SUMMARY
Efficient repair of UV-induced DNA damage requires the precise coordination of nucleotide excision repair (NER) with numerous other biological processes. To map this crosstalk, we generated a differential genetic interaction map centered on quantitative growth measurements of >45,000 double mutants before and after different doses of UV radiation. Integration of genetic data with physical interaction networks identified a global map of 89 UV-induced functional interactions amongst 62 protein complexes, including a number of links between the RSC complex and several NER factors. We show that RSC is recruited to both silenced and transcribed loci following UV damage where it facilitates efficient repair by promoting nucleosome remodeling. Finally, a comparison of the response to high versus low levels of UV shows that the degree of genetic rewiring correlates with dose of UV and reveals a network of dose-specific interactions. This study makes available a large resource of UV-induced interactions, and it illustrates a methodology for identifying dose-dependent interactions based on quantitative shifts in genetic networks.
INTRODUCTION
Helix-distorting DNA lesions, such as those caused by exposure to ultraviolet (UV) radiation, are sensed and repaired by the nucleotide excision repair (NER) pathway (Prakash and Prakash, 2000). Following damage recognition, the lesion is excised, the resulting gap is filled in by a DNA polymerase, and finally the remaining nick is sealed by a DNA ligase (Prakash and Prakash, 2000). The NER machinery, however, does not work in isolation. Increasing evidence points to the precise coordination of NER with several other biological processes such as the cell-cycle checkpoint (Sertic et al., 2012) and chromatin remodeling (Gong et al., 2006; Luijsterburg et al., 2012; Sarkar et al., 2010; Yu et al., 2005). Thus, a critical next step in defining the UV damage response will require an understanding of how distinct cellular processes cooperate with NER to promote the efficient repair of UV-induced lesions.
Large-scale screens for genetic interactions, facilitated by high-throughput techniques such as synthetic genetic arrays (SGA) or diploid synthetic lethal analysis by microarray (dSLAM), have been used with great success to rapidly map functional synergies among most genes in the yeast genome (Costanzo et al., 2010; Pan et al., 2007; Schuldiner et al., 2005; Schuldiner et al., 2006). However, it has become increasingly clear that many gene functional relationships are condition-dependent (St Onge et al., 2007) and identifying genetic networks that are essential to responding to an external stimulus will require a differential methodology. To this end we have recently developed an interaction mapping technique termed differential epistasis mapping (Bandyopadhyay et al., 2010) which enables the detection of quantitative changes in genetic interaction following an environmental change. Such differential genetic interactions have been shown to specifically highlight functional connections relevant to stress conditions with both high power and sensitivity (Guenole et al., 2012).
Towards the goal of defining the crosstalk between NER and other cellular processes following UV irradiation, we constructed a large differential epistasis network by measuring changes in genetic interactions in response to two doses of UV. The genetic data reveal a novel link between the NER machinery and the RSC chromatin remodeling complex. We find that, unlike chromatin remodeling complexes previously implicated in NER (Gong et al., 2006; Sarkar et al., 2010), RSC is recruited to sites of UV-induced lesions in both silenced and transcribed loci, where it helps to promote efficient repair. Finally, we leverage measurements made across multiple doses of UV to pinpoint a network of 79 dose-specific interactions, which, strikingly, are observed only at low or high doses but not both. This study makes available a large resource of UV-induced differential interactions, which we expect will prove indispensable for modeling the response to UV at the level of single genes, protein complexes and global processes.
RESULTS
A UV-based differential genetic interaction map
To map the functional connections between genes and pathways that underlie the response to UV-induced DNA damage, we measured changes in genetic interactions between a set of 37 query genes (Table S1) and 1397 array genes (Table S2). Query genes were chosen to represent a majority of the core NER factors and many known chromatin-remodeling complexes, while array genes were drawn from numerous functional categories. Using SGA technology (Tong and Boone, 2006), >45,000 double mutant combinations were generated and growth rates were measured in untreated (UT) conditions as well as in response to two doses of UV radiation: a ‘low’ dose of 20 J/m2and a ‘high’ dose of 80 J/m 2 (Methods and Figure 1A).
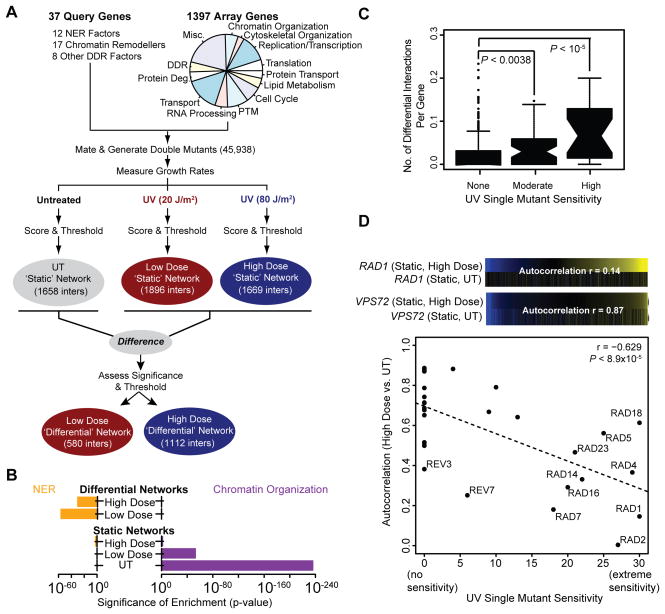
Figure 1. A UV-induced differential genetic network.
(A) Outline of the genetic interaction screen. The functional categories represented by the array genes are shown in the pie chart (Misc – Miscellaneous, DDR – DNA Damage Response, Protein Deg. – Protein degradation, PTM – Post-translational modifications). See Table S2 for more details. (B) The significance of enrichment for interactions with genes annotated to Nucleotide Excision Repair (NER) or Chromatin Organization (see Table S7 for process definitions) is shown for each network. Enrichment p-values were calculated as previously described (Bandyopadhyay et al., 2010). (C) Each gene considered in this study is binned according to its UV-induced single mutant sensitivity (Begley et al., 2004) and the distribution of the number of high dose differential interactions for all genes in a bin (# of significant differential interactions/# of tested differential interactions) is summarized as a box-and-whisker plot. Significance is assessed using a Mann-Whitney U test. (D) For each query gene, the Pearson’s correlation between it’s high dose static interaction profile and static untreated profile (‘Autocorrelation’) is plotted against the gene’s UV-induced single mutant sensitivity (Begley et al., 2004). The high dose and untreated static profiles are shown for two query genes: RAD1 and VPS72. See also Figure S1.
