be viewed mainly as a proof-of-principle study which can be built upon by ourselves and others. As a result of this work, we now know that we can readily access and process lungs in batches and easily cut several dozen regular cubes of tissue from each lung. This knowledge, along with the ability to use the data presented here in power calculations, will allow researchers to design larger-scale experiments which provide more reliable estimates of between-strain or between-genotype differences. Second, if we are to determine how well this model recapitulates chronic infection in humans, a detailed exploration of the chemical environment in EVPL and how this coevolves with infecting microbes over time (days to weeks) is required. For instance, we do not yet know whether the reported chemical similarities between human and pig lung are maintained in this ex vivo system or whether the ASM needs to be modified when it is used in conjunction with tissue (for example, ASM contains iron, but if this is also supplied in abundance by the lung tissue, then overall levels of bioavailable iron may be unrealistically high, and this could explain why siderophore gene expression appeared to be switched off in our experiment). Further, the oxygen regime in infected lungs is likely to be an important factor, and infection foci may become less aerobic over the course of infection (81, 82). Future work could address how and when oxygen levels change inside sections of EVPL and how this affects the growth and virulence of P. aeruginosa. Finally, a careful comparison of results obtained in EVPL with those obtained from live animal models, with clinical data from CF patients where applicable, will help us to determine whether the differences we observe between genotypes in this model are likely to be meaningful in vivo.
Role of QS and fitness consequences of lasR mutation in EVPL.
3-Oxo-C12-HSL accumulated to nanomolar levels after 24 h of infection with WT PAO1. It is hard to know how this corresponds to the level of expression in CF lungs, since studies using various assay methods report concentrations ranging from femtomolar to micromolar in CF secretions and tissues (83–88). We detected significant differences between WT PAO1 and lasR mutants, which do not respond to 3-oxo-C12-HSL. lasR mutants produced no detectable 3-oxo-C12-HSL and significantly less protease and pyocyanin than the WT; further, expression of one of the phenazine biosynthetic operons, phzA1, was significantly reduced. Consistent with these results, lasR mutant-infected tissue exhibited qualitatively less tissue damage. We therefore conclude that P. aeruginosa “senses a quorum” in EVPL. Moreover, our results are consistent with reports that P. aeruginosa isolates from CF patients undergoing periods of acute exacerbation overproduce various QS-dependent exoproducts (8, 55, 89) and that in P. aeruginosa mouse infection models, areas of tissue with higher N-acylhomoserine lactone (AHL) concentrations exhibit more severe pathological changes (86), and lasR mutants cause less tissue damage than the WT (36). That our lasR mutants showed reduced pyocyanin production is interesting, because in standard laboratory medium in vitro, lasR mutants have been reported to produce significantly more pyocyanin than the WT (90, 91). In contrast, other studies have shown that among P. aeruginosa clones isolated from CF patients, lasR mutation is often associated with a loss of pyocyanin production in vitro (55; see also reference 92).
A key finding from our study is that lasR mutants grew as well as or better than the WT in EVPL. This is noteworthy because there has been considerable debate about the evolutionary dynamics of lasR mutants in chronic infection. There are at least three possible explanations for the presence of lasR QS-blind mutants in chronic P. aeruginosa infections. First, loss of QS response could be adaptive, conferring a growth or persistence advantage in the context of an established infection. Second, lasR mutants may act as social “cheats” and persist because they take advantage of coinfecting QS-proficient genotypes, whose QS-dependent exoproducts may benefit any cell in the vicinity, regardless of its own level of production (22, 93–96). Third, QS-blind mutants may be maladaptive but arise due to recurrent mutation and persist at low frequencies due to stochastic evolutionary drift. It is difficult to choose which of these alternatives (if any) is correct, because there is very little quantitative data on the frequency of QS-blind mutants within chronically infected hosts and how this changes (or not) over time.
Generally, the first hypothesis, that QS-blind mutants have a fitness advantage, has had little support, because loss of lasR function reduces the ability of P. aeruginosa to establish acute infections (31–36). We also found that in ASM in the absence of pig lung, lasR mutants were less fit than the WT. These observations, combined with demonstrations that lasR mutants can act as WT-exploiting cheats in some situations in vitro (22, 26, 97, 98) and in acute burn wound infections (32, 35), have led some researchers to give the second hypothesis serious consideration. This has led to the idea of deliberately introducing cheating mutants to trigger population collapse or to act as “Trojan horses” for carrying useful alleles (e.g., antibiotic susceptibility) into infectious populations (e.g. see reference 99).
