Significance
Phytochrome A (phyA) is the photoreceptor in Arabidopsis that mediates the far-red light high radiance response. Its partner FHY1 (FAR-RED ELONGATED HYPOCOTYL 1) is involved in each step of phyA signaling, including phyA nuclear translocation, interaction with transcription factors, and association with gene promoters. Although there is evidence that an FHY1-independent nuclear phyA signaling branch is present, whether phyA and FHY1 in fact act separately is currently under debate. This study identifies phyA and FHY1 unique direct target genes and in particular analyzes the phyA-independent FHY1 nuclear actions involved in regulating gene transcription. Significant light has been shed on the molecular mechanisms through which phyA or FHY1 performs their own functions in response to the far-red light signal.
Abstract
To incorporate the far-red light (FR) signal into a strategy for optimizing plant growth, FAR-RED ELONGATED HYPOCOTYL1 (FHY1) mediates the nuclear translocation of the FR photoreceptor phytochrome A (phyA) and facilitates the association of phyA with the promoters of numerous associated genes crucial for the response to environmental stimuli. However, whether FHY1 plays additional roles after FR irradiation remains elusive. Here, through the global identification of FHY1 chromatin association sites through ChIP-seq analysis and by the comparison of FHY1-associated sites with phyA-associated sites, we demonstrated that nuclear FHY1 can either act independently of phyA or act in association with phyA to activate the expression of distinct target genes. We also determined that phyA can act independently of FHY1 in regulating phyA-specific target genes. Furthermore, we determined that the independent FHY1 nuclear pathway is involved in crucial aspects of plant development, as in the case of inhibited seed germination under FR during salt stress. Notably, the differential presence of cis-elements and transcription factors in common and unique FHY1- and/or phyA-associated genes are indicative of the complexity of the independent and coordinated FHY1 and phyA pathways. Our study uncovers previously unidentified aspects of FHY1 function beyond its currently recognized role in phyA-dependent photomorphogenesis.
Light is one of the most important environmental cues in plant growth and development. Arabidopsis has thus evolved several photoreceptors to perceive different wavelengths in the visible light spectrum (1). Among them, phytochrome A (phyA) is the primary photoreceptor to mediate the far-red light (FR) and early red light (R) responses (2, 3). PhyA therefore plays a predominant role in plant adaptation to a shade environment where the R/FR ratio decreases. Upon FR and R irradiation phyA will shuttle between the inactive R-absorbing Pr form and the active FR-absorbing Pfr form. These two forms display distinct biological activities, nuclear translocation rates, degradation rates, and affinities for various signaling intermediates (2, 4, 5).
Both FR and R trigger the localization of cytosolic phyA to the nucleus (6), albeit through different mechanisms. Two essential partners for phyA nuclear translocation, FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and its less abundant homolog FHY1-LIKE (FHL), influence this process at two levels; first, after the initial burst of phyA nuclear importation under R, FHY1 is gradually phosphorylated by action of the Pfr form of phyA (7). This event inhibits the nuclear translocation of FHY1 and consequently prevents further nuclear accumulation of phyA. Under FR, however, persistent phyA nuclear translocation is observed with nonphosphorylated FHY1 (8); second, the nucleo-cytoplasmic shuttling of FHY1/FHL is also reduced under R owing to the low dissociation rate of the Pfr–FHY1/FHL complex. The FR-driven phyA conversion to its Pr form benefits the recycling of FHY1/FHL for continuous phyA nuclear transportation (4).
The mechanism through which nuclear phyA subsequently regulates a considerable number of FR-modulated genes has been intensively investigated (9, 10). It was recently demonstrated that the nuclear phyA–FHY1 complex is recruited to the promoter regions of FR-responsive genes CHS (chalcone synthase) and NAC019 [no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF), cup-shaped cotyledon (CUC) 019] through transcription factors to coregulate gene transcription (8, 9). Furthermore, genomic analysis suggests that the “phyA-promoter association” model is a universal mechanism of regulation for thousands of phyA-associated genes (9). A wide range of transcription factors may act downstream of phyA for the purpose of recognizing diverse cis-elements that are responsive to multiple internal or external stimuli (9). Therefore, light and other signals are integrated within the cell through phyA.
The possibility of an FHY1-independent phyA nuclear signaling pathway has been suggested due to the fact that the phenotype of the fhy1 mutant can be rescued by the expression of nuclear phyA (phyA-NLS-GFP) (11). Thus, it is possible that FHY1 is not necessary for phyA-promoter association. On the other hand, whether nuclear FHY1 acts independently of phyA to regulate gene expression remains unknown.
In this study we performed FHY1 ChIP-sequencing (ChIP-seq) analysis to identify FHY1-associated genes. Strikingly, comparison of the FHY1- and known phyA-associated genes demonstrated that they uniquely associate with most of their respective genes, suggesting that phyA and FHY1 perform their own functions in the nucleus upon FR exposure in addition to functioning coordinately. We also demonstrated that FHY1 independently modulates salt-resistant seed germination under FR by the association and regulation of its unique target gene AFP4 (ABI five binding protein 4), indicating that the independent FHY1 pathway is involved in specific developmental processes in the plant. With the analysis of downstream cis-elements being preferentially associated by phyA and FHY1 and several transcription factors involved in light signaling, we propose that in addition to the coordinate performance of phyA and FHY1, they also act independently of one another with distinct molecular partners in multiple aspects of plant development.
