Significance
Hexaploid bread wheat is generally more salt tolerant than its tetraploid progenitor. However, the physiological bases and the relative contributions of immediate effects of polyploidization and subsequently acquired adaptive changes in the salt tolerance of hexaploid wheat remained elusive. This study compared a large suite of morphophysiological traits in synthetic and natural hexaploid wheats, and their tetraploid and diploid progenitors, under normal and salt-stressed conditions, and studied subgenome-specific expression of a critical salt-tolerance gene, HKT1;5. Results have thrown light on the major physiological bases underlying salt tolerance of hexaploid wheat and revealed polyploidy-induced alteration in gene regulation under salt stress. These allopolyploidization-induced immediate molecular and physiological changes showed evolutionary perseverance and hence bear implications for polyploid crop improvement.
Keywords: transcriptional rewiring, Na+ homeostasis, salinity tolerance
Abstract
Hexaploid bread wheat (Triticum aestivum L., genome BBAADD) is generally more salt tolerant than its tetraploid wheat progenitor (Triticum turgidum L.). However, little is known about the physiological basis of this trait or about the relative contributions of allohexaploidization and subsequent evolutionary genetic changes on the trait development. Here, we compared the salt tolerance of a synthetic allohexaploid wheat (neo-6x) with its tetraploid (T. turgidum; BBAA) and diploid (Aegilops tauschii; DD) parents, as well as a natural hexaploid bread wheat (nat-6x). We studied 92 morphophysiological traits and analyzed homeologous gene expression of a major salt-tolerance gene High-Affinity K+ Transporter 1;5 (HKT1;5). We observed that under salt stress, neo-6x exhibited higher fitness than both of its parental genotypes due to inheritance of favorable traits like higher germination rate from the 4x parent and the stronger root Na+ retention capacity from the 2x parent. Moreover, expression of the D-subgenome HKT1;5 homeolog, which is responsible for Na+ removal from the xylem vessels, showed an immediate transcriptional reprogramming following allohexaploidization, i.e., from constitutive high basal expression in Ae. tauschii (2x) to salt-induced expression in neo-6x. This phenomenon was also witnessed in the nat-6x. An integrated analysis of 92 traits showed that, under salt-stress conditions, neo-6x resembled more closely the 2x than the 4x parent, suggesting that the salt stress induces enhanced expressivity of the D-subgenome homeologs in the synthetic hexaploid wheat. Collectively, the results suggest that condition-dependent functionalization of the subgenomes might have contributed to the wide-ranging adaptability of natural hexaploid wheat.
Polyploidy or whole genome duplication (WGD) is a pervasive, driving force in plant and vertebrate evolution, which has fascinated biologists for more than a century (1, 2). The common occurrence of WGD suggests an evolutionary advantage of having multiple genomes at least in certain circumstances, which might have enabled the polyploid organisms to be better adapted to some adverse environmental conditions than their diploid progenitors (3, 4). Polyploidy can instantaneously develop novel features that allow them to invade new territories or expand their parental niche (3). Polyploids may also exhibit higher evolvability than their diploid progenitors, which allows them to adapt to capricious environmental conditions (4). Thus, polyploidy has been demonstrated as a process that may lead to saltational speciation especially when novel ecological niches are available for colonization.
Although abrupt genome duplication often produces adverse effects on physiology at both cellular and organismal levels (4, 5), it has been shown that polyploidy in plants may result in favorable physiological consequences such as increased photosynthetic capacity and enhanced tolerance to biotic and abiotic stresses, which can be immediately penetrant upon WGD or emerge in the due course of evolution. For example, Coate et al. (6) showed that tolerance to chronic light stress was significantly increased in a newly formed allotetraploid of soybean (Glycine dolichocarpa) compared with its diploid progenitors. Neoallopolyploid tobacco (Nicotiana miersii × Nicotiana attenuata) showed an innovation of defense against herbivores (7). Both natural and artificial Arabidopsis tetraploids exhibit higher potassium uptake and salinity tolerance than their diploid counterparts (8). These studies have suggested a direct and instantaneous effect of WGD on stress tolerance, which can facilitate physiological differentiation of the neopolyploid from its diploid or lower-ploidal progenitors and establishment as a new species. Nevertheless, our understanding of the physiological bases underlying the instantaneous effects of WGD and their evolutionary consequences are still limited in major crops.
Salinity is a severe problem, affecting more than 800 million hectares of land worldwide that accounts for more than 6% of the global land mass (9). It is well known that hexaploid bread wheat generally shows higher salt tolerance than its tetraploid progenitor, Triticum turgidum (9). However, little is known about the physiological differences between hexaploid and tetraploid wheats and the relative importance of nascent allopolyploidization vs. subsequent evolutionary changes in conferring salt tolerance to hexaploid wheat.
The higher ploidal level and complex genome composition of hexaploid wheat provided it greater physiological and ecological plasticity than its tetraploid and diploid progenitors (10, 11). Owning to its remarkable adaptability, hexaploid wheat has spread rapidly throughout the globe after its inception in the Middle East and became established as a primary grain crop that feeds roughly one-third of the world’s population (10). Hexaploid wheat is a relatively young species that formed nearly 10,000 y ago by hybridization of a domesticated form of allotetraploid wheat (T. turgidum, genome BBAA) and a diploid goat grass Aegilops tauschii (genome DD) (10, 12). Due to the short evolutionary history and the still extant progenitor species, early stages in the evolutionary history of allohexaploid wheat can be largely recapitulated by reconstituting genomically equivalent neoallohexaploid plants via artificial hybridization and induced WGD.