Our result is not consistent with the “social cheat” hypothesis. Rather, it adds weight to the first hypothesis: that loss of LasR function enhances growth in chronic infection. The chemical environment in chronically infected, damaged tissues may confer a growth advantage on lasR-null mutants; D'Argenio et al. (100) report that the relative growth of WT and lasR-null monocultures in vitro is dependent on medium composition and that in some media, lasR-null mutants outgrow the WT. In addition, Duan and Surette (101) show that changes to medium composition can change the way that the QS system reacts to cell density. Moreover, in one of the few studies to track the frequency of lasR-null P. aeruginosa in human patients over the course of infection, Köhler et al. (50) interpret their data as showing that lasR mutants are cheats, but they report that patients colonized only by lasR mutants had bacterial loads similar to those of patients colonized only by the WT, and this strongly suggests that these mutants are not impaired in chronic persistence. For a detailed discussion of the evidence for adaptive loss of LasR, we refer the reader to the review by Heurlier et al. (102), and for a detailed discussion of social cheating, see Ghoul et al. (51).
We could not detect production of the siderophores pyoverdine and pyochelin in EVPL. While siderophores are necessary for acute infections of mice (58) and waxmoth larvae (27), their role in chronic CF lung infection is unclear because tissue damage and low oxygen levels may render iron more accessible (103–106). Studies of CF sputum samples have shown that siderophores are sometimes, but not always, present at detectable levels (61, 62).
Future directions.
Further optimization of EVPL could render it a realistic, ethical, and high-throughput model for studying the evolutionary ecology and pathology of chronic lung infection. As discussed above, several questions regarding the biological realism of the model remain to be answered, but the methodology we have developed for processing and handling lung tissue will allow us and others to address these in detail. The model as presented here produced more extensive and less localized tissue damage than seen in postmortem CF lungs, but since we inoculated with a high dose of bacteria and used aerobic culture conditions, this is not surprising. Now that we have demonstrated the potential of EVPL, the next steps in developing the model will be to find conditions that produce stable and long-lived infections (e.g., by titrating the number of cells inoculated, culturing under microaerobic or anaerobic conditions, and simulating key aspects of the host immune response). In this study, we used alveolar tissue to minimize between-cube variation in structure, but future work could focus on dissecting regular sections of bronchiole, since these are the foci of infection in human CF lungs (107). A comparison between lung cubes obtained from healthy pigs and lung cubes taken from pigs genetically engineered to express human CF mutations (114) may also help to validate the system. In the future, EVPL could especially enhance research into interactions between microbes in multispecies infections, which are the norm in CF (13, 15, 108–110) and are increasingly recognized as important in other respiratory diseases, such as chronic obstructive pulmonary disease (COPD) and asthma (111–113).
Supplementary Material
ACKNOWLEDGMENTS
We thank Kin-Chow Chang for advice on setting up the explant system, Kevin Foster, Kendra Rumbaugh, Alan Smyth, Keith Turner, and two anonymous reviewers for comments on the manuscript, James Gurney for lab help, Vishakha Sovani and Jeni Luckett for histopathology advice, the Histopathology Department at the Queen's Medical Centre (Nottingham University Hospitals NHS Trust) for sample preparation, Ashleigh Griffin for use of the SpectraMax M2, and A. Holmes and Son Butchers (Coalville, Leicestershire, United Kingdom) for supplying lungs.
This work was funded by NERC (NE/J007064/1) and by a Royal Society University Research Fellowship awarded to S.P.D.
F.H. and S.P.D. conceived and designed the study, F.H. and A.M. developed the dissection and infection protocol, F.H. carried out experimental work and analyzed the data, S.H. made the phzA1-luxCDABE reporter and contributed to manuscript preparation, and F.H. and S.P.D. wrote the article.
We have no conflict of interest to declare.
Footnotes
Published ahead of print 27 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01554-14.