Results
Identification of FHY1 Chromatin Association Sites by ChIP-seq.
To determine whether phyA and FHY1 associate with the same genes upon FR irradiation, 4-d-old etiolated 35S: GFP-FHY1 fhy1-1 transgenic seedlings (12) were exposed to 3 h FR, the same conditions used for detection of the phyA-associated genes (9). The treated plants were used for ChIP-seq analysis with an anti-FHY1 antibody that exhibits high specificity in the detection of FR-induced FHY1-association events (8). FHY1-associated DNA samples from three distinct biological replicates and an input DNA sample (as a negative control) were subjected to high-throughput Solexa (Illumina) sequencing. A total of 33, 47, 39, and 89 million reads were uniquely mapped to the Arabidopsis genome from three FHY1 and input libraries, respectively, by using the read aligner Bowtie (http://bowtie.cbcb.umd.edu). The three FHY1 ChIP-seq replicates displayed excellent repeatability, with Pearson correlation coefficients (13) higher than 0.95 (Fig. S1A). Through an optimized peak calling method devised for weaker signals caused by the indirect binding between protein and DNA (9), we identified 3,866 FHY1 chromatin association sites that were reproducible in all three biological replicates. The reliability of our ChIP-seq approach was confirmed by the isolation of FHY1 chromatin association sites detected on two known FHY1-associated genes, CHS and NAC019 (Fig. S1B). Their positions were consistent with that of G-boxes, DNA elements that are critical for phyA-dependent CHS and NAC019 expression (8, 9).
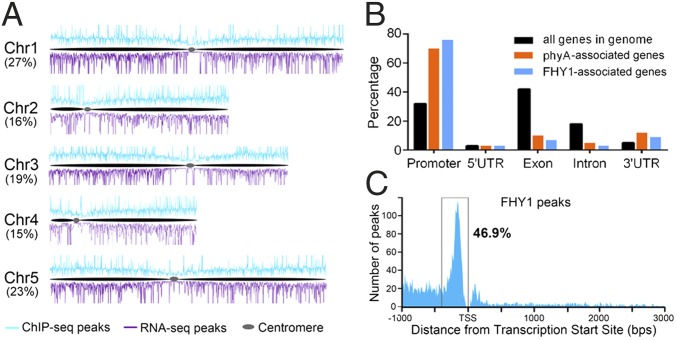
The percentage of FHY1 chromatin association sites on each chromosome was proportional to chromosome size, suggesting that the sites were evenly distributed across the chromosomes (Fig. 1A). However, FHY1 association with DNA did exhibit preferences. First, only approximately 10% of FHY1 association sites were intergenic, consistent with the observation that FHY1 association sites are rarely found in centromeric regions (Fig. 1A). Second, more than 80% of intragenic FHY1 association sites were found in the promoter or the 5′ UTR (Dataset S1), whose proportions only account for 35% of all genic regions in the Arabidopsis genome. Consequently, FHY1 associated with either the promoter or 5′ UTR on 79% of its associated genes (Fig. 1B), whereas the transcription factor FHY3 only binds the promoter or 5′ UTR on 55% of its target genes (14). Last, the precise locations of FHY1 chromatin association sites further revealed that 46.9% of FHY1 association specifically occurs within a 400-bp region upstream of the transcription start site (Fig. 1C). These results suggest that FHY1 preferentially associates with transcriptional regulatory regions. Notably, the distribution patterns of FHY1 chromatin association sites are similar to that of phyA (9), both highly enriched in gene promoter regions. Therefore, we suggest a role for FHY1 as a transcriptional regulator with characteristics similar to those of phyA.
Fig. 1.
Genome-wide distribution of FHY1 chromatin association sites. (A) Distribution of FHY1 chromatin association sites and FHY1-regulated genes across five chromosomes. ChIP-seq peaks and RNA-seq peaks are indicative of the FHY1 chromatin association sites and the FHY1-regulated genes, respectively. The numbers in brackets indicate the percentage of ChIP-seq peaks from each chromosome. (B) Comparison of the chromatin association site distribution patterns between FHY1 and phyA over genic regions. Distribution of individual gene regions in all genomic genes is shown as a control. The promoter region is defined as the 1,000-bp region that precedes the transcription start site (TSS). (C) FHY1 chromatin association sites are highly concentrated within a 400-bp window upstream of the TSS.
FHY1 and phyA Modulate Most of Their Direct Target Genes Independently.
