Significance
The nonenzymatic RNA polymerization introduces backbone heterogeneity with a mixture of 2′–5′ and 3′–5′ linkages. RNA polymerase II (pol II) is a key modern enzyme responsible for synthesizing 3′–5′–linked RNA with high fidelity. It is unclear how pol II selectively recognizes the 3′–5′ over 2′–5′ linkage. Here, we systematically investigated how phosphodiester linkages of nucleic acids govern pol II transcriptional efficiency and fidelity. We revealed pol II has an asymmetric (strand-specific) recognition of phosphodiester linkage, which may reflect a universal principle of template-dependent genetic information transfer. Our results elucidate essential contributions of the phosphodiester linkage to pol II transcription and provide important understanding on nucleic acid recognition and genetic information transfer during molecular evolution.
Keywords: 2′–5′ phosphodiester linkage, trigger loop, synthetic nucleic acid analogues
Abstract
Nonenzymatic RNA polymerization in early life is likely to introduce backbone heterogeneity with a mixture of 2′–5′ and 3′–5′ linkages. On the other hand, modern nucleic acids are dominantly composed of 3′–5′ linkages. RNA polymerase II (pol II) is a key modern enzyme responsible for synthesizing 3′–5′–linked RNA with high fidelity. It is not clear how modern enzymes, such as pol II, selectively recognize 3′–5′ linkages over 2′–5′ linkages of nucleic acids. In this work, we systematically investigated how phosphodiester linkages of nucleic acids govern pol II transcriptional efficiency and fidelity. Through dissecting the impacts of 2′–5′ linkage mutants in the pol II catalytic site, we revealed that the presence of 2′–5′ linkage in RNA primer only modestly reduces pol II transcriptional efficiency without affecting pol II transcriptional fidelity. In sharp contrast, the presence of 2′–5′ linkage in DNA template leads to dramatic decreases in both transcriptional efficiency and fidelity. These distinct effects reveal that pol II has an asymmetric (strand-specific) recognition of phosphodiester linkage. Our results provided important insights into pol II transcriptional fidelity, suggesting essential contributions of phosphodiester linkage to pol II transcription. Finally, our results also provided important understanding on the molecular basis of nucleic acid recognition and genetic information transfer during molecular evolution. We suggest that the asymmetric recognition of phosphodiester linkage by modern nucleic acid enzymes likely stems from the distinct evolutionary pressures of template and primer strand in genetic information transfer during molecular evolution.
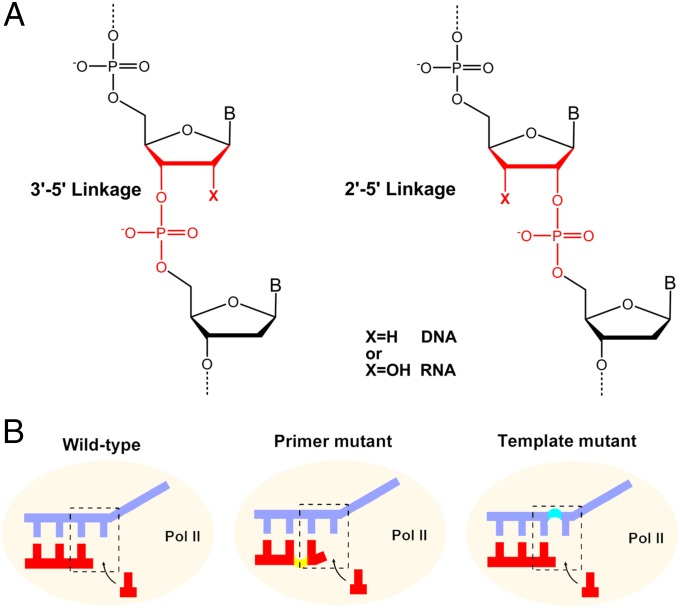
The remarkable capacity of RNA as the functional catalyst as well as the genetic information carrier provides a strong support for the RNA world hypothesis, in which RNA is proposed to play a key role in the early evolution of primitive life before DNA and protein (enzyme) evolved (1, 2). One critical issue for RNA replication in early life is backbone heterogeneity because RNA contains both 2′-OH and 3′-OH, in which both can form phosphodiester bond during RNA polymerization. Indeed, previous studies revealed that nonenzymatic RNA replication would lead to a mixture of 2′–5′ and 3′–5′ linkages (Fig. 1A) (3–9). The 2′–5′–linked nucleic acids can form duplex structures by themselves or by pairing with natural nucleic acids (10–15). The 2′–5′ linkage substitutions in some functional RNAs retain molecular recognition and catalytic properties (16–19). These discoveries suggested that the 2′–5′ linkage in nucleic acids could be an important alternative linkage during early evolution.
Fig. 1.
Nucleic acid structures with 3′–5′ and 2′–5′ phosphodiester linkage. (A) Scheme of 3′–5′– and 2′–5′–linked DNA and RNA. (B) Wild-type, primer mutant, and template mutant scaffolds for RNA pol II transcription.
This backbone heterogeneity problem is largely eliminated during evolution. Modern nucleic acids are dominated by 3′–5′ linkages. The 2′–5′ RNA linkages are only present in a few specific processes. One example is the formation of lariat RNA intron during splicing, where the 2′-OH of an adenosine attacks the 5′ exon–intron junction to release the 5′ exon and form a lariat RNA containing both 3′–5′ and 2′–5′ linkages (20–22). Another example is the generation of 2′–5′ oligomers of adenosine by 2′–5′–oligoadenylate synthetase (OAS) during the IFN antiviral response, which can in turn activate latent RNase (RNaseL) to degrade viral and cellular RNA and thus limit the viral infection (23, 24). However, in the genetic information storage and transfer, the phosphodiester linkages of nucleic acids are unified in a 3′–5′ linked form. RNA polymerase II (pol II), an essential contemporary enzyme, is responsible for synthesizing precursor mRNA in all eukaryotic cells (25, 26). All of these RNA transcripts contain the 3′–5′ linkage backbone. This result leads to several essential questions: How do contemporary enzymes, such as RNA pol II, selectively recognize the 3′–5′ linkages over the 2′–5′ linkages? How does this modern enzyme achieve high fidelity of phosphodiester linkage during the RNA polymerization reaction? As a contemporary enzyme, does RNA pol II still tolerate the phosphodiester linkage alteration (i.e., 2′–5′ linkage)? How would pol II transcription be affected when the phosphodiester linkage configuration of primer strand or template strand is changed? The answers to these questions remain elusive.
In this work, we systematically investigated how phosphodiester linkages of nucleic acids in the pol II active site govern the pol II transcription activity. Using site-specific alteration from a 3′–5′ to a 2′–5′ linkage configuration in either RNA or DNA, we quantitatively dissected the effects of phosphodiester linkage on pol II substrate incorporation, elongation, and proofreading. We also used the α-amanitin transcription inhibition assays to unveil the molecular mechanisms for pol II trigger loop conformational change upon the phosphodiester linkage alteration. Unexpectedly, we revealed strand-specific (asymmetric) contributions of 3′–5′ phosphodiester linkage in ensuring effective and accurate transcription, which may serve as an inherent feature for template-dependent genetic information transfer. This study provided important understanding on the molecular basis of RNA pol II recognition of phosphodiester backbone as well as novel insights into genetic information storage and transfer during molecular evolution.