REFERENCES
- 1.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194–222. 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher RC. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23:146–158. 10.1183/09031936.03.00057003. [DOI] [PubMed] [Google Scholar]
- 3.Govan J, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eberl L, Tummler B. 2004. Pseudomonas aeruginosa and Burkholderia cepacia in cystic fibrosis: genome evolution, interactions and adaptation. Int. J. Med. Microbiol. 294:123–131. 10.1016/j.ijmm.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 5.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 10:841–851. 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 6.Hoboth C, Hoffmann R, Eichner A, Henke C, Schmoldt S, Imhof A, Heesemann J, Hogardt M. 2009. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 200:118–130. 10.1086/599360. [DOI] [PubMed] [Google Scholar]
- 7.Huse HK, Kwon T, Zlosnik JEA, Speert DP, Marcotte EM, Whiteley M. 2010. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. mBio 1(4):e00199–10. 10.1128/mBio.00199-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, Brockhurst MA, Winstanley C. 2011. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 183:1674–1679. 10.1164/rccm.201009-1430OC. [DOI] [PubMed] [Google Scholar]
- 9.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492. 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilder CN, Allada G, Schuster M. 2009. Instantaneous within-patient diversity of Pseudomonas aeruginosa quorum-sensing populations from cystic fibrosis lung infections. Infect. Immun. 77:5631–5639. 10.1128/IAI.00755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MOA, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl. Acad. Sci., U. S. A. 108:7481–7486. 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjarnsholt T, Jensen PØ, Jakobsen TH, Phipps R, Nielsen AK, Rybtke MT, Tolker-Nielsen T, Givskov M, Høiby N, Ciofu O, Scandinavian Cystic Fibrosis Study Consortium 2010. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One 5:e10115. 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153:917–923. 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 14.Moser C, Van Gennip M, Bjarnsholt T, Jensen PØ, Lee B, Hougen HP, Calum H, Ciofu O, Givskov M, Molin S, Høiby N. 2009. Novel experimental Pseudomonas aeruginosa lung infection model mimicking long-term host-pathogen interactions in cystic fibrosis. APMIS 117:95–107. 10.1111/j.1600-0463.2008.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, Conrad D. 2012. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 6:471–474. 10.1038/ismej.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. 2008. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4:e1000184. 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benahmed MA, Elbayed K, Daubeuf F, Santelmo N, Frossard N, Namer IJ. 2014. NMR HRMAS spectroscopy of lung biopsy samples: comparison study between human, pig, rat, and mouse metabolomics. Magn. Reson. Med. 71:35–43. 10.1002/mrm.24658. [DOI] [PubMed] [Google Scholar]
- 18.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. 2012. The pig: a model for human infectious diseases. Trends Microbiol. 20:50–57. 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, López CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG, Inflammation and Host Response to Injury Large Scale Collaborative Research Program 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 110:3507–3512. 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams PP, Gallagher JE. 1978. Preparation and long-term cultivation of porcine tracheal and lung organ cultures by alternate exposure to gaseous and liquid medium phases. In Vitro 14:686–696. 10.1007/BF02616165. [DOI] [PubMed] [Google Scholar]
- 21.Williams PP, Gallagher JE. 1978. Cytopathogenicity of Mycoplasma hyopneumoniae in porcine tracheal ring and lung explant organ cultures alone and in combination with monolayer cultures of fetal lung fibroblasts. Infect. Immun. 20:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diggle SP, Griffin AS, Campbell GS, West SA. 2007. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450:411–414. 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 23.Kümmerli R, Griffin A, West SA, Buckling A, Harrison F. 2009. Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc. Biol. Sci. 276:3531–3538. 10.1098/rspb.2009.0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189:8079–8087. 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer KL, Mashburn LM, Singh PK, Whiteley M. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187:5267–5277. 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popat R, Crusz SA, Messina M, Williams P, West SA, Diggle SP. 2012. Quorum-sensing and cheating in bacterial biofilms. Proc. Biol. Sci. 279:4765–4771. 10.1098/rspb.2012.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison F, Browning LE, Vos M, Buckling A. 2006. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 4:21. 10.1186/1741-7007-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Racey D, Inglis RF, Harrison F, Oliver A, Buckling A. 2010. The effect of elevated mutation rates on the evolution of cooperation and virulence of Pseudomonas aeruginosa. Evolution 64:515–521. 10.1111/j.1558-5646.2009.00821.x. [DOI] [PubMed] [Google Scholar]