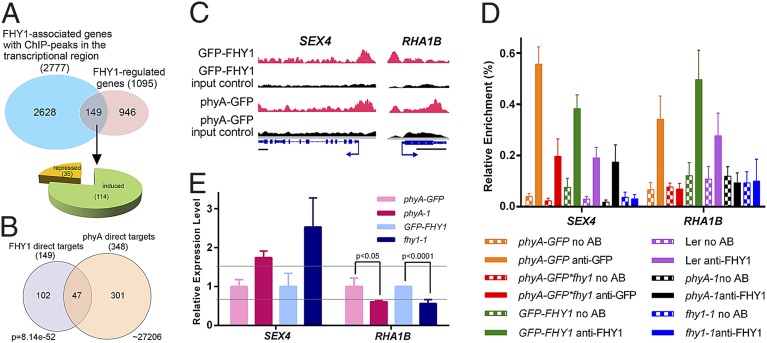
The 3,866 FHY1 chromatin association sites were assigned to 3,352 FHY1-associated genes, of which 2,777 were associated by FHY1 through their promoter or 5′ UTR (Dataset S2). To determine which of the FHY1-associated genes are transcriptionally regulated by FHY1 (thereafter defined as FHY1 direct target genes), we performed an RNA-seq analysis. The 35S: GFP-FHY1 fhy1-1 transgenic line and the fhy1-1 mutant (15) were grown under the same light conditions used for ChIP-seq (D4d+FR3h) for RNA preparation and sequencing. Three biologically replicates exhibited a high value (>0.997) of Pearson correlation coefficients (Fig. S1C). A total of 1,095 FHY1-regulated genes were subsequently identified (Datasets S2 and S3) and were found to be evenly distributed across five Arabidopsis chromosomes (Fig. 1A). Comparison of the FHY1 ChIP-seq and RNA-seq data further defined 149 FHY1 direct target genes (Fig. 2A and Dataset S2), accounting for 5.4% of total FHY1-associated genes. This percentage is lower than that of phyA (13.6%) (9), suggesting that FHY1 is more dependent on other transcriptional regulators for its coregulation activity in comparison with phyA. The promoters of 946 of 1,095 FHY1-regulated genes did not associate with FHY1, indicating that their expression may be indirectly influenced by FHY1-dependent phyA nuclear localization. Of the FHY1 direct target genes, 77% (114 genes) are FHY1-induced (Fig. 2A and Dataset S2).
Fig. 2.
FHY1 and phyA coregulate common target genes by associating with the same position on the promoter. (A) Identification of FHY1 direct target genes by compiling the FHY1 ChIP-seq data and RNAseq data. (B) Identification of common direct target genes of FHY1 and phyA. The P value of the Venn diagram was calculated using the hypergeometric distribution. (C) Integrative Genomics Viewer (IGV) shows that FHY1 and phyA ChIP-seq peaks are located at the same position on common direct target promoters. For each gene, phyA or FHY1 peaks in ChIP samples (red), in input control sample (black), and the gene structure are shown in the top, middle and bottom row. (Scale bar, 500 bp.) (D) Both FHY1 and phyA associate with the ChIP-seq peak-covered promoter region. All materials were grown under dark for 4 d and exposed to FR for 3 h for ChIP. (E) RT-PCR shows that both FHY1 and phyA are crucial to the expression of common target genes. Four-day-old dark grown seedlings were exposed to 3 h FR for RNA preparation. All transcripts were normalized to UBQ1. The relative expression levels in phyA-1 and fhy1-1 lines were expressed as the ratio to phyA-GFP and GFP-FHY1 lines, respectively. The gray lines indicate the ±1.5-fold change. All error bars in D and E represent ±SD (n = 3) of three biological replicates.
To further investigate the diverging mechanisms through which FHY1 and phyA regulate their target genes, we compared the direct target genes of FHY1 and of phyA (Fig. 2B). Only 47 phyA/FHY1 common target genes were found to be transcriptionally regulated by both phyA and FHY1. Most of these genes serve as transcriptional regulators or enzymes involved in light response, photosynthesis, hormone signaling, and other metabolic processes (Table S1), whereas 102 and 301 direct target genes were found to be unique to FHY1 and phyA, respectively (Dataset S4).
FHY1 and phyA Coordinately Regulate Their Common Direct Target Genes Through Distinct Molecular Mechanisms.
We next verified the coassociation of FHY1 and phyA on two common target genes, SEX4 (AT3G52180) and RHA1B (AT4G11360). SEX4 is a phosphatase involved in the diurnal cycle-controlled starch accumulation, and the flowering is slightly delayed in sex4 mutant (16). RHA1B is a ubiquitin ligase induced by plant defense elicitors flg22 and chitin (17, 18). The colocalization of phyA and FHY1 on SEX4 and RHA1B promoters was confirmed by ChIP–quantitative PCR (ChIP-qPCR) using primers that amplify the region detected by ChIP-seq (Fig. 2 C and D, orange and green bars). We next validated FHY1 association by demonstrating that FHY1 association with the promoters of SEX4 and RHA1B was not due to FHY1 overexpression. FHY1 ChIP-qPCR analysis revealed enrichment of FHY1 at these two promoters in wild-type seedlings but not in the fhy1-1 mutant after FR irradiation. It is worth noting, however, that the association is weaker in wild-type plants than in the overexpression line (Fig. 2D, purple and blue bars). Furthermore, FHY1 association with promoters seemed to be FR-inducible (Fig. 2D and Fig. S2A). This result is consistent with the observation that FHY1 accumulates in the nucleus upon FR irradiation (8).