Measurements were first analyzed to assign each double mutant a ‘static’ interaction score in each condition separately (SUT, SLow, SHigh) which reflects the extent to which the double mutant either grew better (S>0; positive interaction or epistasis) or worse than expected (S<0; negative interaction or synthetic sick). To assess shifts in interaction following UV irradiation, the difference in static scores between treated and untreated conditions (SLow–SUT and SHigh–SUT) was computed for each gene pair and then assigned a p-value (PHigh–UT, PLow–UT) using a null distribution of differences observed when comparing replicate measurements made in the same condition (Methods). Using previously established static (S≥2.0 or S≤−2.5; Methods) or differential interaction thresholds (PHigh–UT, PLow–UT≤0.001; FDR ≈ 7%, see Figure S1A), we thus obtained three static genetic networks and two differential genetic networks (Figure 1A; Table S3 for raw data). Quality control metrics were monitored through this process, ensuring the high quality of these datasets (Figures S1B–D).
All three static networks were enriched for interactions involving genes that function in chromatin organization, as has been noted previously for other genetic interaction data (Bandyopadhyay et al., 2010; Guenole et al., 2012). In contrast, the two differential networks exhibited no such enrichment (Figure 1B), as strong signals present in both conditions are effectively ‘cancelled out’ in the differential mode of analysis (Ideker and Krogan, 2012). Instead, the two differential networks were highly enriched for interactions involving genes functioning specifically in the NER pathway (Figure 1B). In addition, we found that the number of differential interactions per gene (Figure 1C and Figure S1E), as well as the extent to which a gene’s static interaction profile was disrupted by UV treatment, is correlated with the UV sensitivity of the corresponding gene deletion strain (Figure 1D and Figure S1F). The static and differential networks thus provide complementary maps of cellular organization, with the differential networks highlighting genes most relevant to the UV damage response.
Differential genetic data link the RSC complex to NER
To identify novel genes and pathways operating in the UV damage response, we integrated all of our genetic data with existing protein-protein interaction data to construct a global map of gene modules and their UV-induced differential interactions. Past work has indicated that sets of genes enriched for static genetic and physical interactions (i.e., modules) often encode for components of the same pathway or complex (Bandyopadhyay et al., 2008; Srivas et al., 2011). On the other hand, differential genetic interactions tend to occur between genes belonging to distinct complexes and represent UV-induced crosstalk or synergy between the two complexes (Bandyopadhyay et al., 2010). Using a previously described workflow (Srivas et al., 2011), we organized our data into a set of 89 functional interactions amongst 62 modules (Figure 2A and Tables S4–5; Methods).
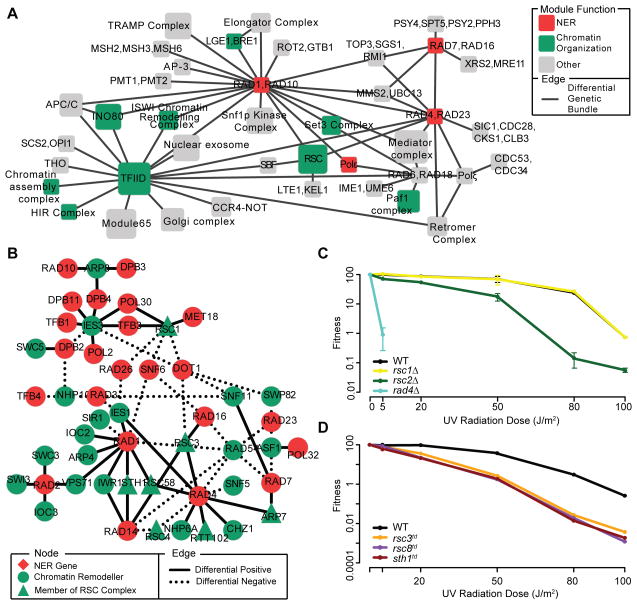
Figure 2. Differential genetic data links RSC to NER.
(A) A map of multi-genic modules spanned by bundles of UV-induced differential interactions. Node size scales with the number of genes in each module, while the node color indicates its function (see Table S7 for a list of process definitions). Modules that overlap with known protein complexes have been labeled accordingly, otherwise a generic name has been provided. For the sake of clarity only a portion of the module map has been shown here. See Table S5 for the complete list of module-module interactions (B) Differential genetic interactions (PLow–UT or PHigh–UT≤0.03) seen between chromatin remodelers and components of the NER pathway (see Table S7 for a list of process definitions). (C,D) Survival curves for (C) non-essential or (D) essential rsc single mutants following exposure to UV radiation across multiple doses. Survival curves were generated through quantification of the spot dilution assay in Figure S2A and one additional replicate (data not shown). Fitness was calculated by counting the number of colonies present in the most dilute spot containing individual colonies and then dividing the count in UV-treated conditions by the count in untreated conditions. All data represent the mean ± 1 s.e.m. of 2 independent replicates. See also Figures S2 and S3.
Detailed inspection of this module map both recapitulates current understanding and suggests many new hypotheses. For example, we observed a link between RAD6/RAD18 and the translesion synthesis polymerase Polζ. This is consistent with past work which has shown that monoubiquitnation of PCNA by the Rad6p-Rad18p dimer leads to the direct activation of Polζ-dependent bypass of DNA lesions through translesion synthesis (Prakash et al., 2005). We also observed crosstalk between the single-stranded DNA endonucleases RAD1/RAD10 and the mismatch repair genes MSH2/MSH3/MSH6; several studies have implicated a joint role for these complexes in ensuring genetic fidelity during mitotic recombination (Saparbaev et al., 1996; Sugawara et al., 1997).
