Abstract
A wide variety of phytochemicals are consumed for their perceived health benefits. Many of these phytochemicals have been found to alter numerous cell functions, but the mechanisms underlying their biological activity tend to be poorly understood. Phenolic phytochemicals are particularly promiscuous modifiers of membrane protein function, suggesting that some of their actions may be due to a common, membrane bilayer-mediated mechanism. To test whether bilayer perturbation may underlie this diversity of actions, we examined five bioactive phenols reported to have medicinal value: capsaicin from chili peppers, curcumin from turmeric, EGCG from green tea, genistein from soybeans, and resveratrol from grapes. We find that each of these widely consumed phytochemicals alters lipid bilayer properties and the function of diverse membrane proteins. Molecular dynamics simulations show that these phytochemicals modify bilayer properties by localizing to the bilayer/solution interface. Bilayer-modifying propensity was verified using a gramicidin-based assay, and indiscriminate modulation of membrane protein function was demonstrated using four proteins: membrane-anchored metalloproteases, mechanosensitive ion channels, and voltage-dependent potassium and sodium channels. Each protein exhibited similar responses to multiple phytochemicals, consistent with a common, bilayer-mediated mechanism. Our results suggest that many effects of amphiphilic phytochemicals are due to cell membrane perturbations, rather than specific protein binding.
Biologically active plant phenols have a broad range of pharmacological effects—including anticarcinogenic, antimicrobial, antioxidant, and anti-inflammatory activity.1−11 Despite widespread popularity in Western medicine, and thousands of scientific publications devoted to the activity of these compounds each year, their molecular mechanisms of action remain poorly understood. Phenolic phytochemicals modulate numerous unrelated proteins and biological pathways but few binding sites have been identified. In the case of membrane proteins, a given protein may be modulated by structurally unrelated plant phenols that can have synergistic effects12−14 suggestive of a common, nonsaturating mechanism. Conversely, a given phytochemical may modulate the function of many different membrane proteins—at similar concentrations (e.g., Table 1 and Supporting Information Table S1). While the many actions of phytochemicals could result from direct interactions with numerous different targets, the presence of binding sites having similar affinities on such a wide variety of targets seems unlikely. We propose a more parsimonious mechanism for the biological activity of many phytochemicals.
Table 1. Membrane Proteins Known to Be Affected by Phytochemicalsa.
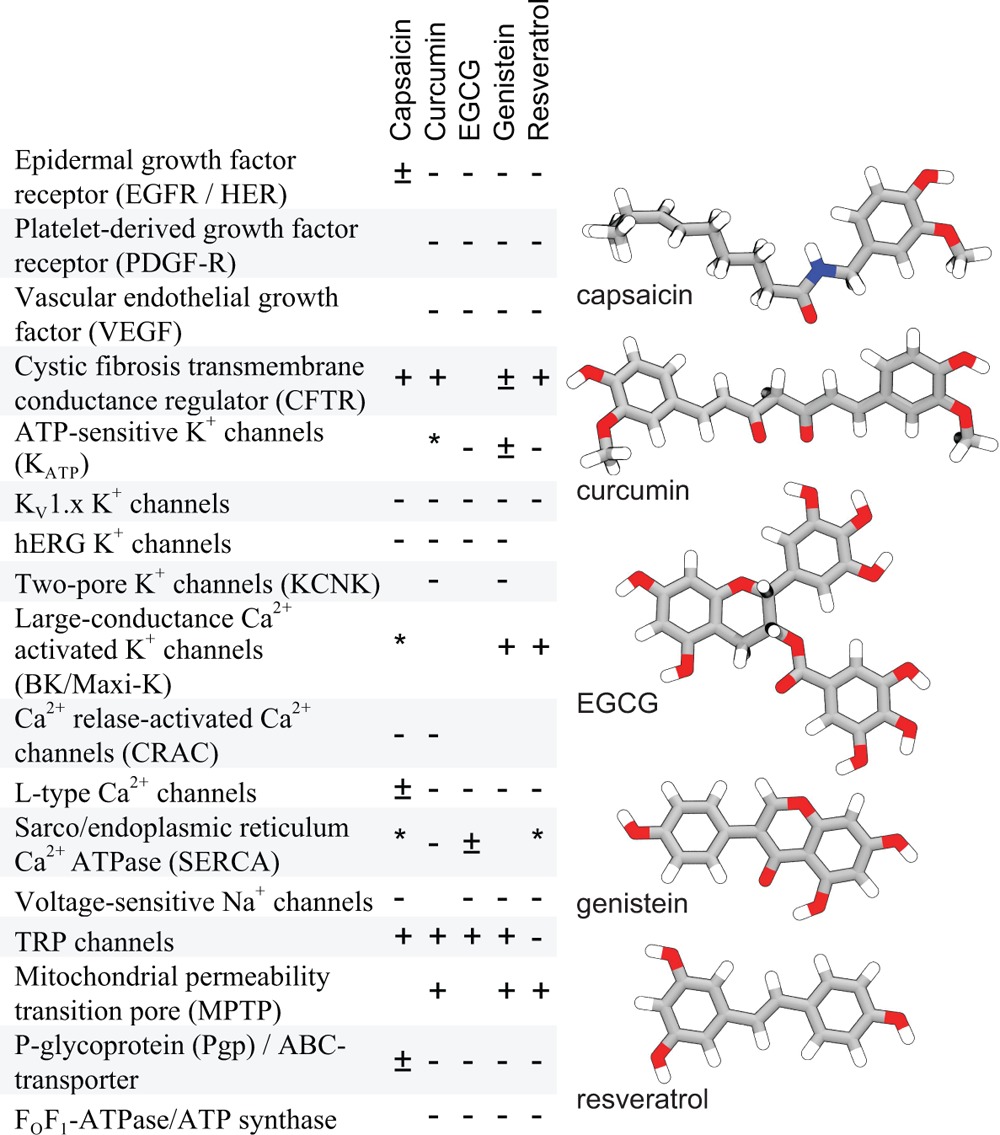
(+) indicates activation or up-regulation, (−) indicates inhibition or down-regulation, (*) indicates “interaction”, (±) indicates biphasic dose response curve or both activation and inhibition reported. For a more extensive listing and references see Table S1 in the Supporting Information.
The common feature of membrane proteins—that they are embedded in a lipid bilayer—leads to a unifying hypothesis for many of the diverse effects of phenolic phytochemicals. These phytochemicals tend to be amphiphilic; they can adsorb to lipid bilayer/solution interfaces and thereby alter bilayer properties, which can lead to changes in membrane protein function.15,16 We therefore propose that, rather than acting through discrete binding sites, physical alteration of membrane properties underlies many of the diverse actions of phenolic phytochemicals.
To test whether the phytochemicals’ bilayer-modifying effects constitute a general mechanism underlying their alteration of membrane protein function, we examined the membrane localization and bilayer-modifying effects of five extensively studied and structurally diverse phenolic phytochemicals—capsaicin (chili peppers), curcumin (turmeric), epigallocatechin gallate (EGCG; green tea), genistein (soybeans), and resveratrol (grapes). The chosen compounds modulate numerous biological pathways and alter the functions of hundreds of different proteins, including many membrane proteins1−11 (Table 1 and Supporting Information Table S1). With a few notable exceptions, such as the binding of capsaicin to TRPV117,18 and the high affinity binding of EGCG to the 67-kDa laminin receptor,19 there is little evidence for direct binding to any of their numerous effector proteins.
We used a combination of molecular dynamics (MD) simulations and a gramicidin-based assay to quantify the compounds’ bilayer-modifying potency. The MD simulations predict and gramicidin experiments verify that all the compounds tested indeed are potent modifiers of bilayer properties. This means that the phytochemicals have the potential to indiscriminately modulate membrane protein function, in the absence of direct binding, through their bilayer-modifying effects. We explored the implications of this membrane-perturbation by testing the compounds’ ability to alter the function of four membrane proteins: the mechanosensitive channel of large conductance (MscL), KV2.1 potassium channels, voltage-dependent sodium channels (NaV), and the membrane-anchored metalloprotease ADAM17. Our results show that membrane-perturbing phytochemicals are indiscriminate modifiers of a wide range of membrane proteins, thus providing a mechanism for their diverse actions—that they alter membrane protein function by altering lipid bilayer properties.
Results and Discussion
Phytochemicals Alter Bilayer Properties
We cataloged the phytochemicals’ effects on membranes—where they localize in the bilayer and what properties they alter. The tested phytochemicals have high octanol/water partition coefficients (logP varies between 3.1 and 4.120), meaning that they readily partition into and permeate through lipid bilayers. A patchwork of previous studies involving MD simulations, NMR, fluorescence spectroscopy, and calorimetry have shown that some of these compounds partition into and cross bilayers and change bilayer properties.21−32 These phenolic compounds would be expected to reside in the bilayer/solution interface and alter bilayer properties such as area per lipid, bilayer thickness and lipid tail order. Polyphenolic compounds have also been found to weaken bilayer integrity through their increase in membrane area33,34 and at high concentrations, they can disrupt bilayers, rupture lipid vesicles, and induce cell lysis.25,35
To develop a consistent description of the phytochemicals’ membrane effects, we used coarse-grained (CG) MD simulations based on the Martini force field36 to characterize how the five phytochemicals modify the structure and dynamics of CG 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayers, at a 1:10 phytochemical:POPC molar ratio (Figure 1). The phytochemicals localize to the bilayer/solution interface (lipid headgroup and backbone region) (Figure 1b); EGCG with its numerous hydroxyl groups resides slightly closer to the bulk aqueous phase, whereas capsaicin with its hydrophobic tail reaches further into the bilayer hydrophobic core. They have rather modest effects on the bulk bilayer properties (less than 3% increases in the average area per lipid or decreases in bilayer thickness) with little effect on average lipid order or bilayer compressibility (Supporting Information Figure S1 and Table S2). The location in the bilayer and the changes in bulk bilayer properties were similar in atomistic simulations (see Supporting Information Methods) with the notable exception of EGCG, which became positioned deeper in the membrane in the atomistic simulations (Supporting Information Figure S3 and Table S2). All the phytochemicals produced significant changes in the bilayer pressure profile (Figure 1c and Supporting Information Figure S1), which may modulate protein function.37 The changes vary among the compounds: EGCG, genistein, and resveratrol primarily perturb the pressure profile in the interfacial region, whereas curcumin and capsaicin shift the profile closer to the center of the bilayer. From the calculated changes in bilayer properties, including the lateral pressure profile, we estimated the bilayer bending modulus, the lipid spontaneous curvature and bilayer elastic ratio, see Supporting Information Methods. The phytochemicals had rather modest effects on these parameters, Supporting Information Table S2.