Interestingly, we noticed that phyA and FHY1 associate with SEX4 promoters independently of each other, because both phyA and FHY1 can still associate with the promoter in absence of FHY1 and phyA, respectively. By contrast, phyA and FHY1 were indispensable for the other protein to associate with the RHA1B promoter (Fig. 2D, red and black bars). Regardless of the different coassociation patterns, both phyA and FHY1 repressed the expression of SEX4 but induced RHA1B transcription (Fig. 2E and Fig. S2B). Considering the gene function of SEX4 and RHA1B, it seemed that under FR, phyA and FHY1 coordinately mediate the enhancement of the plant’s innate immune system at the cost of delayed flowering, in the interest of survival.
FHY1 and phyA Associate with Their Unique Direct Target Genes for Independent Transcriptional Regulation.
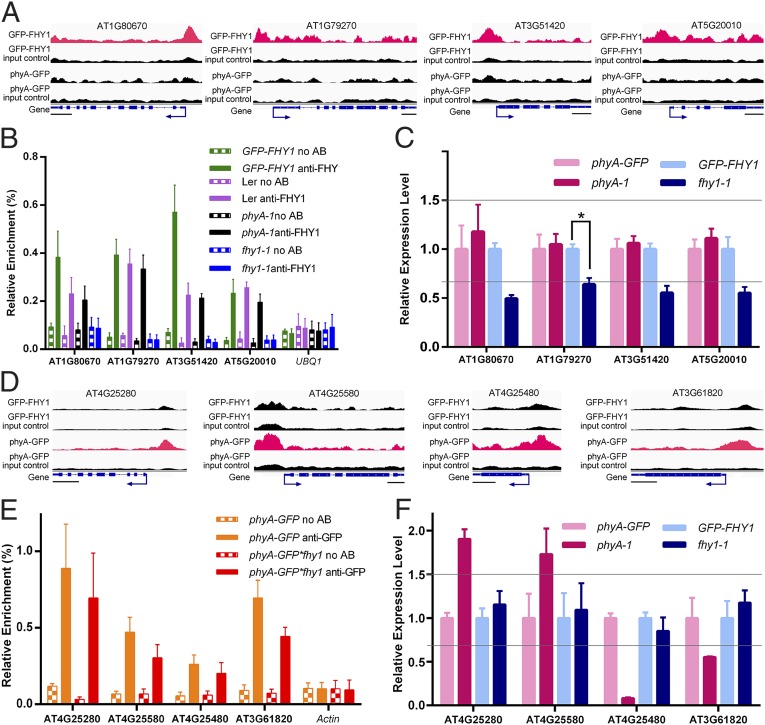
To substantiate the hypothesis that nuclear FHY1 possesses functions independent of the phyA signaling pathway, we examined the association of FHY1 with its unique direct target genes. As shown in Fig. 3A, the presence of phyA on the promoters of four randomly selected target genes unique to FHY1 was undetectable or much weaker than that of FHY1 signals compared with their respective input control background. This suggested that phyA rarely associates with target genes unique to FHY1. ChIP-qPCR assays further demonstrated a phyA-independent FHY1 association with these promoters, because FR-enriched FHY1 signals were detected in the phyA-1 mutant line (Fig. 3B, purple and black bars, and Fig. S2A). Moreover, FHY1, but not phyA, regulated the expression of these four genes (Fig. 3C and Fig. S2B), confirming the results of the RNA-seq assay (Dataset S2).
Fig. 3.
FHY1 and phyA can associate and regulate their target genes independently of one another. (A and D) IGV shows that ChIP-seq peaks specifically enrich in the GFP-FHY1 line (A) or the phyA-GFP line (D). (Scale bars, 500 bp.) (B and E) ChIP-PCRs verify that FHY1 (B) and phyA (E) associate with their unique direct target promoters independently. UBQ1 in B and Actin in E are used as negative controls. (C and F) RT-PCR analyses demonstrated that phyA (C) and FHY1 (F) do not transcriptionally regulate unique direct target genes of the other protein. *P < 0.05. Transcripts were normalized to UBQ1. The gray lines indicate ±1.5-fold change. All materials used in B, C, E, and F were treated as described in Fig. 2. All error bars in this figure represent ±SD (n = 3) of three biological replicates.
To determine whether an FHY1-independent nuclear phyA signaling pathway does in fact exist (11), we looked at four randomly selected phyA unique target genes with promoters that showed enrichment only for phyA according to the ChIP-seq data (Fig. 3D). A high enrichment of phyA at these promoters was detected, and this phyA association with the promoters was independent of FHY1 (Fig. 3E). FHY1 was not involved in the transcriptional regulation of these four phyA unique direct target genes in the transgenic lines, in accordance with the RT-qPCR (Fig. 3F).
The PhyA-Independent FHY1-Mediated Nuclear Pathway Inhibits Salt-Resistant Seed Germination Under FR.