We reasoned that, being a very young species, the wide adaptability of hexaploid wheat is unlikely to be acquired solely by accumulation of genetic variations in the course of evolution and/or domestication. Therefore, a major role of instantaneous effects of allohexaploidization and their evolutionary perseverance on the wheat adaptability is expected. To test how allopolyploidization has immediately affected the fitness of hexaploid wheat under salt stress conditions, we compared salt responses of a synthetic allohexaploid wheat (neo-6x; genome BBAADD), its parental genotypes, Ae. tauschii (genome DD) (2x) and T. turgidum L. (genome BBAA) (4x) with a natural hexaploid wheat (nat-6x). We measured in parallel 92 morphophysiological traits closely related to carbon/nitrogen metabolism, photosynthesis, and salt tolerance in these genotypes and assayed homeolog-specific expression of a critical salt-tolerance gene, High-Affinity K+ Transporter 1;5 (HKT1;5) (13), under both normal and salt-stressed conditions. Our results have suggested major physiological bases for the acquisition and evolution of salt tolerance in hexaploid wheat, which were likely contingent upon an immediate transition, following allohexaploidization, of regulatory control of the D-subgenome homeolog of the HKT1;5 gene that is known to play an important role for conferring salt tolerance in natural hexaploid wheat (13). Collectively, our results have provided insights into the immediate and long-term physiological consequences of allohexaploidization in wheat, which might have practical implications for genetic improvement of salt tolerance in wheat and other crops.
Results
Enhanced Fitness of Hexaploid Wheat Compared with Its Tetraploid and Diploid Progenitors Under Salt Stress.
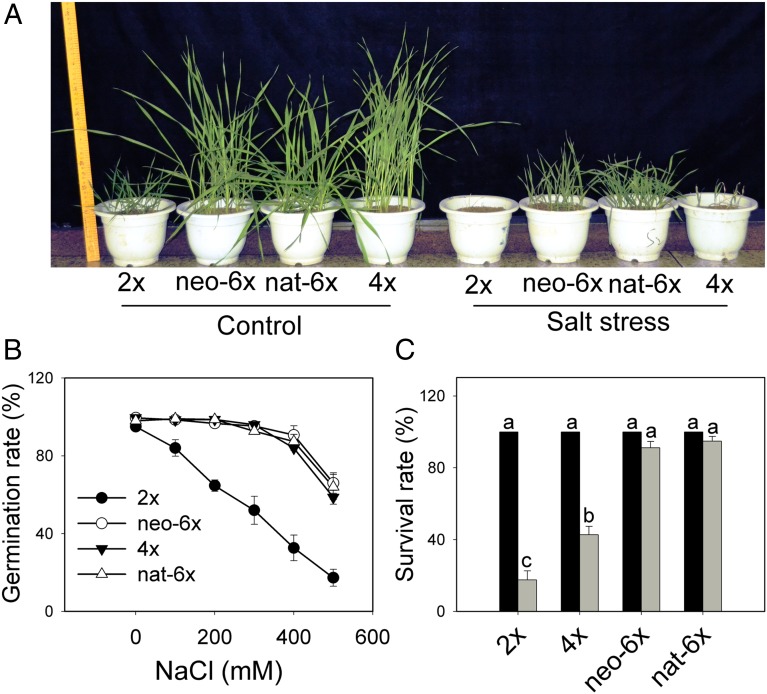
To assess the behavior under salt stress, we subjected plants of a synthetic allohexaploid wheat (neo-6x), its tetraploid (T. turgidum, 4x) and diploid (Ae. tauschii, 2x) parents, and a natural allohexaploid wheat (Triticum aestivum, nat-6x) to a constant salt stress: from seed germination to seedling growth for 30 d. We defined this treatment as type I salt stress (Fig. 1A and SI Materials and Methods), which mimics the natural saline habitats. We quantified germination and survival rates for each of the four genotypes under normal and salt-stress conditions. We found that under normal conditions, both germination and survival rates of the four genotypes were similar (Fig. 1 B and C). However, under salt stress, the germination rate of 4x was similar to those of neo-6x and nat-6x (Fig. 1B), but its survival rate was significantly lower than the hexaploid wheats, as most of the emerged seedlings of 4x died within 30 d (Fig. 1C). The latter observation can be largely attributed to its poor Na+ retention within roots, as detailed below. Under salt-stress conditions the germination rate of Ae. tauschii (2x) was much lower (Fig. 1B and Fig. S1). Collectively, neo-6x and nat-6x showed much higher survival rates than both 2x and 4x (Fig. 1 A and C). Because only a small number of 2x and 4x plants survived under the type I salt-stress treatment, we performed another stress treatment by subjecting the 30-d-old seedlings (starting from sprouting) of the four genotypes to salt stress for a set duration, which we defined as type II salt stress (SI Materials and Methods). We found that when seedlings were subjected to type II salt stress for 32 d, 2x, nat-6x, and neo-6x all showed better growth than 4x (Fig. S2). Furthermore, the salt stress strongly hampered spikelet development in 4x, but did not affect this trait in both synthetic and natural hexaploid wheats (Fig. S3). As 2x required vernalization for flowering, it was not suitable to include 2x in this comparison, as it had involved a cold treatment. Observations of both types of salt stress taken together, it was clear that hexaploid wheat (both neo-6x and nat-6x) had a higher fitness than both of its tetraploid and diploid progenitors under salt stress, most likely by inheriting the higher germination rate from the former (as observed in type I salt-stress treatment) and the stronger salt tolerance of the latter (as observed in type II salt-stress treatment).