Figure 1.
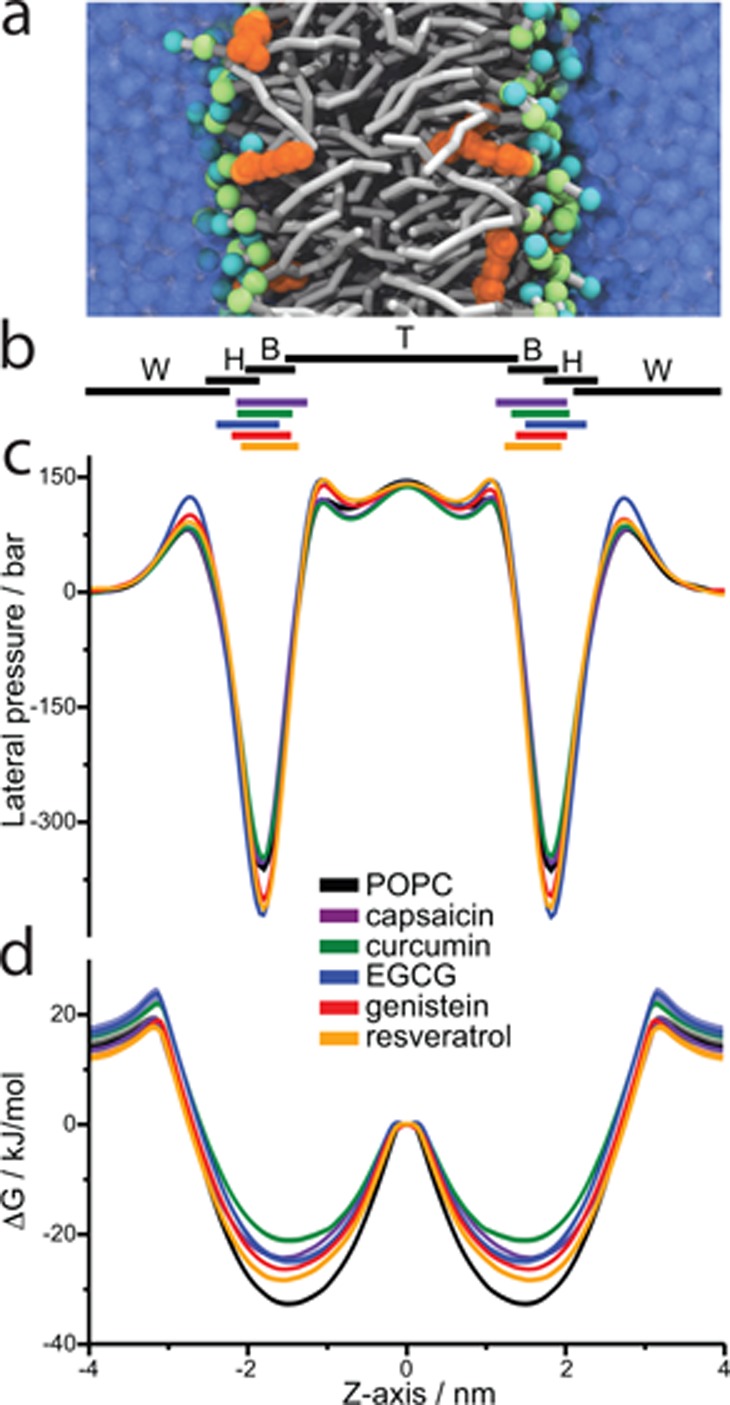
Phytochemicals partition into phospholipid bilayers and alter their properties. The phytochemicals’ effects on 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) CG bilayers were explored using Martini simulations at 1:10 phytochemical/lipid molar ratio. (a) Simulation snapshot showing resveratrol (orange) in a CG POPC bilayer (tails and backbone in gray and the head groups in cyan and green). (b) Lateral density, indicated as density width at half-maximum height of the distributions for water (W), POPC lipid head groups (H), backbone (B), tails (T), and for the phytochemicals (see also Supporting Information Figure S1e). (c) Lateral pressure profile. (d) Symmetrized potential of mean force (PMF) for translocating a probe of radius 0.9 nm through the bilayer. The bilayer normal is set to the Z-axis with zero at the center of the bilayer.
To further estimate how the compounds alter the energy required to perturb the bilayer, we moved a 0.9 nm radius spherical probe (with high affinity for the lipid head and linker region and no preferential interaction with the phytochemicals) across the bilayer and calculated the potential of mean force (PMF) in the absence and presence of the phytochemicals (Figure 1e and Supporting Information Figure S2). Based on this estimate, the phytochemicals reduced the energy required to perturb the bilayer (allowed the bead to pass through more easily) by lowering the transition barriers for crossing the bilayer by 5–10 kJ mol–1. This suggests that the phytochemicals reduce the energetic cost of bilayer adaptations perpendicular to the plane of the bilayer. Therefore, they would alter the conformational equilibria of membrane proteins with conformational changes that are associated with a bilayer perturbation. Integrating the PMF for moving the probe across the bilayer yielded estimates of the work required for the associated bilayer deformation. Relative to pure POPC, curcumin and EGCG were the most potent (reducing the work by 67 ± 5% and 54 ± 8%, respectively), followed by capsaicin (38 ± 3%), genistein (29 ± 3%), and resveratrol (14 ± 4%).
Using a calibrated gramicidin-based assay, we found that these bilayer-modifying effects are sufficient to alter membrane protein function. Gramicidin (gA) is a small antibacterial protein (15 amino acids long) that forms monovalent cation-conducting channels by the transmembrane dimerization of nonconducting subunits residing in opposing bilayer leaflets. Because the channel length is less than the bilayer thickness, the bilayer has to perturb around the gA channel as it forms—meaning that the gA monomer/dimer equilibrium is coupled to the energetic cost of deforming the bilayer. Changes in gA channel activity (quantified as changes in single channel lifetime or time-averaged activity) thus serve as measures for changes in lipid bilayer properties, as sensed by a bilayer-spanning channel.16,38
To estimate what change in bilayer bulk properties would be required to observe a significant change in gA channel activity, we used the continuum elastic model of Nielsen, Goulian, and Andersen39,40 to predict the changes in bilayer properties (bilayer elasticity) that would result in a 10-fold increase in gA function (∼6 kJ mol–1 reduction in the dimerization energy); see Supporting Information Methods. An isolated reduction in the hydrophobic thickness by ∼1%, the bilayer spring constant by ∼4%, the area compressibility by ∼5%, or the bilayer bending modulus by ∼10% would each result in a 6 kJ mol–1 reduction in the dimerization energy. Based on the changes in lipid bilayer properties estimated from the MD simulations, at a 0.1 mol-fraction of each phytochemical, the gA free energy of dimerization is predicted to decrease by ∼12 kJ mol–1, in general agreement with our experimental results, see below.
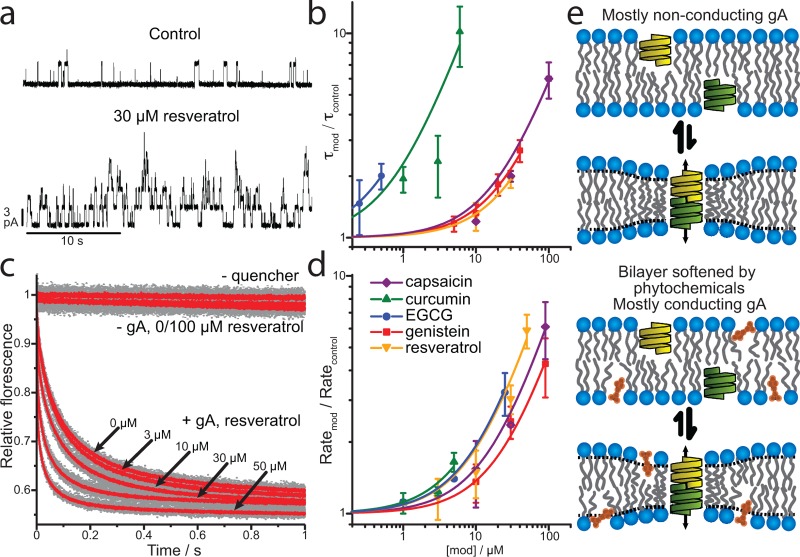
We probed for bilayer-modifying effects in planar bilayers (Figure 2a, b) using gA single-channel electrophysiology41 and in large unilamellar vesicles (LUVs) (Figure 2c, d) using a gramicidin-based fluorescence assay that monitors changes in channel activity in LUVs.42 These two assays are complementary: the single-channel electrophysiology assay allows for determining single-channel appearance frequencies and lifetimes, but planar bilayers contain organic solvent (in this case n-decane); the gramicidin-based fluorescence assay allows for determining changes in the average number of conducting channels in the hydrocarbon-free LUV membrane. Focusing on the concentration ranges where most in vitro experiments with the chosen phytochemicals were done, cf. the studies cited in Table 1 and Supporting Information Table S1, we found that all five compounds increased channel activity—doubling gA single channel lifetimes at 0.5–32 μM aqueous drug concentration and the rate of quencher influx through gA channels (gA activity) at 9–27 μM (Figure 2). EGCG and curcumin were the most potent bilayer-modifying compounds followed closely by capsaicin, genistein, and resveratrol. Maximum concentrations in the gA experiments were limited by bilayer stability. Higher concentrations resulted in frequent breakage of planar bilayers and leak of quencher into vesicles. This is an expected consequence of the nominal aqueous phase concentrations, which translate into significant concentrations in the bilayer due to the compounds high partition coefficients into the membrane. Using octanol/water partition coefficients to estimate membrane/electrolyte partitioning suggests membrane concentrations will be three-to-four orders of magnitude higher than the aqueous concentrations. At the maximum concentrations used in this study, the predicted mole-fractions ranged from ∼0.01 to ∼0.05, close to the 0.1 mol-fraction used in MD simulations. Consistent with previous studies and our MD simulations, the phytochemicals reduce the energetic cost of bilayer deformation associated with gA channel formation. Even at low micromolar nominal concentrations, the phytochemicals alter bilayer properties sufficiently to change the function of gA channels in planar bilayers and lipid vesicles (Figure 2b, d).