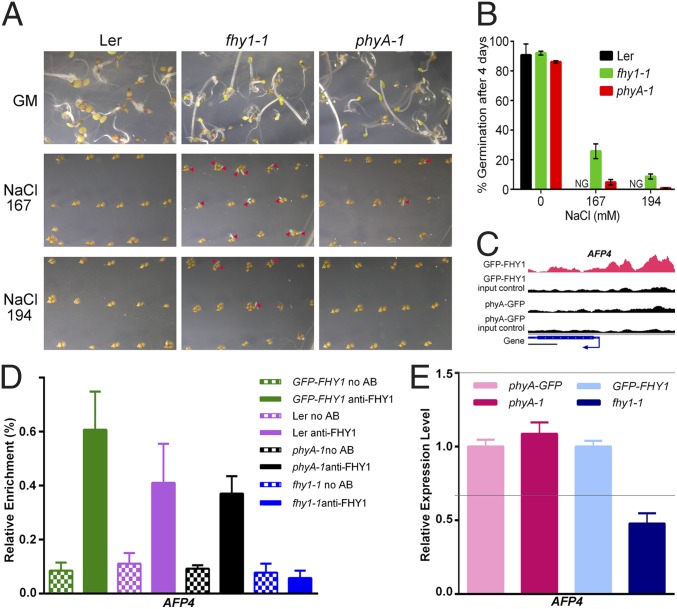
The knockout line of an FHY1 unique direct target gene, AFP4 (AT3G02140), was reported to be mildly resistant to salt stress in the seed germination stage of development (19). Intriguingly, the fhy1-1 mutant also exhibited an increased seed germination rate in comparison with wild-type (Ler) and the phyA-1 mutant under FR (Fig. 4 A and B) when the same salt treatment was applied (19). Therefore, FHY1, like AFP4, seems to play a negative role in salt-tolerant seed germination under FR, whereas phyA involvement in this developmental process is negligible.
Fig. 4.
FHY1 modulates seed germination under FR independently of phyA through association and regulation of a unique direct target gene, whose knockout line is reported to be resistant to salt. (A) The fhy1-1 mutant exhibits a higher germination rate compared with wild-type (Ler) and the phyA-1 mutant upon salt treatment under FR. Seeds were spread on Murashige and Skoog plates without or with NaCl (167 mM and 194 mM) and grown under FR for 4 d. Arrowheads mark germinated seeds. (B) Statistics of seed germination are shown in A. NG, not germinated. (C) FHY1 but not phyA associates with the promoter of AFP4. (Scale bar, 500 bp.) (D) ChIP-PCR shows that FHY1 associates with AFP4 promoter independent of phyA. (E) FHY1 but not phyA is crucial for AFP4 expression under FR. Transcripts were normalized to UBQ1. The gray lines indicate ±1.5-fold change. Materials used in D and E were treated as described in Fig. 2. All error bars in this figure represent ±SD (n = 3) of three biological replicates.
The enrichment of FHY1 on the AFP4 promoter region was confirmed by both the ChIP-seq (Fig. 4C) and the ChIP-qPCR (Fig. 4D, green and purple bars) results. Furthermore, the presence of FHY1 was essential for the typical expression of AFP4 under FR (Fig. 4E and Fig. S2B). PhyA was not enriched in the AFP4 promoter (Fig. 4C) and was not required either for the FHY1-AFP4 promoter association (Fig. 4D, black bars) or for the transcriptional regulation of AFP4 (Fig. 4E and Fig. S2B). The above results suggested that FHY1 itself is crucial for the FR-responsive plant development through the phyA-independent FHY1 nuclear pathway.
Shared and Unique Associated Genes of phyA and FHY1 Feature Three Different Modes of Regulation.
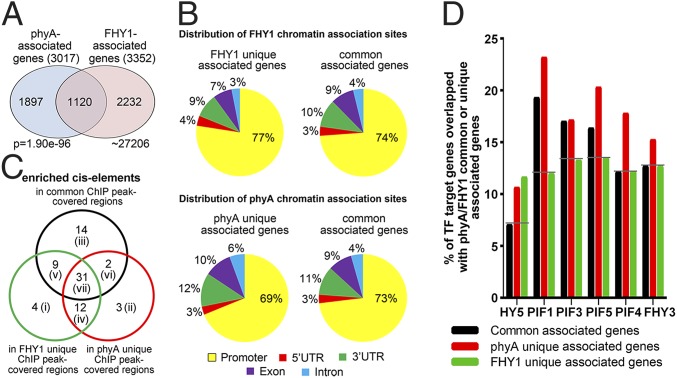
We next sought to identify the factors that influence the association of phyA and FHY1 with chromatin, either coordinately or uniquely. As shown in Fig. 5A and Dataset S4, approximately 37% of phyA- and 33% of FHY1-associated genes resided in the shared group (1,120). The remaining genes were identified as unique associated genes of either FHY1 or phyA (Fig. 5A). The distribution patterns of phyA and FHY1 chromatin association sites on their unique associated genes were similar to the patterns on shared associated genes (Fig. 5B), where they were found to be enriched in the promoter regions with concentrated cis-elements.