Fig. 1.
Effects of salt stress on survivorship and germination rate of four genotypes: a synthetic allohexaploid wheat (neo-6x), its diploid parent, Aegilops tauschii (2x), its tetraploid parent, Tritium turgidum (4x), and a natural hexaploid bread wheat, T. aestivum cv. Chinese Spring (nat-6x). (A) An example of plant growth of four genotypes subjected to continuous salt stress (150 mM NaCl) for 30 d from seed germination (type I salt stress). (B) The germination rate was determined and the values are means (±SE) of three biological replicates. (C) Survival rate (%) of the four genotypes under normal (black bars) and salt-stressed (gray bars) conditions. The values were means (±SE) of six biological replicates. Means followed by different letters among genotypes under the same treatment (normal or stress) were significantly different according to a least significant difference (LSD) test (P < 0.05).
The D Subgenome Positively Contributes to the Strong Capacity for Na+ Retention by Hexaploid Wheat Roots Under Salt Stress, Which Is Concomitant with Regulatory Transition of a Critical Salt-Tolerance Gene.
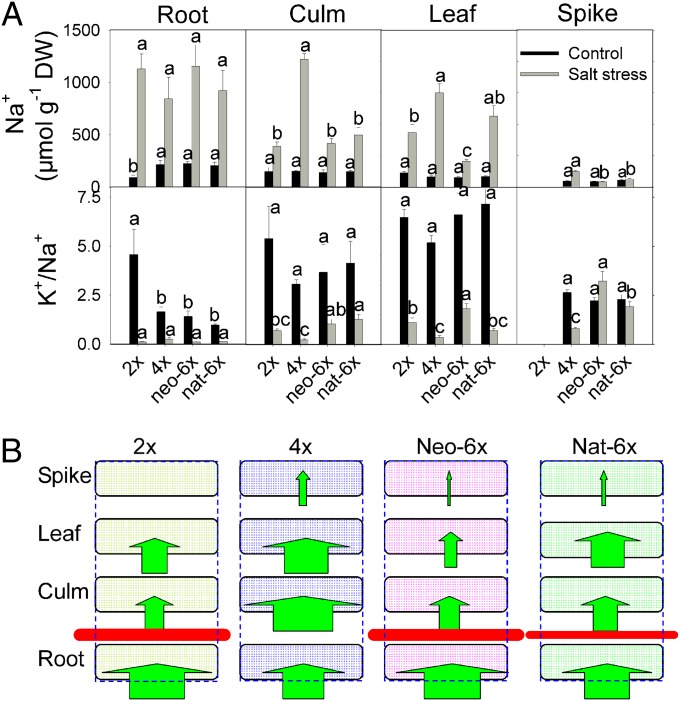
Low Na+ and high K+/Na+ ratio in the cytoplasm are essential to maintain ion homeostasis under both normal and stress conditions. Under normal conditions, Na+ content was similar in the four genotypes in roots, leaves, culms, and spikes; however, roots of 2x showed a lower Na+ content than the other three genotypes and exhibited a reverse trend for K+/Na+ ratio (Fig. 2A). Under type II salt stress, there was no difference in root Na+ content and K+/Na+ ratios of the four genotypes. In contrast, culms and leaves of neo-6x, nat-6x, and 2x showed much lower Na+ content and higher K+/Na+ ratios than 4x (Fig. 2A). In spikes, nat-6x and neo-6x (spikes were not examined for 2x, as explained above) also had much lower Na+ contents and higher K+/Na+ ratios than 4x (Fig. 2A). We further compared differences in Na+ content among the four organs for a given genotype under salt stress and found that Na+ content in 4x was much lower in roots than in culms, suggesting a poor Na+ retention capacity of roots in the 4x (Fig. 2B). In contrast, Na+ content was much higher in roots than in the other organs in the other three genotypes, suggesting a strong Na+ retention capacity of roots in these genotypes (Fig. 2B). Collectively, these results suggested that 2x, neo-6x, and nat-6x all possessed a mechanism whereby Na+ was efficiently sequestered within roots that interrupted its transportation to the above ground organs, but 4x did not possess this mechanism. Thus, immediately after allohexaploidization, neo-6x has acquired a stronger capacity from 2x to retain Na+ within roots, which might have attributed to its strong salt tolerance, and this trait has been largely maintained in natural hexaploid wheat.
Fig. 2.
