Significance
Hypoxia-inducible factor 1α (HIF-1α) is required for adaptive changes to low oxygen levels, which include a reduced rate of cell division. However, many cell types continue to proliferate under hypoxic conditions. Here, we show that cyclin-dependent kinases 1 and 2 physically and functionally interact with HIF-1α, inhibiting and promoting its degradation by lysosomes, respectively. Cancer cells that proliferate under hypoxia failed to do so when treated with lysosome inhibitors. Our studies reveal that HIF-1α levels are coupled to phases of the cell cycle through lysosomal degradation and identify a novel role for the lysosome as a regulator of cell-cycle progression under hypoxic conditions.
Keywords: cell proliferation, chaperone mediated autophagy, lysosome
Abstract
Hypoxia-inducible factor 1 (HIF-1) is a transcription factor that mediates adaptive responses to oxygen deprivation. In addition, the HIF-1α subunit has a nontranscriptional role as a negative regulator of DNA replication through effects on minichromosome maintenance helicase loading and activation. However, some cell types continue to replicate under hypoxic conditions. The mechanism by which these cells maintain proliferation in the presence of elevated HIF-1α levels is unclear. Here we report that HIF-1α physically and functionally interacts with cyclin-dependent kinase 1 (Cdk1) and Cdk2. Cdk1 activity blocks lysosomal degradation of HIF-1α and increases HIF-1α protein stability and transcriptional activity. By contrast, Cdk2 activity promotes lysosomal degradation of HIF-1α at the G1/S phase transition. Blocking lysosomal degradation by genetic or pharmacological means leads to HIF-1α–dependent cell-cycle arrest, demonstrating that lysosomal degradation of HIF-1α is an essential step for the maintenance of cell-cycle progression under hypoxic conditions.
Hypoxia elicits a variety of adaptive cellular and systemic responses, which include changes in angiogenesis, red blood cell production, metabolism, and autophagy (1). Many of these changes are mediated through the transcriptional activity of hypoxia-inducible factor 1 (HIF-1) (2). HIF-1 is a heterodimer composed of HIF-1α and HIF-1β subunits (3). HIF-1 activity is regulated by cellular O2 availability through O2-dependent hydroxylation reactions. Proline hydroxylation targets HIF-1α for ubiquitination by the von Hippel–Lindau ubiquitin ligase complex and subsequent proteasomal degradation (4–6), whereas asparagine hydroxylation inhibits binding of the HIF-1α transactivation domain to the coactivator p300 (7). Under hypoxic conditions, both proline and asparagine hydroxylation are inhibited, leading to increased stability of HIF-1α, enhanced binding of coactivators, and increased transcription of HIF-1 target genes. Among the hundreds of target genes regulated by HIF-1 are GLUT1, encoding glucose transporter 1 (8); PDK1, encoding pyruvate dehydrogenase kinase 1 (9, 10); VEGF, encoding vascular endothelial growth factor (11); BNIP3, encoding Bcl-2/adenovirus E1B 19-kDa protein-interacting protein 3 (12); PLOD2, encoding procollagen lysyl hydroxylase 2 (13); and P4HA1, encoding procollagen prolyl 4-hydroxylase α1 (14). HIF-2, which is composed of HIF-2α and HIF-1β subunits, is regulated by oxygen in a similar manner, although HIF-2 has a more limited tissue distribution and in some cases regulates distinct target genes (15). Mechanisms by which HIF-1α is regulated in an O2-independent manner have also been identified (16–26). In addition to proteasome-dependent pathways for HIF-1α degradation, we identified a pathway by which HIF-1α can be targeted for lysosomal degradation through chaperone-mediated autophagy (27), which subsequently was confirmed by others (28–30).
Because an inadequate supply of oxygen will only be exacerbated by an increase in cell number, inhibition of proliferation is a fundamental adaptive response to hypoxia. This effect has been shown to be dependent on HIF-1α in multiple cell types, including various cancer cell lines (31–34), fibroblasts (34), lymphocytes (34), and hematopoietic stem cells (35). Forced overexpression of HIF-1α is sufficient to arrest the mammalian cell cycle in G1 phase (31, 36). We have recently shown that HIF-1α functions in a transcription-independent manner to inhibit DNA replication. HIF-1α binds to the minichromosome maintenance (MCM) proteins (31, 32), which normally assemble as a hexamer during G1 phase and are kept in a loaded but inactivated state by the proteins Cdc6 and Cdt1 (37). During the G1/S-phase transition, phosphorylation of this complex by cyclin-dependent kinase 2 (Cdk2) causes nuclear export of Cdc6 and subsequent phosphorylation and activation of the MCM helicase by Cdc7 (38, 39). MCM complex activation leads to DNA replication during S phase, with Cdk1 activity subsequently coordinating the events of G2/M phase. HIF-1α promotes interaction of Cdc6 with the MCM helicase, leading to enhanced loading of the MCM helicase onto chromatin, but the presence of HIF-1α blocks phosphorylation and activation of the MCM complex (31). However, because some fraction of cells proceed through the cell cycle during hypoxia, we hypothesized that cells possess a mechanism to selectively degrade HIF-1α during S phase of the cell cycle.
In this article, we report that lysosomal degradation of HIF-1α is regulated by the activity of Cdk1 and Cdk2, which physically interact with HIF-1α. Overexpression of Cdk1 increased HIF-1α protein levels, whereas Cdk2 overexpression decreased HIF-1α levels. Pharmacological or genetic inhibition of lysosome function led to cell-cycle arrest, which was rescued by knockdown of HIF-1α and HIF-2α. This study establishes that Cdk-dependent regulation of HIF-1α lysosomal degradation is essential for DNA replication under hypoxic conditions.
Results
Cdk1 and Cdk2 Interact with HIF-1α.
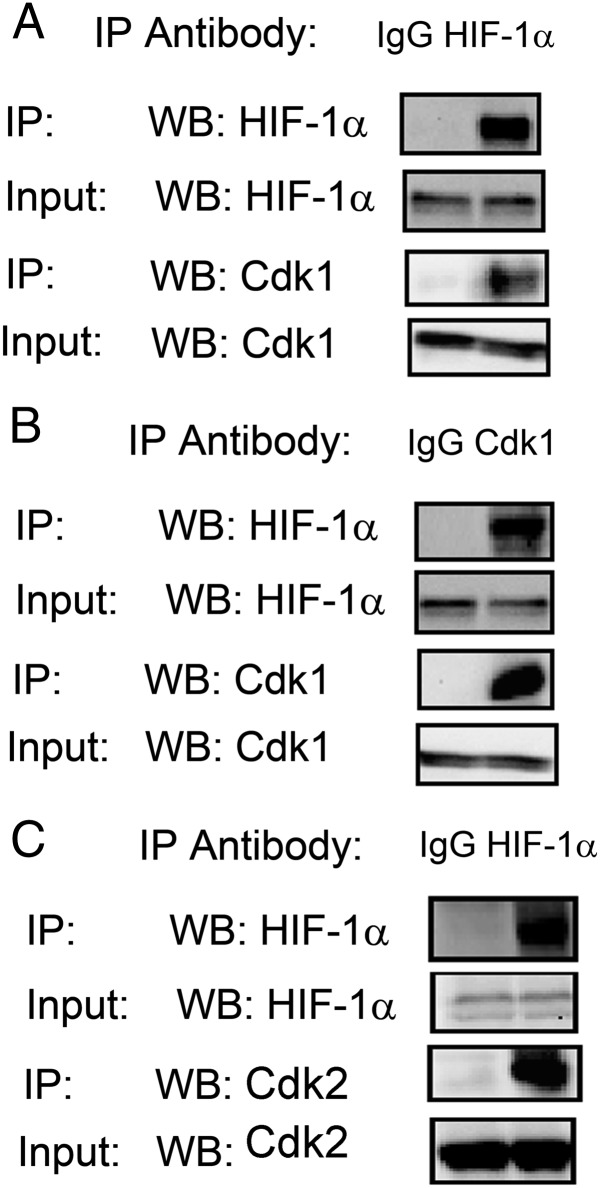
We hypothesized that HIF-1α may be subject to cell-cycle phase-specific regulation and therefore analyzed binding of HIF-1α to Cdk1 and Cdk2. We detected Cdk1 after immunoprecipitation of endogenous HIF-1α from lysates of hypoxic HeLa human cervical carcinoma cells (Fig. 1A), and HIF-1α was detected after immunoprecipitation of Cdk1 (Fig. 1B). Similarly, we detected Cdk2 after immunoprecipitation of HIF-1α (Fig. 1C). Thus, HIF-1α interacts with both Cdk1 and Cdk2.