Fig. 5.
The preferences of FHY1 and phyA when associating with genes. (A) The common and unique associated genes of phyA and FHY1. The P value of the Venn diagram was calculated using the hypergeometric distribution. (B) Both FHY1 (Upper) and phyA (Lower) prefer to associate with the promoter regions of their unique genes as well as the promoters of common genes. (C) Different cis-elements are enriched in the coordinate and unique associations of phyA and FHY1. Enriched cis-elements located within common and unique ChIP peak-covered regions indicate coordinate and unique association sites of phyA and FHY1, respectively. (D) FHY1 and phyA prefer different transcription factors when associating with common or unique genes. Target genes of these transcription factors were overlapped with phyA/FHY1 common associated genes (black), phyA unique associated genes (red), and FHY1 unique associated genes (green) with different degrees.
To identify the cis-elements that might determine the coordinate or unique association of phyA and FHY1, we focused on the ChIP peak-covered regions rather than the 1,000-bp promoter regions for the screening of enriched cis-elements. By using the PLACE database (20), a total of 75 cis-elements exhibited a high frequency of association (>1.5 fold of that on random genomic DNA), suggesting that they play a role in mediating phyA or FHY1 associations with the DNA. These cis-elements were classified into seven different groups (Fig. 5C and Dataset S5). Among them, the groups i, ii, and iii specifically mediated the FHY1 unique association, the phyA unique association, and the coordinate association, respectively. The other four groups of cis-elements represented a more complicated mechanism for mediating the diverse association patterns of phyA and FHY1. Although the group v, vi, and vii cis-elements, as well as the group iii factors, were able to mediate the coordinate association, they also participated in the unique associations. This is probably because the recruitment of FHY1 or phyA on these factors was obstructed by other factor(s). Moreover, the group iv cis-elements mediated unique associations of both phyA and FHY1. The seven different groups of cis-elements allowed us to postulate that distinct underlying molecular events might occur on each group of cis-elements.
Because of the absence of a DNA binding domain (1, 21), both phyA and FHY1 rely on transcription factors for their cis-element associations and transcriptional coregulatory activities. We thus compared the common associated genes, phyA unique associated genes, and FHY1 unique associated genes with the target genes of several transcription factors known to be involved in light signaling (14, 22–26). Each of these transcription factors was required for both coordinate and unique associations, but with different degrees of participation (Fig. 5D). Unique promoter associations of both phyA and FHY1 preferred HY5, for example, rather than other transcription factors. In another case, phytochrome interacting factor (PIF)1, PIF3, and PIF5 were more involved in the coordinate association and the phyA unique association, whereas the transcription factors PIF4 and FHY3 mediated the phyA unique association most frequently. FHY1 associated with fewer targets of PIFs and FHY3 than phyA did, suggesting that the FHY1-DNA association relies more on other transcription factors compared with the phyA-DNA association. Collectively, the coordinate and unique associations prefer different cis-elements and transcription factors.
We next investigated whether the coordinate and unique associations of phyA and FHY1 occur in response to different cellular events. A WEGO analysis revealed that the common associated genes were more enriched in light and stress signaling (Fig. S3), consistent with the conventional recognition of phyA and FHY1 functions. PhyA and FHY1 unique associated genes, however, seemed to be playing a greater role in the transport, metabolism, and cell growth processes (Fig. S3 and Dataset S2).
Discussion
Substantial research has been performed to elucidate the combined actions of the FR photoreceptor phyA and its partner FHY1 in transcriptional regulation in response to FR exposure. FHY1 mediates not only phyA nuclear translocation through physical interaction (4, 6, 7) but also the assembly of the phyA/FHY1/transcription factors complex with DNA (8, 9, 27). On the basis of our data, along with that of previous studies (8, 9), it seems that both phyA and FHY1 coregulate the activity of their common target promoters in the phyA signaling complex, suggesting a coordinated action in transcriptional regulation.
However, we observed that although phyA and FHY1 share a pool of both associated genes and direct target genes, distinct molecular mechanisms may be adopted for the coordinate association of phyA and FHY1 on the target chromatin (Fig. 6, Right). Because the group iii cis-elements only mediate the phyA/FHY1 coassociation, the assembly of phyA/FHY1/transcription factor would require the prior interaction of phyA with FHY1 (Fig. 6, TF8 case). By contrast, for the group v, vi, and vii cis-elements, transcription factors physically interact with phyA or FHY1 for their unique associations, as well as the coordinate association if the subsequent recruitment of the other protein occurs favorably (Fig. 6, TF6, TF7, and TF9 cases). FHY1 and phyA coassociations on RHA1B and SEX4 corroborate the group iii and vii models, respectively.
Fig. 6.
A working model illustrating that phyA and FHY1 can either uniquely or coordinately associate with gene promoters. Overall, transcription factors binding to diverse cis-elements exhibit different affinity with phyA or FHY1, thus leading to the distinct association patterns of phyA and FHY1 on the promoters. The numbers (i–vii) correspond to different cis-element categories labeled in Fig. 5C. X and Y, unknown factors that are involved in the unique associations.
