Abstract
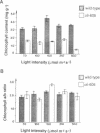
Olive (oli) is a recessive nuclear mutation of Antirrhinum majus which reduces the level of chlorophyll pigmentation and affects the ultrastructure of chloroplasts. The oli-605 allele carries a Tam3 transposon insertion which has allowed the locus to be isolated. The oli gene encodes a large putative protein of 153 kDa which shows homology to the products of two bacterial genes necessary for tetrapyrrole-metal chelation during the synthesis of bacteriochlorophyll or cobyrinic acid. We therefore propose that the product of the oli gene is necessary for a key step of chlorophyll synthesis: the chelation of magnesium by protoporphyrin IX. Somatic reversion of the oli-605 allele produces chimeric plants which indicate that the oli gene functions cell-autonomously. Expression of oli is restricted to photosynthetic cells and repressed by light, suggesting that it may be involved in regulating the rate of chlorophyll synthesis in green tissues.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aagaard J., Sistrom W. R. Control of synthesis of reaction center bacteriochlorophyll in photosynthetic bacteria. Photochem Photobiol. 1972 Feb;15(2):209–225. doi: 10.1111/j.1751-1097.1972.tb06240.x. [DOI] [PubMed] [Google Scholar]
- Apel K. Phytochrome-induced appearance of mRNA activity for the apoprotein of the light-harvesting chlorophyll a/b protein of barley (Hordeum vulgare). Eur J Biochem. 1979 Jun;97(1):183–188. doi: 10.1111/j.1432-1033.1979.tb13101.x. [DOI] [PubMed] [Google Scholar]
- Apel K., Santel H. J., Redlinger T. E., Falk H. The protochlorophyllide holochrome of barley (Hordeum vulgare L.). Isolation and characterization of the NADPH:protochlorophyllide oxidoreductase. Eur J Biochem. 1980 Oct;111(1):251–258. doi: 10.1111/j.1432-1033.1980.tb06100.x. [DOI] [PubMed] [Google Scholar]
- Armstrong G. A., Alberti M., Leach F., Hearst J. E. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet. 1989 Apr;216(2-3):254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- Becerril J. M., Duke S. O. Protoporphyrin IX Content Correlates with Activity of Photobleaching Herbicides. Plant Physiol. 1989 Jul;90(3):1175–1181. doi: 10.1104/pp.90.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel A. J., Marrs B. L. Transcriptional regulation of several genes for bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata in response to oxygen. J Bacteriol. 1983 Nov;156(2):686–694. doi: 10.1128/jb.156.2.686-694.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollivar D. W., Bauer C. E. Nucleotide Sequence of S-Adenosyl-l-Methionine: Magnesium Protoporphyrin Methyltransferase from Rhodobacter capsulatus. Plant Physiol. 1992 Jan;98(1):408–410. doi: 10.1104/pp.98.1.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D., Carpenter R., Sommer H., Hartley N., Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993 Jan 15;72(1):85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Castelfranco P. A., Weinstein J. D., Schwarcz S., Pardo A. D., Wezelman B. E. The Mg insertion step in chlorophyll biosynthesis. Arch Biochem Biophys. 1979 Feb;192(2):592–598. doi: 10.1016/0003-9861(79)90130-9. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Carpenter R. A semi-dominant allele, niv-525, acts in trans to inhibit expression of its wild-type homologue in Antirrhinum majus. EMBO J. 1988 Apr;7(4):877–883. doi: 10.1002/j.1460-2075.1988.tb02891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Carpenter R., Martin C. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell. 1986 Oct 24;47(2):285–296. doi: 10.1016/0092-8674(86)90451-4. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Romero J. M., Doyle S., Elliott R., Murphy G., Carpenter R. floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell. 1990 Dec 21;63(6):1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Coomber S. A., Chaudhri M., Connor A., Britton G., Hunter C. N. Localized transposon Tn5 mutagenesis of the photosynthetic gene cluster of Rhodobacter sphaeroides. Mol Microbiol. 1990 Jun;4(6):977–989. doi: 10.1111/j.1365-2958.1990.tb00670.x. [DOI] [PubMed] [Google Scholar]