Fig. 1.
Cdks interact with HIF-1α. (A and B) Antibody against HIF-1α (A) or Cdk1 (B), or IgG control, was used for immunoprecipitation (IP) of lysates prepared from HeLa cells that were exposed to 1% O2 for 6 h. Western blot (WB) assays of the immunoprecipitates were performed using antibody against HIF-1α or Cdk1. (C) Anti–HIF-1α antibody or IgG was used for IP of lysates prepared from HeLa cells that were exposed to 1% O2 for 6 h. WB assays of the immunoprecipitates were performed using antibody against HIF-1α or Cdk2.
Cdk1 Is a Positive Regulator of HIF-1α.
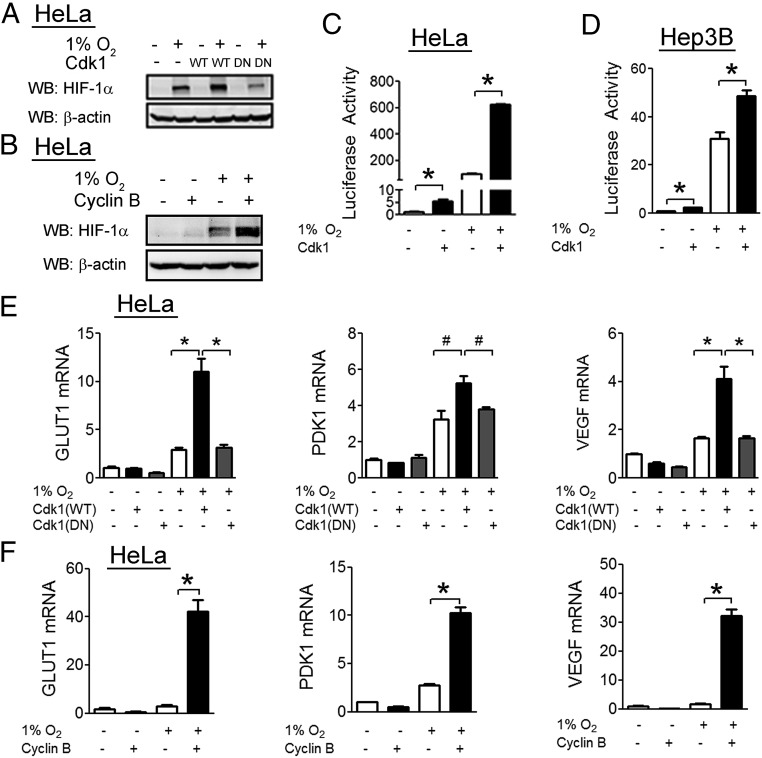
To examine the effect of Cdk1 activity on HIF-1α, we overexpressed wild-type Cdk1 or a catalytically inactive Cdk1(D146N) mutant in HeLa cells. Cells transfected with wild-type Cdk1 showed increased induction of HIF-1α upon exposure to hypoxia, whereas cells transfected with Cdk1(D146N) showed no increase (Fig. 2A). The kinase activity of Cdk1 is dependent upon binding of cyclin B and overexpression of cyclin B also led to increased HIF-1α protein levels under hypoxic conditions (Fig. 2B).
Fig. 2.
Cdk1 is a positive regulator of HIF-1α. (A) HeLa cells were transfected with either empty vector (−) or vector encoding wild-type (WT) or catalytically-inactive (DN) Cdk1, then exposed to either 20% or 1% O2 for 6 h. Cell lysates were analyzed by WB with the indicated antibody. (B) HeLa cells were transfected with empty vector (−) or vector encoding cyclin B (+), then exposed to 20% O2 (−) or 1% O2 (+) for 6 h. Cell lysates were analyzed by WB. (C and D) HeLa (C) and Hep3B (D) cells were transfected with HIF-dependent firefly luciferase reporter p2.1, control Renilla luciferase reporter pSV-RL, and either empty vector (−) or vector encoding Cdk1 (+). Cells were exposed to 20% O2 (−) or 1% O2 (+) for 24 h, and the ratio of firefly:Renilla luciferase activity was determined. (E) HeLa cells were transfected with empty vector or vector encoding Cdk1 that was either WT or DN. Cells were exposed to 20% or 1% O2 for 24 h, RNA was isolated, and RT-qPCR was performed to quantify the indicated mRNA. (F) HeLa cells were transfected with either empty vector or vector encoding cyclin B, and mRNA analyses were performed. All results in bar graphs are presented as mean ± SEM (n = 4). *P < 0.01; #P < 0.05.
To determine the effect of Cdk1 on HIF-1 transcriptional activity, cells were cotransfected with p2.1, a reporter plasmid that contains a 68-bp hypoxia response element from the human ENO1 gene upstream of a basal SV40 promoter and firefly luciferase coding sequences, and pSV-RL, a control reporter that contains Renilla luciferase coding sequences downstream of the SV40 promoter only (40). The ratio of firefly:Renilla luciferase activity serves as a measure of HIF transcriptional activity. In both HeLa cells (Fig. 2C) and Hep3B human hepatocellular carcinoma cells (Fig. 2D), Cdk1 overexpression was associated with a significant increase in HIF-1 transcriptional activity. We also analyzed expression of three different HIF-1 target genes by reverse transcription and quantitative real-time PCR (RT-qPCR). Cdk1 enhanced induction of GLUT1, PDK1, and VEGF mRNA expression in HeLa cells under hypoxic conditions (Fig. 2E), whereas the inactive D146N mutant had no effect. Similarly, overexpression of the Cdk1 activator cyclin B significantly increased the hypoxia-induced expression of all three HIF-1 target genes analyzed (Fig. 2F).
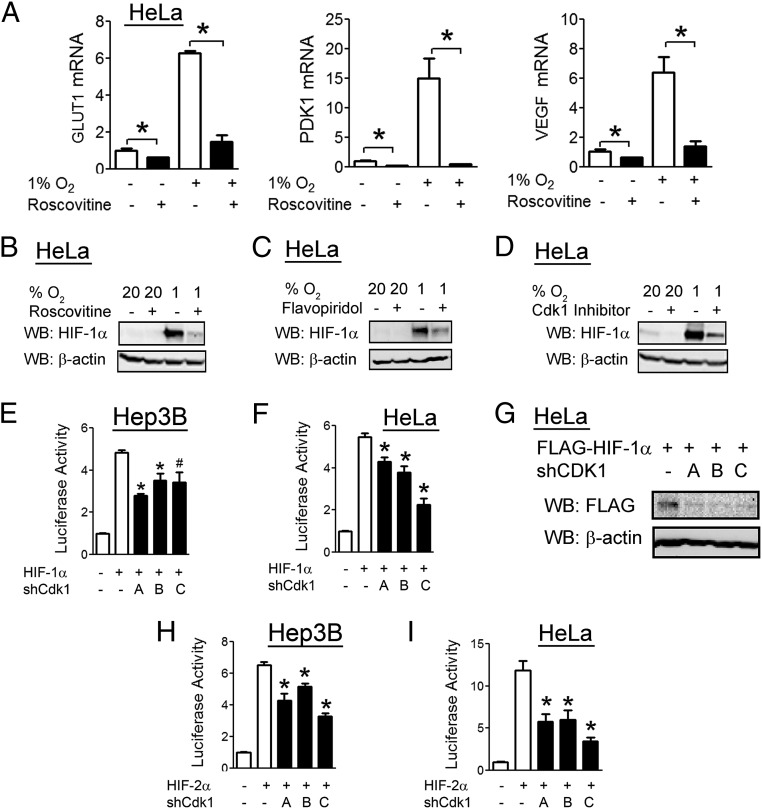
To examine the effect of Cdk1 inhibition on HIF-1, we used both pharmacological and genetic approaches. Treatment of HeLa cells with the Cdk inhibitor roscovitine (20 µM) impaired the hypoxic induction of all three HIF-1 target genes examined (Fig. 3A). Roscovitine treatment also inhibited the induction of HIF-1α protein levels in response to hypoxia (Fig. 3B). Flavopiridol (300 nM), a Cdk inhibitor that is chemically unrelated to roscovitine, also decreased hypoxic induction of HIF-1α protein levels (Fig. 3C), as did a Cdk1-specific inhibitor (Fig. 3D).
Fig. 3.