- 29.Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. 2011. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 55:2655–2661. 10.1128/AAC.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papaioannou E, Wahjudi M, Nadal-Jimenez P, Koch G, Setroikromo R, Quax WJ. 2009. Quorum-quenching acylase reduces the virulence of Pseudomonas aeruginosa in a Caenorhabditis elegans infection model. Antimicrob. Agents Chemother. 53:4891–4897. 10.1128/AAC.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutter EI, Duong J, Purighalla S, Storey DG. 2012. Lethality and cooperation of Pseudomonas aeruginosa quorum sensing mutants in Drosophila melanogaster infection models. Microbiology 158:2125–2132. 10.1099/mic.0.054999-0. [DOI] [PubMed] [Google Scholar]
- 32.Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, West SA. 2009. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19:341–345. 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 33.Rumbaugh KP, Griswold JA, Hamood AN. 1999. Contribution of the regulatory gene lasR to the pathogenesis of Pseudomonas aeruginosa infection of burned mice. J. Burn Care Rehabil. 20:42–49. 10.1097/00004630-199901001-00008. [DOI] [PubMed] [Google Scholar]
- 34.Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. 1999. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 67:5854–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rumbaugh KP, Trivedi U, Watters C, Burton-Chellew MN, Diggle SP, West SA. 2012. Kin selection, quorum sensing and virulence in pathogenic bacteria. Proc. Biol. Sci. 279:3584–3588. 10.1098/rspb.2012.0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang HB, DiMango E, Bryan R, Gambello M, Iglewski BH, Goldberg JB, Prince A. 1996. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect. Immun. 64:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kukavica-Ibrulj I, Levesque RC. 2008. Animal models of chronic lung infection with Pseudomonas aeruginosa: useful tools for cystic fibrosis studies. Lab. Anim. 42:389–412. 10.1258/la.2007.06014e. [DOI] [PubMed] [Google Scholar]
- 38.Stoltz DA, Meyerholz DK, Pezzulo AA, Ramachandran S, Rogan MP, Davis GJ, Hanfland RA, Wohlford-Lenane C, Dohrn CL, Bartlett JA, Nelson GA, Chang EH, Taft PJ, Ludwig PS, Estin M, Hornick EE, Launspach JL, Samuel M, Rokhlina T, Karp PH, Ostedgaard LS, Uc A, Starner TD, Horswill AR, Brogden KA, Prather RS, Richter SS, Shilyansky J, McCray PB, Zabner J, Welsh MJ. 2010. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci. Transl. Med. 28:29ra31. 10.1126/scitranslmed.3000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saiman L, Cacalano G, Prince A. 1990. Pseudomonas cepacia adherence to respiratory epithelial cells is enhanced by Pseudomonas aeruginosa. Infect. Immun. 58:2578–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu H, Thuruthyil SJ, Willcox MD. 2000. Invasive strains of Pseudomonas aeruginosa are able to cause epithelial cell cytotoxicity that is dependent on bacterial cell density. Clin. Exp. Ophthalmol. 28:201–204. 10.1046/j.1442-9071.2000.00289.x. [DOI] [PubMed] [Google Scholar]
- 41.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. 2012. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 4:159ra147 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nunes SF, Murcia PR, Tiley LS, Brown IH, Tucker AW, Maskell DJ, Wood JLN. 2010. An ex vivo swine tracheal organ culture for the study of influenza infection. Influenza Other Respir. Viruses 4:7–15. 10.1111/j.1750-2659.2009.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann N, Rasmussen TB, Jensen PØ, Stub C, Hentzer M, Molin S, Ciofu O, Givskov M, Johansen HK, Høiby N. 2005. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect. Immun. 73:2504–2514. 10.1128/IAI.73.4.2504-2514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedersen SS, Shand GH, Hansen BL, Hansen GN. 1990. Induction of experimental chronic Pseudomonas aeruginosa lung infection with P. aeruginosa entrapped in alginate microspheres. APMIS 98:203–211. 10.1111/j.1699-0463.1990.tb01023.x. [DOI] [PubMed] [Google Scholar]