Figure 2.
Phytochemicals perturb phospholipid bilayers as sensed by gramicidin channels. (a, b) Phytochemical effect on gA channel lifetime measured using single-channel electrophysiology in planar DOPC/n-decane bilayers. (a) Representative current traces. (b) Changes to gA lifetime with the addition of phytochemicals. The solid lines are f([mod]) = 1 + [mod]/D fits to the results. The phytochemicals double the gA lifetime at (concentrations in μM) 20.7 ± 1.3 capsaicin, 0.8 ± 0.1 curcumin, 0.5 ± 0.01 EGCG, 25.6 ± 1.5 genistein, and 32.3 ± 0.7 resveratrol. (c, d) Phytochemical effect on gA channel activity measured with a gA permeable quencher rate of influx into fluorescent vesicles doped with gA. (d) Changes to gA activity with the addition of phytochemicals. The solid lines are f([mod]) = 1 + [mod]/D fits to the results. The phytochemicals double gA induced quencher influx rates at (concentrations in μM) 18.0 ± 0.6 capsaicin, 8.6 ± 1.0 curcumin, 11.2 ± 0.2 EGCG, 26.7 ± 1.4 genistein, and 11.3 ± 0.9 resveratrol. (e) Schematic depicting increased gA channel activity following the addition of phytochemicals that partition into the bilayer/solution interface.
Phytochemicals Modify Membrane Protein Function
Bilayer-mediated alterations of membrane protein function arise because the proteins are coupled to the bilayer through hydrophobic interactions.16 When membrane proteins undergo conformational changes that involve the protein/bilayer boundary, they perturb the adjacent bilayer, effectively coupling changes in bilayer properties to changes in membrane protein function (the energetics and kinetics of their conformational equilibria). The direction and magnitude of changes in the conformational equilibrium (protein function) depend on both the protein (the change in conformation) in question and the properties of the host bilayer.15,16,37,43−45 For example, for a protein in which the hydrophobic length of the active state is longer than that of the inactive state, the protein activity will increase when the average bilayer thickness increases (within a given range), and vice versa. Softening the bilayer (decreasing bilayer elasticity) will shift the conformational equilibrium toward the state with more hydrophobic mismatch. Hence, bilayer softening could increase or decrease protein activity depending on the specific protein–bilayer match. A quantitative treatment is given in the Supporting Information Membrane Protein–Lipid Bilayer Coupling section.
Our results suggest that many phenolic phytochemicals are likely to modify membrane protein function by partitioning into the bilayer/solution interface and thereby alter bilayer properties, akin to their effects on gA (Figure 2e). To test this hypothesis, we explored whether the five phytochemicals’ bilayer-modifying effects are sufficient to alter the function of four transmembrane proteins in different membrane environments: the bacterial mechanosensitive channel of large conductance (MscL); the voltage-dependent potassium channel KV2.1; neuronal voltage-dependent sodium channels (NaV); and the ADAM17 disintegrin-type metalloproteinase.
MscL serves as a last resort emergency release valve in case of severe osmotic shock46 and is a commonly used model for mechanosensation. The channel activates with increasing bilayer tension, opening a large (∼3 nS), nonselective pore. As MscL senses bilayer tension, it should be sensitive to changes in bilayer properties. Indeed, MscL’s gating threshold is affected by bilayer thicknesses and the asymmetric insertion of amphiphiles or lysophosphatidylcholine (LPC) into the bilayer.47,48 The effects on MscL gating were determined in asolectin lipid vesicles using a calcein fluorescence dequenching assay.49 The G22C MscL mutant was incorporated in fluorophore-filled vesicles and the channels were activated by adding the positively charged sulfhydryl reagent [2-(trimethylammonium)ethyl] methanethiosulfonate bromide (MTSET), which reacts with Cys22 and thereby weakens the gate—allowing for channel activation without applied tension.50 Channel activation was monitored by the increase in fluorescence as calcein exited LUVs through open MscL channels (Figure 3a, control). All the phytochemicals tested modified MscL function, reducing MscL activity to 50% of control (for both maximal release and efflux rate) at 2–130 μM (nominal concentration) (Figure 3b, c), with curcumin being the most potent. The phytochemicals inhibited channel activation similarly, reducing both the initial rate and steady-state fluorescence intensity (Figure 3).
Figure 3.
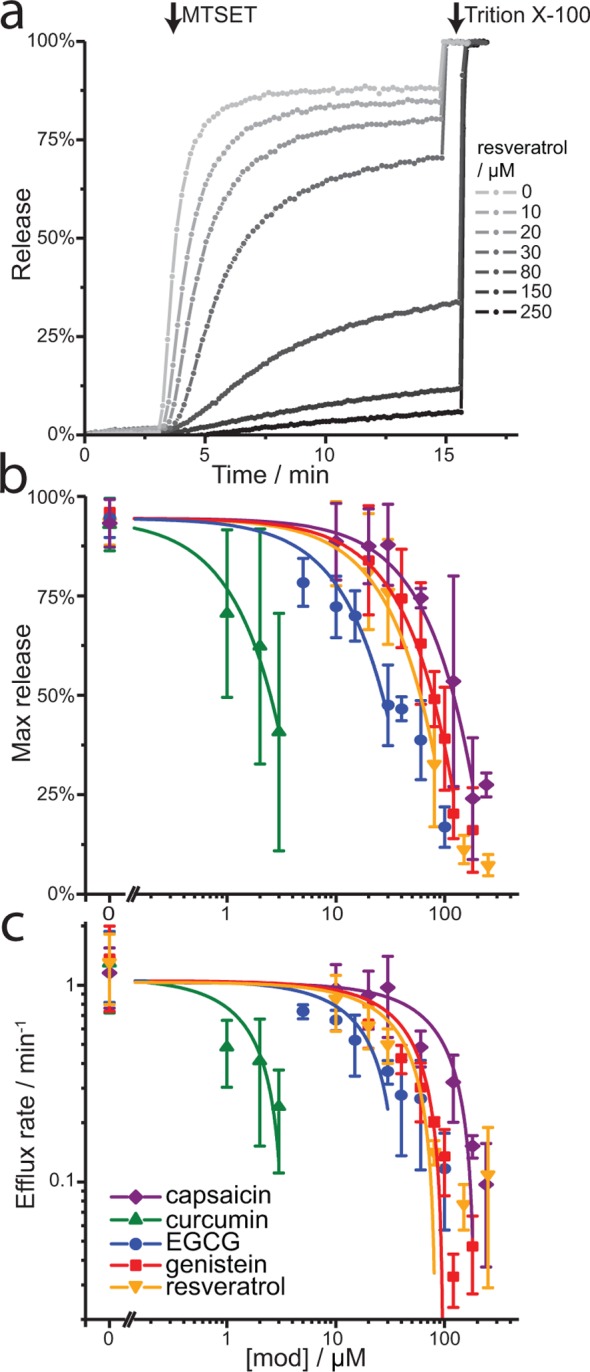
Phytochemicals inhibit mechanosensitive channels. MscL channels were reconstituted into calcein-loaded vesicles. Channel activation was initiated by exposure to MTSET and the release of calcein through open MscL channels is monitored as an increase in fluorescence. (a) Representative calcein release curves. (b, c) Changes in MscL activity after addition of phytochemicals, avg ± standard deviation (SD), n = 3. The solid lines are f([mod]) = f(control) – [mod]/D fits to the results (excluding saturating concentration). (b) The phytochemicals produce a 50% reduction in the max release at (concentrations in μM) 127 ± 5 capsaicin, 2.7 ± 0.2 curcumin, 28 ± 2 EGCG, 82 ± 2 genistein, and 63 ± 2 resveratrol μM concentration and half the efflux rate (c) at 92 ± 12 capsaicin, 1.6 ± 0.4 curcumin, 18 ± 5 EGCG, 46 ± 9 genistein, and 40 ± 9 resveratrol.
Voltage-dependent potassium channels (KV) are transmembrane proteins that are modulated by changes in lipid bilayer properties.51−55 We explored the phytochemicals’ effects on KV2.1 channels heterologously expressed in the plasma membrane of CHO-K1 cells using voltage-clamp electrophysiology (Figure 4). Four of the five phytochemicals induced similar changes in KV2.1 function. Capsaicin, curcumin, EGCG, and genistein significantly inhibited peak KV2.1 currents (Figure 4c) with little effect on time course of activation or the conductance–voltage relation (Figure 4d, e). This common signature in their mode of action suggests that the active compounds alter channel function via a common mechanism.