Cdk1 inhibition decreases HIF-1α protein levels and transcriptional activity. (A and B) HeLa cells were treated with vehicle or roscovitine (20 µM), exposed to 20% or 1% O2 for 24 h, and RT-qPCR was performed to quantify the indicated mRNA (A) or cell lysates were analyzed by WB with the indicated antibody (B). (C and D) HeLa cells were treated with vehicle, flavopiridol (300 nM) (C), or Cdk1 inhibitor (D) and exposed to 20% or 1% O2 for 24 h, and cell lysates were analyzed by WB. (E and F) Hep3B (E) and HeLa (F) cells were cotransfected with FLAG–HIF-1α vector, p2.1 HIF reporter, pSV-RL control reporter, and either empty vector (−) or one of three shRNA expression vectors (A, B, or C) targeting different Cdk1 mRNA sequences. At 48 h posttransfection, luciferase activities were determined. (G) HeLa cells were transfected with FLAG–HIF-1α vector and either empty vector or one of three shRNA vectors targeting different Cdk1 mRNA sequences. At 48 h posttransfection, cell lysates were analyzed by WB. (H and I) Hep3B (H) and HeLa (I) cells were transfected with HIF-2α vector, p2.1, pSV-RL, and either empty vector or one of three shRNA vectors targeting different Cdk1 mRNA sequences. At 48 h posttransfection, luciferase activities were determined. All results in bar graphs are presented as mean ± SEM (n = 4). *P < 0.01; #P < 0.05.
As pharmacological treatments may be confounded by off-target effects, we generated three shRNA vectors targeting different nucleotide sequences within Cdk1 mRNA, which were designated A, B, and C. Knockdown of Cdk1 with each shRNA vector led to decreased HIF-1 transcriptional activity in luciferase reporter assays in Hep3B cells (Fig. 3E) and HeLa cells (Fig. 3F). Cdk1 knockdown led to decreased HIF-1α protein levels in HeLa cells (Fig. 3G). Cdk1 knockdown also led to decreased HIF-2 transcriptional activity in both Hep3B cells (Fig. 3H) and HeLa cells (Fig. 3I). Taken together, the data in Fig. 3 demonstrate that Cdk1 is a positive regulator of HIF-1 and HIF-2 activity.
Regulation of HIF-1α by Cdk1 Is Proteasome-Independent and Lysosome-Dependent.
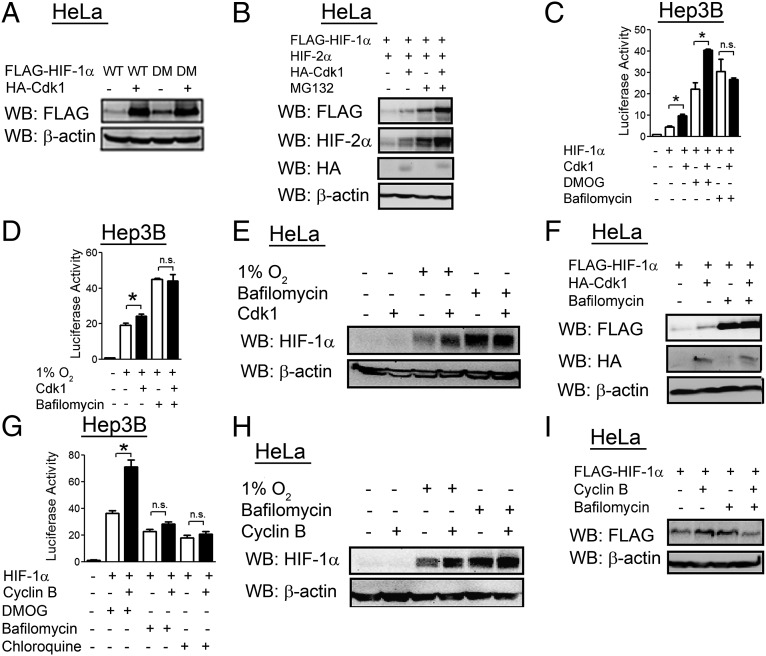
As proline hydroxylation is a major regulator of HIF-1α protein levels, we tested the hypothesis that Cdk1 might regulate hydroxylation-dependent proteasomal degradation of HIF-1α. However, Cdk1 overexpression led to increased levels of a double mutant HIF-1α protein, which contained Pro → Ala mutations in both hydroxylation sites (P402A/P564A), to a similar extent as wild-type HIF-1α (Fig. 4A). Cdk1 increased HIF-1α and HIF-2α levels in the presence or absence of the proteasome inhibitor MG132 (Fig. 4B). Cdk1 overexpression also increased HIF transcriptional activity in the presence of the hydroxylase inhibitor dimethyloxalylglycine (DMOG) (Fig. 4C). These data indicate that Cdk1 regulation of HIF-1 is independent of proline hydroxylation and proteasomal degradation.
Fig. 4.
Regulation of HIF-1α by Cdk1 is proteasome-independent and lysosome-dependent. (A) HeLa cells were cotransfected with expression vector encoding FLAG epitope-tagged HIF-1α that was either wild-type (WT) or P402A/P564A double-mutant (DM), and either empty vector (−) or vector encoding HA-tagged Cdk1 (+). At 24 h posttransfection, cell lysates were analyzed by WB. (B) HeLa cells were cotransfected with FLAG–HIF-1α vector, HIF-2α vector, and empty vector or Cdk1 vector as indicated. At 24 h posttransfection, cells were treated with vehicle or MG132 (20 μM) for 6 h, and cell lysates were analyzed by WB. (C) Hep3B cells were cotransfected with p2.1, pSV-RL, FLAG–HIF-1α vector, and either empty vector or Cdk1 vector. At 24 h posttransfection, cells were treated with vehicle, bafilomycin (10 nM), or DMOG (500 nM) for an additional 24 h, and luciferase activities were determined. (D) Hep3B cells were cotransfected with p2.1, pSV-RL, and empty vector or Cdk1 vector. Cells were exposed to 1% O2 for 24 h as indicated, and luciferase activities were determined. (E and H) HeLa cells were transfected with empty vector or vector encoding either Cdk1 (E) or cyclin B (H). At 24 h posttransfection, cells were exposed to 1% O2 or treated with bafilomycin (10 nM) for 24 h, and cell lysates were analyzed by WB. (F and I) HeLa cells were cotransfected with FLAG–HIF-1α vector and empty vector or vector encoding either Cdk1 (F) or cyclin B (I). At 24 h posttransfection, cells were treated with vehicle or bafilomycin (10 nM) for an additional 24 h, and cell lysates were analyzed by WB. (G) Hep3B cells were cotransfected with p2.1, pSV-RL, FLAG–HIF-1α vector, and either empty vector or cyclin B vector. At 24 h posttransfection, cells were treated with vehicle, DMOG (500 nM), bafilomycin (10 nM), or chloroquine (50 µM) for an additional 24 h, and luciferase activities were determined. All results in bar graphs are presented as mean ± SEM (n = 4). *P < 0.01; n.s., not significant.
We next investigated whether Cdk1 regulated lysosomal degradation of HIF-1α. Cdk1 overexpression increased HIF-1 transcriptional activity in cells in which HIF-1α was overexpressed (Fig. 4C) or induced by hypoxia (Fig. 4D), but Cdk1 had no effect on HIF-1 activity in cells that were treated with the lysosome inhibitor bafilomycin. Cdk1 overexpression increased HIF-1α protein levels in hypoxic cells (Fig. 4E) and in cells transfected with a HIF-1α expression vector (Fig. 4F), but had no effect on HIF-1α levels when cells were treated with bafilomycin. Similarly, cyclin B overexpression increased HIF-1 transcriptional activity in cells treated with DMOG, but had no effect in cells treated with bafilomycin or chloroquine, which is a lysosome inhibitor that is chemically unrelated to bafilomycin (Fig. 4G). HIF-1α protein levels were increased by cyclin B overexpression in hypoxic cells (Fig. 4H) and in cells overexpressing FLAG–HIF-1α (Fig. 4I), but cyclin B did not increase HIF-1α levels in cells treated with bafilomycin. Taken together, the data in Fig. 4 indicate that Cdk1 increases levels of HIF-1α by protecting it from lysosomal degradation.
Cdk2 Activity Decreases HIF-1α Protein Levels.