Effects of salt stress on the Na+ content, K+/Na+ ratio, and Na+ influx among organs in the neo-6x, its diploid (2x), and tetraploid (4x) parents, and nat-6x. To take these measurements, the seedlings for all genotypes were first grown under normal conditions for 30 d and then subjected to salt stress (150 mM NaCl) for 32 d (type II stress). The values are means (±SE) of four biological replicates, and each replicate consisted of a pool of five plants. (A) Na+ content and K+/Na+ ratio; means followed by different letters among genotypes of the same treatment are significantly different according to LSD (P < 0.05). (B) Na+ influx among organs under salt stress; green arrows indicate Na+ influx. The width of arrows denotes relative concentrations of Na+ in a given organ, according to data in Fig. 2A. The horizontal red bars denote Na+ retention by roots, and difference in retention capacities of the individual genotypes was depicted by the relative thickness of the bars.
To investigate the possible molecular basis for this physiological phenomenon, we examined expression of the HKT1;5 gene, which encodes one of a few characterized Na+ transporters in wheat. HKT1;5 functions to remove excess Na+ from the xylem vessels, thereby keeping Na+ below toxic levels in the photosynthetic tissues by avoiding its transport to the leaves (9, 13). Hexaploid wheat has two HKT1;5 homeologs, HKT1;5-B and HKT1;5-D, which map to chromosomes 4B and 4D, respectively (14). The A subgenome of tetraploid and hexaploid wheats, which was contributed by Triticum urartu (genome AuAu), lacks the HKT1;5 gene (14). However, the cultivated diploid einkorn wheat Triticum monococcum (genome AmAm) was shown to possess a copy of the HKT1;5 gene (13). Moreover, HKT1;5-B, albeit present, may have lost its function for salt tolerance because tetraploid wheat (T. turgidum) is Na+ sensitive, and introgression of HKT1;5-A from T. monococcum (genome AmAm) to a tetraploid wheat was shown to provide it with Na+ tolerance similar to that of hexaploid wheat (13). These prior studies have established that HKT1;5-D plays a key role in mediating Na+ tolerance in natural hexaploid wheat (Fig. S4A). We coupled quantitative RT-PCR (qRT-PCR) and the TaqMan SNP genotyping assay to quantify the total as well as B and D subgenome-specific expression of HKT1;5. We made the following observations: (i) HKT1;5-D showed constitutive high expression levels in both leaves and roots of 2x, which did not show significant alteration in response to salt stress (Fig. S4B). This suggests that the high level of salt tolerance of 2x, if conditioned by this gene, was due to its constitutive high basal expression rather than stress induction. (ii) HKT1;5-B showed similar constitutive high expression in 4x as HKT1;5-D in 2x, except that it showed further up-regulation in leaves under salt stress (Fig. S4B). Given that 4x was Na+ sensitive, our results were therefore consistent with the earlier suggestion that HKT1;5-B was not functional with respect to salt tolerance (13). (iii) HKT1;5-D and HKT1;5-B showed markedly lower expression in both leaves and roots of both neo-6x and nat-6x than 2x and 4x, respectively, under normal conditions, but both genes showed significant up-regulation in roots but not leaves of neo-6x and nat-6x under salt stress (Fig. S4B). Taken together, our results, on the one hand, were consistent with previous findings showing that HKT1;5-D plays a major role in conditioning Na+ tolerance in natural hexaploid wheat; and on the other hand, our results demonstrated that in contrast to the constitutive high expression pattern of HKT1;5-D observed in Ae. tauschii, expression of this gene in hexaploid wheat has undergone a regulatory transition, i.e., from constitutive high basal expression to induced high expression upon salt stress. Importantly, this immediately rewired expression pattern of HKT1;5-D following allohexaploidization showed evolutionary conservation in natural hexaploid wheat (nat-6x) (Fig. S4B).
Hexaploid Wheat Showed Transgressive Performance in Photosynthesis and Nitrogen Metabolism Relative to Its Tetraploid and Diploid Progenitors Under Salt Stress.
We measured 10 parameters related to photosynthesis for each of the four genotypes under both normal and salt stress (type II) conditions (Fig. S5). Under normal conditions, nat-6x and neo-6x showed either no significant differences or lower values than the 2x and 4x for the 10 measured parameters (Fig. S5). Under salt stress, we observed that nat-6x and neo-6x had (i) higher stomatal conductance (gs) and net photosynthetic rate (PN) than both 2x and 4x (Fig. S5); (ii) much higher chlorophyll and starch contents than 4x but similar to 2x (Fig. S5); and (iii) much lower electrolyte leakage rates than 4x but similar to 2x (Fig. S5), suggesting more severe salt damage to membranes in 4x than in the other three genotypes. Higher PN in neo-6x and nat-6x might be due to their higher stomata conductance (gs) and chlorophyll accumulation, which, respectively, promote CO2 assimilation and water uptake (Fig. S5). Collectively, it is evident that hexaploid wheat (both neo-6x and nat-6x) had either inherited the advantageous photosynthetic traits from its progenitors or showed transgressive expression of the traits, culminating in the higher photosynthetic capacity in hexaploid wheat.