Figure 4.
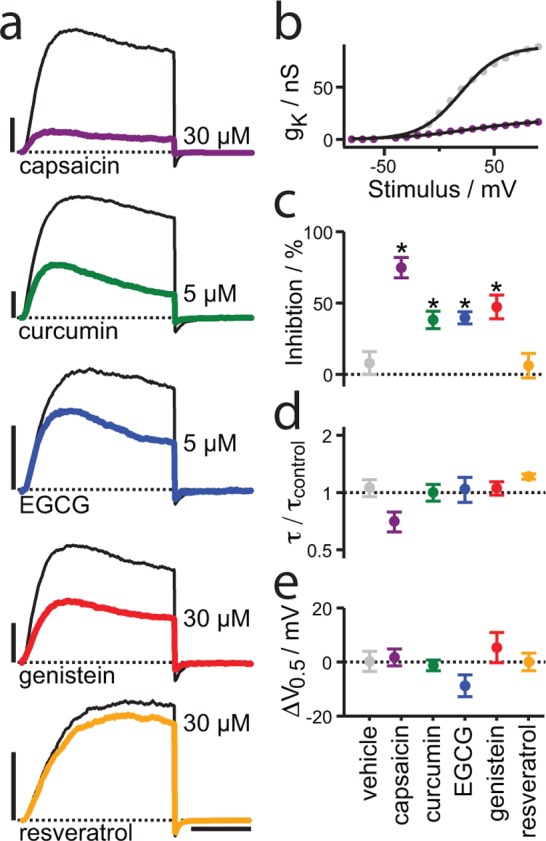
Phytochemicals inhibit voltage-dependent potassium channels. (a) Representative KV2.1 current traces from 100 ms steps to +20 mV, returning to the holding potential of −100 mV. Black lines, control. Colored lines, during application of indicated phytochemical. Abscissa bar, 40 ms; ordinate, 1 nA. (b) Conductance–voltage relation. Gray circles, KV2.1 control; purple circles, 30 μM capsaicin. Lines are fitted Boltzmann relations. (c–e) Phytochemical effects on KV2.1 currents. Same concentrations as panel a. Circles indicate mean, bars standard error. Asterisks indicate significant difference from DMSO vehicle treatments, P < 0.05 two-tailed, Mann–Whitney U-test, n = 4–6. (c) Inhibition of peak KV2.1 current at +20 mV. (d) Ratio of KV2.1 activation time constant at +20 mV in phytochemical versus control. (e) Shift of conductance–voltage relation midpoint by phytochemicals.
Voltage-dependent sodium channels (NaV) are also regulated by changes in lipid bilayer properties.56−58 NaV channels are structurally related to KV channels, yet share little sequence homology or drug sensitivity. The phytochemical effects on endogenous NaV were determined by whole-cell electrophysiology in neuronal ND7/23 cells (Figure 5). The phytochemicals produced voltage-dependent inhibition of peak Na+ current (INa) (Figure 5c). Except for curcumin, inhibition was greater when the test pulse was preceded by a prepulse to V1/2 (−69 ± 3 mV), a voltage at which approximately half the channels were in the inactivated state. Effects on NaV steady-state inactivation were tested using a double-pulse protocol (Figure 5d, e). Capsaicin, genistein, and resveratrol acted similarly, shifting the voltage-dependence of steady-state inactivation toward more hyperpolarized potentials.
Figure 5.
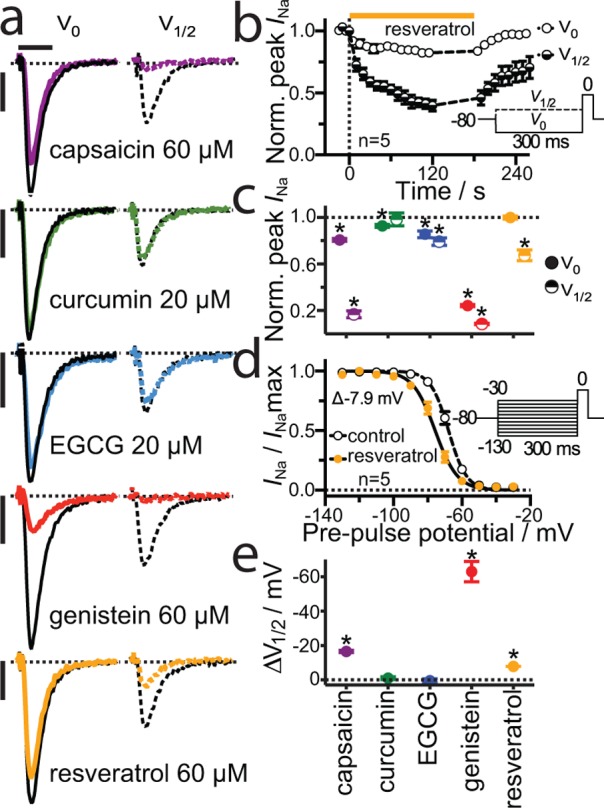
Phytochemicals inhibit sodium channels. (a) Representative NaV current traces from an experiment with an alternating two-pulse protocol. A test pulse to 0 mV, to elicit peak Na+ current (INa) was preceded by a 300 ms prepulse to holding potentials of either V0 (−130 mV) or V1/2 (see insert in panel (b) V1/2 = −69 ± 3 mV). Black lines, control. Colored lines, results obtained during application of the listed concentration of the indicated phytochemical. Abscissa bar, 1 ms; ordinate, 1 nA. (b) Inhibition of peak INa during wash-in and wash-out of resveratrol. (c) Inhibition of peak INa after 120 s treatment with the different compounds. (d, e) Shift in steady-state inactivation tested using a double-pulse protocol in which a test pulse to 0 mV was preceded by a 300 ms conditioning prepulse to potentials ranging from −130 mV to −30 mV. (d) Shift in the voltage-dependence of steady-state inactivation. The results from each experiment were fitted with a standard Boltzmann equation to calculate the individual V1/2 values. (e) Shift in V1/2 caused by the phytochemicals. For all panels, phytochemicals were tested at the concentrations shown in panel a (avg ± sem). Asterisks denote significant difference from control, p < 0.05 two-tailed, Student’s t-test, n = 4–6.
ADAM17 is a single transmembrane-helix membrane-anchored disintegrin-type metalloproteinase that cleaves membrane proteins to release peptides from cells in a process known as ectodomain shedding.59,60 It cleaves a variety of membrane proteins, including the pro-tumor necrosis factor α (TNFα) and the pro-transforming growth factor α (TGFα).61 ADAM17-dependent ectodomain shedding is a regulated process that can be rapidly activated by several intracellular signaling pathways through a mechanism that requires its transmembrane domain,59,60 raising the question of whether its function might be sensitive to changes in bilayer properties. To address this question, we tested whether treatment with the phytochemicals stimulated shedding of TGFα from mouse fibroblasts expressing ADAM17. At the low micromolar concentrations tested in the preceding experiments, capsaicin, EGCG and genistein had no effect on ADAM17-driven TGFα shedding (Figure 6); curcumin and resveratrol could not be tested as they interfered with the colorimetric detection assay. Higher concentrations of capsaicin induced the shedding of TGFα, whereas EGCG and genistein did not, even at the highest concentrations tested. Of all the membrane proteins tested, ADAM17 was the least sensitive to the phytochemicals.
Figure 6.
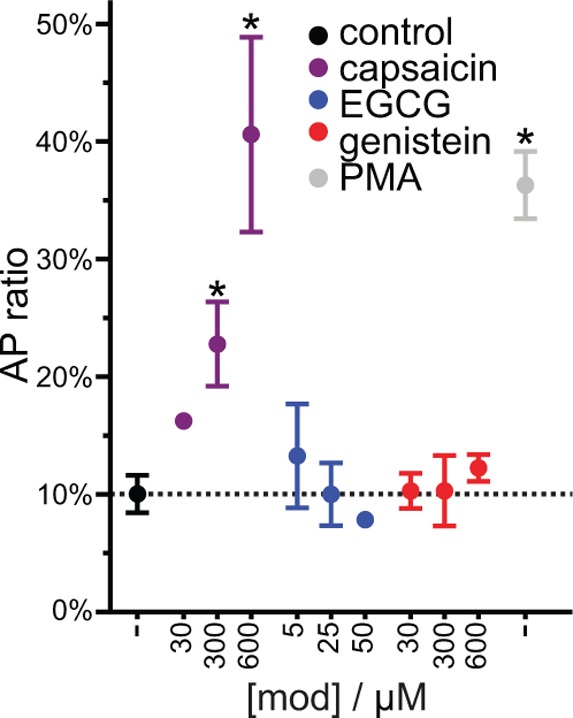
Phytochemicals’ effect on ADAM17-mediated shedding of TGFα. ADAM17-mediated shedding of alkaline phosphatase (AP)-tagged TGFα was measured and shown as an AP-ratio. TGFα shedding was sensitive to capsaicin but not to EGCG or genistein at concentrations between 5 and 600 μM, as indicated. Phorbol-12-myristate-13-acetate (PMA), 25 ng/mL, was used as a positive control for activation of ADAM17. Asterisks indicate significant difference from control, p < 0.05 two-tailed, Student’s t-test, avg ± sem, n = 1–5.