Our map also revealed a significant number of complexes involved in chromatin organization (P = 0.031, Fisher’s exact test; Figure 2A). This observation was not expected given the lack of enrichment for interactions with genes annotated to this function in the differential networks (Figure 1B), thus suggesting a NER-specific role for these complexes. For example, the INO80 chromatin remodeling complex was found to interact with Rad1p-Rad10p dimer, which is consistent with a recent finding that INO80 is required for efficient repair of UV-induced lesions at a heterochromatic locus (Sarkar et al., 2010). Another chromatin remodeling complex that was featured prominently in our map with links to two different NER modules (RAD4/RAD23 and RAD1/RAD10) was RSC (Remodel the Structure of Chromatin). RSC is a highly conserved chromatin remodeling complex with DNA-dependent ATPase activity (Cairns et al., 1996), but as of yet has no known role in the UV damage response. We observed multiple interactions between components of the RSC chromatin remodeling complex (RSC1, RSC3, RSC58, ARP7) and several NER factors, including RAD1 and RAD4 (Figure 2B) as well as more moderate differential positive interactions between RSC3 and RAD14/RAD16 (P=0.01). Moreover, we found that deletion of non-essential genes encoding RSC subunits (RSC2 and HTL1, but not RSC1; Figure 2C and Figures S2A,B) as well as depletion of RSC subunits encoded by essential genes (RSC3, RSC8 and STH1; Figure 2D and Figure S2A) led to increased UV sensitivity. Together these observations support the hypothesis that RSC is required during the UV damage response.
Rsc2 is required for NER at both transcribed and silenced loci
The UV sensitivity of RSC-deficient strains could be caused by a checkpoint or NER defect. To distinguish between these possibilities, we first examined checkpoint activation in wildtype (WT) and rsc2Δ cells following exposure to UV. Analysis of the phosphorylation levels of Rad53 (a central effector of the checkpoint response) revealed efficient checkpoint activation, with Rad53 becoming phosphorylated within 30 minutes after UV exposure, and remaining phosphorylated for at least 3 hours in both WT and rsc2Δ cells (Figure S2C). Furthermore, FACS analysis revealed no major differences in the cell cycle profile between WT and rsc2Δ following UV exposure (Figure S2C), indicating normal checkpoint activation following UV irradiation in rsc2Δ cells. A similar phenotype has been noted in rsc2Δ mutants following exposure to the DNA alkylating agent, methyl methanesulfonate (Chambers et al., 2012). To monitor the efficiency of NER in WT and rsc2Δ cells, we used a sensitive qPCR-based assay (Methods) to measure the rate of repair within the highly transcribed MATa locus and the non-transcribed HMLα locus. At both loci, UV lesions were rapidly repaired in WT cells (~80% lesions removed by 3 hours; Figures 3A,B), whereas the rate of repair in the NER deficient rad14Δ mutants (Guzder et al., 1993) was significantly reduced (PMATa = 1.1×10−5 and PHMLα = 1.8×10−9; F test). The rsc2Δ cells, while not as deficient in repair as rad14Δ cells (PMATa = 0.024 and PHMLα = 0.013; F test), had nearly twice as many lesions present as WT at both loci three hours after UV exposure, indicating that RSC contributes to an efficient NER response. Importantly, both WT and rsc2Δ cells were found to accumulate equivalent amounts of UV-induced lesions, suggesting that the difference in repair rates is not due to differences in DNA damage susceptibility in these cells (Figure S2D).
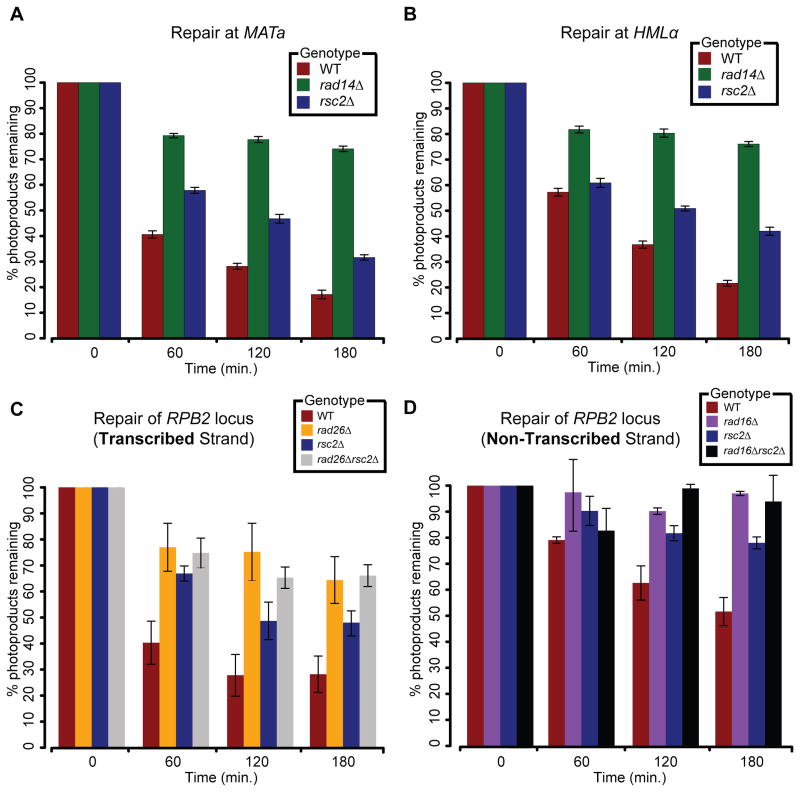
Figure 3. RSC is required for efficient TCR- and GGR-NER.
(A,B) Rate of photoproduct removal at the (A) MATa and (B) HMLα loci measured in G1-synchronized cells using a sensitive qPCR-based assay (Methods). (C,D) Rate of photoproduct removal on the (C) transcribed and (D) non-transcribed strands of the RPB2 locus measured in G1-synchronized cells using a strand specific repair assay (Methods). All data represent the mean ± 1 s.e.m. of at least 3 independent replicates.