We further observed that the salt stress greatly reduced the accumulation of most of the 15 measured amino acids related to carbon/nitrogen metabolism in 4x, but the reductions were to a much lesser extent in the 2x, neo-6x, and nat-6x (Fig. S6 and Dataset S1). In particular, the extent of reduction for the three critical amino acids (Ser, Arg, and Asp) were significantly smaller in neo-6x and nat-6x than in both 2x and 4x (Fig. S6). Specifically, the salt stress had (i) increased the Ser and Arg content in nat-6x and neo-6x, decreased it in 4x, but did not change it in 2x; and (ii) had no effect on the Asp content in nat-6x and neo-6x, but decreased it in both 2x and 4x (Fig. S6). Taken together, for each of the 15 measured amino acids under the salt stress, hexaploid wheat (neo- or nat-) either inherited traits related to carbon/nitrogen metabolism under salt stress from its diploid progenitor or showed transgressive expression, similar to the situation of photosynthetic traits. Under salt-stressed conditions, elevated concentrations of amino acids in the hexaploid wheat relative to their tetraploid and diploid progenitors not only plays an osmotic role (Dataset S1) but also serves as a source of carbon and nitrogen, both of which may help to cope with the salt stress. Notably, these traits were also expressed immediately following allohexaploidization and maintained during evolution of the natural hexaploid wheat.
Global Biological Effects of the Newly Added D Subgenome in the Synthetic Hexaploid Wheat Were Dramatically Augmented Under Salt Stress.
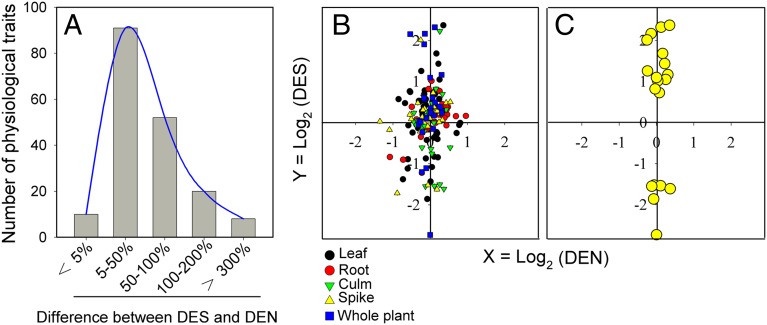
Given the multifaceted differences between hexaploid and tetraploid wheats, especially under salt-stressed conditions, we were interested in testing the biological consequences of adding the D subgenome in hexaploid wheat evolution. Because the exact founding parents of natural hexaploid wheats cannot be determined, the synthetic hexaploid wheat represents a tractable system for this analysis. Thus, in theory, if there were no effects of the D subgenome addition on a given morphophysiological trait in a newly synthesized hexaploid wheat (neo-6x) for which the exact parental genotypes are known and available for comparison, the neo-6x and its tetraploid parent (4x) are expected to show the same values. Conversely, if there were penetrable effects of the D-subgenome addition, the value of a given trait in neo-6x is expected to be different from that in 4x. We defined the effect of D subgenome on a physiological or morphological trait under normal and salt-stress conditions as DEN (D-subgenome effect under normal condition) and DES (D-subgenome effect under salt stress), respectively (Materials and Methods), and calculated these parameters for each of the 92 morphophysiological traits (detailed in Dataset S2) (Figs. 1 and 2, Figs. S1, S2, S3, S5, S6, and Dataset S3). We observed that for many of the measured traits, DES was statistically different (χ2 test, P < 0.05) from DEN (Fig. 3A), indicating that the salt stress substantially changed the effects of the D subgenome within the same hexaploid genetic background. To test how this D-subgenome–specific response to salt stress was manifested, we compared DES and DEN of each trait, and plotted Log2DEN (x axial) and Log2DES (y axial) for all of the 92 traits (Fig. 3B and Dataset S2). Thus, the icons aggregating to the y axis indicated that the traits had small differences between neo-6x and 4x under normal conditions, whereas the icons aggregating to x axis indicated that the traits had small differences between neo-6x and 4x under salt stress. We found that the majority of the icons were close to the y axis but separated away from the x axis, indicating that the difference between neo-6x and 4x for many of the 92 measured traits was small under normal conditions, but was significantly augmented under salt stress (Fig. 3B). In particular, this trend was even more pronounced for several key traits, for example, survival rate, chlorophyll content, photosynthesis, Na+ metabolism, free amino acids, and starch accumulation (Fig. 3C).
Fig. 3.
Augmented effects by the D subgenome of hexaploid wheat on physiological and morphological traits under salt stress. (A) Number of traits showing differences in the D-subgenome effect between normal conditions (DEN) and salt-stress conditions (DES); for a given trait, DEN = ratio of neo-6x's value/4x's value under normal conditions, and DES = ratio of neo-6x's value/4x's value under salt stress conditions. (B) DEN was plotted against DES on a logarithmic scale; icons represent given traits. For a given physiological performance, x = Log2(DEN) and y = Log2(DES). The icons aggregated to the y axis indicate that the physiological traits have small differences between neo-6x and 4x under normal conditions; the icons aggregated to the x axis indicate small differences between neo-6x and 4x under salt stress. (C) Distribution of several key physiological traits including survivorship, photosynthesis, chlorophyll content, Na+ metabolism, free amino acid (Tyr), and accumulation of starch. Detailed information of all measured traits is given in Dataset S2.