The other remarkable finding of this study is the existence of independent nuclear pathways for phyA and FHY1 on their own associated genes (Fig. 6, Left). The greater number of unique direct target genes compared with the number of shared targets implies a more extensive adoption of separate actions for both phyA and FHY1 in the nucleus. It remains possible that transcription factors with the ability to directly interact with both phyA and FHY1 still mediate the unique phyA or FHY1 associations because of the potential concealment of the binding site caused by the protein–protein interaction (Fig. 6, TF5 case). Overall, our data imply that the specificity of cis-elements and the affinity of their corresponding transcription factors for phyA and FHY1 interaction could be decisive factors for the different coordination or unique phyA/FHY1 working patterns on associated gene promoters.
In absence of FHY1, other signaling components may contribute to the phyA–DNA association, especially when PIF5, PIF4, and FHY3 are involved because these transcription factors cannot directly interact with phyA. FHL, the low-abundant homolog of FHY1, is one of the possible candidates because it mediates the phyA nuclear accumulation (6) and facilitates the interaction between phyA and transcription factors (27), just like FHY1 behaves. The existence of FHL might account for some, but not all, of the FHY1-indepednent phyA nuclear action, considering that FHY1 only associates and regulates 13.5% of phyA direct targets (47 of 348 genes).
Although phyA could be dispensable for the FR-induced FHY1–DNA association, it might indirectly facilitate this event. For example, phyA can promote FHY1 and FHL to reside in the light-induced nuclear speckles (6), the possible sites for regulation of transcription in light signaling (28). In addition, phyA might stabilize related transcription factors or other FHY1 binding proteins to facilitate the formation of FHY1 regulatory complexes on DNA under FR. Notably, we are not sure whether all of the FHY1-DNA associations depend on FR because only limited FHY1-associated genes were tested in darkness in our study. It remains possible that the FHY1-DNA association occurs in darkness but is hard to be detected owing to the low abundance of nuclear FHY1 in dark. The underlying mechanism of the FR-induced phyA-independent FHY1–DNA association awaits further investigation.
We also checked whether FHY1 and phyA are direct targets of themselves. We found that both phyA and FHY1 did not associate with their own promoters but induced their own expressions (Dataset S2) (9). This result suggests that phyA and FHY1 possibly enhance the FR signaling through an indirect regulation mechanism. Interestingly, the FHY1 promoter is directly associated and repressed by phyA (9), indicating a feedback regulation of FHY1 under FR. The phyA signaling is known to repress the FHY1 transcription by repressing FHY3/FAR1 transcription (29) and inducing the HY5 inhibition on the FHY1 transcription (30). Our ChIPseq data revealed an additional mechanism in the feedback regulation of FHY1 by phyA.
On the basis of the cis-elements identified in our study, we predict that the transcription factor ATBPC3, involved in the meristem maintenance, is able to mediate the FHY1 unique association. Additionally, AG, a transcription factor that functions in flower development, may facilitate the phyA unique association. Furthermore, several transcription factors in ABA the response pathway (ATHB1, ATHB5, and GBF4) are likely to mediate the coassociation of phyA and FHY1 (Dataset S5). Multiple transcription factors beyond those involved in light signaling may be responsible for the coordinate or unique association of phyA and FHY1 with DNA, indicating that the FR signal is coordinated with other plant responses through the photoreceptor phyA and its partner FHY1.
To our knowledge this is the first report that reveals a role for FHY1 as an independent transcriptional regulator in plant development. It plays a predominant role in the negative control of salt-resistant seed germination compared with phyA under FR. Given that the FHY1 unique target gene AFP4 encodes an ABI5 binding protein (19), there may be cross-talk between the FHY1 nuclear pathway and the abscissic acid (ABA) signaling pathway in the cell. It is possible that in the fhy1 mutant the reduced AFP4 expression weakens ABA signaling so that the inhibition of seed germination that results from salt stress-induced ABA accumulation (31) is relieved. Interestingly, the nuclear phyA pathway positively regulates ABA response in the root elongation inhibition (9). In this case, FR and ABA signals (two stimuli that inhibit plant growth) are integrated to prevent plants from developing under less-than-ideal growth conditions. The example of AFP4 suggests that the phyA-independent FHY1 nuclear pathway enables FHY1 to influence FR-responsive plant development by itself. This study thus not only suggests a correlation between the FHY1/phyA nuclear behaviors and particular cellular events but also provides valuable data for further research in regard to the independent roles of phyA and FHY1 in various aspects of plant growth and development under FR light. The result of this study suggests that both coordinate and unique associations of phyA and FHY1 play a role in the transcriptional regulation of target genes and require the involvement of transcription factors and various types of cis-elements, many of which remain to be characterized.
Materials and Methods
Plant Materials and Growth Conditions.
The wild-type Arabidopsis thaliana used in this study was of the Landsberg erecta (Ler) ecotype. The phyA-1 (32) and fhy1-1 (15) mutants and the 35S: GFP-FHY1 fhy1-1 (12) and PphyA: phyA-GFP phyA201 (33) transgenic lines have been described previously. For the phyA-GFP*fhy1-1 line, phyA-GFP phyA201 was crossed with fhy1-1. Genotyping and hygromycin (20 µg/mL) screening were performed for the fhy1 and phyA-GFP homozygotes, respectively, in the T3 generation. The growth conditions and light sources were used as described in ref. 8.