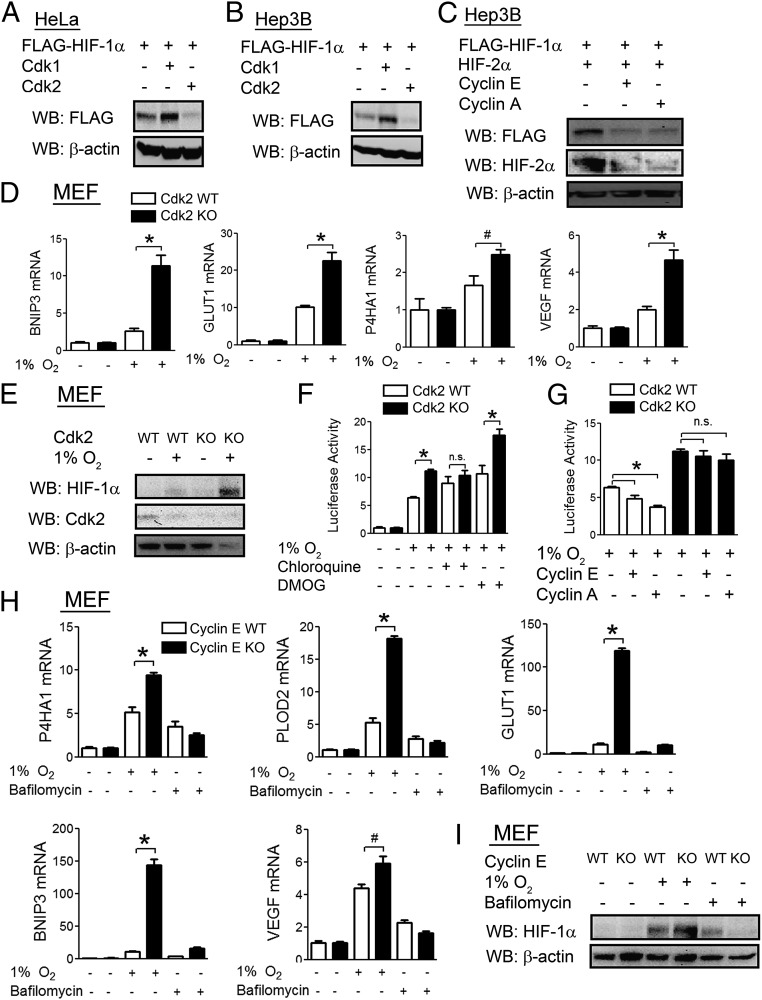
As Cdk2 was also found to interact with HIF-1α, we analyzed the effect of Cdk2 overexpression on HIF-1α protein levels. Whereas Cdk1 increased HIF-1α protein levels in both HeLa (Fig. 5A) and Hep3B (Fig. 5B) cells, Cdk2 had the opposite effect. Overexpression of the Cdk2 binding partner cyclin E or cyclin A also led to decreased levels of both HIF-1α and HIF-2α (Fig. 5C). Mouse embryo fibroblasts (MEFs) from Cdk2 knockout (KO) mice (41) showed enhanced expression of HIF-1 target genes in response to hypoxia compared with MEFs from wild-type mice (Fig. 5D), which was associated with increased induction of HIF-1α protein levels (Fig. 5E). Cdk2 KO increased HIF-1 transcriptional activity in hypoxic or DMOG-treated MEFs but had no effect in chloroquine-treated cells (Fig. 5F). Overexpression of either cyclin E or cyclin A led to decreased HIF-1 transcriptional activity in wild-type MEFs, but not in Cdk2 KO MEFs (Fig. 5G), implicating Cdk2:cyclin E and Cdk2:cyclin A complexes as negative regulators of HIF-1α. We further analyzed the role of cyclin E, which activates Cdk2 at the G1/S phase transition, using MEFs from cyclin E KO mice that lack expression of both cyclin E1 and cyclin E2 (42). Cyclin E KO MEFs had increased expression of multiple HIF-1 target genes, including P4HA1, PLOD2, GLUT1, BNIP3, and VEGF, but this effect was lost with bafilomycin treatment (Fig. 5H). Cyclin E KO MEFs had a corresponding increase in HIF-1α levels in response to hypoxia compared with wild-type MEFs, but not in the presence of bafilomycin (Fig. 5I). Taken together, the data in Fig. 5 indicate that Cdk2 activity stimulates the lysosomal degradation of HIF-1α.
Fig. 5.
Cdk2 down-regulates HIF-1α protein levels. (A and B) HeLa (A) and Hep3B (B) cells were cotransfected with FLAG–HIF-1α vector and either empty vector or vector encoding Cdk1 or Cdk2. At 24 h posttransfection, cell lysates were analyzed by WB. (C) Hep3B cells were cotransfected with FLAG–HIF-1α vector and either empty vector or vector encoding cyclin E or cyclin A. At 24 h posttransfection, cells lysates were analyzed by WB. (D and E) Mouse MEFs from Cdk2 WT or KO mice were exposed to 20% or 1% O2 for 24 h, and RT-qPCR (D) or WB (E) assays were performed. (F) Cdk2 WT or KO MEFs were cotransfected with p2.1 and pSV-RL. At 24 h posttransfection, cells were treated with vehicle, DMOG (500 nM), or chloroquine (50 µM), then exposed to 1% O2 for an additional 24 h, and luciferase activities were determined. (G) Cdk2 WT or KO MEFs were cotransfected with p2.1, pSV-RL, and either empty vector or vector encoding cyclin E or cyclin A. At 24 h posttransfection, cells were exposed to hypoxia for an additional 24 h, and luciferase activities were determined. (H and I) MEFs from Cyclin E1/Cyclin E2 WT or KO mice were exposed to 20% or 1% O2 or treated with bafilomycin (10 nM) for 24 h, and RT-qPCR (H) or WB (I) assays were performed. All results in bar graphs are presented as mean ± SEM (n = 4). *P < 0.01; #P < 0.05; n.s., not significant.
Cdk2 Promotes HIF-1 Transcriptional Activity in Cancer Cell Lines.
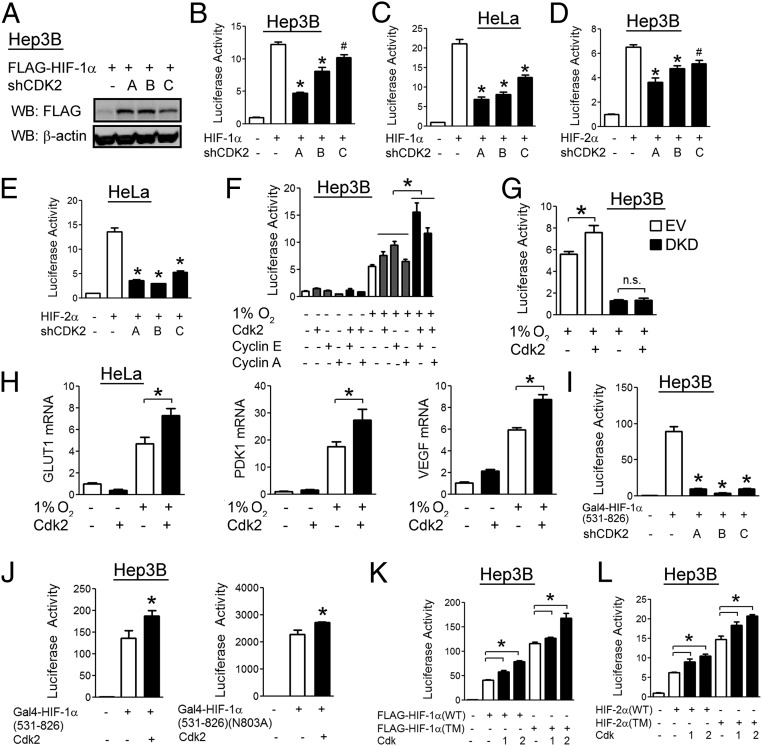
In Hep3B cells transfected with a HIF-1α expression vector, knockdown of Cdk2 with each of three different shRNA vectors led to increased HIF-1α protein levels under nonhypoxic conditions, which was consistent with a role for Cdk2 in stimulating HIF-1α degradation (Fig. 6A). Surprisingly, under similar conditions, Cdk2 knockdown was also associated with decreased HIF-1 transcriptional activity in both Hep3B (Fig. 6B) and HeLa (Fig. 6C) cells. Cdk2 knockdown also decreased HIF-2 transcriptional activity (Fig. 6 D and E). Overexpression of Cdk2, cyclin E, or cyclin A led to an increase in hypoxia-induced HIF-1 transcriptional activity and the effect was even greater when overexpression of Cdk2 was combined with either of its cyclin partners (Fig. 6F). Using Hep3B cells stably transfected with vectors encoding shRNAs against both HIF-1α and HIF-2α (38), we found that the stimulatory effect of Cdk2 on HIF reporter activity was dependent on HIF-1α and HIF-2α (Fig. 6G), excluding the possibility that Cdk2 had nonspecific effects on gene transcription. We also found that overexpression of Cdk2 led to increased induction of HIF target genes in response to hypoxia (Fig. 6H), which is consistent with a role for Cdk2 as a positive regulator of HIF transcriptional activity in HeLa cells.