NER is composed of two distinct pathways, Global Genome Repair (GGR) and Transcription Coupled Repair (TCR), which remove, respectively, lesions throughout the entire genome or on the transcribed strand of expressed loci only (Prakash and Prakash, 2000). While both the INO80 and SWI/SNF complexes have been implicated in NER previously, neither was found to have a role in promoting efficient TCR (Gong et al., 2006; Sarkar et al., 2010). Strikingly, deletion of RSC2 or STH1 (catalytic core of the RSC complex) in combination with either RAD26 (a component of TCR) or RAD16 (a component of GGR) revealed a UV-dependent synthetic sick relationship (Figures S3A-G). We also observed a differential negative interaction between RSC2 and RAD14 (a component of both GGR and TCR; Figures S3A,D). Together with our finding that RSC mediates efficient repair in a variety of genomic contexts (Figures 3A,B), these data suggest that RSC may affect both pathways of NER.
To assess RSC’s role in both GGR and TCR, we employed an assay in which we measured the rate of photoproduct removal at the non-transcribed and transcribed strands of the RPB2 locus (Methods). Critically, all experiments were performed in G1-arrested cells in which lesion removal is dependent solely on NER (Gong et al., 2008; Gong et al., 2006; Sarkar et al., 2010; Smerdon and Lieberman, 1978) and any effect of DNA replication or replication fork stalling/collapse due to photoproduct induction can be ruled out. As expected, in rad26Δ and rad16Δ cells in which, respectively, TCR (Aboussekhra and Al-Sharif, 2005) and GGR (Verhage et al., 1994) are completely abrogated, we observed virtually no repair over the duration of the experiment, neither at the transcribed (Figure 3C) nor non-transcribed strands (Figure 3D). In rsc2Δ cells, repair at both strands was compromised compared to wild type, but not completely abolished, with respectively, ~1.8- and ~1.5-times more photoproducts present at three hours after UV irradiation. This suggests that RSC is required, but not essential, for GG- and TC-NER. Finally, the rsc2Δrad16Δ and rsc2Δrad26Δ double mutants displayed a reduction in the rate of repair similar to that of the corresponding NER-deficient single mutants suggesting a potential epistatic effect and providing further evidence for a role for RSC in both NER pathways. Importantly, the reduced rate of repair seen at the transcribed strand of RPB2 in rsc2Δ cells was not due to transcriptional misregulation, as the expression level of RPB2 was comparable in WT and rsc2Δ cells (Figure S3H).
RSC is recruited to sites of UV lesions via Rad4 and promotes nucleosome remodeling
We considered that RSC might affect NER indirectly by regulating the expression of one or more NER factors, or by acting directly at sites of UV lesions. To test the former hypothesis, we obtained previously published gene expression data generated in rsc mutants in nominal conditions (Lenstra et al., 2011). Analysis of these data found no changes in the expression of NER factors, while only nine genes annotated to the broader DNA damage response appeared to be differentially expressed in a rsc mutant compared to wild type (Figure S4A). However, none of these nine genes were found previously to be differentially expressed following exposure to UV (Wade et al., 2009). Finally, we randomly selected 11 core NER genes, and measured their expression levels via qPCR in both wild type and rsc2Δ cells following UV exposure (Methods). None of these genes’ change in UV-induced expression was found to be dependent on Rsc2 (Figure S4B). Together, this suggests that Rsc2 does not affect NER indirectly through transcriptional regulation of NER factors.
We next asked if RSC might be acting directly at UV lesions. Using a modified ChIP protocol that allows the analysis of protein occupancy in the presence of UV photoproducts (Methods) (Sarkar et al., 2010), we monitored Rsc2-Myc recruitment to MATa and HMLα following UV irradiation (Figures 4A,B). In wild-type cells, Rsc2-Myc accumulates at both loci almost immediately after irradiation, reaches maximal occupancy at 30 minutes post-irradiation and then decreases during the remainder of the experiment. Rsc2 recruitment was UV damage-dependent as we observed little enrichment of Rsc2-Myc at either locus in untreated conditions (Figure S4C). Finally, we also observed strong recruitment of Sth1-Myc to both loci in a UV-dependent fashion, providing further evidence for the recruitment of the RSC complex to damaged chromatin (Figure S4D).
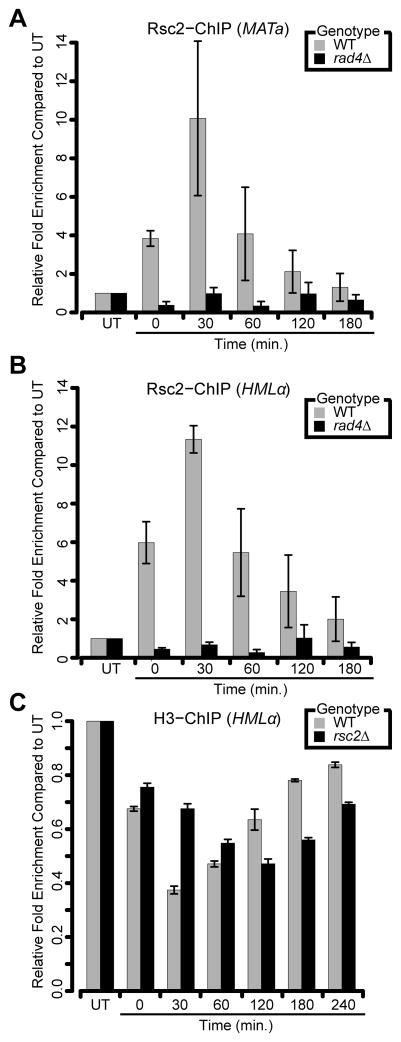
Figure 4. RSC promotes proper nucleosome remodeling following UV-induced damage.