Discussion
We have shown that five widely consumed and extensively studied phenolic phytochemicals (capsaicin, curcumin, EGCG, genistein, and resveratrol) modify membrane protein function at nominal concentrations similar to those where they alter lipid bilayer properties. The generality of the phytochemicals’ effects against five membrane proteins (gA, MscL, KV2.1, NaV, and ADAM17) in six different membrane environments (two synthetic lipid systems, an extract of native lipids, and three cell types), leads us to conclude they modulate membrane protein function through a common mechanism, namely changes in lipid bilayer physical properties—and that the regulation of membrane protein function by changes in lipid bilayers properties provides a general mechanism for altering membrane protein (and cell) function. The generality of these bilayer-mediated effects does not exclude that these compounds also may alter cellular (and membrane protein) function by more conventional cell signaling pathways, as has been observed for capsaicin in primary sensory neurons.62−64
A consistent physical model emerges when our results are combined with prior studies:15,44,65−67 molecules that alter bilayer properties shift the energetics and kinetics of membrane protein conformational equilibria, thereby altering protein function. Our MD simulations show that the phytochemicals localize to the bilayer/solution interface (Figure 1) and produce changes in bilayer properties. Though the compounds have rather modest effects on bilayer bulk properties (Supporting Information Figure S1 and Table S2), they all altered the lateral pressure profile (Figure 1c) and made the bilayer easier to bend and perturb (Figure 1d). Curcumin and EGCG were the most potent, and the unique structure of each phytochemical produced subtly different changes in bilayer properties (Figure 1 and Supporting Informatio Figures S1 and S2) and protein function. Not surprisingly, the absolute magnitude of the bilayer-mediated changes in membrane protein function that are produced by a given phytochemical varies with the specific membrane protein in question, as well as the bilayer in which it is embedded.
The phytochemicals modify gA channel activity at low micromolar aqueous concentrations (the membrane concentration will be three-to-four orders of magnitude higher than the aqueous concentrations), with curcumin and EGCG being somewhat more potent (based on the aqueous concentrations) than the other compounds tested. These results confirm the predictions from the MD simulations. Considering the rather modest effects observed in the MD simulations, the changes in gA channel function are striking—reflecting the sensitivity of functional assays to changes in membrane properties. That is, changes in protein function reflect the absolute changes in the bilayer contribution to the free energy of the conformational transition.58 Of the four other membrane proteins tested, the three that are known to be sensitive to their bilayer environment (MscL, KV2.1, and NaV) were affected by the phytochemicals at concentrations comparable to those where they alter gA channel function. ADAM17, which is a single-span membrane protein whose catalytic activity is not known to depend on its bilayer environment, was affected only by capsaicin at concentrations so high (0.3–0.6 mM) that bilayer stability may be questioned.
The dose-dependence of the changes in MscL function is consistent with predictions based on the MD simulations and the gA results. There is considerable structural information about MscL’s closed↔open transition. The bilayer-spanning domain of the open state of MscL has a larger radius and a smaller hydrophobic length than the closed state.48,68 The bilayer softening predicted by the MD simulations, and observed in the gA experiments,67 would tend to reduce the energetic cost of bilayer thinning, but the accumulation of material at the bilayer/solution interface would tend to increase the cost of the area expansion in the interface, as suggested by the more shallow energy well in Figure 1d. These structural changes suggest that amphiphiles at the bilayer/solution interface will stabilize the closed conformation of the MscL channel, an effect we observed experimentally for all five phytochemicals.
As further evidence for the generality of the phytochemicals’ effects, each membrane protein’s response characteristics were similar for multiple phytochemicals. In a wide variety of membrane proteins, all the phenolic phytochemicals produced the same qualitative change in function (Table 1 and Supporting Information Table S1). For the proteins examined here, several functional parameters were measured, and the response to any one phytochemical was predictive of results obtained with the others. All the phytochemicals increased gA lifetimes and increased channel frequency without affecting the maximum conductance. All the phytochemicals reduced MscL activity, both reducing efflux rate and max release. Four of five phytochemicals decreased peak KV2.1 conductance, without altering kinetics or the voltage dependence. Three of five phytochemicals increased NaV steady-state inactivation, without altering activation or inactivation kinetics, a demonstrated response to membrane softening by amphiphiles.56,58,67 The unique signature of the phytochemicals on this host of targets is explained parsimoniously by a common mechanism of action. We suggest that the signature response of each protein to multiple phytochemical represents its ‘amphiphile response,’ and that many other phenolic compounds would produce similar responses.
The present study examined how a number of structurally diverse amphiphiles alter the function of diverse membrane proteins, and provides evidence for lipid bilayer regulation of membrane protein function. Specifically, we showed that when structurally diverse phenolic phytochemicals adsorb to the bilayer/solution interface to an extent where they alter lipid bilayer properties they also alter membrane protein function. Even at quite low aqueous concentrations, significant accumulation can occur at the bilayer/solution interface, which is sufficient to produce measurable changes in lipid bilayer properties that result in alterations of membrane protein function. Though the changes in bilayer bulk properties are subtle, they may cause considerable (>kBT) changes in the energetics of membrane protein conformational rearrangements and thus protein function.16,58 Our results suggest that the phytochemicals tested here would alter the function of many other membrane proteins, going beyond the proteins summarized in Supporting Information Table S1. The imposed changes in protein function, however, will depend on the conformational transitions involved in the normal function of the protein in question, as well as the specific lipid environment. Fundamentally, these compounds alter the lipid bilayer contribution to the free energy differences of conformational transitions. The existence of the lipid bilayer contribution to the thermodynamics of protein conformational transitions implies that a protein’s lipid environment is likely to be adjusted to allow for optimal function; this in turn suggests that even modest changes in bilayer properties will alter protein function—as reported here with the different proteins tested in varying membrane environments (synthetic model membranes, native lipid extracts and different eukaryotic cell membranes). This mechanism is general, which leads us to suggest that any membrane protein functional change produced by an amphiphilic molecule should be tested for sensitivity to known bilayer-modifiers to determine whether the action is due to general bilayer perturbation, cf. ref (16).
The tested phytochemicals are just a few specific examples of amphiphiles that partition into the membrane and thereby become promiscuous modifiers of membrane protein function. In the absence of evidence for direct, specific interactions, it thus becomes prudent to assume that amphiphiles’ effects on membrane protein function could involve changes in lipid bilayer properties. Bilayer effects should be ruled out before specific binding interactions of an amphiphile with a membrane protein are implicated.
Methods
Molecular Dynamics Simulations
Simulations were performed using the Martini coarse-grain force field69,70 and the GROMACS simulation package71,72 with the standard Martini simulation setup. Topologies of the phytochemicals and their derivation from atomistic resolution simulations are described in Supporting Information and Figure S5. The temperature (310 K) and pressure (1 atm) of the systems were controlled using a weak-coupling algorithm.73 Phytochemical partitioning and perturbation of lipid bilayer properties were extracted from simulations of CG 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayers containing phytochemicals at a 1:10 phytochemical to POPC molar ratio. At least 0.5 μs equilibration was used prior to collecting the 2 μs trajectories used for analysis. To quantify the phytochemicals’ effect to the bilayer deformation energy the potential of mean force (PMF) of dragging a large object (radius 0.9 nm) across a CG POPC bilayer was determined (see details in Supporting Information).
Selected Phenolic Phytochemicals
The phytochemicals capsaicin, curcumin, EGCG, genistein, and resveratrol were purchased from Sigma at the highest available purity and stocks diluted in DMSO unless otherwise stated. All concentrations reported are nominal. The hydrophobic phytochemicals readily partition into bilayers and onto plastics. Consequently, because of the finite aqueous to lipid phase volume ratios and differences in the assays, the actual aqueous concentrations will be lower than the added (or nominal) concentrations and will vary among the assays.45,74 Phytochemical concentrations used for the MscL, KV2.1, NaV, and ADAM17 experiments were chosen to be sufficient to modify membrane properties; all concentrations exceeded that required for a doubling of gA activity in the fluorescence assays. Concentrations used are generally similar to those reported to alter function of many membrane proteins (Table 1 and Supporting Table S1). Higher concentrations are used in many cellular studies (e.g., in Supporting Table S1) but had frank bilayer-destabilizing effects in our experimental preparations, and for that reason, they were not studied further.
gA Single-Channel Electrophysiology
Lipid bilayers were formed using 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 2% (w/v) in n-decane. Synthesized gA analogues were prepared and purified as described previously.75 Single-channel experiments were performed using the bilayer punch method76 at 25 ± 1 °C using a Dagan 3900A Integrating patch clamp, with 200 mV applied potential in 1 M NaCl, 10 mM HEPES, pH 7.0. The current signal was low-pass Bessel filtered at 2–5 kHz, digitized at 20 kHz and digitally filtered at 500 Hz. Single-channel current transition amplitudes and lifetimes were determined as described previously76,77 and the average channel lifetimes were determined by fitting a single exponential distribution to the lifetime histograms.77 Detailed descriptions of the phytochemicals’ effects at the singe-channel level can be found regarding capsaicin,56 curcumin,74 EGCG78,79 genistein,80 and resveratrol (Supporting Information Figure S4).
gA Based Fluorescence Assay
The phytochemicals’ bilayer-modifying potency was determined using a gramicidin-based fluorescence assay as described previously.42,81 Fluorophore-loaded large unilamellar lipid vesicles (LUVs) were made of 1,2-dierucoyl-sn-glycero-3-phosphocholine (DC22:1PC) using a mixture of hydration, sonication, freeze–thawing, and extrusion. We use long-chain lipids to increase the bilayer thickness sufficiently to shift the gramicidin monomer↔dimer equilibrium toward the nonconducting monomers, which is necessary in order to detect changes in the monomer↔dimer equilibrium. The LUVs were doped with 260 nM gramicidin (gA) from Bacillus brevis 24 h before use and incubated at 12 °C in the dark. The phytochemicals were incubated for 10 min at 25 °C with the LUV suspension. The vesicle-entrapped fluorophore, 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS), is quenched by a gramicidin permeable quencher (Tl+). The time course of fluorescence quenching was measured using a SX.20 Stopped-Flow Spectrometer from Applied Photophysics, excitation was set at 352 nm and the fluorescence emission above 455 nm was recorded. The quencher fluorescence was normalized to the initial buffer value and a stretched exponential82 was fit to the first 2–100 ms and the rate at 2 ms was calculated. Each phytochemical was measured in four to six experiments from at least two different vesicle preparations.