Fig. 6.
Cdk2 enhances HIF-1α transactivation function in cancer cells. (A) Hep3B cells were cotransfected with FLAG–HIF-1α vector and empty vector or vector encoding one of three shRNAs targeting Cdk2. At 48 h posttransfection, cell lysates were analyzed by WB. (B–E) Hep3B (B and D) and HeLa (C and E) cells were cotransfected with p2.1, pSV-RL, FLAG–HIF-1α vector, and the indicated shRNA vector. At 48 h posttransfection, luciferase activities were determined. (F) Hep3B cells were cotransfected with p2.1, pSV-RL, and the indicated expression vector. At 24 h posttransfection, cells were exposed to 20% or 1% O2 for an additional 24 h, and luciferase activities were determined. (G) Hep3B cells stably transfected with either shEV (white) or shRNAs against both HIF-1α and HIF-2α [double knockdown (DKD); black] were cotransfected with p2.1, pSV-RL, and Cdk2 vector as indicated. At 24 h posttransfection, cells were exposed to 20% or 1% O2 for an additional 24 h and luciferase activities were determined. (H) HeLa cells were transfected with empty vector or Cdk2 vector followed by exposure to 20% or 1% O2 for an additional 24 h, and qRT-PCR was performed. (I) Hep3B cells were cotransfected with vector encoding Gal4–HIF-1α(531–826), firefly luciferase reporter pG5E1bLuc, pSV-RL, and the indicated shRNA vector. At 48 h posttransfection, luciferase activities were determined. (J) Hep3B cells were cotransfected with vector encoding Gal4–HIF-1α(531–826) or Gal4–HIF-1α(531–826)(N803A), pG5E1bLuc, pSV-RL, and the indicated expression vector. At 48 h posttransfection, luciferase activities were determined. (K and L) Hep3B cells were cotransfected with p2.1, pSV-RL, vector encoding wild-type (WT) or triple mutant (TM) HIF-1α (K), or HIF-2α (L), and the indicated expression vector. At 24 h posttransfection, luciferase activities were determined. All results in bar graphs are presented as mean ± SEM (n = 4). *P < 0.01; #P < 0.05; n.s., not significant.
To examine the effect of Cdk2 activity on HIF-1 transactivation domain function, HeLa cells were cotransfected with reporter plasmid pG5-E1b-Luc, which contains five Gal4 binding sites upstream of the E1b gene promoter and firefly luciferase coding sequences, and an expression vector encoding the Gal4 DNA-binding domain either alone (Gal4-EV) or fused to HIF-1α(531–826), which encompasses the HIF-1α transactivation domain (43). Cdk2 knockdown led to decreased HIF-1α transactivation domain function (Fig. 6I), whereas overexpression led to the opposite effect (Fig. 6J, Left). Transactivation domain function is O2-regulated through hydroxylation of asparagine 803 (N803), which prevents interaction with the HIF coactivators p300 and CBP (7). However, Cdk2 overexpression led to increased activity of HIF-1α(531–826/N803A), indicating that Cdk2 does not modulate asparagine hydroxylation (Fig. 6J, Right). The effect of Cdk2 overexpression on HIF transcriptional activity was maintained even when a HIF-1α construct containing mutations in all three hydroxylation sites (P402A/P564A/N803A) (Fig. 6K) or the analogous triple-mutant HIF-2α construct (Fig. 6L) was expressed. Thus, Cdk2 can stimulate HIF-1 and HIF-2 transcriptional activity in a hydroxylation-independent manner.
Lysosome Function Promotes Cell-Cycle Progression by Degrading HIF-1α.
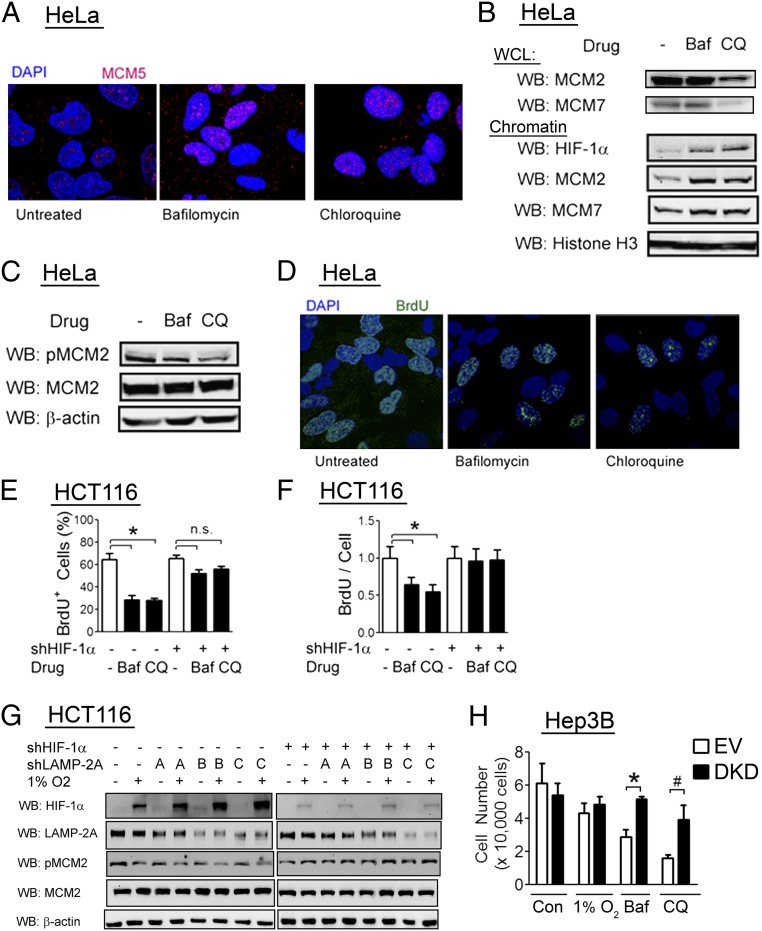
We previously described nontranscriptional effects of HIF-1α protein, which enhanced chromatin loading of the MCM complex but impaired MCM helicase activation, thereby blocking DNA replication (31). Induction of HIF-1α levels with a lysosome inhibitor (bafilomycin or chloroquine) led to increased MCM chromatin loading as determined by immunofluorescence (Fig. 7A) and immunoblot analysis of chromatin-enriched fractions (Fig. 7B, Lower), despite no increase of total MCM levels in whole cell lysates (Fig. 7B, Upper). Lysosome inhibitors decreased MCM2 phosphorylation (Fig. 7C), which is a marker of MCM helicase activation, as well as DNA replication, as determined by BrdU incorporation (Fig. 7D). We next analyzed HCT116 colorectal carcinoma cells stably transfected with either empty shRNA vector or shRNA vector targeting HIF-1α. Consistent with results observed in HeLa cells, treatment with bafilomycin or chloroquine led to a decrease in the percentage of BrdU+ cells that were replicating DNA (Fig. 7E) as well as a decreased number of replication sites in dividing cells (Fig. 7F), and these effects were abolished by HIF-1α knockdown.
Fig. 7.
Lysosome activity promotes DNA replication via HIF-1α degradation. (A–C) HeLa cells were treated with vehicle, bafilomycin (10 nM), or chloroquine (50 µM) for 24 h. Cells were fixed, stained with anti-MCM5 antibody, and analyzed by confocal microscopy (A), or whole cell lysates (WCLs) and chromatin fractions were prepared and analyzed by WB (B and C). (D) HeLa cells were treated with vehicle, bafilomycin, or chloroquine for 24 h, and exposed to a 5-min BrdU pulse before fixation and confocal imaging with anti-BrdU antibody. (E and F) HCT116 cells stably transfected with empty vector or HIF-1α shRNA vector were treated with vehicle (−), bafilomycin (Baf), or chloroquine (CQ). The percentage of BrdU-positive cells (E) and BrdU incorporation per replicating cell (F) were determined. Results are shown as mean ± SEM (n = 3). *P < 0.01; n.s., not significant. (G) HCT116 cells stably transfected with either empty vector or one of three shRNA vectors targeting LAMP-2A were transfected with empty vector or HIF-1α shRNA vector, exposed to 1% O2 for 24 h, and lysates were analyzed by WB. (H) Hep3B cells stably transfected with either empty vector or vectors encoding shRNAs against HIF-1α and HIF-2α (DKD) were treated with vehicle control (Con), Baf, or CQ, or exposed to 1% O2. After 36 h of treatment, cells were counted. Results are shown as mean ± SEM (n = 4). *P < 0.01; #P < 0.05; n.s., not significant.