- Crouzet J., Levy-Schil S., Cameron B., Cauchois L., Rigault S., Rouyez M. C., Blanche F., Debussche L., Thibaut D. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J Bacteriol. 1991 Oct;173(19):6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V., Furtek D. B., Nelson O. E. Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac). Proc Natl Acad Sci U S A. 1984 Jun;81(12):3825–3829. doi: 10.1073/pnas.81.12.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuesler T. P., Wright L. A., Castelfranco P. A. Properties of Magnesium Chelatase in Greening Etioplasts: METAL ION SPECIFICITY AND EFFECT OF SUBSTRATE CONCENTRATIONS. Plant Physiol. 1981 Feb;67(2):246–249. doi: 10.1104/pp.67.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRANICK S. Magnesium protoporphyrin monoester and protoporphyrin monomethyl ester in chlorophyll biosynthesis. J Biol Chem. 1961 Apr;236:1168–1172. [PubMed] [Google Scholar]
- Gorchein A. Control of magnesium-protoporphyrin chelatase activity in Rhodopseudomonas spheroides. Role of light, oxygen, and electron and energy transfer. Biochem J. 1973 Aug;134(4):833–845. doi: 10.1042/bj1340833d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchein A. Magnesium protoporphyrin chelatase activity in Rhodopseudomonas spheroides. Studies with whole cells. Biochem J. 1972 Mar;127(1):97–106. doi: 10.1042/bj1270097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough S. Defective synthesis of porphyrins in barley plastids caused by mutation in nuclear genes. Biochim Biophys Acta. 1972 Nov 24;286(1):36–54. doi: 10.1016/0304-4165(72)90086-4. [DOI] [PubMed] [Google Scholar]
- Griffiths W. T. Protochlorophyll and protochlorophyllide as precursors for chlorophyll synthesis in vitro. FEBS Lett. 1974 Dec 15;49(2):196–200. doi: 10.1016/0014-5793(74)80510-7. [DOI] [PubMed] [Google Scholar]
- Griffiths W. T. Some observations on chlorophyll(ide) synthesis by isolated etioplasts. Biochem J. 1975 Jan;146(1):17–24. doi: 10.1042/bj1460017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel E., Klein S. Light dependent formation of -aminolevulinic acid in etiolated leaves of higher plants. Biochem Biophys Res Commun. 1972 Oct 17;49(2):364–370. doi: 10.1016/0006-291x(72)90419-6. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Herrin D. L., Battey J. F., Greer K., Schmidt G. W. Regulation of chlorophyll apoprotein expression and accumulation. Requirements for carotenoids and chlorophyll. J Biol Chem. 1992 Apr 25;267(12):8260–8269. [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Kiley P. J., Kaplan S. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides light-harvesting B800-850-alpha and B800-850-beta genes. J Bacteriol. 1987 Jul;169(7):3268–3275. doi: 10.1128/jb.169.7.3268-3275.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Mayerhofer R., Koncz-Kalman Z., Nawrath C., Reiss B., Redei G. P., Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990 May;9(5):1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Coen E. S., Doyle S., Carpenter R. Pigmentation mutants produced by transposon mutagenesis in Antirrhinum majus. Plant J. 1991 Jul;1(1):59–69. [PubMed] [Google Scholar]
- Lötscher H. R., Winterhalter K. H., Carafoli E., Richter C. The energy-state of mitochondria during the transport of Ca2+. Eur J Biochem. 1980 Sep;110(1):211–216. doi: 10.1111/j.1432-1033.1980.tb04857.x. [DOI] [PubMed] [Google Scholar]