Results
To systematically dissect the role of phosphodiester linkage on RNA pol II transcription, we constructed a well-defined in vitro transcription system with canonical 3′–5′ linkage backbone (referred to as “wild-type” system) or the scaffold containing a site-specific 2′–5′ phosphodiester linkage in either RNA primer or DNA template strand (referred to as “primer mutant” or “template mutant,” respectively) (Fig. 1B). Direct comparison of pol II transcription from the wild-type, primer linkage mutant, and template linkage mutant systems would allow us to quantitatively and systematically investigate the impact of the phosphodiester linkage alteration on pol II transcription. Such information gained from this defined system would also reveal the potential imprints of pol II as a contemporary RNA polymerase during phosphodiester linkage evolution from the RNA world.
Impact of the 2′–5′ Primer Linkage Mutant on pol II Transcription.
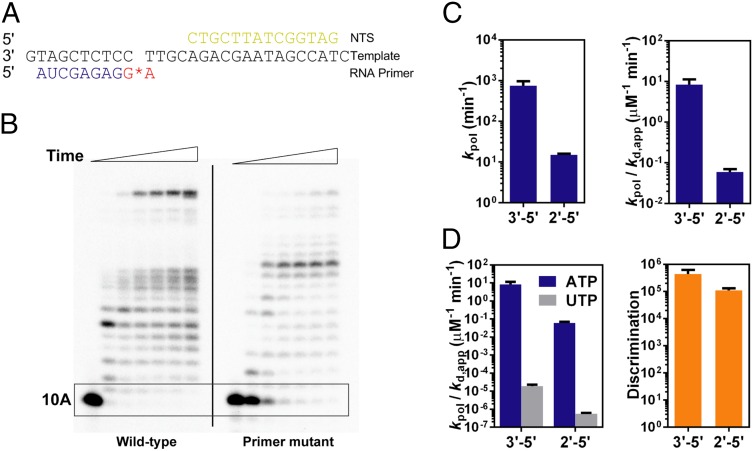
We first investigated the impact of 2′–5′ phosphodiester linkage alteration at 3′-RNA primer terminus (between −1 and −2 position) on pol II transcription (Fig. 2A). As shown in Fig. 2B, the first nucleotide incorporation right after the 2′–5′ linkage RNA primer is greatly compromised (the boxed area in Fig. 2B, Right). In contrast, pol II can extend efficiently from a wild-type RNA primer (Fig. 2B, Left). We observed long RNA transcripts with prolonged incubation indicating that a 2′–5′ linkage in RNA primer is not an absolute blockage for pol II transcription and the rate limiting step for pol II elongation is the first nucleotide addition immediately after the 2′–5′ linkage.
Fig. 2.
The 2′–5′ phosphodiester linkage in RNA primer end reduces RNA pol II transcriptional efficiency but does not affect fidelity. (A) Scaffold of RNA, template DNA, and nontemplate DNA for runoff elongation from the 2′–5′–linked primer end. The red star refers to the 2′–5′ linkage. (B) RNA pol II transcription products of 3′–5′–linked (left part of the gel) and 2′–5′–linked (right part of the gel) scaffolds in the presence of 25 μM NTP. Time points are 0, 30 s, 5 min, 20 min, 1 h, 2 h, and 4 h from left to right. The boxed area shows the major difference with linkage alteration. (C) Catalytic constants (kpol) and specificity constants (kpol/Kd,app) of pol II transcription from the 3′–5′ primer (blue bars) and the 2′–5′ primer (gray bars). (D) Specificity constants for both correct incorporation (ATP) and incorrect incorporation (UTP) (Left) and the discrimination power (Right).
We then quantitatively measured the effect of the 2′–5′ phosphodiester linkage alteration at 3′-RNA primer terminus on transcriptional efficiency by using pre-steady state single turnover incorporation assays. These assays allowed us to determine the kinetic parameters kpol (catalytic rate constant for substrate incorporation), Kd,app (apparent substrate dissociation constant), and substrate specificity (kpol/Kd,app, as a measurement of enzymatic efficiency). A single 2′–5′ linkage in the RNA primer leads to a 50-fold decrease in kpol, a 2.8-fold decrease in Kd,app, and an ∼140-fold decrease in substrate specificity (kpol/Kd,app), in comparison with transcription from the wild-type scaffold (Fig. 2C and Table 1). These results highlighted an important contribution of 3′–5′ linkage orientation in RNA primer to pol II transcriptional efficiency.
Table 1.
Selected kinetic data for effects of phosphodiester linkage alteration in RNA and DNA on pol II transcription
| Linkage | Incorporation | kpol, min−1 | Kd,app, μM | kpol/Kd,app, μM−1·min−1 | Relative efficiency* | Discrimination† | |
| Wild type | ATP | 750 ± 210 | 90 ± 20 | 8.3 ± 3.0 | 1 | (4.4 ± 1.8) × 105 | |
| UTP | 0.015 ± 0.003 | 800 ± 60 | (1.9 ± 0.4) × 10−5 | — | 1 | ||
| Primer mutant‡ | ATP | 15 ± 1 | 250 ± 40 | 0.06 ± 0.01 | 0.007 ± 0.003 | — | (1.1 ± 0.2) × 105 |
| UTP | (4.5 ± 0.2) × 10−4 | 800 ± 90 | (5.6 ± 0.7) × 10−7 | — | 0.029 ± 0.007 | ||
| Template mutant‡ | ATP | 17 ± 1 | 530 ± 60 | 0.032 ± 0.004 | 0.004 ± 0.003 | — | 910 ± 200 |
| UTP | 0.13 ± 0.01 | 3,700 ± 700 | (3.5 ± 0.7) × 10−5 | — | 1.8 ± 0.5 | ||
We further tested whether the transcriptional fidelity can be affected by the phosphodiester linkage alteration in the primer. Strikingly, we found that the phosphodiester linkage alteration in the RNA primer does not affect pol II transcriptional fidelity, although it does slow down both the matched ATP and mismatched UTP incorporation. As shown in Fig. 2D and Table 1, pol II has strong discrimination power [(4.4 ± 1.8) × 105] for incorporation of matched ATP over mismatched UTP to a wild-type 3′-RNA terminus (3′–5′ linkage). Unexpectedly, pol II still maintains very high transcriptional fidelity with strong discrimination power [(1.1 ± 0.2) × 105] even in the presence of a 2′–5′ phosphodiester linkage in RNA primer.
To further investigate whether pol II can recognize the 2′–5′ phosphodiester linkage alteration in the RNA primer via backtracking and proofreading cleavage activity, we measured transcription factor IIS (TFIIS)-stimulated cleavage rates of pol II elongation complex containing either a wild-type RNA primer or a 2′–5′ RNA primer linkage mutant. Similar to the result from pol II elongation complex with wild-type RNA, we observed a very weak cleavage for pol II elongation complex with a 2′–5′ RNA primer linkage mutant (Fig. S1). This result implies that the majority of pol II elongation complex is resistant to TFIIS cleavage and located in a posttranslocation state despite the presence of a 2′–5′ phosphodiester linkage at its 3′-RNA terminus. The 2′–5′ phosphodiester linkage alteration in the primer does not affect pol II translocation. Taken together, we found out that substitution of a 3′–5′ linkage with a 2′–5′ linkage in RNA primer plays a negligible role in pol II transcriptional fidelity and pol II translocation.