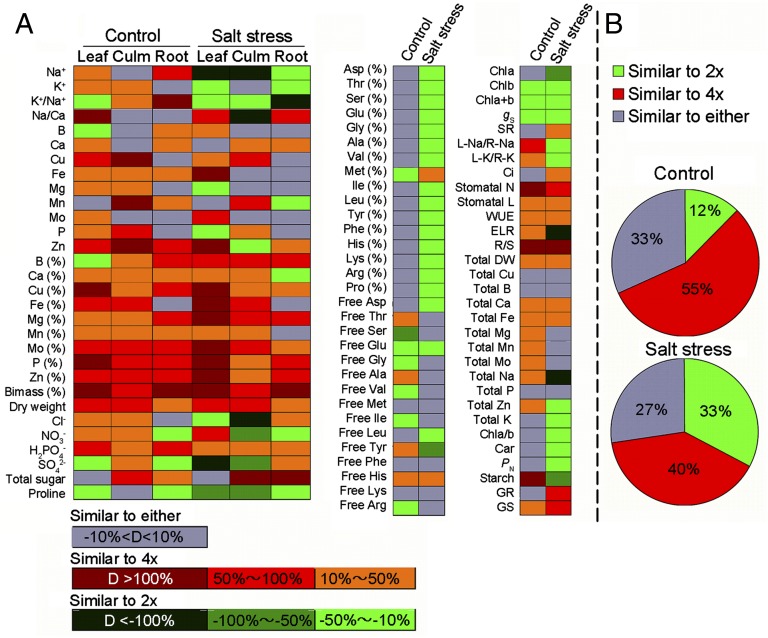
We next explored the possibility that the augmented effects of the D subgenome by the salt stress might have transformed neo-6x, being similar to its diploid parent 2x for the measured physiological traits under salt stress compared with the normal conditions. Indeed, we found that, under normal conditions, the majority of the traits of neo-6x were more similar to 4x than to 2x, as only 19 of 153 data points (traits × organs) were similar to 2x; in contrast, under the salt stress, 31 more data points were converted from being more similar to 4x (or to both) to being more similar to 2x (Fig. 4 A and B). Furthermore, this physiological conversion was organ specific (Fig. 4A).
Fig. 4.
Differentiation in physiological traits between neo-6x and its 2x and 4x parents in leaf, culm, or root. Difference between 2x and neo-6x was calculated by D2 = (2x − neo-6x)/neo-6x × 100%, and difference between 4x and neo-6x was calculated by D4 = (4x − neo-6x)/neo-6x × 100%. Thus, for a given physiological trait, the difference (D = |D2|−|D4|) between D2 and D4 indicates whether neo-6x was more similar to 2x or to 4x. Thus, |D2|>|D4| indicates neo-6x is more similar to 4x than 2x (red rectangles); conversely, |D2|<|D4| indicates neo-6x is more similar to 2x than 4x (green rectangles). For those traits in which D is in the range of −10% to +10%, it denotes neo-6x is similar to either 2x or 4x (gray rectangles). (A) All measured traits in three organs (leaf, culm, and root, Leftmost panel) and in one organ (leaf, Center and Rightmost panels). (B) Relative proportions of the above three situations. Chl, chlorophyll; Car, carotenoid; ELR, electrolyte leakage rate; WUE, water use efficiency; gs, stomatal conductance; PN, net photosynthetic rate; Ci, internal CO2 concentration; DW, dry weight; stomatal N, stomatal number; stomatal L, stomatal length; L-Na/R-Na, leaf Na+ content /root Na+ content; L-K/R-K, leaf K+ content/root K+ content; R/S, root DW/shoot DW; GR, germination rate; GS, germination speed; and SR, survival rate. Delineations of all physiological performances are shown in Dataset S2.
Discussion
Stronger Salt Tolerance of Hexaploid Wheat Was Likely Acquired Immediately Following Allohexaploidization and Preserved During Subsequent Evolution.
Although it is generally believed that an adaptive trait like tolerance to biotic and abiotic stresses has evolved over evolutionary time under selective constraints, speciation via polyploidy might be an exception to this general rule. This is because WGD per se may generate profound physiological alterations that are heritable, provided that euploidy is maintained and intersubgenome rearrangements are restrained (4, 5, 15). Notably, most stabilized neo- and natural allopolyploid plant species meet these criteria (15–17). An elegant example supporting the de novo creativity of biological effects by WGD itself is the recent demonstration that autopolyploid Arabidopsis plants are generally more salt tolerant than their diploid progenitors due to enhanced potassium uptake (8). In the case of allopolyploidy, both WGD and hybridity (merging of divergent genomes of different species) are involved, thus, in theory even greater biological effects are expected (3, 4, 18, 19). We show here that to a great extent the salt-tolerant trait of hexaploid wheat can be attributed to allopolyploidization followed by evolutionary perseverance. This was supported by our observations that for most of the measured morphophysiological traits directly related to salt tolerance, including survivorship, photosynthesis, osmotic regulation, Na+ removal, amino acid and NO3− accumulation, were highly similar between the synthetic and natural hexaploid wheats, but substantially different from their progenitor species. In this respect, our findings are different from that of Maherali et al. (20), who showed that natural tetraploid Chamerion angustifolium exhibited greater drought tolerance than both its newly formed tetraploid counterpart and diploid progenitors. However, our results are consistent with the findings of Chao et al. (8) in Arabidopsis, who demonstrated that enhanced salt tolerance is an immediate consequence of the WGD, which was manifested by both synthetic and natural tetraploids. Nevertheless, our results are also distinct from the findings in Arabidopsis (8), as the enhanced salt tolerance observed in hexaploid wheat is largely a consequence of hybridity rather than WGD, i.e., a combination of the higher germination capability of tetraploid and the stronger Na+ retention ability of the diploid progenitors under salt stress, respectively.