- 45.Kemp MW, Massey RC. 2007. The use of insect models to study human pathogens. Drug Discov. Today Dis. Models 4:105–110. 10.1016/j.ddmod.2007.06.007. [DOI] [Google Scholar]
- 46.Wiles S, Hanage WP, Frankel G, Robertson B. 2006. Modelling infectious disease—time to think outside the box? Nat. Rev. Microbiol. 4:307–312. 10.1038/nrmicro1386. [DOI] [PubMed] [Google Scholar]
- 47.Hoffmann N. 2007. Animal models of chronic Pseudomonas aeruginosa lung infection in cystic fibrosis. Drug Discov. Today Dis. Models 4:99–104. 10.1016/j.ddmod.2007.11.008. [DOI] [Google Scholar]
- 48.Huang S, Wiszniewski L, Constant S. 2011. The use of in vitro 3D cell models in drug development for respiratory diseases, p 169–190 In Kapetanović IM. (ed), Drug discovery and development—present and future. InTech, Rijeka, Croatia. 10.5772/28132. [DOI] [Google Scholar]
- 49.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. 2013. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67:43–63. 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 50.Köhler T, Buckling A, van Delden C. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. U. S. A. 106:6339–6344. 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghoul M, Griffin AS, West SA. 2014. Toward an evolutionary definition of cheating. Evolution 68:318–331. 10.1111/evo.12266. [DOI] [PubMed] [Google Scholar]
- 52.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4:597–607. 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 53.Bjarnsholt T, Givskov M. 2007. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Phil. Trans. R. Soc. B Biol. Sci. 362:1213–1222. 10.1098/rstb.2007.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Defoirdt T, Brackman G, Coenye T. 2013. Quorum sensing inhibitors: how strong is the evidence? Trends Microbiol. 21:619–624. 10.1016/j.tim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Fothergill JL, Panagea S, Hart CA, Walshaw MJ, Pitt TL, Winstanley C. 2007. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 7:45. 10.1186/1471-2180-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunter RC, Klepac-Ceraj V, Lorenzi MM, Grotzinger H, Martin TR, Newman DK. 2012. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am. J. Respir. Cell. Mol. Biol. 47:738–745. 10.1165/rcmb.2012-0088OC. [DOI] [PubMed] [Google Scholar]
- 57.Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. 1988. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56:2515–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buckling A, Harrison F, Vos M, Brockhurst MA, Gardner A, West SA, Griffin A. 2007. Siderophore-mediated cooperation and virulence in Pseudomonas aeruginosa. FEMS Microbiol. Ecol. 62:135–141. 10.1111/j.1574-6941.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 60.Harrison EF. 2007. Cooperative behaviour in Pseudomonas aeruginosa: ecology, evolution and pathology. D.Phil. thesis. University of Oxford, Oxford, United Kingdom. [Google Scholar]
- 61.Haas B, Kraut J, Marks J, Zanker SC, Castignetti D. 1991. Siderophore presence in sputa of cystic fibrosis patients. Infect. Immun. 59:3997–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin L, Reid D, Sharples K, Lamont I. 2011. Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals 24:1059–1067. 10.1007/s10534-011-9464-z. [DOI] [PubMed] [Google Scholar]
- 63.Winson MK, Swift S, Fish L, Throup JP, Jørgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GSAB. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185–192. 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 64.Jiricny N, Diggle SP, West SA, Evans BA, Ballantyne G, Ross-Gillespie A, Griffin AS. 2010. Fitness correlates with the extent of cheating in a social bacterium. J. Evol. Biol. 23:738–747. 10.1111/j.1420-9101.2010.01939.x. [DOI] [PubMed] [Google Scholar]
- 65.Dumas Z, Ross-Gillespie A, Kümmerli R. 2013. Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc. Biol. Sci. 280:20131055. 10.1098/rspb.2013.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fox J, Weisberg S. 2011. An R companion to applied regression, 2nd ed. Sage, Thousand Oaks, CA. [Google Scholar]
- 67.Development Core Team R. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 68.Bakkal S, Robinson SM, Ordonez CL, Waltz DA, Riley MA. 2010. Role of bacteriocins in mediating interactions of bacterial isolates taken from cystic fibrosis patients. Microbiology 156:2058–2067. 10.1099/mic.0.036848-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harrison F, Paul J, Massey RC, Buckling A. 2008. Inter-specific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa. ISME J. 2:49–55. 10.1038/ismej.2007.96. [DOI] [PubMed] [Google Scholar]