Impact of the 2′–5′ Template Linkage Mutant on pol II Transcription.
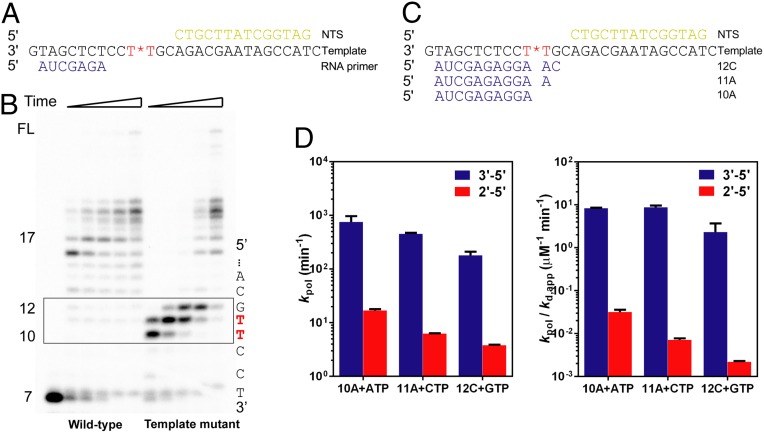
To test the impact of the 2′–5′ linkage in template strand on pol II transcription, we first performed a runoff assay, which allows RNA pol II to elongate in the presence of all four NTPs (Fig. 3A). As shown in Fig. 3B, RNA pol II is stalled near the 2′–5′ linkage site. In contrast, pol II can elongate along the wild-type template (3′–5′ linkage) and produce long transcripts. A single 2′–5′ linkage change in the template strand leads to three strong pausing sites during RNA pol II transcription, which refer to the 10-, 11-, and 12-mer products. RNA pol II can eventually bypass these pausing sites with prolonged incubation. To further confirm that all three pausing bands were directly caused by 2′–5′ linkage and not due to nucleotide misincorporation, we also performed transcription assays with the scaffolds containing primers with varied lengths that mimic stepwise bypass of the 2′–5′ linkage site in template strand (Fig. S2). The results from these scaffolds revealed significant pausing and slow extension along the DNA template containing a 2′–5′ linkage. Taken together, all three pausing bands as observed in Fig. 3B are due to the presence of 2′–5′ linkage site.
Fig. 3.
Transcriptional efficiency of RNA pol II is greatly reduced by the linkage alteration in the DNA template. (A) Scaffold of RNA, template DNA, and nontemplate DNA for runoff elongation through the 2′–5′ linkage site in the template. The red star refers to the 2′–5′ linkage in the DNA. (B) RNA pol II transcription products of 3′–5′–linked (left part of the gel) and 2′–5′–linked (right part of the gel) scaffolds in the presence of 25 μM NTP. Time points are 30 s, 2 min, 5 min, 20 min, and 1 h from left to right. Numbers on the left refer to lengths of RNA elongation products; positions of these RNA products on the DNA template are partly shown on the right. The boxed area shows the major difference with linkage alteration. (C) Scaffolds used to study every transcriptionally slowed down step. Three primers refer to three stalled positions in the DNA template. The red star refers to the 2′–5′ linkage. (D) Catalytic constants (kpol) and specificity constants (kpol/Kd,app) of pol II transcription through 3′–5′ linkage (blue) and 2′–5′ linkage (red).
To further quantitatively investigate the effects of the 2′–5′ linkage alteration in template on RNA pol II transcription, we assembled three scaffolds for kinetic studies, which are in correspondence with the three pausing sites shown in Fig. 3C (referred to as the 10A, 11A, and 12C scaffolds). The 10A scaffold mimics the state in which pol II encounters the 2′–5′ linkage site, whereas the 11A and 12C scaffolds refer to the following two consecutive bypass steps of the 2′–5′ linkage site. For the 10A scaffold, the capacity of nucleotide incorporation was greatly reduced for transcription from the 2′–5′ template linkage mutant comparing with that of the natural 3′–5′ linkage, with an ∼44-fold decrease in kpol, an approximately sixfold decrease in Kd,app, and an ∼260-fold decease in substrate specificity. This linkage change can cause further effects on the following elongation beyond the 2′–5′ linkage position as revealed by 11A and 12C scaffolds (Fig. 3D and Table S1). For the 11A scaffold, kpol was reduced by ∼70-fold, and kpol/Kd,app was greatly decreased by over 1,200-fold. A similar pattern was observed for the 12C scaffold, where kpol/Kd,app was reduced by ∼1,000-fold. Collectively, one single 2′–5′ linkage alteration in the template strand can cause three pausing sites during transcription, which can result in at least over 3 × 108-fold of total decrease in pol II enzymatic specificity (kpol/Kd,app).
To further investigate the impact of 2′–5′ linkage in template strand on pol II transcriptional fidelity, we systematically and quantitatively dissected the three fidelity checkpoints: nucleotide incorporation, extension, and proofreading step, which also provide us with a comprehensive understanding on the role of correct phosphodiester linkage in the DNA template in ensuring pol II transcriptional fidelity.
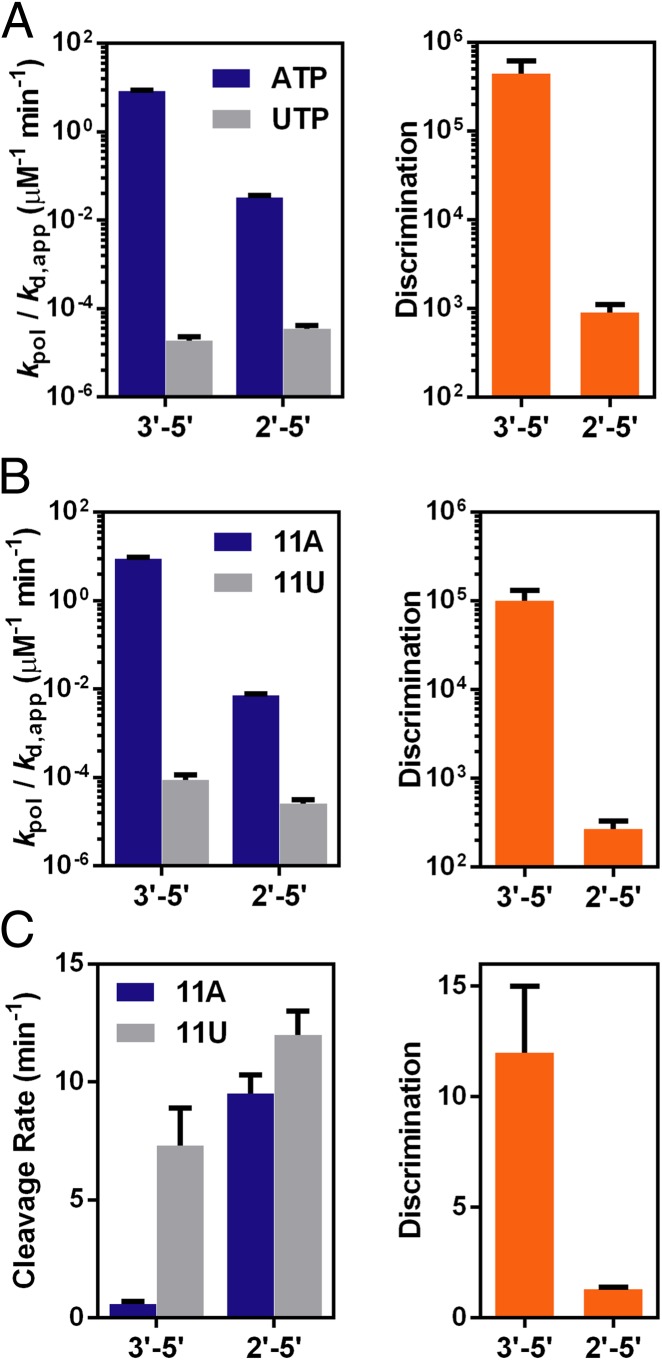
For the first fidelity checkpoint step, nucleotide selection and incorporation, pol II discrimination power for the 2′–5′ template linkage mutant is greatly reduced by ∼500-fold. The discrimination of ATP over UTP drops from ∼105 for a wild-type template to ∼900 for the 2′–5′ template linkage mutant (Fig. 4A and Table S2). Such decrease in pol II fidelity is mainly caused by inefficient correct ATP incorporation on the 2′–5′ linkage template. The specificity constant of ATP incorporation is reduced by ∼260-fold due to linkage alteration, whereas that of the mismatched UTP incorporation remains essentially unchanged (approximately twofold difference). kpol of UTP incorporation on the 2′–5′ linkage template (0.13 ± 0.01 min−1) is even ∼10-fold faster than that on the 3′–5′ linkage template (0.015 ± 0.003 min−1) (Table S2).