Salt tolerance is a complex trait involving a cascade of primary and secondary stress responses as well as whole plant coordination (21). Our integrated analysis of 92 morphophysiological traits indicated that at least three physiological bases are involved: First, immediately after allohexaploidization, the synthetic hexaploid wheat inherited the higher germination capacity of its tetraploid wheat progenitor T. turgidum. Germination is the most critical period in the life cycle of salt-stress–affected plants, a period that is extremely sensitive to high salinity and critical for seedling survival and growth. Lower germination capacity may have been a major cause for the lower survivorship of Ae. tauschii under the salt-stress conditions. Second, immediately after allohexaploidization, the synthetic hexaploid wheat inherited the stronger ability to retain Na+ within roots from Ae. tauschii. This is consistent with observations of the two earlier studies (22, 23), which compared leaf Na+ content in a large number of synthetic/natural hexaploid and tetraploid wheat genotypes, and found that in general both synthetic and natural hexaploid wheats contained lower leaf Na+ content than tetraploid wheats, implicating, although untested in these studies, lower capacity to retain Na+ by roots of tetraploid than hexaploid wheats. Third, immediately after allohexaploidization, the synthetic hexaploid wheat showed transgressive performance in carbon and nitrogen metabolism over both its diploid and tetraploid parents. Importantly, characteristics of all three physiological bases are highly similar between the synthetic and the natural hexaploid wheats, suggesting their evolutionary perseverance. This is consistent with the broadly shared regulatory shift (from constitutive to inductive) in the expression control of the D-subgenome homeolog of a critical Na+-tolerance gene, HKT1;5 between natural and synthetic hexaploid wheats (9, 13).
Generally Augmented Biological Effect of the D Subgenome in the Synthetic Hexaploid Wheat Under Salt Stress.
It has been repeatedly shown that allopolyploidization induces reorchestration of homeologous gene expression, and the extent of these changes often augment with time, leading to genetic diploidization of the resultant polyploid (24–26). In hexaploid wheat, the three subgenomes, B, A, and D, are known to express differentially due to inherent parental expression divergence, novel interactions of parental cis- and trans-regulatory factors, de novo epigenetic remodeling, and biased physical retention of homeologs (11, 27, 28). In cotton (Gossypium hirsutum), an allotetraploid species (genome AADD), it was found that the homeologs manifest extensive partitioning of labor across development (29) and in response to abiotic stresses (30). Together, it appears to be the rule that the subgenomes of a given allohexaploid species have redundant but also distinct functions, which may represent a major advantage of allopolyploidy over its diploid progenitors (5, 19). However, it remained uninvestigated whether the subgenomes may function differentially under different conditions to such an extent that it may culminate in overall alternation in “more phenotypic or physiological resemblance to one of the progenitors.” Here, based on an integrated analysis of the 92 morphophysiological traits, we showed that, under unstressed normal conditions, major traits in the synthetic hexaploid wheat (neo-6x) were more similar to its tetraploid parent (4x) than its diploid parent (2x), as expected for a BBAA:DD = 2:1 genome contribution. However, under salt-stress conditions, differences between neo-6x and 4x for these morphophysiological traits were dramatically augmented, and many of these traits of neo-6x became more similar to 2x than to 4x, suggesting that the salt stress has enhanced functionality of the D subgenome in the synthetic hexaploid wheat, giving it the property of salt tolerance. This remarkable ability to immediately reorchestrate functionality of the subgenomes in response to different growing conditions is clearly a unique property of allopolyploids. That said, magnitude of adaptability to ambient conditions is expected to increase with the ploidy level, which is consistent with the enhanced adaptability of hexaploid wheat than its tetraploid progenitor (10). Further studies are required to explore the molecular mechanisms underlying the condition-dependent physiological alternation in allopolyploid crops and assess specificity as well as generality of the phenomenon in response to diverse stress conditions.
Materials and Methods
Plant Material and Physiological Measurements.
A synthetic allohexaploid wheat genotype (Allo-960), their tetraploid wheat and diploid goat-grass parents, and a natural hexaploid bread wheat (T. aestivum, cv. Chinese Spring) were used in this study (detailed in SI Materials and Methods). Stress treatments were applied with sodium chloride (NaCl), and 92 morphological and physiological parameters of four organs were measured (Dataset S2), as detailed in SI Materials and Methods.
The TaqMan SNP Genotyping Assay and qRT-PCR Analysis.