As both bafilomycin and chloroquine are nonspecific inhibitors of lysosome function, we generated HCT116 cells stably transfected with any of three shRNA vectors targeting different sequences within LAMP-2A to specifically investigate the role of chaperone-mediated autophagy in cell-cycle regulation. Hypoxic induction of HIF-1α protein was increased in LAMP-2A knockdown cells, and the magnitude of the increase was inversely proportional to the magnitude of the decrease in LAMP-2A and phospho-MCM2 levels (Fig. 7G). Importantly, the inhibitory effect of LAMP-2A knockdown on MCM2 phosphorylation was lost with simultaneous knockdown of HIF-1α (Fig. 7G) in HCT116 cells, which express predominantly HIF-1α, unlike HeLa cells, which express abundant amounts of both HIF-1α and HIF-2α (31). We conclude that chaperone-mediated autophagy regulates the cell cycle through its effects on HIF-1α.
Consistent with prior results (44), we found that Hep3B cells do not have impaired proliferation in the presence of hypoxia (Fig. 7H), unlike both HCT116 and HeLa cells. This could be due to a mechanism that renders the DNA replication complex insensitive to the effects of HIF-1α or due to degradation of HIF-1α before the onset of DNA replication in this cell line. We tested the hypothesis that lysosomal degradation of HIF-1α before DNA replication was an essential step in this process. Consistent with this hypothesis, we found that treatment with bafilomycin or chloroquine had a significant effect on Hep3B cell proliferation, in contrast to hypoxia (Fig. 7H). Using Hep3B cells with stable knockdown of both HIF-1α and HIF-2α (45), we showed that the effect of either lysosome inhibitor on proliferation was dependent on induction of HIF-1α and HIF-2α levels (Fig. 7H), demonstrating that Hep3B cells are sensitive to the inhibitory effects of HIF-1α and HIF-2α on cell proliferation. We conclude that lysosomal degradation of HIF-1α is an essential step in the proliferation of mammalian cells under hypoxic conditions.
Discussion
Oxygen availability is a critical and dynamic regulator of cell proliferation. Hypoxia induces reversible cell-cycle arrest in a HIF-dependent manner (31–36), but suppresses senescence, which is the irreversible loss of replicative potential, in a HIF-independent manner (46). In the present study, we have delineated a molecular mechanism by which Cdks regulate HIF-1. Both Cdk1 and Cdk2 interact with HIF-1α. Overexpression of wild-type Cdk1 increased HIF-1α protein levels, HIF-1 transcriptional activity, and HIF target gene expression, whereas a catalytically inactive mutant did not. These effects were also observed with overexpression of the Cdk1-activating protein, cyclin B. In contrast, overexpression of Cdk2 decreased HIF-1α protein levels, as did overexpression of cyclin E or cyclin A. However, whereas Cdk2 activity led to decreased HIF target gene expression in MEFs, in two cancer cell lines, Cdk2 activity led to increased HIF-1 transcriptional activity. The effects of Cdk1 and Cdk2 on HIF-1 were maintained in the presence of proteasome or hydroxylase inhibitors but abolished by inhibitors of lysosomal degradation. While this manuscript was in preparation, another group reported that Cdk1 promoted tumor growth through increased HIF-1α protein levels and that Cdk1 phosphorylated HIF-1α at serine residue 668, which inhibited its proteasomal degradation (47). However, our data demonstrate that Cdk1 and Cdk2 regulate lysosomal degradation of HIF-1α. Additional studies are required to determine if Cdk2 phosphorylates HIF-1α (on a residue other than Ser-688) to increase its lysosomal degradation.
Progression through the cell cycle is a highly controlled process, dependent on cell-cycle phase-specific synthesis and degradation of multiple regulatory proteins. Cell-cycle phase-specific proteasomal degradation of proteins is generally mediated by two ubiquitin ligase complexes: the Skp1-Cullin-F box complex, which is active during the G1/S and G2/M transitions, and the anaphase-promoting complex/cyclosome complex, which is active during the metaphase–anaphase transition (37). We have previously demonstrated that several MCM subunits bind to and promote the proteasomal degradation of HIF-1α independent of their helicase activity (32). Induction of cellular quiescence led to decreased levels of MCM proteins and increased HIF-1α levels, whereas stimulation of cellular proliferation led to an MCM-dependent decrease in HIF-1 activity (32). However, MCM protein levels do not vary with particular phases of the cell cycle. The results of the present study indicate that under conditions of cell proliferation, degradation of HIF-1α to allow DNA replication is mediated by the lysosome. This is the first report, to our knowledge, implicating lysosome function as a regulator of cell-cycle progression.
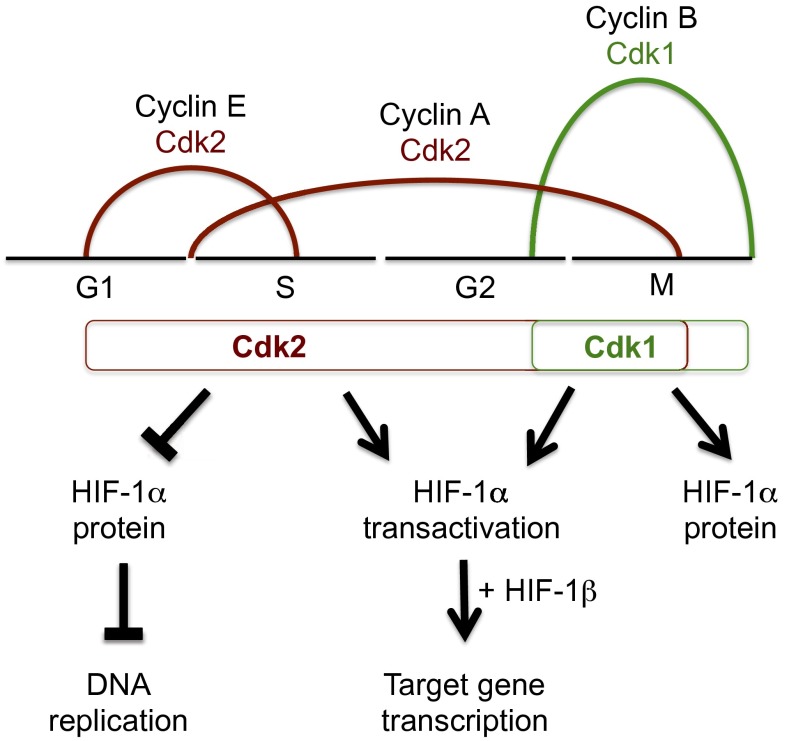
Our results suggest the following model (Fig. 8): Cdk2–cyclin E, which is active in late G1/early S phase, stimulates HIF-1α degradation by chaperone-mediated autophagy to overcome the nontranscriptional inhibitory effect of HIF-1α on DNA replication. In cancer cells, Cdk2–cyclin E activity also stimulates HIF-1 transactivation function to compensate for the effect of reduced HIF-1α protein levels on HIF-1 target gene transcription. Because Cdk1–cyclin B is only active in late-G2/M phase, it is not involved in the regulation of DNA replication by HIF-1α and serves mainly to counteract the negative effect of Cdk2–cyclin A on HIF-1α stability and to further increase HIF-1 target gene transcription in late-G2/early M phase. During late M phase, Cdk1–cyclin B is unopposed, allowing for high levels of HIF-1 transcriptional activity. In addition, high levels of HIF-1α at the beginning of G1 may promote the initial steps of MCM helicase loading, although MCM helicase activation requires the subsequent degradation of HIF-1α at the G1/S phase transition. This mechanism likely underlies past observations that HIF-1α activity fluctuates when arrested cells are induced to proceed through the cell cycle (32, 47).
Fig. 8.
Cdk activity couples HIF-1α levels to the cell cycle. Cdk2:cyclin E and Cdk2:cyclin A complexes target HIF-1α for lysosomal degradation during the G1/S phase transition and throughout the S phase, respectively, thereby promoting DNA replication and cell-cycle progression. In cancer cell lines, Cdk2 has a positive effect on HIF-1α transactivation domain function, thereby promoting HIF-1 target gene expression. Cdk1:cyclin B complexes promote HIF-1α stabilization and HIF-1 target gene expression at the conclusion of the S phase by blocking lysosomal degradation of HIF-1α. Cell-cycle phases are coupled to HIF-1α levels through Cdk activity, thereby allowing cell proliferation while simultaneously inducing the adaptive HIF-1 transcriptional response.