MscL Fluorescence Assay
The phytochemicals’ effect on MscL function was measured using a calcein fluorescence assay.49 When entrapped in vesicles at high concentration calcein self-quenches, such that the fluorescence increases when calcein is released from the vesicles. Calcein-loaded asolectin LUVs were made by extrusion, E. coli G22C MscL mutant at 1:50 protein to lipid (w/w) ratio was incorporated and external calcein was removed using a Sephadex G50 size-exclusion column. Calcein fluorescence was monitored using a Varian Cary Eclipse fluorometer excitation at 495 nm and emission recorded at 515 nm. Phytochemicals were added to the vesicle suspension and incubated for 3–5 min. [2-(Trimethylammonium)ethyl] methanethiosulfonate bromide (MTSET; 1 mM) was then added to activate the G22C mutant MscL and release calcein from the vesicles. At the end of the experiment 0.5% (v/v) Triton X-100 (Triton) was added to dissolve all vesicles and measure the maximal fluorescence. All data were normalized using initial fluorescence as 0% calcein release and fluorescence after Triton addition as 100% release. Maximum release was recorded right before Triton addition. Control vesicles without MscL were prepared and only nominal calcein release was observed (at the relevant time scales) both with and without added phytochemicals, except for the very highest phytochemical concentrations tested which was presumably due to the phytochemicals’ destabilization of the vesicles (data not shown).
KV2.1 Electrophysiology
A CHO-K1 cell line stably transfected with rat KV2.1 in a tetracycline-inducible vector83 was maintained in Ham’s F12 media containing 10% fetal bovine serum, 1% penicillin-streptomycin, 1 μg/mL blasticidin, and 25 μg/mL zeocin at 37 °C in a 5% CO2 atmosphere. To induce KV2.1 expression for electrophysiological recording, 1 μg/mL tetracycline was added to the maintenance media for 1 h. Cells were harvested by scraping in a phosphate buffered saline solution containing EDTA, pelleted at 1000 g for 2 min, resuspended in CHO-SFMII media (Invitrogen) and rotated at room temperature (RT) until use. Standard whole cell patch clamp recordings were used to measure currents from KV2.1 channels (IK). Aliquots of the CHO cell suspension were added to a recording chamber and rinsed with external solution before sealing. The external (bath) solution contained (in mM): 50 HEPES, 20 KOH, 155 NaCl, 2 CaCl2, 2 MgCl2, 0.1 MgEDTA, pH 7.3. After sealing, the external solution was replaced with one containing 5 μM tetrodotoxin, 0.1% DMSO ± phytochemicals. The internal (pipet) solution contained (in mM): 50 KF, 70 KCl, 35 KOH, 5 EGTA, 50 HEPES, and adjusted to pH 7.3 with HCl. Pipettes were pulled from thin wall borosilicate glass (Sutter), coated with Sylgard (Dow), heat-cured and fire polished. Pipette tip resistances with these solutions were <3 MΩ. Recordings were at RT (22–24 °C). Voltage clamp was achieved with an Axon200B amplifier and digitized with a HEKA ITC-18 controlled by Patchmaster software. Recordings were low-pass Bessel filtered at 10 kHz by the amplifier and smoothed with a 1 kHz Gaussian filter for presentation. Series resistance compensation was used to constrain maximal voltage error to less than 10 mV. Holding potential was −100 mV. KV2.1 currents were activated by step depolarization to potentials illustrated in the figures. P/5 leak subtraction was used from −100 mV. Solution exchange was accomplished by flowing at least 200 μL of desired solution through a recording chamber containing less than 100 μL solution. Experiments varying external K+ concentrations while measuring IK reversal indicated >90% solution exchange. Data analysis was conducted using IgorPro (Wavemetrics), which uses a Levenberg–Marquardt algorithm for least-squares fitting. Conductances were determined from peak currents using a calculated K+ reversal potential of −52 mV. Boltzmann and time constant fitting procedures are as described previously.84 Inactivation rates varied widely and were influenced by solution flow. They were not analyzed in detail.
NaV Electrophysiology
Endogenous TTX-sensitive Na+ currents were recorded from the mammalian neuronal cell line ND7/23 as previously described.58 In brief, cells were grown on 12 mm coverslips and whole cell voltage-clamp recordings were performed at RT (22–24 °C) using a patch-clamp amplifier (Molecular Devices) with a sampling rate of 50 kHz and a 10 kHz low-pass filter. External bath solution contained (in mM): 130 NaCl, 10 HEPES, 3.25 KCl, 2 MgCl2, 2 CaCl2, 20 TEACl, and 5 d-glucose, adjusted to pH 7.4 (with NaOH), and 310 mOsm/kg H2O (with sucrose). Recording pipettes were pulled from borosilicate glass capillaries (Sutter Instruments, Novato, CA) using a P-97 puller (Sutter Instruments). Pipettes had a tip resistance of 1.5–2.5 MΩ when filled with following pipet solution (in mM): 120 CsF, 10 NaCl, 10 HEPES, 10 EGTA, 10 TEACl, 1 CaCl2, and 1 MgCl2, adjusted to pH 7.3 (with CsOH), and 310 mOsm/kg H2O (with sucrose). Access resistance was further decreased using 70–80% series resistance compensation. Liquid–junction potentials were not corrected; capacitative current transients were electronically canceled with the internal amplifier circuitry. Stock solutions of phytochemicals were prepared in DMSO and further diluted to the final working concentration with external bath solution prior to the experiment. Control solutions had the same amount of DMSO as the drug solutions and did not exceed 0.14%. Cells were superfused using a pressurized perfusion system (ALA Scientific Instruments) with a 200 μm-diameter perfusion pipet positioned in close proximity of the cell.
Ectodomain Shedding Assay
Shedding of alkaline phosphatase (AP)-tagged transforming growth factor α (TGFα) was monitored as described previously.85 Briefly immortalized WT-mouse embryonic fibroblasts (mEFs) on six-well plates were transiently transfected with AP-tagged TGFα and treated with or without capsaicin, EGCG, genistein, or the positive control phorbol-12-myristate-13-acetate (PMA) the day after transfection at the indicated concentrations. AP activity in the supernatant (conditioned for 45 min) and cell lysates was measured by colorimetry. Three identical wells were prepared, and the ratio between the AP activity in the supernatant and the cell lysate plus supernatant was calculated for normalization. Curcumin and resveratrol could not be tested as they interfered with the colorimetric detection assay.
Acknowledgments
This work was financially supported by The Netherlands Organization for Scientific Research (NWO): H.I.I. by a Rubicon grant, A.K. by an ERC-Starting grant (208814), and S.J.M., D.H.J., and X.P. by an ECHO grant (700.58.005). P.T. and J.T.S. were supported by U.S. National Institutes of Health (NIH) grant 5P30GM092328-02 and U.S. American Heart Association (AHA) grant 10SDG4220047. O.S.A., E.A.H., H.I.I., and N.B.R. were supported by U.S. NIH grant R01 GM021342 and U.S. American Recovery and Reinvestment Act (ARRA) supplement GM0213420-35S1. H.C.H. and K.F.H. were supported by U.S. NIH grant GM58055.