(A,B) Analysis of Rsc2-Myc recruitment to either (A) MATa or (B) HMLαfollowing exposure to UV radiation. (C) Analysis of histone H3 occupancy at the HMLα locus following UV exposure in G1-synchronized cells. All data represent the mean ± 1 s.e.m of at least 3 independent replicates. See also Figure S4.
Rad4 is a core NER factor responsible for the initial damage recognition step and subsequent recruitment of other NER factors (Jansen et al., 1998). Previous work has shown an important role for Rad4 in mediating the recruitment of chromatin remodelers to sites of UV-induced lesions (Gong et al., 2006; Sarkar et al., 2010). Thus, we asked whether Rad4 may play a similar role in targeting RSC to damaged chromatin. Using our modified ChIP assay, we observed that Rad4 was recruited to both loci in a UV-dependent manner (Figure S4D), indicating that both Rad4 and RSC are present at the same sites of UV damage. Moreover, deletion of RAD4 completely abrogated Rsc2-Myc accumulation at both loci (Figures 4A,B). Together, these results suggest that Rad4 is required for the recruitment of RSC to sites of UV damage.
RSC possesses the capability to perturb nucleosome structures (Cairns et al., 1996; Cairns et al., 1999; Saha et al., 2002), suggesting that it may affect nucleosome remodeling dynamics at UV lesions to promote NER. To test this hypothesis, we monitored levels of histone H3 at the HMLα locus following UV treatment. In the wild-type strain, we observed a rapid loss of histone H3 within the first 30 minutes, followed by a gradual restoration over the next three hours (Figure 4C). These results are consistent with the rates of repair seen earlier (Figure 3), in which the majority of repair occurs within the first hour and is completed within three hours. In rsc2Δ mutants, we observed a delay in both the initial loss of H3 around UV lesions, as well as restoration of H3 occupancy at later time points (Figure 4C). As there was minimal difference in histone H3 levels between WT and rsc2Δ in untreated conditions, these results suggest that RSC promotes proper nucleosome remodeling at sites of UV lesions (Figure S4E).
Comparing network rewiring across low and high levels of UV
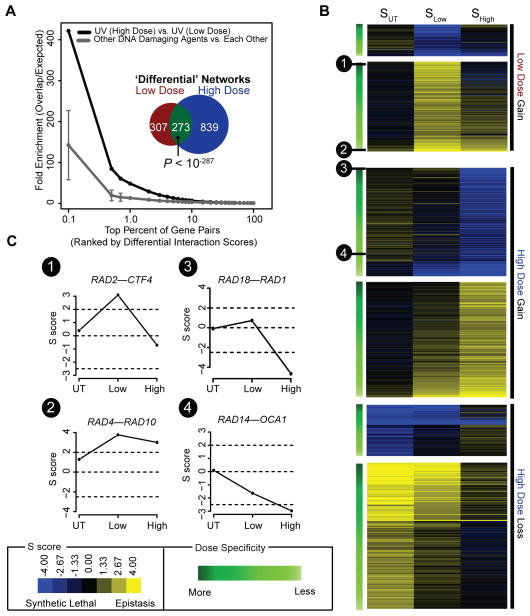
As this is the first study to measure genetic interactions in response to varying doses of the same agent, we next compared the effect of low versus high UV dose on the genetic network. We found that both UV-induced differential genetic networks had significant similarity, especially in comparison to networks measured under genotoxic agents other than UV (Figures 5A and Figure S5A). On the other hand, the high dose induced nearly twice the number of genetic interaction changes than the low dose (1112 versus 580, Figure 5A). This observation was clear through analysis of either differential (Figure 5A) or static networks (Figure S5B) and was robust to the choice of threshold used to define significant interactions (Figures S5C,D). Thus, while both UV doses produce overlapping networks and highlight gene functions related to the NER pathway, they also appear to induce a network of dose-specific interactions.
Figure 5. Identifying dose-specific differential interactions.
(A) Overlap between high and low UV dose differential networks (black line) or the average overlap seen amongst three previously published differential networks generated in response to distinct genotoxic agents (dark grey line/‘Other DNA Damaging Agents’; (Guenole et al., 2012)). Fold enrichment is defined as n/r, where n is the number of top-ranked gene pairs (x-axis; ranked by differential p-value) common to a pair of networks and r is the number expected at random. Error bars indicate 1 s.d. The inset shows the overlap between all significant differential interactions (PLow–UT, PHigh–UT≤0.001) uncovered in high dose versus low dose conditions. Significance of overlap was assessed using a one-tailed Fisher’s exact test. (B) Heat map of the static dose profiles (SUT → SLow → SHigh) for all 849 and 307 high and low dose differential interactions. Interactions have been categorized as “Gain of Interaction” or “Loss of Interaction” and then ordered (top to bottom) based on their likelihood of being a dose-specific differential interaction. For more details, see Supplementary Methods. (C) Example static dose profiles are shown for four interactions. See also Figure S5.
To further characterize this space of potential dose-specific interactions, we visualized the underlying static genetic changes between untreated, low dose, and high dose conditions for each of these interactions (Figures 5B). Visualizing the data in this manner revealed a continuum of differential interactions ranging from those interactions which displayed a marked change in interaction exclusively in low or high UV dose compared to untreated (e.g. RAD2-CTF4 and RAD18-RAD1; Figure 5C) to interactions in which a shift in genetic interaction was observed at both UV doses compared to untreated, but where the magnitude of this shift differed between doses (e.g. RAD4-RAD10 and RAD14-OCA1; Figure 5C).
To explicitly identify dose-specific interactions, i.e., interactions with a strong change in one dose only, we developed a bioinformatics pipeline to analyze an interaction’s underlying static dose profile. As described further in the Supplementary Methods, we first defined a set of model dose profiles representing high or low dose specific interactions (Figure S5E), as well as a ‘null’ model profile devoid of change between UV doses. Interactions whose static dose profile more closely aligns with a dose-specific profile compared to the null model are classified as dose-specific interactions. Applied to our data set, our method identified, respectively, 35 and 44 high and low dose specific interactions (Table S6; FDR < 40%).