The method is detailed in SI Materials and Methods. Amplification curves of the TaqMan SNP genotyping assay are shown in Dataset S4, and the primers used in this assay are listed in Dataset S5.
Interactive Effects of D Subgenome Under Salt Stress.
We used mathematical analysis to assess interactive effects of salt stress and functionality of the D subgenome of hexaploid wheat. We defined the following parameters to assess the biological effects of the D subgenome in the synthetic hexaploid wheat: the DEN = ratio of neo-6x's measured value/4x's measured value under normal conditions and the DES = ratio of neo-6x's value/4x's value under salt-stress conditions.
Statistical Analysis.
Statistical analysis was performed using the statistical program SPSS 13.0 and detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. George Fedak (Agriculture and Agri-Food Canada) for providing the initial seeds of the synthetic allohexaploid wheat (Allo-960) and its parental genotypes. This work was supported by the National Natural Science Foundation of China (nos. 31290211 and 31300192), the Program for Introducing Talents to Universities (B07017), and the Joint Center for Plant Genetic Research between Northeast Normal University and Washington State University.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412839111/-/DCSupplemental.
References
- 1.Madlung A. Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Heredity (Edinb) 2013;110(2):99–104. doi: 10.1038/hdy.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hegarty M, et al. Lessons from natural and artificial polyploids in higher plants. Cytogenet Genome Res. 2013;140(2-4):204–225. doi: 10.1159/000353361. [DOI] [PubMed] [Google Scholar]
- 3.Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- 4.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131(3):452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6(11):836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 6.Coate JE, Powell AF, Owens TG, Doyle JJ. Transgressive physiological and transcriptomic responses to light stress in allopolyploid Glycine dolichocarpa (Leguminosae) Heredity (Edinb) 2013;110(2):160–170. doi: 10.1038/hdy.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearse IS, Krügel T, Baldwin IT. Innovation in anti-herbivore defense systems during neopolypoloidy: The functional consequences of instantaneous speciation. Plant J. 2006;47(2):196–210. doi: 10.1111/j.1365-313X.2006.02776.x. [DOI] [PubMed] [Google Scholar]
- 8.Chao D-Y, et al. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science. 2013;341(6146):658–659. doi: 10.1126/science.1240561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 10.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316(5833):1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman M, Levy AA, Fahima T, Korol A. Genomic asymmetry in allopolyploid plants: Wheat as a model. J Exp Bot. 2012;63(14):5045–5059. doi: 10.1093/jxb/ers192. [DOI] [PubMed] [Google Scholar]
- 12.Huang S, et al. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA. 2002;99(12):8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James RA, Davenport RJ, Munns R. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 2006;142(4):1537–1547. doi: 10.1104/pp.106.086538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S, Spielmeyer W, Lagudah ES, Munns R. Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J Exp Bot. 2008;59(4):927–937. doi: 10.1093/jxb/ern033. [DOI] [PubMed] [Google Scholar]
- 15.Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16(7):351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- 16.Doyle JJ, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 17.Gaeta RT, Chris Pires J. Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol. 2010;186(1):18–28. doi: 10.1111/j.1469-8137.2009.03089.x. [DOI] [PubMed] [Google Scholar]
- 18.Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42(1):225–249. [PubMed] [Google Scholar]
- 19.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maherali H, Walden AE, Husband BC. Genome duplication and the evolution of physiological responses to water stress. New Phytol. 2009;184(3):721–731. doi: 10.1111/j.1469-8137.2009.02997.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J-K. Plant salt tolerance. Trends Plant Sci. 2001;6(2):66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 22.Schachtman DP, Lagudah ES, Munns R. The expression of salt tolerance from Triticum tauschii in hexaploid wheat. Theor Appl Genet. 1992;84(5-6):714–719. doi: 10.1007/BF00224174. [DOI] [PubMed] [Google Scholar]
- 23.Pritchard D, et al. K+/Na+ discrimination in synthetic hexaploid wheat lines: Transfer of the trait for K+/Na+ discrimination from Aegilops tauschii into a Triticum turgidum background. Cereal Res Commun. 2002;30(3-4):261–267. [Google Scholar]
- 24.Birchler JA. Insights from paleogenomic and population studies into the consequences of dosage sensitive gene expression in plants. Curr Opin Plant Biol. 2012;15(5):544–548. doi: 10.1016/j.pbi.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Freeling M, et al. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr Opin Plant Biol. 2012;15(2):131–139. doi: 10.1016/j.pbi.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Roulin A, et al. The fate of duplicated genes in a polyploid plant genome. Plant J. 2012;73(1):143–153. doi: 10.1111/tpj.12026. [DOI] [PubMed] [Google Scholar]
- 27.Osborn TC, et al. Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics. 2003;165(3):1569–1577. doi: 10.1093/genetics/165.3.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pont C, et al. Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo- and neoduplicated subgenomes. Plant J. 2013;76(6):1030–1044. doi: 10.1111/tpj.12366. [DOI] [PubMed] [Google Scholar]
- 29.Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA. 2003;100(8):4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong S, Adams KL. Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytol. 2011;190(4):1045–1057. doi: 10.1111/j.1469-8137.2011.03650.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.