Fig. 4.
Transcriptional fidelity of RNA pol II is also reduced by the linkage alteration in the DNA template. (A) Fidelity decreases in the first checkpoint: nucleotide selection and incorporation. (Left) Specificity constants between correct incorporation (ATP) and incorrect incorporation (UTP) and (Right) the discrimination power. (B) Fidelity decreases in the second checkpoint: subsequent extension. (Left) Specificity constants between extension after the correct 3′-terminus (11A) and the incorrect 3′-terminus (11U) and (Right) the discrimination power. (C) Fidelity decreases in the third checkpoint: proofreading. (Left) Rate constants for cleavage of the correct 3′-terminus (11A) and the incorrect 3′-terminus (11U) and (Right) the discrimination power.
To investigate the impact of 2′–5′ linkage in template on the extension step (second checkpoint), we used 11A and 11U scaffolds to evaluate subsequent extension after matched and mismatched 3′-RNA termini, respectively. Consistent with previous studies, the extension after a mismatched 3′-terminus 11U on the 3′–5′ wild-type DNA template is ∼105 less efficient than a matched 11A, which ensured the high transcriptional fidelity (Fig. 4B and Table S2). In sharp contrast, the specificity (kpol/Kd,app) of subsequent extension following 11A was greatly reduced by ∼1,200 fold for the 2′–5′ linked template in comparison with that from a wild-type 3′–5′ linked template, whereas the specificity constant for 11U extension on the 2′–5′ linkage is only about threefold lower than the extension on the 3′–5′ linkage (Fig. 4B and Table S2). As a result, the discrimination power in the second checkpoint step sharply drops to (270 ± 60) for 2′–5′ linked template, which is ∼370-fold lower than that of the natural 3′–5′ linkage (Table S2). Similar to the first checkpoint step, the 2′–5′ linkage template-induced transcriptional fidelity decrease in this extension step is also mainly attributed by a significant decrease in extension efficiency from the matched terminus rather than a mismatched terminus.
We further measured TFIIS-stimulated cleavage rates to determine the impact of 2′–5′ linkage in template on pol II’s proofreading ability (the third checkpoint step) (Fig. 4C). The 2′–5′ linkage alteration in template greatly enhances the cleavage rate even for the fully matched 11A scaffold. The rate constant is about 16-fold faster than that of the wild-type scaffold (Fig. 4C and Table S2). This result suggests that a 2′–5′ linkage in the template can be effectively recognized by pol II and promotes backtracking. For the mismatched 11U scaffold containing 2′–5′ linkage template mutant, the cleavage rate was only slightly faster than that of 11A scaffold containing 2′–5′ linkage template mutant. As a result, the presence of 2′–5′ linkage at template abolishes the discrimination capacity of pol II proofreading [preferential cleavage of mismatched terminus (11U) over matched terminus (11A)] with an ∼10-fold decrease (from ∼12 to ∼1.3) (Fig. 4C). The presence of 2′–5′ linkage also affects the cleavage patterns by pol II (Fig. S3). We observed a minor but detectable amount of n-1 cleavage product for the mismatched 11U scaffold containing 2′–5′ linkage template mutant (Fig. S3B, Right), whereas no n-1 cleavage product was observed for the wild-type 11U scaffold with 3′–5′ linkage (Fig. S3A, Right), suggesting the existence of a pol II pretranslocation state during proofreading for the mismatched 11U scaffold containing 2′–5′ linkage template mutant. In contrast, for the matched 11A scaffold, strong n-1 and n-2 products were observed for both wild type and template mutant (Fig. S3 A and B, Left). Collectively, the linkage alteration from 3′–5′ to 2′–5′ linkage in template not only changes cleavage efficiency and fidelity but also causes different cleavage patterns.
In summary, the 2′–5′ linkage template mutant leads to a significant decrease in transcriptional fidelity for all three checkpoint steps. This is in sharp contrast with the impact of 2′–5′ linkage primer mutant on transcriptional fidelity.
Impact of the 2′–5′ Linkage Mutants on RNA pol II Trigger Loop Conformations.
In RNA pol II, an evolutionarily conserved mobile structural motif called trigger loop (TL) facilitates nucleotide selection and promotes efficient incorporation (27–30). The TL can adapt either closed (active) or open (inactive) conformations, in which closed TL greatly enhances enzymatic efficiency and fidelity of pol II. α-Amanitin, a natural toxin, can trap TL in an open inactive conformation and inhibit pol II transcription (29, 31, 32). Recently, we have used α-amanitin and synthetic nucleotide analogs to profile the TL conformations (33). As aforementioned, the 2′–5′ linkage alteration at either primer or template strand greatly affects the capacity of pol II for nucleotide selection and incorporation. We are interested in obtaining the mechanistic insights into the impact of 2′–5′ linkage on pol II transcription by investigating whether the conformation of TL is affected by the 2′–5′ linkage using α-amanitin transcription inhibition assay.