ChIP-qPCR and ChIP-seq.
ChIP was performed as previously described (9) with the anti-FHY1 (12) or anti-GFP (Clontech) antibody in the indicated materials. The ChIP-DNA samples were subsequently used for qPCR with Power SYBR Green PCR Master Mix (Applied Biosystems) or for ChIP-seq at the Yale Center for Genome Analysis. Input control for the ChIP-seq was the DNA sample before antibody immunoprecipitation in the ChIP. Primer information can be found in Table S2.
RT-qPCR and RNA-seq.
RNA from indicated materials were extracted using RNeasy Plant Mini Kit (Qiagen). RNA-seq was conducted by the Yale Center for Genome Analysis using Illumina HiSeq 2000. For RT-qPCR, cDNA was obtained by using a Superscipt II Reverse Transcriptase Kit (Invitrogen). The qPCR was subsequently performed as for above ChIP-DNA samples. Primer information can be found in Table S2.
ChIP-seq and RNA-seq Data Analysis.
ChIP-seq and RNA-seq data analysis was performed as previously described (9) for identification of FHY1 chromatin association sites and FHY1 regulated genes. The gene ontology (GO) analysis and P values calculation for the Venn diagrams were performed as previously described (9). For cis-elements analysis, the FHY1/phyA common ChIP peak-covered region was defined as the overlapped regions that must cover more than 50% of the phyA ChIP peak. The remaining FHY1 or phyA ChIP peaks were identified as FHY1 or phyA unique ChIP peak-covered regions. High-throughput sequencing data analyzed in this study are available in the GEO database with the accession number GSE58084.
Supplementary Material
Acknowledgments
This work is supported by National Institutes of Health Grant GM47850, by Ministry of Science and Technology of China Grant 2012CB910900, and by Grant PJ00901003 from the Next-Generation BioGreen 21 Program, Rural Development Administration, Republic of Korea.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE58084).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412528111/-/DCSupplemental.
References
- 1.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 2.Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21(11):664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tepperman JM, Hwang YS, Quail PH. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006;48(5):728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- 4.Rausenberger J, et al. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell. 2011;146(5):813–825. doi: 10.1016/j.cell.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18(6):617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiltbrunner A, et al. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47(8):1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y, et al. Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell. 2009;21(2):494–506. doi: 10.1105/tpc.108.061259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F, et al. Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light in Arabidopsis. Plant Cell. 2012;24(5):1907–1920. doi: 10.1105/tpc.112.097733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F, et al. Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell. 2014;26(5):1949–1966. doi: 10.1105/tpc.114.123950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA. 2001;98(16):9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genoud T, et al. FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 2008;4(8):e1000143. doi: 10.1371/journal.pgen.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Y, et al. Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 2005;139(3):1234–1243. doi: 10.1104/pp.105.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang X, et al. Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell. 2011;23(7):2514–2535. doi: 10.1105/tpc.111.085126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desnos T, Puente P, Whitelam GC, Harberd NP. FHY1: A phytochrome A-specific signal transducer. Genes Dev. 2001;15(22):2980–2990. doi: 10.1101/gad.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokolov LN, Dominguez-Solis JR, Allary AL, Buchanan BB, Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA. 2006;103(25):9732–9737. doi: 10.1073/pnas.0603329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro L, et al. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135(2):1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact. 2007;20(8):900–911. doi: 10.1094/MPMI-20-8-0900. [DOI] [PubMed] [Google Scholar]
- 19.Garcia ME, Lynch T, Peeters J, Snowden C, Finkelstein R. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol Biol. 2008;67(6):643–658. doi: 10.1007/s11103-008-9344-2. [DOI] [PubMed] [Google Scholar]
- 20.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeidler M, Zhou Q, Sarda X, Yau CP, Chua NH. The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J. 2004;40(3):355–365. doi: 10.1111/j.1365-313X.2004.02212.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9(1):e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornitschek P, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71(5):699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 24.Oh E, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21(2):403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011;65(3):346–358. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- 26.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14(8):802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang SW, Jang IC, Henriques R, Chua NH. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell. 2009;21(5):1341–1359. doi: 10.1105/tpc.109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Buskirk EK, Decker PV, Chen M. Photobodies in light signaling. Plant Physiol. 2012;158(1):52–60. doi: 10.1104/pp.111.186411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin R, et al. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318(5854):1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell. 2010;22(11):3634–3649. doi: 10.1105/tpc.110.075788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia W, Wang Y, Zhang S, Zhang J. Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J Exp Bot. 2002;53(378):2201–2206. doi: 10.1093/jxb/erf079. [DOI] [PubMed] [Google Scholar]
- 32.Whitelam GC, et al. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5(7):757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim L, et al. Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 2000;22(2):125–133. doi: 10.1046/j.1365-313x.2000.00729.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.