HIF-1α induction leads to an arrest of DNA replication through two main mechanisms. The first is an immediate and nontranscriptional mechanism, which is mediated through binding of HIF-1α to Cdc6 and the MCM complex, leading to inhibition of helicase activation (31). The second mechanism relies on sequestration of c-Myc and subsequent induction of the cell-cycle inhibitors p21 and p27 (33). The mechanism by which certain cancer cell lines maintain proliferation while expressing HIF-1α protein has been a subject of investigation and did not seem to correlate with levels of HIF-1α, p21, or p27 levels (44). Previous studies have suggested that HIF-2α may promote cellular proliferation, with the relative balance between HIF-2α and HIF-1α determining the response to hypoxia (48). Supporting data for this hypothesis are particularly strong in renal cell carcinoma, where cancers expressing HIF-2α have been shown to have higher proliferation rates than those expressing both HIF-1α and HIF-2α (49). However, both HIF-1α and HIF-2α bind to the MCM helicase (32) and overexpression of either protein inhibits proliferation in other cell types (36). We found that Hep3B cells maintained proliferation upon hypoxic induction of HIF-1α and HIF-2α, but lysosome inhibitors led to potent cell-cycle arrest, which was dependent on HIF-1α and HIF-2α. This finding confirms that Hep3B cells are sensitive to the effects of HIF-1α on DNA replication, but use lysosomal degradation of HIF-1α to maintain cell proliferation under hypoxic conditions. The ability to degrade HIF-1α and HIF-2α before DNA replication through chaperone-mediated autophagy may maintain cancer cell proliferation in the hypoxic tumor microenvironment, and partially explain recent results implicating chaperone-mediated autophagy in tumor growth (50).
The effect of Cdk2 on HIF-1α is particularly interesting as it promotes lysosomal degradation of HIF-1α while increasing HIF-1α transactivation domain function in cancer cells. Cdk2 can thereby functionally uncouple the transcriptional effects of HIF-1, which promote metabolic adaptation, from the nontranscriptional effect of HIF-1α on DNA replication, which arrests the cell cycle. This arrangement is highly unusual but remarkably similar to the effect of S-phase kinase-associated protein 2 (Skp2) on the c-Myc transcription factor, whereby Skp2 promotes c-Myc degradation while acting as a cofactor for its transcriptional activity (51, 52). c-Myc is similar to HIF-1α, as it is a transcription factor with a nontranscriptional role in DNA replication, although it functions as an activator rather than an inhibitor of MCM helicase activity (53). Thus, it appears that regulation of HIF-1α by Cdk2 is analogous to regulation of c-Myc by Skp2 and indicates that mammalian cells have evolved mechanisms to differentially regulate the nontranscriptional and transcriptional functions of HIF-1α and c-Myc. A recent study demonstrated that the Hcm1 transcription factor was phosphorylated by Cdk1, which increased Hcm1 transcriptional activity through phosphorylation of the N terminus and targeted it for degradation through phosphorylation of the C terminus, suggesting another instance whereby transcription factor activity and abundance are dissociated (54).
We demonstrated that pharmacological inhibition of Cdk1 was associated with decreased induction of HIF-1α and HIF-1 target genes, which is in agreement with a recent study (47). Previous studies revealed that the Cdk inhibitor flavopiridol decreased HIF-1α levels and HIF target gene expression (55, 56). This effect was noted to be proteasome-independent, although the molecular mechanism was not delineated (55). Cdk inhibitors have long been of interest as anticancer agents (57). Our results lend mechanistic support for the use of Cdk1 inhibitors in tumors with high levels of HIF-1α, given their dual effects as inhibitors of HIF-1α transcriptional activity and of cell-cycle progression.
Materials and Methods
Tissue Culture.
HeLa and Hep3B cell lines and MEFs were cultured in DMEM, and HCT116 cells were cultured in McCoy’s 5A medium, all supplemented with 10% (vol/vol) FBS and penicillin/streptomycin. Cells were maintained at 37 °C in a 5% (vol/vol) CO2, 95% air incubator. Cells were subjected to hypoxia by exposure to 1% O2/5% CO2/balance N2 at 37 °C in a modular incubator chamber (Billups–Rothenberg). DMOG (Sigma–Aldrich) was used at a concentration of 1 mM. Hep3B–shEV and Hep3B–DKD cells were described previously (45). HCT116–shEV and HCT116–shHIF-1α cells were described previously (31). Roscovitine, flavopiridol, and Cdk1 inhibitor were obtained from Sigma–Aldrich.
Reporter Assays.
A total of 20,000 HeLa or Hep3B cells were seeded onto 24-well plates and 48 h after seeding were transfected with plasmid DNA using PolyJet (SignaGen). Control reporter pSV-RL (10 ng), HIF-1–dependent reporter p2.1 (120 ng), and expression vectors were cotransfected. For transactivation assays, pSV-RL (10 ng), pG5-E1b-Luc (100 ng), and expression vectors were cotransfected. The cells were lysed, and luciferase activities were determined with a multiwell luminescence reader (Perkin–Elmer Life Science) using a dual luciferase reporter assay system (Promega).
Immunoprecipitation and Immunoblot Assays.
Cells were lysed in PBS with 0.1% Tween 20, 1 mM DTT, protease inhibitor mixture, 1 mM Na3VO4, and 10 mM NaF, followed by gentle sonication. For immunoprecipitation assays, 2 μg of antibody and 30 µL of protein G-Sepharose beads (GE Healthcare) were incubated with 2 mg of cell lysate overnight at 4 °C. Beads were washed four times in lysis buffer. Proteins were eluted in SDS sample buffer and fractionated by SDS–polyacrylamide gel electrophoresis. The following antibodies were used in immunoblot and immunoprecipitation assays: histone H3 and β-actin (Santa Cruz Biotechnology), HIF-1α (BD Biosciences), FLAG (Sigma), and IgG, Cdk1, Cdk2, Mcm2, Mcm5, Mcm7, phospho-Mcm2, Lamp-2A, and HIF-2α (Novus Biologicals).
RT-qPCR Assays.
Total RNA was extracted from 293T cells using TRIzol (Invitrogen) and treated with DNase I (Ambion). A 1-μg aliquot was used for first-strand synthesis with the iScript cDNA Synthesis System (Bio-Rad). The qPCR assays were performed with iQ SYBR Green Supermix and iCycler Real-Time PCR Detection System (Bio-Rad). Primer sequences are listed in Table S1. The induced expression (E) of each target mRNA, normalized to 18S rRNA in each sample, was calculated based on the threshold cycle (Ct) as E = 2−∆(∆Ct), where ∆Ct = Ct(target) – Ct(18S) and ∆(∆Ct) = ∆Ct(control) – ∆Ct(treatment).
Immunofluorescence Assay.
Cells were plated on gelatin-coated glass-bottomed plates (Live Assay). Posttreatment, samples were washed with ice-cold PBS, fixed with 4% (wt/vol) paraformaldehyde for 20 min at room temperature, permeabilized with 0.05% Triton X-100 for 15 min, washed twice with PBS, and blocked with 10% (vol/vol) goat serum and 1% AlbuMAX (Invitrogen) for 1 h. Samples were incubated with rabbit polyclonal anti-MCM5 (Santa Cruz) and sheep polyclonal anti-BrdU (Abcam) primary antibodies for 1 h, washed, and incubated with Alexa Fluor-conjugated secondary antibodies (Invitrogen) for 1 h (27). Samples were washed and mounted on microscope slides with a drop of SlowFade (Invitrogen) and sealed with Vectashield (Vector Labs). Samples were imaged within 2 d postpreparation using a Nikon A1R confocal microscope with a 60× oil immersion objective and 1.4 numerical aperture. Images were analyzed using Nikon Elements Software (Nikon Instruments).
Chromatin Isolation.
Chromatin fractions were isolated as previously described (31). Briefly, cells were washed with PBS, pelleted, and lysed with cytoskeleton buffer [20 mM Hepes (pH 7.8), 10 mM KCl, 2 mM EDTA, 300 mM sucrose, and 0.5% Triton X-100, supplemented with protease inhibitors and phenylmethylsulfonyl fluoride]. After incubation on ice for 10 min, samples were centrifuged, and the pellet was isolated. This process was repeated twice, after which the pellets were suspended in 10 mM Hepes (pH 7.8), 2 mM EDTA, 0.3 mM EGTA, and 1 mM DTT. Samples were sonicated, and protein concentrations were normalized before immunoblot assays were performed.