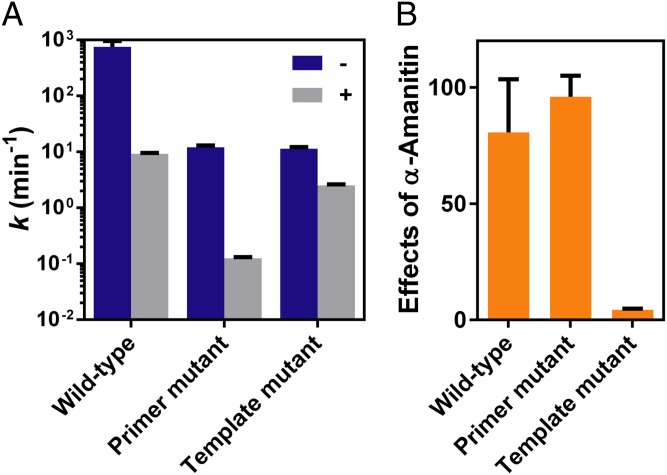
We first investigated effects of the 2′–5′ linkage primer mutant on TL conformation probed by α-amanitin (see the scaffold in Fig. 2A). For the 3′–5′ wild-type system, we expected the TL to be closed during nucleotide incorporation and therefore the catalytic rate would sharply decrease with α-amanitin treatment. Indeed, the observed rate constant drops to 9.3 ± 0.4 min−1, with an ∼80-fold decrease, which is consistent with the reported results (29, 33). Strikingly, the 2′–5′ linkage alteration in the primer does not affect the TL conformation because there is an over 90-fold decrease in the rate constant, from 12 ± 1 min−1 to 0.13 ± 0.01 min−1, with α-amanitin treatment (Fig. 5A). The strong effect of α-amanitin in the system containing a 2′–5′ linkage primer mutant is comparable with that in the 3′–5′–linked primer (Fig. 5B), indicating the TL is likely in a closed and active conformation even in the presence of a 2′–5′ linkage in RNA primer. This active TL ensures accurate nucleotide selection, which is consistent with the high transcriptional fidelity we observed in the pol II transcription from the 2′–5′ linkage primer mutant system (Fig. 2D).
Fig. 5.
Distinct effects of α-amanitin on RNA pol II in the 2′–5′ linkage primer and template. (A) Nucleotide incorporation rates in the absence (–) and presence (+) of α-amanitin. (B) Effects of α-amanitin on nucleotide incorporation. The effects of α-amanitin refer to folds of rate change before and after α-amanitin treatment.
In sharp contrast, for the 2′–5′ linkage template mutant (scaffold 10A in Fig. 3C), the incorporation rate was only slightly decreased by α-amanitin, varying from 11 ± 1 min−1 to 2.5 ± 0.1 min−1, with approximately fivefold reduction (Fig. 5), suggesting that the TL is open and has lost its ability to facilitate nucleotide incorporation opposite the 2′–5′ linkage template. This result suggests that a 2′–5′ linkage alteration in the template is sufficient to prevent the TL closure. This result is also consistent with low transcriptional fidelity in a 2′–5′ linkage DNA template mutant system (Fig. 4A). In addition, the TL adopts an open conformation during incorporation of mismatched nucleotide for all these three types of scaffolds (Fig. S4).
Discussion
The Impacts of Phosphodiester Linkage on pol II Transcription Are Strand-Specific.
Here we systematically investigated the impact of 2′–5′ phosphodiester linkage substitution on pol II transcriptional efficiency, fidelity, and trigger loop conformations. Strikingly, we revealed that the impacts of phosphodiester linkage on pol II transcription are strand-specific.
The 2′–5′ linkage substitution in the primer strand causes a single strong pausing site immediately after the 2′–5′ linkage, with an ∼150-fold reduction of pol II transcription efficiency (Table 1). In contrast, the 2′–5′ linkage substitution at the template strand leads to three consecutive strong pausing sites, with a total of ∼3 × 108-fold decrease in pol II enzymatic specificity (kpol/Kd,app) (Table 1 and Table S1). In both cases, we observed the bypass of long RNA transcripts with prolonged incubation, indicating that the 2′–5′ linkage is not an absolute blockage for pol II transcription. Similarly, early studies revealed that DNA polymerase I and HIV reverse transcriptase can also read through 2′–5′–linked templates (34, 35).
Further investigations of the impact of the phosphodiester linkage on pol II transcriptional fidelity also reveal a striking strand-specific pattern. We found that a 2′–5′ linkage substitution in the primer strand does not affect overall pol II transcriptional fidelity (Table 1). In sharp contrast, a 2′–5′ linkage substitution in the template strand causes a remarkable decrease in pol II transcriptional fidelity (total ∼106-fold decease) during all three fidelity checkpoint steps (Table 1 and Table S2).
To obtain mechanistic insights into how phosphodiester linkage alteration affects RNA pol II transcription, we probed the TL conformation profile upon linkage change using α-amanitin transcription inhibition assay. Again, here we reveal a striking strand-specific pattern of the impact of the phosphodiester linkage alteration on pol II TL conformation profile. We found that TL can still achieve a closed conformation (sensitive to α-amanitin treatment) for the 2′–5′ linkage primer strand mutant. In sharp contrast, TL remains in an open conformation (resistant to α-amanitin treatment) for the 2′–5′ linkage template strand mutant.
The strand-specific pattern of TL conformation profile for primer and template linkage substitution are well correlated with the strand-specific pattern of pol II transcriptional fidelity. The closure of TL is critical for pol II catalytic activity as well as transcriptional fidelity (27–30, 36–39). For the wild-type 3′–5′ linkage system, the high transcriptional fidelity is due to pol II’s capacity to incorporate matched NTP in a fast and TL-dependent manner (in which TL can achieve a closed conformation during nucleotide addition process) and incorporate mismatched NTP in a slow and TL-independent manner (in which TL remains at an open conformation during nucletotide addition). Similarly, for the system with a 2′–5′ linkage substitution in the primer strand, pol II maintains its capacity to incorporate matched NTP in a relatively fast and TL-dependent manner and incorporate mismatched NTP in a slow and TL-independent manner, which results in high transcriptional fidelity. In contrast, for the system with a 2′–5′ linkage substitution in the template strand, the pol II TL is unable to reach a closed conformation, and pol II incorporates both matched and mismatched NTP in a TL-independent manner, which results in low transcriptional fidelity.
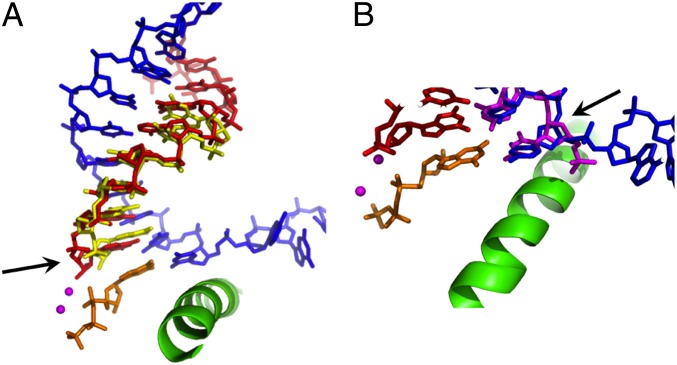
To understand how the 2′–5′ phosphodiester linkage substitution would accommodate at pol II active site, we superimposed the structure of pol II elongation complex (PDB: 2E2H and 2E2J) (28) with recent duplex structures containing a 2′–5′ linkage (PDB: 4MSB or NMR structure) (12, 40, 41). We found that 2′–5′ linkage in the primer strand only locally distorts the RNA terminus at −1 position (Fig. 6A). The incoming nucleotide can still pair with the template base in the canonical position, which can trigger the TL closure. The distortion induced by 2′–5′ linkage increases the distance between 3′-OH and alpha phosphate of incoming NTP, resulting in a decrease of catalytic rate in comparison with the wild-type 3′–5′ linkage system. In contrast, the substitution of a 2′–5′ linkage in template strand would rotate and shift the template bases (Fig. 6B) and in turn push the incoming NTP away from its canonical binding site. As a result, the NTP is misaligned with RNA primer and prevents TL from closing. Hence, the transcription rate is significantly reduced. Future structural studies will reveal the molecular details of how the 2′–5′ phosphodiester linkage substitution would accommodate at pol II active site.