- 70.Hoffman LR, Déziel E, D'Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 103:19890–19895. 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riedel K, Hentzer M, Geisenberger O, Huber B, Steidle A, Wu H, Høiby N, Givskov M, Molin S, Eberl L. 2001. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249–3262. [DOI] [PubMed] [Google Scholar]
- 72.Traverse CC, Mayo-Smith LM, Poltak SR, Cooper VS. 2013. Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc. Natl. Acad. Sci. U. S. A. 110:E250–E259. 10.1073/pnas.1207025110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weaver VB, Kolter R. 2004. Burkholderia spp. alter Pseudomonas aeruginosa physiology through iron sequestration. J. Bacteriol. 186:2376–2384. 10.1128/JB.186.8.2376-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffin AS, West SA, Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430:1024–1027. 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 75.Gutierrez JP, Grimwood K, Armstrong DS, Carlin JB, Carzino R, Olinsky A, Robertson CF, Phelan PD. 2001. Interlobar differences in bronchoalveolar lavage fluid from children with cystic fibrosis. Eur. Respir. J. 17:281–286. 10.1183/09031936.01.17202810. [DOI] [PubMed] [Google Scholar]
- 76.Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW, Rainey PB, Schmieder R, Youle M, Conrad D, Rohwer F. 2012. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am. J. Respir. Cell Mol. Biol. 46:127–131. 10.1165/rcmb.2011-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Narisawa N, Haruta S, Arai H, Ishii M, Igarashi Y. 2008. Coexistence of antibiotic-producing and antibiotic-sensitive bacteria in biofilms is mediated by resistant bacteria. Appl. Environ. Microbiol. 74:3887–3894. 10.1128/AEM.02497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slater FR, Bailey MJ, Tett AJ, Turner SL. 2008. Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol. Ecol. 66:3–13. 10.1111/j.1574-6941.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 79.Hermsen R, Deris JB, Hwa T. 2012. On the rapidity of antibiotic resistance evolution facilitated by a concentration gradient. Proc. Natl. Acad. Sci. U. S. A. 109:10775–10780. 10.1073/pnas.1117716109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perron GG, Lee AEG, Wang Y, Huang WE, Barraclough TG. 2012. Bacterial recombination promotes the evolution of multi-drug-resistance in functionally diverse populations. Proc. Biol. Sci. 279:1477–1484. 10.1098/rspb.2011.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer K, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas J, Randell S, Boucher R, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325. 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65:153–165. 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chambers CE, Visser MB, Schwab U, Sokol PA. 2005. Identification of N-acylhomoserine lactones in mucopurulent respiratory secretions from cystic fibrosis patients. FEMS Microbiol. Lett. 244:297–304. 10.1016/j.femsle.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 84.Favre-Bonté S, Pache J-C, Robert J, Blanc D, Pechère J-C, van Delden C. 2002. Detection of Pseudomonas aeruginosa cell-to-cell signals in lung tissue of cystic fibrosis patients. Microb. Pathog. 32:143–147. 10.1006/mpat.2001.0487. [DOI] [PubMed] [Google Scholar]
- 85.Hooi DS, Bycroft BW, Chhabra SR, Williams P, Pritchard DI. 2004. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect. Immun. 72:6463–6470. 10.1128/IAI.72.11.6463-6470.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Middleton B, Rodgers HC, Cámara M, Knox AJ, Williams P, Hardman A. 2002. Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol. Lett. 207:1–7. 10.1111/j.1574-6968.2002.tb11019.x. [DOI] [PubMed] [Google Scholar]
- 87.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764. 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 88.Wu H, Song Z, Hentzer M, Andersen JB, Heydorn A, Mathee K, Moser C, Eberl L, Molin S, Høiby N, Givskov M. 2000. Detection of N-acylhomoserine lactones in lung tissues of mice infected with Pseudomonas aeruginosa. Microbiology 146:2481–2493. [DOI] [PubMed] [Google Scholar]
- 89.Winstanley C, Fothergill JL. 2009. The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections. FEMS Microbiol. Lett. 290:1–9. 10.1111/j.1574-6968.2008.01394.x. [DOI] [PubMed] [Google Scholar]