These dose specific connections were enriched for interactions with genes annotated to DNA metabolism (P = 0.00012; high dose), DNA recombination (P = 0.00012; high dose), translesion synthesis (P = 0.02459; low dose), and intriguingly components of protein degradation (P = 0.0146; low dose). Consistent with these observations, exposure to genotoxic agents, including UV radiation, has previously been shown to result in increased rates of protein degradation (Burgis and Samson, 2007). Strikingly, we found that all low dose specific interactions involved NER factors involved in downstream repair activities such as DNA incision and gap filling (RAD1, RAD2, RAD10) and not by any of the early sensors. Consistent with this observation, deletion of factors involved in sensing DNA damage was found to have no impact on the rate of damage-induced protein degradation (Burgis and Samson, 2007), suggesting that the signal for increased protein degradation does not originate from the damaged DNA itself.
Although further work will be required to resolve the precise mechanisms underlying these dose specific connections, our study provides an important proof of principle that the framework for analyzing shifts in genetic networks in response to external stimuli (Bandyopadhyay et al., 2010; Bean and Ideker, 2012) can be expanded to understand how such networks are influenced by varying dosage.
DISCUSSION
Here, we have mapped the UV-induced genetic network between most components of the NER pathway and over 1,300 genes spanning a wide range of biological processes (Figure 1A). Unlike previous differential genetic studies (Bandyopadhyay et al., 2010; Guenole et al., 2012), we have for the first time made measurements across multiple doses of treatment and developed a bioinformatics pipeline to specifically identify dose-specific interactions (Figure 5 and Supplementary Methods). While the induction of genetic interactions exclusively at higher doses may be expected, our data also implicate a number of low dose specific connections (Table S6), suggesting the presence of response pathways unique to low versus high doses of DNA damage. Indeed, past genome-wide expression studies have shown the induction of transcriptional programs at low (but not high) concentrations of other genotoxic agents such as methyl methanesulfonate (MMS) and ionizing radiation (Benton et al., 2006). Future studies may combine differential genetic interaction mapping with our new bioinformatics pipeline to the study of other genotoxic agents that have known shifts in the mode of action between high and low doses. For example, moderate to high doses of hydroxyurea inhibit DNA, RNA, and protein synthesis, while at low concentrations, it interferes exclusively with DNA synthesis (Timson, 1969). Similarly, low concentrations of MMS have been shown to activate only the intra-S checkpoint, whereas higher doses also lead to activation of the G1/S checkpoint (Frei and Gasser, 2000). Examining dose-specific genetic networks may help to disentangle the pathways contributing to the distinct modes of action of these compounds.
Combining our differential genetic data with other physical interaction data sets (Collins et al., 2007; Gavin et al., 2006; Krogan et al., 2006) revealed a link between the RSC chromatin remodeling complex and both pathways of NER making RSC the first complex to be linked to TCR (Figure 2B, Figure 3, and Figure S3A–G). We found that Rsc2 is recruited in a UV-dependent manner to both expressed and silenced loci (Figures 4A,B) and that deletion of RSC2 led to altered histone remodeling dynamics at sites of UV damage (Figure 4C). It is worth noting that the remodeling defect observed in rsc2Δ cells is not as severe as that observed in rad4Δ cells, possibly due to the fact that other chromatin remodelers, such as INO80, are also recruited to sites of UV damage in a Rad4-dependent manner (Gong et al., 2006; Sarkar et al., 2010). While Sth1 (and not Rsc2) forms the catalytic core of the RSC complex, in vitro studies have demonstrated that purified Sth1, without other RSC components (including Rsc2), exhibits a severe reduction in remodeling activity (Saha et al., 2002). These results, coupled with our data showing recruitment of Sth1 and Rsc2 to sites of UV-induced lesions (Figure S4D), lead us to conclude that RSC mediates efficient NER by remodeling chromatin at sites of UV damage.
How might RSC be recruited to damaged chromatin? One possibility is that NER factors such as Rad4, Rad23, or Rad26, which are responsible for initial damage recognition (Prakash and Prakash, 2000), may physically interact with RSC to facilitate its recruitment. Although we were not able to demonstrate a physical interaction between Rsc2 and Rad4 or Rad23 (data not shown), the RSC complex contains over 15 different subunits (as defined in the Saccharomyces Genome Database) and thus an interaction with these NER factors might also occur through one of the other subunits. Alternatively, the damage recognition step itself may help to recruit RSC to chromatin. Structural studies of Rad4-DNA complexes have shown that the binding of Rad4 at UV-induced lesions results in the destabilization of the helical structure (Min and Pavletich, 2007) and the formation of a highly kinked structure (Janicijevic et al., 2003). Such structures have been shown to act as a platform for the assembly of an active NER complex and it is tempting to speculate that they may also serve to promote the recruitment of RSC to damaged chromatin.
Acetylation of histone H3 lysine 14 (H3K14) by NER factors Gcn5 and Rad16 is a critical mark for efficient NER (Teng et al., 2002). Past work has shown that Rsc4, one of the RSC subunits, contains a tandem bromodomain that binds specifically to H3K14ac (VanDemark et al., 2007). Moreover, Rsc4 itself is known be acetylated by Gcn5, an event which has been shown to be important for promoting resistance to DNA damage and replicative stress (Charles et al., 2011). This suggests that post-translational modification of either histones or components of RSC may also contribute to the recruitment and regulation of RSC-dependent chromatin remodeling at sites of UV-induced lesions.
Rsc1 and Rsc2 are known to define two mutually exclusive RSC complexes (Cairns et al., 1999) and appear to have overlapping but not identical functions (Cairns et al., 1999; Hillenmeyer et al., 2008; Rossio et al., 2010). For example, while both mutants are sensitive to double-stranded break inducing agents and exhibit defects in non-homologous end joining, these defects could not be rescued by additional copies of the other gene, suggesting similar but non-overlapping functions (Chai et al., 2005; Chambers et al., 2012; Kent et al., 2007). Here, we found that deletion of RSC2 but not RSC1, rendered cells sensitive to UV (Figure 2C). Consistent with this finding, recent work has implicated a role for Rsc2, but not Rsc1, in the replication of UV-damaged DNA (Niimi et al., 2012). However, given that our differential network revealed several connections between RSC1 and DDR factors, we cannot exclude a role for the Rsc1-containing complex in the UV response. For instance, Rsc1 may function in other facets of the UV damage response, such as facilitating the repair of DNA breaks that arise as a consequence of UV-induced replication fork collapse.