Fig. 6.
Superimposition of RNA pol II elongation complex (PDB ID: 2E2J) with 2′–5′ phosphodiester linkage in RNA primer (PDB ID: 4MSB) (A) and DNA template (B). RNA primer is shown in red, and DNA template is shown in blue. The superimposed RNA and DNA with 2′–5′ phosphodiester linkages are shown in yellow (A) and magenta (B), respectively. The misaligned template or primer is highlighted by arrows.
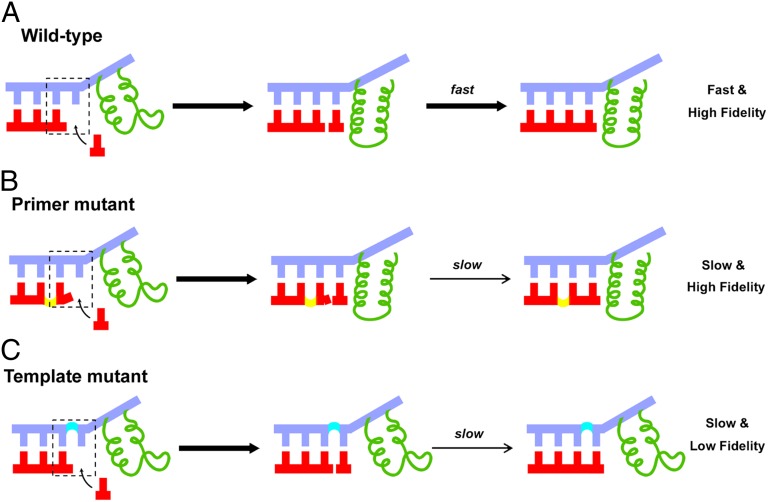
Collectively, the distinct impacts of 2′–5′ phosphodiester linkage substitution on pol II transcriptional efficiency, fidelity, and TL conformation are depicted in Fig. 7. For the wild-type 3′–5′ linkage, pol II can read through DNA template and synthesize RNA via a fast and high transcriptional fidelity manner. The pol II transcription is TL-dependent for correct NTP addition (Fig. 7A). The 2′–5′ linkage substitution at the RNA primer modestly reduces transcriptional efficiency but maintains high transcriptional fidelity. The pol II transcription in this mutant system is TL-dependent (Fig. 7B). In contrast, the 2′–5′ linkage substitution in the DNA template reduced both transcriptional efficiency and fidelity as TL remained open during pol II transcription (Fig. 7C).
Fig. 7.
Phosphodiester linkages in RNA primer and DNA template play two distinct roles during pol II transcription. (A) RNA pol II transcription on the wild-type RNA/DNA scaffold. Nucleotide incorporation has a good alignment with the upstream duplex, and the trigger loop of pol II (colored in green) is in the closed state, ensuring high efficiency and fidelity. (B) RNA pol II transcription after a 2′–5′–linked primer end. The trigger loop of pol II can be closed but the misaligned 3′-terminus lowers the primer extension efficiency. Hence, fidelity is maintained but the overall enzymatic efficiency is reduced. (C) RNA pol II transcription through a 2′–5′–linked position in the DNA template. During incorporation opposite the 2′–5′ linkage, the trigger loop of pol II cannot be closed because of the linkage alteration induced base and sugar shift. Therefore, both transcriptional efficiency and fidelity decreased.
Insights into Molecular Evolution of Phosphodiester Linkage Recognition.
The 2′–5′ linkage is an alternative form of nucleic acid backbone in addition to the 3′–5′ linkage during molecular evolution (42, 43). Previous studies have demonstrated that a mixture of 2′–5′ and 3′–5′ phosphodiester linkage can be obtained during nonenzymatic RNA replication, suggesting a high probability of mixture of phosphodiester linkages in the early evolution of life (3–9). It is conceivable that RNA in early evolution of life may have contained a mixture of 2′–5′ and 3′–5′ phosphodiester linkages. Such backbone heterogeneity may be tolerated by enzymes in the early evolutionary stage of life.
On the other hand, pol II represents the other end of the evolutionary spectrum. It is an advanced enzyme that has specifically evolved based on modern 3′–5′–linked nucleic acid scaffold. Our results provide striking insights into how pol II, as a contemporary enzyme, has evolved to recognize these key structural features of natural nucleic acids. Surprisingly, we found that pol II has a striking strand-specific recognition pattern of phosphodiester linkage. Pol II is very tolerant of a 2′–5′ linkage substitution in primer strand. In sharp contrast, a 2′–5′ linkage substitution in template strand leads to a catastrophic effect on both transcriptional efficiency and fidelity. We are tempted to speculate that the reason for this asymmetric pattern of linkage discrimination may reflect a universal principle of template-dependent genetic information transfer: the template strand and primer strand share distinct roles in supporting template-dependent genetic information transfer. The template strand plays a vital role in genetic information transfer. It provides both genetic coding instruction and structural scaffold support in selecting and positioning the correct incoming NTP through hydrogen bonding, size and shape recognition as well as base stacking. In contrast, the primer strand plays a much minor role. It stabilizes the incoming NTP mainly by base stacking and aligns its terminal hydroxyl group poised for attacking the alpha-phosphate of NTP. Thus, the penalty of 2′–5′phosphodiester linkage in the template strand is significantly higher than that in the primer strand, requiring high evolutionary pressure for evolving enzyme to recognize the desired phosphodiester linkage in the template strand. Therefore, although we fully acknowledge the huge benefits of evolving DNA dedicated for genetic information storage, we speculate that avoiding the high penalty of backbone heterogeneity during genetic information transfer could also be one of the many reasons to evolve DNA from RNA to eliminate 2′–5′phosphodiester linkage in the template strand. Recent discovery of high level of ribonucleotide incorporation in the DNA template (44, 45) suggests that the evolutionary route from a RNA world to a perfect DNA-RNA-protein world is not yet complete.
Although RNA pol II has evolved to possess a strong built-in ability to recognize the 3′–5′ linkage, the bypass of the 2′–5′ linkage still implies tolerance to linkage alteration to some extent. This seems like a common feature for other nucleic acid enzymes (34, 35). These results motivate us, as a future direction, to explore a systematic comparative study of polymerases at different evolutionary stage, which may exhibit a different spectrum of tolerance to 2′–5′ linkage. Indeed, a recent study (46) revealed that oligoadenylate synthetases from the phylogenetically oldest metazoan phylum, Porifera, are able to catalyze formation of oligomers having both 2′–5′ and 3′–5′ linkages, suggesting the existence of a link between primitive (linkage-tolerant) and evolutionarily advanced nucleic acid enzymes (linkage-specific). Future comparative studies of nucleic acid enzymes from different evolutionary stage would give us a clue on how phosphodiester backbone was evolved from a mixture of 2′–5′ linkage and 3′–5′ linkage in early stages of evolution to a dominant 3′–5′ linkage in our modern world, in a similar way as current comparative population genetics that allow us to trace back to the possible migration routes of our ancestors out of Africa.