- Martin C., Carpenter R., Sommer H., Saedler H., Coen E. S. Molecular analysis of instability in flower pigmentation of Antirrhinum majus, following isolation of the pallida locus by transposon tagging. EMBO J. 1985 Jul;4(7):1625–1630. doi: 10.1002/j.1460-2075.1985.tb03829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet J. E., Klein P. G., Klein R. R. Chlorophyll regulates accumulation of the plastid-encoded chlorophyll apoproteins CP43 and D1 by increasing apoprotein stability. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4038–4042. doi: 10.1073/pnas.87.11.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsat B., Monfort A., Chatellard P., Stutz E. Mapping and sequencing of an actively transcribed Euglena gracilis chloroplast gene (ccsA) homologous to the Arabidopsis thaliana nuclear gene cs(ch-42). FEBS Lett. 1992 Jun 1;303(2-3):181–184. doi: 10.1016/0014-5793(92)80514-h. [DOI] [PubMed] [Google Scholar]
- Pardo A. D., Chereskin B. M., Castelfranco P. A., Franceschi V. R., Wezelman B. E. ATP requirement for mg chelatase in developing chloroplasts. Plant Physiol. 1980 May;65(5):956–960. doi: 10.1104/pp.65.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Scolnik P. A., Marrs B. L. Genetic research with photosynthetic bacteria. Annu Rev Microbiol. 1987;41:703–726. doi: 10.1146/annurev.mi.41.100187.003415. [DOI] [PubMed] [Google Scholar]
- Sganga M. W., Aksamit R. R., Cantoni G. L., Bauer C. E. Mutational and nucleotide sequence analysis of S-adenosyl-L-homocysteine hydrolase from Rhodobacter capsulatus. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6328–6332. doi: 10.1073/pnas.89.14.6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. P., Cohen S. N., Clark W. G., Marrs B. L. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J Bacteriol. 1983 May;154(2):580–590. doi: 10.1128/jb.154.2.580-590.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. J., Weinstein J. D. Further characterization of the magnesium chelatase in isolated developing cucumber chloroplasts : substrate specificity, regulation, intactness, and ATP requirements. Plant Physiol. 1991 Apr;95(4):1189–1196. doi: 10.1104/pp.95.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. J., Weinstein J. D. In vitro assay of the chlorophyll biosynthetic enzyme Mg-chelatase: resolution of the activity into soluble and membrane-bound fractions. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5789–5793. doi: 10.1073/pnas.88.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. M., Bauer C. E. Rhodobacter capsulatus genes involved in early steps of the bacteriochlorophyll biosynthetic pathway. J Bacteriol. 1990 Sep;172(9):5001–5010. doi: 10.1128/jb.172.9.5001-5010.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H. C., Marrs B. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J Bacteriol. 1976 May;126(2):619–629. doi: 10.1128/jb.126.2.619-629.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. A., Bauer C. E., Williams J. C., Marrs B. L. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol Gen Genet. 1989 Jul;218(1):1–12. doi: 10.1007/BF00330558. [DOI] [PubMed] [Google Scholar]
- Zhu Y. S., Hearst J. E. Regulation of expression of genes for light-harvesting antenna proteins LH-I and LH-II; reaction center polypeptides RC-L, RC-M, and RC-H; and enzymes of bacteriochlorophyll and carotenoid biosynthesis in Rhodobacter capsulatus by light and oxygen. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7613–7617. doi: 10.1073/pnas.83.20.7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsebo K. M., Hearst J. E. Genetic-physical mapping of a photosynthetic gene cluster from R. capsulata. Cell. 1984 Jul;37(3):937–947. doi: 10.1016/0092-8674(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Zucconi A. P., Beatty J. T. Posttranscriptional regulation by light of the steady-state levels of mature B800-850 light-harvesting complexes in Rhodobacter capsulatus. J Bacteriol. 1988 Feb;170(2):877–882. doi: 10.1128/jb.170.2.877-882.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R. G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989 Apr 1;180(3):535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]