To facilitate access to our resource, all data have been made available as Cytoscape session files on a Supplementary Website (http://chianti.ucsd.edu/~rsrivas/srivas_2013/). Using Cytoscape (Shannon et al., 2003) one can query for genes of interest and easily browse the higher-level analysis of interactions between complexes (Figure 2A). Data on UV single mutant sensitivity, as well as gene-level data from the Saccharomyces Genome Database (Cherry et al., 2012), have been added to the session files, allowing for a wealth of information from different fields to be cross-referenced. We anticipate that this resource will be invaluable in increasing our understanding of the UV damage response.
Experimental Procedures
Differential Genetic Experiments
Double mutants were constructed using the SGA technology (Tong and Boone, 2006), except that in the final step double mutants were replica pinned on the prescribed media and exposed to UV-C radiation (20 J/m2 and 80 J/m2) or mock-treatment. Static and differential scores were computed as previously described (Bandyopadhyay et al., 2010; Collins et al., 2006). A list of all strains used is provided in Table S8.
Constructing the Module Map
A list of multi-genic modules was obtained from a previous study (Guenole et al., 2012). This list was filtered to include only those modules that contained at least two genes for which we had screened genetic interactions resulting in a list of 123 modules. The sum of the absolute value of differential scores for gene pairs spanning two modules was then compared to a null distribution of summed differential scores for equal-sized random samples of gene pairs. This analysis was performed separately for low dose and high dose differential networks, after which a threshold of P < 0.012 was used to generate the module map (Figure 2A; Tables S4,5).
Transcription Analysis
Mid-log phase cultures of cells were exposed to UV-C radiation (100 J/m2) or mock-treatment and allowed to recover in YPD media for 60 minutes at 30°C. Cell were then lysed and RT-qPCR was performed as previously described (Chen et al., 2013). For a list of primers used, see Table S9. Measurements were normalized against the housekeeping gene GCN4 and final fold changes were computed using the Pfaffl method (Pfaffl, 2001).
Repair Assays
Photoproduct removal rates at MATa, HMLα and RPB2 were determined as described previously (den Dulk et al., 2006; Sarkar et al., 2010). To test for a difference in the rate of photoproduct removal between two strains, a standard linear model was built in which ‘% photoproducts remaining’ was regressed against factors ‘Time’, ‘Strain’ (categorical variable representing the genotype of the strain, e.g. wildtype or rsc2Δ) and an interaction term (‘Time’ * ‘Strain’). The significance of the interaction term, which represents the difference in the rate of photoproduct removal between strains, was then assessed using an F test. All statistical analysis was performed in R (version 2.11.1).
Cell Cycle Profiling Experiments
Exponentially growing cells were exposed to UV (70 J/m2), and then released into fresh YPD medium. FACS analysis was performed at different time points following UV irradiation using a BD LSRII instrument and WinMDI software. Rad53 phosphorylation status was monitored via Western blot analysis using an anti-Rad53 antibody (Santa Cruz Biotechnology) as previously described (Guenole et al., 2012).
Measuring Protein Occupancy at UV-induced Lesions by ChIP-qPCR
ChIP-qPCR experiments were performed as described previously (Sarkar et al., 2010) using either an anti-histone H3 (Abcam #AB1791) or anti-Myc (Cell Signaling #9B11) antibody. Briefly, cells were exposed to UV radiation (200 J/m2) and harvested at the indicated time points (Figure 4). Cells were then processed as for conventional ChIP, except that after immunoprecipitation (‘IP’) and reversal of DNA-protein cross-links, the DNA was treated with 5 μgD. melanogaster 6-4 photolyase and 50 ng Escherichia coli CPD photolyase for 1 hr at room temperature to remove all unrepaired UV-lesions and permit equal amplification of all DNA. In addition, an aliquot of each extract was taken prior to immunoprecipitation and treated with photolyase enzymes (‘Input’), and a no antibody control IP was performed (‘No Antibody’). IP, Input, and No Antibody DNA were subsequently analyzed by RT-qPCR using primers targeting MATa or HMLα (see Table S9 for primer sequences). Absolute enrichment for histone H3, Rsc2, Sth1, or Rad4 at these loci was calculated by comparing IP threshold cycle values (Ct) to no antibody values using input as a reference. Finally, relative fold enrichment was defined as the ratio of absolute enrichment of UV-treated samples to that of untreated samples (UT).
Additional experimental details have been provided in the Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Differential genetic analysis of 45,938 double mutants in response to UV.
Genetic data link RSC to global genome and transcription-coupled repair.
RSC facilitates repair via nucleosome remodeling at sites of UV-induced lesions.
High and low dose UV induce a set of dose-specific genetic interactions.