Key Structural Features Governing pol II Transcriptional Efficiency and Fidelity.
Finally, our findings also revealed the key contribution of the 3′–5′ phosphodiester linkage backbone on pol II transcriptional efficiency and fidelity. Previous studies on molecular recognition of RNA pol II have focused on the contributions of functional groups in nucleic acids, such as nucleotide bases, 2′- and 3′-OH of ribose (28–30, 47–50). Recently, we used synthetic nucleic acid analogs to reveal the importance of chemical interactions and the intrinsic structural features of nucleic acids in controlling pol II transcriptional fidelity. To this end, we revealed the distinct contributions of hydrogen bonds between nucleobases in controlling individual fidelity checkpoint steps using hydrogen bond-deficient nucleotide analogues (51). Recently, we revealed that the sugar backbone is a dominant factor in controlling all three fidelity checkpoint steps of pol II transcriptional fidelity using sugar-ring disrupted mutant (52). Our work herein revealed asymmetric roles of the phosphodiester linkage in RNA primer and DNA template in maintaining pol II’s high transcriptional fidelity and efficiency. A general theme from these studies is to use synthetic nucleic acid chemistry to chemically modify the specific functional groups or motifs of nucleic acids. This chemical mutation approach allows us to systematically dissect the individual roles of chemical interactions and intrinsic structural motifs of nucleic acids in controlling pol II transcriptional fidelity, which cannot be achieved by conventional biochemical and genetic approaches (50–53).
Materials and Methods
Materials.
RNA pol II was purified from Saccharomyces cerevisiae as previously described (28, 54). The DNA template and nontemplate oligonucleotides were purchased from Integrated DNA Technologies. RNA primers were purchased from TriLink Biotechnologies and radiolabeled using [γ-32P] ATP and T4 Polynucleotide Kinase (NEB). The 2′–5′–linked oligos were gel purified and purchased from Gene Link Inc. The pol II elongation complexes for transcription assays were assembled using established methods (51, 55).
In Vitro Transcription Assays.
The pol II elongation complexes for transcription assays were assembled using established methods (51, 55). Briefly, an aliquot of 5′-32P–labeled RNA was annealed with a 1.5-fold amount of template DNA and twofold amount of nontemplate DNA to form RNA/DNA scaffold in elongation buffer [20 mM Tris⋅HCl (pH = 7.5), 40 mM KCl, 5 mM MgCl2]. An aliquot of annealed scaffold of RNA/DNA was then incubated with a fourfold excess amount of pol II at room temperature for 10 min to ensure the formation of a pol II elongation complex. The pol II elongation complex is ready for in vitro transcription upon mixing with equal volumes of NTP solution of various concentrations. The quenched products were analyzed by denaturing PAGE and visualized using a storage phosphor screen and Pharos FX imager (Bio-Rad).
Single Turnover Nucleotide Incorporation Assays.
The assay was carried out as previously described (51, 55). Briefly, nucleotide incorporation assays were conducted by preincubating 50 nM scaffold with 200 nM pol II for 10 min in elongation buffer at 22 °C. The preincubated enzyme:scaffold complex was then mixed with an equal volume of solution containing 40 mM KCl, 20 mM Tris⋅HCl (pH = 7.5), 10 mM DTT, 10 mM MgCl2, and twofold concentrations of various nucleotides. Final reaction concentrations after mixing were 25 nM scaffold, 100 nM pol II, 5 mM MgCl2, and various nucleotide concentrations in elongation buffer. Reactions were quenched at various times by addition of one volume of 0.5 M EDTA (pH = 8.0).
TFIIS Cleavage Assays.
Recombinant TFIIS was purified as described (51, 55). Cleavage reactions were performed by preincubating pol II with various scaffolds as previously described with slight modification. The solution was then mixed with an equal volume of solution containing TFIIS and MgCl2 in elongation buffer. Final reaction conditions were 100 nM pol II, 25 nM scaffold, 1.5 μM TFIIS, and 5 mM MgCl2. Reactions were quenched at various time points by addition of one volume of 0.5 M EDTA (pH = 8.0). Products were separated by denaturing PAGE as described above.
Inhibition Assays of α-Amanitin.
In these assays, we used high concentration of α-amanitin (500 mg/L) to make sure that the trigger loop of pol II was fully inhibited (29, 33). Briefly, the transcriptional scaffolds were preincubated with a fourfold amount of pol II at room temperature for 5 min before addition of α-amanitin. The mixture was then allowed to be further incubated for another 5 min before activation by NTPs. Final reaction conditions were 100 nM pol II, 25 nM scaffold, 500 mg/L α-amanitin, and 1 mM NTP. Reactions were quenched at various time points by addition of one volume of 0.5 M EDTA (pH = 8.0). Products were separated by denaturing PAGE as described above.
Data Analysis.
Nonlinear-regression data fitting was performed using Prism 6. The time dependence of product formation was fit to a one-phase association, Eq. 1 to determine the observed rate (kobs). The substrate concentration dependence was fit to a hyperbolic Eq. 2 to obtain values for the maximum rate of NTP incorporation (kpol) and apparent Kd (Kd,app) governing NTP binding essentially as described (Fig. S5).
| [1] |
| [2] |
The specificity constant was determined by kpol/Kd,app. Discrimination was calculated as the ratio of specificity constants governing two different nucleotide incorporation events as described (51, 55).
Supplementary Material
Acknowledgments
D.W. acknowledges the National Institutes of Health (Grant GM102362), Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research, and start-up funds from Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, for support. X.H. acknowledges the Hong Kong Research Grants Council (Grant AoE/M-09/12).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406234111/-/DCSupplemental.