Statistical Analysis.
Data are presented as mean ± SEM, except where otherwise noted. Differences between two conditions were analyzed using Student's t test.
Supplementary Material
Acknowledgments
We are grateful to Dr. Mariano Barbacid (Centro Nacional de Investigaciones Oncológicas) for the gift of Cdk2 KO and wild-type MEFs and Dr. Peter Sicinski (Dana–Farber Cancer Institute) for the gift of cyclin E1/E2 double-KO and wild-type MEFs. We thank Karen Padgett (Novus Biologicals) for providing IgG, Cdk1, Cdk2, Mcm2, Mcm5, Mcm7, phospho-Mcm2, Lamp-2A, and HIF-2α antibodies. D.M.G. is supported by funding from the National Cancer Institute (K99CA181352). G.L.S. is an American Cancer Society Research Professor and the C. Michael Armstrong Professor at the Johns Hopkins University School of Medicine.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412840111/-/DCSupplemental.
References
- 1.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iyer NV, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 α. Genes Dev. 1998;12(2):149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola P, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 6.Ivan M, et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 7.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295(5556):858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 8.Ebert BL, Firth JD, Ratcliffe PJ. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct Cis-acting sequences. J Biol Chem. 1995;270(49):29083–29089. doi: 10.1074/jbc.270.49.29083. [DOI] [PubMed] [Google Scholar]
- 9.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Forsythe JA, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16(9):4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci USA. 2000;97(16):9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilkes DM, et al. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol Cancer Res. 2013;11(5):456–466. doi: 10.1158/1541-7786.MCR-12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Gilkes DM, et al. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73(11):3285–3296. doi: 10.1158/0008-5472.CAN-12-3963. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92(3):967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YV, et al. RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol Cell. 2007;25(2):207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YV, et al. Calcineurin promotes hypoxia-inducible factor 1α expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J Biol Chem. 2007;282(51):37064–37073. doi: 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo W, et al. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J Biol Chem. 2010;285(6):3651–3663. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh MY, Darnay BG, Powis G. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1α, leading to its oxygen-independent degradation. Mol Cell Biol. 2008;28(23):7081–7095. doi: 10.1128/MCB.00773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbi ME, Hu H, Kshitiz, Gilkes DM, Semenza GL. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J Biol Chem. 2013;288(29):20768–20775. doi: 10.1074/jbc.M113.476903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montagner M, et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487(7407):380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- 22.Baek JH, et al. Spermidine/spermine N(1)-acetyltransferase-1 binds to hypoxia-inducible factor-1α (HIF-1α) and RACK1 and promotes ubiquitination and degradation of HIF-1α. J Biol Chem. 2007;282(46):33358–33366. doi: 10.1074/jbc.M705627200. [DOI] [PubMed] [Google Scholar]
- 23.Ravi R, et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α. Genes Dev. 2000;14(1):34–44. [PMC free article] [PubMed] [Google Scholar]
- 24.Flugel D, Gorlach A, Michiels C, Kietzmann T. Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1α and mediated its destabilization in a VHL-independent manner. Mol Cell Biol. 2007;27(9):3252–3265. doi: 10.1128/MCB.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D, Yao Y, Lu L, Costa M, Dai W. Plk3 functions as an essential component of the hypoxia regulatory pathway by direct phosphorylation of HIF-1α. J Biol Chem. 2010;285(50):38944–38950. doi: 10.1074/jbc.M110.160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang X, et al. PIASy stimulates HIF1α SUMOylation and negatively regulates HIF1α activity in response to hypoxia. Oncogene. 2010;29(41):5568–5578. doi: 10.1038/onc.2010.297. [DOI] [PubMed] [Google Scholar]
- 27.Hubbi ME, et al. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J Biol Chem. 2013;288(15):10703–10714. doi: 10.1074/jbc.M112.414771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira JV, et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9(9):1349–1366. doi: 10.4161/auto.25190. [DOI] [PubMed] [Google Scholar]
- 29.Egger ME, Huang JS, Yin W, McMasters KM, McNally LR. Inhibition of autophagy with chloroquine is effective in melanoma. J Surg Res. 2013;184(1):274–281. doi: 10.1016/j.jss.2013.04.055. [DOI] [PubMed] [Google Scholar]
- 30.Yan ZW, Hou JK, He W, Fan L, Huang Y. Chloroquine enhances cobalt chloride-induced leukemic cell differentiation via the suppression of autophagy at the late phase. Biochem Biophys Res Commun. 2013;430(3):926–932. doi: 10.1016/j.bbrc.2012.12.052. [DOI] [PubMed] [Google Scholar]
- 31.Hubbi ME, et al. A nontranscriptional role for HIF-1α as a direct inhibitor of DNA replication. Sci Signal. 2013;6(262):ra10. doi: 10.1126/scisignal.2003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubbi ME, Luo W, Baek JH, Semenza GL. MCM proteins are negative regulators of hypoxia-inducible factor 1. Mol Cell. 2011;42(5):700–712. doi: 10.1016/j.molcel.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koshiji M, et al. HIF-1α induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23(9):1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goda N, et al. Hypoxia-inducible factor 1α is essential for cell cycle arrest during hypoxia. Mol Cell Biol. 2003;23(1):359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takubo K, et al. Regulation of the HIF-1α level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7(3):391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 36.Hackenbeck T, et al. HIF-1 or HIF-2 induction is sufficient to achieve cell cycle arrest in NIH3T3 mouse fibroblasts independent from hypoxia. Cell Cycle. 2009;8(9):1386–1395. doi: 10.4161/cc.8.9.8306. [DOI] [PubMed] [Google Scholar]
- 37.Costa A, Hood IV, Berger JM. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha P, et al. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18(5):2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei M, et al. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11(24):3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semenza GL, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 41.Martín A, et al. Cdk2 is dispensable for cell cycle inhibition and tumor suppression mediated by p27(Kip1) and p21(Cip1) Cancer Cell. 2005;7(6):591–598. doi: 10.1016/j.ccr.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Geng Y, et al. Cyclin E ablation in the mouse. Cell. 2003;114(4):431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 43.Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1α. Modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272(31):19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 44.Box AH, Demetrick DJ. Cell cycle kinase inhibitor expression and hypoxia-induced cell cycle arrest in human cancer cell lines. Carcinogenesis. 2004;25(12):2325–2335. doi: 10.1093/carcin/bgh274. [DOI] [PubMed] [Google Scholar]
- 45.Hubbi ME, Gilkes DM, Baek JH, Semenza GL. Four-and-a-half LIM domain proteins inhibit transactivation by hypoxia-inducible factor 1. J Biol Chem. 2012;287(9):6139–6149. doi: 10.1074/jbc.M111.278630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leontieva OV, et al. Hypoxia suppresses conversion from proliferative arrest to cellular senescence. Proc Natl Acad Sci USA. 2012;109(33):13314–13318. doi: 10.1073/pnas.1205690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warfel NA, Dolloff NG, Dicker DT, Malysz J, El-Deiry WS. CDK1 stabilizes HIF-1α via direct phosphorylation of Ser668 to promote tumor growth. Cell Cycle. 2013;12(23):3689–3701. doi: 10.4161/cc.26930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11(4):335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordan JD, et al. HIF-α effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14(6):435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kon M, et al. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med. 2011;3(109):ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11(5):1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 52.von der Lehr N, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11(5):1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 53.Dominguez-Sola D, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448(7152):445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 54.Landry BD, Mapa CE, Arsenault HE, Poti KE, Benanti JA. Regulation of a transcription factor network by Cdk1 coordinates late cell cycle gene expression. EMBO J. 2014;33(9):1044–1060. doi: 10.1002/embj.201386877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melillo G, et al. Flavopiridol, a protein kinase inhibitor, down-regulates hypoxic induction of vascular endothelial growth factor expression in human monocytes. Cancer Res. 1999;59(21):5433–5437. [PubMed] [Google Scholar]
- 56.Newcomb EW, et al. Flavopiridol downregulates hypoxia-mediated hypoxia-inducible factor-1α expression in human glioma cells by a proteasome-independent pathway: Implications for in vivo therapy. Neuro-oncol. 2005;7(3):225–235. doi: 10.1215/S1152851704000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cicenas J, Valius M. The CDK inhibitors in cancer research and therapy. J Cancer Res Clin Oncol. 2011;137(10):1409–1418. doi: 10.1007/s00432-011-1039-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.