Acknowledgments
The authors thank C. Logie for generously providing yeast strains; A. Guénolé and I. Stulemeijer for assistance with the screen; P. Jaeger, G. Hannum, G. Bean, and M. Hofree for insightful discussions; and J.P. Shen for a critical reading of the manuscript. This work was supported by grants from the U.S. National Institutes of Health to T.I. (ES014811, GM084279), the Netherlands Organization for Scientific Research (NWO-VIDI and TOP-GO) and Human Frontiers Science Program (CDA grant) to H.v.A, and a Programme Grant from Cancer Research (UK) to S.S. and P.M.c.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboussekhra A, Al-Sharif IS. Homologous recombination is involved in transcription-coupled repair of UV damage in Saccharomyces cerevisiae. The EMBO journal. 2005;24:1999–2010. doi: 10.1038/sj.emboj.7600665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS computational biology. 2008;4:e1000065. doi: 10.1371/journal.pcbi.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean GJ, Ideker T. Differential analysis of high-throughput quantitative genetic interaction data. Genome biology. 2012;13:R123. doi: 10.1186/gb-2012-13-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Molecular cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Benton MG, Somasundaram S, Glasner JD, Palecek SP. Analyzing the dose-dependence of the Saccharomyces cerevisiae global transcriptional response to methyl methanesulfonate and ionizing radiation. BMC genomics. 2006;7:305. doi: 10.1186/1471-2164-7-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgis NE, Samson LD. The protein degradation response of Saccharomyces cerevisiae to classical DNA-damaging agents. Chemical research in toxicology. 2007;20:1843–1853. doi: 10.1021/tx700126e. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Molecular cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes & development. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AL, Brownlee PM, Durley SC, Beacham T, Kent NA, Downs JA. The two different isoforms of the RSC chromatin remodeling complex play distinct roles in DNA damage responses. PloS one. 2012;7:e32016. doi: 10.1371/journal.pone.0032016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles GM, Chen C, Shih SC, Collins SR, Beltrao P, Zhang X, Sharma T, Tan S, Burlingame AL, Krogan NJ, et al. Site-specific acetylation mark on an essential chromatin-remodeling complex promotes resistance to replication stress. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10620–10625. doi: 10.1073/pnas.1019735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Licon K, Otsuka R, Pillus L, Ideker T. Decoupling epigenetic and genetic effects through systematic analysis of gene position. Cell reports. 2013;3:128–137. doi: 10.1016/j.celrep.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic acids research. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Molecular & cellular proteomics : MCP. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome biology. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dulk B, Sun SM, de Ruijter M, Brandsma JA, Brouwer J. Rad33, a new factor involved in nucleotide excision repair in Saccharomyces cerevisiae. DNA repair. 2006;5:683–692. doi: 10.1016/j.dnarep.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes & development. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell cycle. 2008;7:1067–1074. doi: 10.4161/cc.7.8.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nature structural & molecular biology. 2006;13:902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- Guenole A, Srivas R, Vreeken K, Wang ZZ, Wang S, Krogan NJ, Ideker T, van Attikum H. Dissection of DNA Damage Responses Using Multiconditional Genetic Interaction Maps. Molecular cell. 2012 doi: 10.1016/j.molcel.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder SN, Sung P, Prakash L, Prakash S. Yeast DNA-repair gene RAD14 encodes a zinc metalloprotein with affinity for ultraviolet-damaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5433–5437. doi: 10.1073/pnas.90.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Krogan NJ. Differential network biology. Molecular systems biology. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicijevic A, Sugasawa K, Shimizu Y, Hanaoka F, Wijgers N, Djurica M, Hoeijmakers JH, Wyman C. DNA bending by the human damage recognition complex XPC-HR23B. DNA repair. 2003;2:325–336. doi: 10.1016/s1568-7864(02)00222-7. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Verhage RA, Brouwer J. Preferential binding of yeast Rad4.Rad23 complex to damaged DNA. The Journal of biological chemistry. 1998;273:33111–33114. doi: 10.1074/jbc.273.50.33111. [DOI] [PubMed] [Google Scholar]
- Kent NA, Chambers AL, Downs JA. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. The Journal of biological chemistry. 2007;282:27693–27701. doi: 10.1074/jbc.M704707200. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Molecular cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, Mullenders LH, Dantuma NP. DDB2 promotes chromatin decondensation at UV-induced DNA damage. The Journal of cell biology. 2012;197:267–281. doi: 10.1083/jcb.201106074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- Niimi A, Chambers AL, Downs JA, Lehmann AR. A role for chromatin remodellers in replication of damaged DNA. Nucleic acids research. 2012;40:7393–7403. doi: 10.1093/nar/gks453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Ooi SL, Wang X, Sookhai-Mahadeo S, Meluh P, Boeke JD. dSLAM analysis of genome-wide genetic interactions in Saccharomyces cerevisiae. Methods. 2007;41:206–221. doi: 10.1016/j.ymeth.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annual review of biochemistry. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutation research. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- Rossio V, Galati E, Ferrari M, Pellicioli A, Sutani T, Shirahige K, Lucchini G, Piatti S. The RSC chromatin-remodeling complex influences mitotic exit and adaptation to the spindle assembly checkpoint by controlling the Cdc14 phosphatase. The Journal of cell biology. 2010;191:981–997. doi: 10.1083/jcb.201007025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes & development. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M, Prakash L, Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Kiely R, McHugh PJ. The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. The Journal of cell biology. 2010;191:1061–1068. doi: 10.1083/jcb.201006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Sertic S, Pizzi S, Lazzaro F, Plevani P, Muzi-Falconi M. NER and DDR: classical music with new instruments. Cell cycle. 2012;11:668–674. doi: 10.4161/cc.11.4.19117. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA- reapir synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivas R, Hannum G, Ruscheinski J, Ono K, Wang PL, Smoot M, Ideker T. Assembling global maps of cellular function through integrative analysis of physical and genetic networks. Nature protocols. 2011;6:1308–1323. doi: 10.1038/nprot.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nature genetics. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Yu Y, Waters R. The Saccharomyces cerevisiae histone acetyltransferase Gcn5 has a role in the photoreactivation and nucleotide excision repair of UV-induced cyclobutane pyrimidine dimers in the MFA2 gene. Journal of molecular biology. 2002;316:489–499. doi: 10.1006/jmbi.2001.5383. [DOI] [PubMed] [Google Scholar]
- Timson J. Hydroxyurea: comparison of cytotoxic and antimitotic activities against human lymphocytes in vitro. British journal of cancer. 1969;23:337–339. doi: 10.1038/bjc.1969.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Molecular cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R, Zeeman AM, de Groot N, Gleig F, Bang DD, van de Putte P, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Molecular and cellular biology. 1994;14:6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade SL, Poorey K, Bekiranov S, Auble DT. The Snf1 kinase and proteasome-associated Rad23 regulate UV-responsive gene expression. The EMBO journal. 2009;28:2919–2931. doi: 10.1038/emboj.2009.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.