References
- 1.Gilbert W. Origin of life: The RNA world. Nature. 1986;319(6055):618. [Google Scholar]
- 2.Joyce GF. RNA evolution and the origins of life. Nature. 1989;338(6212):217–224. doi: 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- 3.Bowler FR, et al. Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation. Nat Chem. 2013;5(5):383–389. doi: 10.1038/nchem.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekland EH, Bartel DP. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature. 1996;382(6589):373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- 5.Ertem G, Ferris JP. Synthesis of RNA oligomers on heterogeneous templates. Nature. 1996;379(6562):238–240. doi: 10.1038/379238a0. [DOI] [PubMed] [Google Scholar]
- 6.Inoue T, Orgel LE. Oligomerization of (guanosine 5′-phosphor)-2-methylimidazolide on poly(C). An RNA polymerase model. J Mol Biol. 1982;162(1):201–217. doi: 10.1016/0022-2836(82)90169-3. [DOI] [PubMed] [Google Scholar]
- 7.Usher DA, McHale AH. Nonenzymic joining of oligoadenylates on a polyuridylic acid template. Science. 1976;192(4234):53–54. doi: 10.1126/science.1257755. [DOI] [PubMed] [Google Scholar]
- 8.Usher DA, McHale AH. Hydrolytic stability of helical RNA: A selective advantage for the natural 3′,5′-bond. Proc Natl Acad Sci USA. 1976;73(4):1149–1153. doi: 10.1073/pnas.73.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris JP, Ertem G. Oligomerization of ribonucleotides on montmorillonite: reaction of the 5′-phosphorimidazolide of adenosine. Science. 1992;257(5075):1387–1389. doi: 10.1126/science.1529338. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty JP, Rizzo CJ, Breslow R. Oligodeoxynucleotides that contain 2',5′' linkages: Synthesis and hybridization properties. J Am Chem Soc. 1992;114(15):6254–6255. [Google Scholar]
- 11.Jin R, et al. Comparative spectroscopic, calorimetric, and computational studies of nucleic acid complexes with 2′,5″-versus 3′,5″-phosphodiester linkages. Proc Natl Acad Sci USA. 1993;90(22):10568–10572. doi: 10.1073/pnas.90.22.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson H, Jung K-E, Switzer C, Wang AH-J. DNA with 2'-5′ phosphodiester bonds forms a duplex structure in the A-type conformation. J Am Chem Soc. 1995;117(2):837–838. [Google Scholar]
- 13.Giannaris PA, Damha MJ. Oligoribonucleotides containing 2′,5′-phosphodiester linkages exhibit binding selectivity for 3′,5′-RNA over 3′,5′-ssDNA. Nucleic Acids Res. 1993;21(20):4742–4749. doi: 10.1093/nar/21.20.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prakash TP, Jung K-E, Switzer C. RNA recognition by the 2'-structural isomer of DNA. Chem Commun. 1996;(15):1793–1794. [Google Scholar]
- 15.Sheppard TL, Breslow RC. Selective binding of RNA, but not DNA, by complementary 2',5′-linked DNA. J Am Chem Soc. 1996;118(40):9810–9811. [Google Scholar]
- 16.Burlina F, Fourrey J, Lefort V, Favre A. Cleavage activity of a hammerhead ribozyme domain containing 2′,5′-phosphodiester linkages. Tetrahedron Lett. 1999;40(24):4559–4562. [Google Scholar]
- 17.Engelhart AE, Powner MW, Szostak JW. Functional RNAs exhibit tolerance for non-heritable 2′-5′ versus 3′-5′ backbone heterogeneity. Nat Chem. 2013;5(5):390–394. doi: 10.1038/nchem.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flynn-Charlebois A, Prior TK, Hoadley KA, Silverman SK. In vitro evolution of an RNA-cleaving DNA enzyme into an RNA ligase switches the selectivity from 3′-5′ to 2′-5′. J Am Chem Soc. 2003;125(18):5346–5350. doi: 10.1021/ja0340331. [DOI] [PubMed] [Google Scholar]
- 19.Torelli AT, Krucinska J, Wedekind JE. A comparison of vanadate to a 2′-5′ linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13(7):1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3(7):a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller W. The RNA lariat: A new ring to the splicing of mRNA precursors. Cell. 1984;39(3 Pt 2):423–425. doi: 10.1016/0092-8674(84)90449-5. [DOI] [PubMed] [Google Scholar]
- 22.Carlomagno T, et al. Structural principles of RNA catalysis in a 2′-5′ lariat-forming ribozyme. J Am Chem Soc. 2013;135(11):4403–4411. doi: 10.1021/ja311868t. [DOI] [PubMed] [Google Scholar]
- 23.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell. 2003;12(5):1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 24.Hovanessian AG, Justesen J. The human 2′-5’oligoadenylate synthetase family: Unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie. 2007;89(6-7):779–788. doi: 10.1016/j.biochi.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Cramer P. RNA polymerase II structure: From core to functional complexes. Curr Opin Genet Dev. 2004;14(2):218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Cramer P. Structure and function of RNA polymerase II. Adv Protein Chem. 2004;67:1–42. doi: 10.1016/S0065-3233(04)67001-X. [DOI] [PubMed] [Google Scholar]
- 27.Larson MH, et al. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc Natl Acad Sci USA. 2012;109(17):6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127(5):941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan CD, Larsson K-M, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30(5):547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang X, et al. RNA polymerase II trigger loop residues stabilize and position the incoming nucleotide triphosphate in transcription. Proc Natl Acad Sci USA. 2010;107(36):15745–15750. doi: 10.1073/pnas.1009898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15(8):811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 32.Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: Alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci USA. 2002;99(3):1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, et al. Dissecting the chemical interactions and substrate structural signatures governing RNA polymerase II trigger loop closure by synthetic nucleic acid analogues. Nucleic Acids Res. 2014;42(9):5863–5870. doi: 10.1093/nar/gku238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorsch JR, Bartel DP, Szostak JW. Reverse transcriptase reads through a 2′-5′linkage and a 2′-thiophosphate in a template. Nucleic Acids Res. 1995;23(15):2811–2814. doi: 10.1093/nar/23.15.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha S, Kim PH, Switzer C. 2′,5′-linked DNA is a template for polymerase-directed DNA synthesis. J Am Chem Soc. 2004;126(1):40–41. doi: 10.1021/ja034986z. [DOI] [PubMed] [Google Scholar]
- 36.Kireeva ML, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30(5):557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27(3):406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Feig M, Burton ZF. RNA polymerase II with open and closed trigger loops: active site dynamics and nucleic acid translocation. Biophys J. 2010;99(8):2577–2586. doi: 10.1016/j.bpj.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17(1):99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Szostak JW. The free energy landscape of pseudorotation in 3′-5′ and 2′-5′ linked nucleic acids. J Am Chem Soc. 2014;136(7):2858–2865. doi: 10.1021/ja412079b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheng J, et al. Structural insights into the effects of 2′-5′ linkages on the RNA duplex. Proc Natl Acad Sci USA. 2014;111(8):3050–3055. doi: 10.1073/pnas.1317799111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benner SA. Synthetic biology: Act natural. Nature. 2003;421(6919):118. doi: 10.1038/421118a. [DOI] [PubMed] [Google Scholar]
- 43.Joyce GF. Forty years of in vitro evolution. Angew Chem Int Ed Engl. 2007;46(34):6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- 44.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6(10):774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50(3):437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Päri M, et al. Enzymatically active 2′,5′-oligoadenylate synthetases are widely distributed among Metazoa, including protostome lineage. Biochimie. 2014;97:200–209. doi: 10.1016/j.biochi.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 47.Yuzenkova Y, et al. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93(4):627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 49.Svetlov V, Nudler E. Basic mechanism of transcription by RNA polymerase II. Biochim Biophys Acta. 2013;1829(1):20–28. doi: 10.1016/j.bbagrm.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S, Wang D. Understanding the molecular basis of RNA polymerase II transcription. Isr J Chem. 2013;53(6-7):442–449. doi: 10.1002/ijch.201300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kellinger MW, Ulrich S, Chong J, Kool ET, Wang D. Dissecting chemical interactions governing RNA polymerase II transcriptional fidelity. J Am Chem Soc. 2012;134(19):8231–8240. doi: 10.1021/ja302077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu L, Plouffe SW, Chong J, Wengel J, Wang D. A chemical perspective on transcriptional fidelity: Dominant contributions of sugar integrity revealed by unlocked nucleic acids. Angew Chem Int Ed Engl. 2013;52(47):12341–12345. doi: 10.1002/anie.201307661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benner SA, Sismour AM. Synthetic biology. Nat Rev Genet. 2005;6(7):533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324(5931):1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19(8):831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.