Glossary
Abbreviations
- EGCG
epigallocatechin gallate
- MD
molecular dynamics
- CG
coarse-grained
- PMF
potential of mean force
- KV2.1
voltage-dependent potassium channels
- MscL
mechanosensitive channel of large conductance
- NaV
voltage-dependent sodium channels
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DC22:1PC
1,2-dierucoyl-sn-glycero-3-phosphocholine
- LPC
lysophosphatidylcholine
- LUVs
large unilamellar lipid vesicles
- gA
gramicidin
- MTSET
[2-(trimethylammonium)ethyl] methanethiosulfonate bromide
- ANTS
8-aminonaphthalene-1,3,6-trisulfonic acid
- TGFα
transforming growth factor α
- PMA
phorbol-12-myristate-13-acetate
- AP
alkaline phosphatase
Supporting Information Available
Additional methods for the MD simulations, providing details on both the force field and the simulation protocols; extended listing with references of membrane proteins shown to be affected by the five phytochemicals; the phytochemicals effects on membrane properties, as measured with CG and atomistic MD simulation; further details on how the phytochemicals reduce the energy required to perturb the bilayer; details on the effects of resveratrol on gA channel activity; CG mapping schema for the phytochemicals. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
H.I.I., J.T.S., and O.S.A. designed the overall experimental plan. X.P., D.H.J., H.I.I., and S.J.M. were responsible for the MD simulations and analysis. E.A.H., H.I.I., N.B.R., and O.S.A. conducted the gramicidin experiments and analysis. M.Z., D.Y., and A.K. conducted the MscL experiments and analysis. P.T. and J.T.S. conducted the KV2.1 experiments and analysis. K.F.H. and H.C.H. conducted the NaV experiments and analysis. K.H., T.M., and C.B. conducted the ADAM17 experiments and analysis. H.I.I., J.T.S., and O.S.A. wrote the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Joe B.; Vijaykumar M.; Lokesh B. R. (2004) Biological properties of curcumin—cellular and molecular mechanisms of action. Crit. Rev. Food Sci. Nutr. 44, 97–111. [DOI] [PubMed] [Google Scholar]
- Shishodia S.; Sethi G.; Aggarwal B. B. (2005) Curcumin: Getting back to the roots. Ann. N.Y. Acad. Sci. 1056, 206–217. [DOI] [PubMed] [Google Scholar]
- Shimizu M.; Weinstein I. B. (2005) Modulation of signal transduction by tea catechins and related phytochemicals. Mutat. Res. 591, 147–160. [DOI] [PubMed] [Google Scholar]
- Khan N.; Afaq F.; Saleem M.; Ahmad N.; Mukhtar H. (2006) Targeting multiple signaling pathways by green tea polyphenol (−)-epigallocatechin-3-gallate. Cancer Res. 66, 2500–2505. [DOI] [PubMed] [Google Scholar]
- Nagle D. G.; Ferreira D.; Zhou Y.-D. (2006) Epigallocatechin-3-gallate (EGCG): chemical and biomedical perspectives. Phytochemistry 67, 1849–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramassamy C. (2006) Emerging role of polyphenolic compounds in the treatment of neurodegenerative diseases: A review of their intracellular targets. Eur. J. Pharmacol. 545, 51–64. [DOI] [PubMed] [Google Scholar]
- Deorukhkar A.; Krishnan S.; Sethi G.; Aggarwal B. B. (2007) Back to basics: How natural products can provide the basis for new therapeutics. Expert Opin. Investig. Drugs 16, 1753–1773. [DOI] [PubMed] [Google Scholar]
- Goel A.; Kunnumakkara A. B.; Aggarwal B. B. (2008) Curcumin as “Curecumin”: From kitchen to clinic. Biochem. Pharmacol. 75, 787–809. [DOI] [PubMed] [Google Scholar]
- Harikumar K. B.; Aggarwal B. B. (2008) Resveratrol: A multitargeted agent for age-associated chronic diseases. Cell Cycle 7, 1020–1035. [DOI] [PubMed] [Google Scholar]
- Chacko S. M.; Thambi P. T.; Kuttan R.; Nishigaki I. (2010) Beneficial effects of green tea: A literature review. Chin. Med. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S.; Phromnoi K.; Yadav V.; Chaturvedi M.; Aggarwal B. (2010) Targeting inflammatory pathways by flavonoids for prevention and treatment of cancer. Planta Med. 76, 1044–1063. [DOI] [PubMed] [Google Scholar]
- Verma S. P.; Salamone E.; Goldin B. (1997) Curcumin and genistein, plant natural products, show synergistic inhibitory effects on the growth of human breast cancer MCF-7 cells induced by estrogenic pesticides. Biochem. Biophys. Res. Commun. 233, 692–696. [DOI] [PubMed] [Google Scholar]
- Khafif A.; Schantz S. P.; Chou T. C.; Edelstein D.; Sacks P. G. (1998) Quantitation of chemopreventive synergism between (−)-epigallocatechin-3-gallate and curcumin in normal, premalignant and malignant human oral epithelial cells. Carcinogenesis 19, 419–424. [DOI] [PubMed] [Google Scholar]
- Zheng J.; Ramirez V. D. (2000) Inhibition of mitochondrial proton F0F1-ATPase/ATP synthase by polyphenolic phytochemicals. Br. J. Pharmacol. 130, 1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen O. S.; Koeppe R. E. II. (2007) Bilayer thickness and membrane protein function: An energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 36, 107–130. [DOI] [PubMed] [Google Scholar]
- Lundbæk J. A.; Collingwood S. A.; Ingólfsson H. I.; Kapoor R.; Andersen O. S. (2010) Lipid bilayer regulation of membrane protein function: Gramicidin channels as molecular force probes. J. R. Soc. Interface 7, 373–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina M. J.; Schumacher M. A.; Tominaga M.; Rosen T. A.; Levine J. D.; Julius D. (1997) The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature 389, 816–824. [DOI] [PubMed] [Google Scholar]
- Cao E.; Liao M.; Cheng Y.; Julius D. (2013) TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana H.; Koga K.; Fujimura Y.; Yamada K. (2004) A receptor for green tea polyphenol EGCG. Nat. Struct. Mol. Biol. 11, 380–381. [DOI] [PubMed] [Google Scholar]
- chemicalize.org was used to predict octanol water partition coefficients (log Poct/wat). ChemAxon: http://www.chemaxon.com (accessed May 2013).
- Meddings J. B.; Hogaboam C. M.; Tran K.; Reynolds J. D.; Wallace J. L. (1991) Capsaicin effects on non-neuronal plasma membranes. Biochim. Biophys. Acta 1070, 43–50. [DOI] [PubMed] [Google Scholar]
- Aranda F. J.; Villalain J.; Gomez-Fernandez J. C. (1995) Capsaicin affects the structure and phase organization of phospholipid membranes. Biochim. Biophys. Acta 1234, 225–234. [DOI] [PubMed] [Google Scholar]
- Jaruga E.; Sokal A.; Chrul S.; Bartosz G. (1998) Apoptosis-independent alterations in membrane dynamics induced by curcumin. Exp. Cell Res. 245, 303–312. [DOI] [PubMed] [Google Scholar]
- Lambert J. W.; Sum A. K. (2006) Molecular dynamics study of the properties of capsaicin in an 1-octanol/water system. J. Phys. Chem. B 110, 2351–2357. [DOI] [PubMed] [Google Scholar]
- Tamba Y.; Ohba S.; Kubota M.; Yoshioka H.; Yoshioka H.; Yamazaki M. (2007) Single GUV method reveals interaction of tea catechin (−)-epigallocatechin gallate with lipid membranes. Biophys. J. 92, 3178–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung W. C.; Chen F. Y.; Lee C. C.; Sun Y.; Lee M. T.; Huang H. W. (2008) Membrane-thinning effect of curcumin. Biophys. J. 94, 4331–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirk T. W.; Brown E. F.; Sum A. K.; Friedman M. (2008) Molecular dynamics study on the biophysical interactions of seven green tea catechins with lipid bilayers of cell membranes. J. Agric. Food Chem. 56, 7750–7758. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Lee C. C.; Hung W. C.; Chen F. Y.; Lee M. T.; Huang H. W. (2008) The bound states of amphipathic drugs in lipid bilayers: Study of curcumin. Biophys. J. 95, 2318–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Hung W.-C.; Chen F.-Y.; Lee C.-C.; Huang H. W. (2009) Interaction of tea catechin (−)-epigallocatechin gallate with lipid bilayers. Biophys. J. 96, 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J.; Fritz M.; Brender J. R.; Smith P. E.; Lee D. K.; Ramamoorthy A. (2009) Determining the effects of lipophilic drugs on membrane structure by solid-state NMR spectroscopy: The case of the antioxidant curcumin. J. Am. Chem. Soc. 131, 4490–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Chu S.; Hagerman A. E.; Lorigan G. A. (2011) Probing the interaction of polyphenols with lipid bilayers by solid-state NMR spectroscopy. J. Agric. Food Chem. 59, 6783–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan M.; Zubovski Y.; Venable R. M.; Pastor R. W.; Nagle J. F.; Tristram-Nagle S. (2012) Structure and elasticity of lipid membranes with genistein and daidzein bioflavinoids using X-ray scattering and MD simulations. J. Phys. Chem. B 116, 3918–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S. A.; Disalvo E. A.; Gawrisch K.; Borovyagin V.; Toone E.; Schiffman S. S.; Needham D.; McIntosh T. J. (1994) Increased adhesion between neutral lipid bilayers: Interbilayer bridges formed by tannic acid. Biophys. J. 66, 1943–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh N. W.; Porter N. A.; McIntosh T. J.; Simon S. A. (1996) The interaction of polyphenols with bilayers: Conditions for increasing bilayer adhesion. Biophys. J. 71, 3261–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaruga E.; Salvioli S.; Dobrucki J.; Chrul S.; Bandorowicz-Pikula J.; Sikora E.; Franceschi C.; Cossarizza A.; Bartosz G. (1998) Apoptosis-like, reversible changes in plasma membrane asymmetry and permeability, and transient modifications in mitochondrial membrane potential induced by curcumin in rat thymocytes. FEBS Lett. 433, 287–293. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; Tieleman D. P. (2013) Perspective on the Martini model. Chem. Soc. Rev. 42, 6801. [DOI] [PubMed] [Google Scholar]
- Cantor R. S. (1997) Lateral pressures in cell membranes: A mechanism for modulation of protein function. J. Phys. Chem. B 101, 1723–1725. [Google Scholar]
- Andersen O. S., Sawyer D. B., and Koeppe R. E. II (1992) Modulation of channel function by the host bilayer, in Biomembrane Structure and Function (Easwaran K. R. K., and Gaber B., Eds.), pp 227–244. Adenine Press, Schenectady, NY. [Google Scholar]