- 90.Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50:29–43. 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 91.Dekimpe V, Déziel E. 2009. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155:712–723. 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 92.Beatson S, Whitchurch C, Sargent J, Levesque R, Mattick J. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605–3613. 10.1128/JB.184.13.3605-3613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Darch SE, West SA, Winzer K, Diggle SP. 2012. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 109:8259−8263. 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Diggle SP. 2010. Microbial communication and virulence: lessons from evolutionary theory. Microbiology 156:3503–3512. 10.1099/mic.0.045179-0. [DOI] [PubMed] [Google Scholar]
- 95.Pai A, Tanouchi Y, You L. 2012. Optimality and robustness in quorum sensing (QS)-mediated regulation of a costly public good enzyme. Proc. Natl. Acad. Sci. U. S. A. 109:19810–19815. 10.1073/pnas.1211072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. 2007. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38:53–77. 10.1146/annurev.ecolsys.38.091206.095740. [DOI] [Google Scholar]
- 97.Sandoz KM, Mitzimberg SM, Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 104:15876–15881. 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilder CN, Diggle SP, Schuster M. 2011. Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J. 5:1332–1343. 10.1038/ismej.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brown SP, West SA, Diggle SP, Griffin AS. 2009. Social evolution in micro-organisms and a Trojan horse approach to medical intervention strategies. Phil. Trans. R. Soc. B Biol. Sci. 364:3157–3168. 10.1098/rstb.2009.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Déziel E, Smith EE, Nguyen H, Ernst RK, Larson Freeman TJ, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64:512–533. 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duan K, Surette MG. 2007. Environmental regulation of Pseudomonas aeruginosa PAO1 las and rhl quorum-sensing systems. J. Bacteriol. 189:4827–4836. 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heurlier K, Denervaud V, Haas D. 2006. Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:93–102. 10.1016/j.ijmm.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 103.Hunter RC, Asfour F, Dingemans J, Osuna BL, Samad T, Malfroot A, Cornelis P, Newman DK. 2013. Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. mBio. 4(4):e00557–13. 10.1128/mBio.00557-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reid DW, Lam QT, Schneider H, Walters EH. 2004. Airway iron and iron-regulatory cytokines in cystic fibrosis. Eur. Respir. J. 24:286–291. 10.1183/09031936.04.00104803. [DOI] [PubMed] [Google Scholar]
- 105.Stites SW, Plautz MW, Bailey K, O'Brien-Ladner AR, Wesselius LJ. 1999. Increased concentrations of iron and isoferritins in the lower respiratory tract of patients with stable cystic fibrosis. Am. J. Respir. Crit. Care Med. 160:796–801. 10.1164/ajrccm.160.3.9811018. [DOI] [PubMed] [Google Scholar]
- 106.Stites SW, Walters B, O'Brien-Ladner AR, Bailey K, Wesselius LJ. 1998. Increased iron and ferritin content of sputum from patients with cystic fibrosis or chronic bronchitis. Chest 114:814–819. 10.1378/chest.114.3.814. [DOI] [PubMed] [Google Scholar]
- 107.Bjarnsholt T, Jensen PØ, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Høiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547–558. 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 108.Lynch SV, Bruce KD. 2013. The cystic fibrosis airway microbiome. Cold Spring Harb. Perspect. Med. 3:a009738. 10.1101/cshperspect.a009738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moore JE, Shaw A, Millar BC, Downey DG, Murphy PG, Elborn JS. 2005. Microbial ecology of the cystic fibrosis lung: does microflora type influence microbial loading? Br. J. Biomed. Sci. 62:175–178. [DOI] [PubMed] [Google Scholar]
- 110.Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD. 2003. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 41:3548–3558. 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beck JM, Young VB, Huffnagle GB. 2012. The microbiome of the lung. Transl. Res. 160:258–266. 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, Lynch SV. 2010. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS 14:9−59. 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Matkovic Z, Miravitlles M. 2013. Chronic bronchial infection in COPD. Is there an infective phenotype? Respir. Med. 107:10–22. 10.1016/j.rmed.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rogers CS, Abraham WM, Brogden KA, Engelhardt JF, Fisher JT, McCray PB, McLennan G, Meyerholz DK, Namati E, Ostedgaard LS, Prather RS, Sabater JR, Stoltz DA, Zabner J, Welsh MJ. 2008. The porcine lung as a potential model for cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 295:L240–L263. 10.1152/ajplung.90203.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.