- Nielsen C.; Goulian M.; Andersen O. S. (1998) Energetics of inclusion-induced bilayer deformations. Biophys. J. 74, 1966–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C.; Andersen O. S. (2000) Inclusion-induced bilayer deformations: Effects of monolayer equilibrium curvature. Biophys. J. 79, 2583–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen O. S.; Bruno M. J.; Sun H.; Koeppe R. E. II. (2007) Single-molecule methods for monitoring changes in bilayer elastic properties. Methods Mol. Biol. 400, 543–570. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Andersen O. S. (2010) Screening for small molecules’ bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay Drug Dev. Technol. 8, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner S. M.; Shyamsunder E. (1991) Is the mechanism of general anesthesia related to lipid membrane spontaneous curvature?. Ann. N.Y. Acad. Sci. 625, 685–697. [DOI] [PubMed] [Google Scholar]
- Lee A. G. (2004) How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87. [DOI] [PubMed] [Google Scholar]
- Bruno M. J.; Koeppe R. E. II; Andersen O. S. (2007) Docosahexaenoic acid alters bilayer elastic properties. Proc. Natl. Acad. Sci. U.S.A. 104, 9638–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levina N.; Tötemeyer S.; Stokes N. R.; Louis P.; Jones M. A.; Booth I. R. (1999) Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: Identification of genes required for MscS activity. EMBO J. 18, 1730–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B.; Adler J.; Kung C. (1990) Mechanosensitive ion channels of E. coli activated by amphipaths. Nature 348, 261–263. [DOI] [PubMed] [Google Scholar]
- Perozo E.; Kloda A.; Cortes D. M.; Martinac B. (2002) Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 9, 696–703. [DOI] [PubMed] [Google Scholar]
- Kocer A.; Walko M.; Feringa B. L. (2007) Synthesis and utilization of reversible and irreversible light-activated nanovalves derived from the channel protein MscL. Nat. Protoc. 2, 1426–1437. [DOI] [PubMed] [Google Scholar]
- Yoshimura K.; Batiza A.; Kung C. (2001) Chemically charging the pore constriction opens the mechanosensitive channel MscL. Biophys. J. 80, 2198–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitko U.; Juranka P. F.; Morris C. E. (2006) Membrane stretch slows the concerted step prior to opening in a Kv channel. J. Gen. Physiol. 127, 687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balijepalli R. C.; Delisle B. P.; Balijepalli S. Y.; Foell J. D.; Slind J. K.; Kamp T. J.; January C. T. (2007) Kv11.1 (ERG1) K+ channels localize in cholesterol and sphingolipid enriched membranes and are modulated by membrane cholesterol. Channels (Austin) 1, 263–272. [DOI] [PubMed] [Google Scholar]
- Rana O. R.; Zobel C.; Saygili E.; Brixius K.; Gramley F.; Schimpf T.; Mischke K.; Frechen D.; Knackstedt C.; Schwinger R. H. G.; Schauerte P.; Saygili E. (2008) A simple device to apply equibiaxial strain to cells cultured on flexible membranes. Am. J. Physiol. Heart. Circ. Physiol. 294, H532–40. [DOI] [PubMed] [Google Scholar]
- Finol-Urdaneta R. K.; McArthur J. R.; Juranka P. F.; French R. J.; Morris C. E. (2010) Modulation of KvAP unitary conductance and gating by 1-alkanols and other surface active agents. Biophys. J. 98, 762–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D.; del Mármol J.; MacKinnon R. (2012) Mechanistic basis for low threshold mechanosensitivity in voltage-dependent K+ channels. Proc. Natl. Acad. Sci. U.S.A. 109, 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundbæk J. A.; Birn P.; Tape S. E.; Toombes G. E.; Søgaard R.; Koeppe R. E. II; Gruner S. M.; Hansen A. J.; Andersen O. S. (2005) Capsaicin regulates voltage-dependent sodium channels by altering lipid bilayer elasticity. Mol. Pharmacol. 68, 680–689. [DOI] [PubMed] [Google Scholar]
- Morris C. E.; Juranka P. F. (2007) Nav channel mechanosensitivity: Activation and inactivation accelerate reversibly with stretch. Biophys. J. 93, 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova R.; Herold K. F.; Sanford R. L.; Greathouse D. V.; Hemmings H. C.; Andersen O. S. (2011) Thiazolidinedione insulin sensitizers alter lipid bilayer properties and voltage-dependent sodium channel function: Implications for drug discovery. J. Gen. Physiol. 138, 249–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnarborg S. W.; Hinkle C. L.; Stevenson M.; Russell W. E.; Raska C. S.; Peschon J. J.; Castner B. J.; Gerhart M. J.; Paxton R. J.; Black R. A.; Lee D. C. (2002) Tumor necrosis factor-α converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J. Biol. Chem. 277, 12838–12845. [DOI] [PubMed] [Google Scholar]
- Le Gall S. M.; Maretzky T.; Issuree P. D. A.; Niu X. D.; Reiss K.; Saftig P.; Khokha R.; Lundell D.; Blobel C. P. (2010) ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J. Cell Sci. 123, 3913–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon J. J.; Slack J. L.; Reddy P.; Stocking K. L.; Sunnarborg S. W.; Lee D. C.; Russell W. E.; Castner B. J.; Johnson R. S.; Fitzner J. N.; Boyce R. W.; Nelson N.; Kozlosky C. J.; Wolfson M. F.; Rauch C. T.; Cerretti D. P.; Paxton R. J.; March C. J.; Black R. A. (1998) An essential role for ectodomain shedding in mammalian development. Science 282, 1281–1284. [DOI] [PubMed] [Google Scholar]
- Liu L.; Oortgiesen M.; Li L.; Simon S. A. (2001) Capsaicin inhibits activation of voltage-gated sodium currents in capsaicin-sensitive trigeminal ganglion neurons. J. Neurophysiol. 85, 745–758. [DOI] [PubMed] [Google Scholar]
- Cao X.; Cao X.; Xie H.; Yang R.; Lei G.; Li F.; Li A.; Liu C.; Liu L. (2007) Effects of capsaicin on VGSCs in TRPV1–/– mice. Brain Res. 1163, 33–43. [DOI] [PubMed] [Google Scholar]
- Xu Y. P.; Zhang J. W.; Li L.; Ye Z. Y.; Zhang Y.; Gao X.; Li F.; Yan X. S.; Liu Z. G.; Liu L. J.; Cao X. H. (2012) Complex regulation of capsaicin on intracellular second messengers by calcium dependent and independent mechanisms in primary sensory neurons. Neurosci. Lett. 517, 30–35. [DOI] [PubMed] [Google Scholar]
- Bienvenüe A.; Marie J. S. (1994) Modulation of protein function by lipids. Curr. Top. Membr. 40, 319–354. [Google Scholar]
- Marsh D. (2008) Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta 1778, 1545–1575. [DOI] [PubMed] [Google Scholar]
- Lundbæk J. A.; Koeppe R. E. II; Andersen O. S. (2010) Amphiphile regulation of ion channel function by changes in the bilayer spring constant. Proc. Natl. Acad. Sci. U.S.A. 107, 15427–15430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollila O. H. S.; Louhivuori M.; Marrink S. J.; Vattulainen I. (2011) Protein shape change has a major effect on the gating energy of a mechanosensitive channel. Biophys. J. 100, 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink S. J.; De Vries A. H.; Mark A. E. (2004) Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B 108, 750–760. [Google Scholar]
- Marrink S. J.; Risselada H. J.; Yefimov S.; Tieleman D. P.; De Vries A. H. (2007) The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824. [DOI] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447. [DOI] [PubMed] [Google Scholar]
- Pronk S.; Pall S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; Hess B.; Lindahl E. (2013) GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.; Postma J. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690. [Google Scholar]
- Ingólfsson H. I.; Koeppe R. E. II; Andersen O. S. (2007) Curcumin is a modulator of bilayer material properties. Biochemistry 46, 10384–10391. [DOI] [PubMed] [Google Scholar]
- Greathouse D. V.; Koeppe R. E. II; Providence L. L.; Shobana S.; Andersen O. S. (1999) Design and characterization of gramicidin channels. Meth. Enzymol. 294, 525–550. [DOI] [PubMed] [Google Scholar]
- Andersen O. S. (1983) Ion movement through gramicidin A channels. Single-channel measurements at very high potentials. Biophys. J. 41, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin J. T.; Koeppe R. E. II; Andersen O. S. (1990) Energetics of gramicidin hybrid channel formation as a test for structural equivalence. Side-chain substitutions in the native sequence. J. Mol. Biol. 211, 221–234. [DOI] [PubMed] [Google Scholar]
- Adachi S.; Nagao T.; Ingólfsson H. I.; Maxfield F. R.; Andersen O. S.; Kopelovich L.; Weinstein I. B. (2007) The inhibitory effect of (−)-epigallocatechin gallate on activation of the epidermal growth factor receptor is associated with altered lipid order in HT29 colon cancer cells. Cancer Res. 67, 6493–6501. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Koeppe R. E. II; Andersen O. S. (2011) Effects of green tea catechins on gramicidin channel function and inferred changes in bilayer properties. FEBS Lett. 585, 3101–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang T. C.; Koeppe R. E. II; Andersen O. S. (2003) Genistein can modulate channel function by a phosphorylation-independent mechanism: Importance of hydrophobic mismatch and bilayer mechanics. Biochemistry 42, 13646–13658. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Sanford R. L.; Kapoor R.; Andersen O. S. (2010) Gramicidin-based fluorescence assay; for determining small molecules potential for modifying lipid bilayer properties. J. Vis. Exp 2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberan-Santos M. N.; Bodunov E. N.; Valeur B. (2005) Mathematical functions for the analysis of luminescence decays with underlying distributions 1. Kohlrausch decay function (stretched exponential). Chem. Phys. 315, 171–182. [Google Scholar]
- Trapani J. G.; Korn S. J. (2003) Control of ion channel expression for patch clamp recordings using an inducible expression system in mammalian cell lines. BMC Neurosci. 4, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack J. T.; Aldrich R. W. (2006) Binding of a gating modifier toxin induces intersubunit cooperativity early in the Shaker K channel’s activation pathway. J. Gen. Physiol. 128, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T.; Yang G.; Ouerfelli O.; Overall C. M.; Worpenberg S.; Hassiepen U.; Eder J.; Blobel C. P. (2009) Characterization of the catalytic activity of the membrane-anchored metalloproteinase ADAM15 in cell-based assays. Biochem. J. 420, 105–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.