Abstract
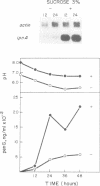
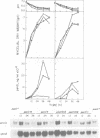
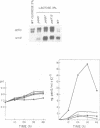
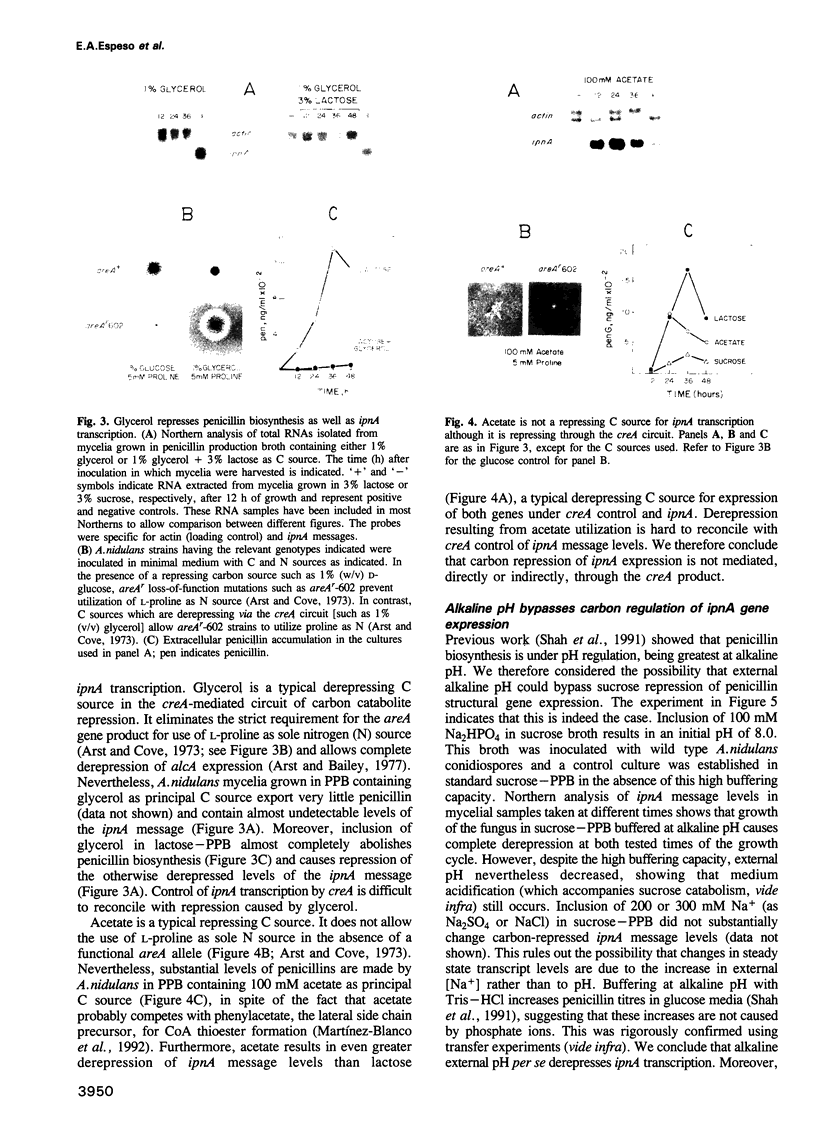
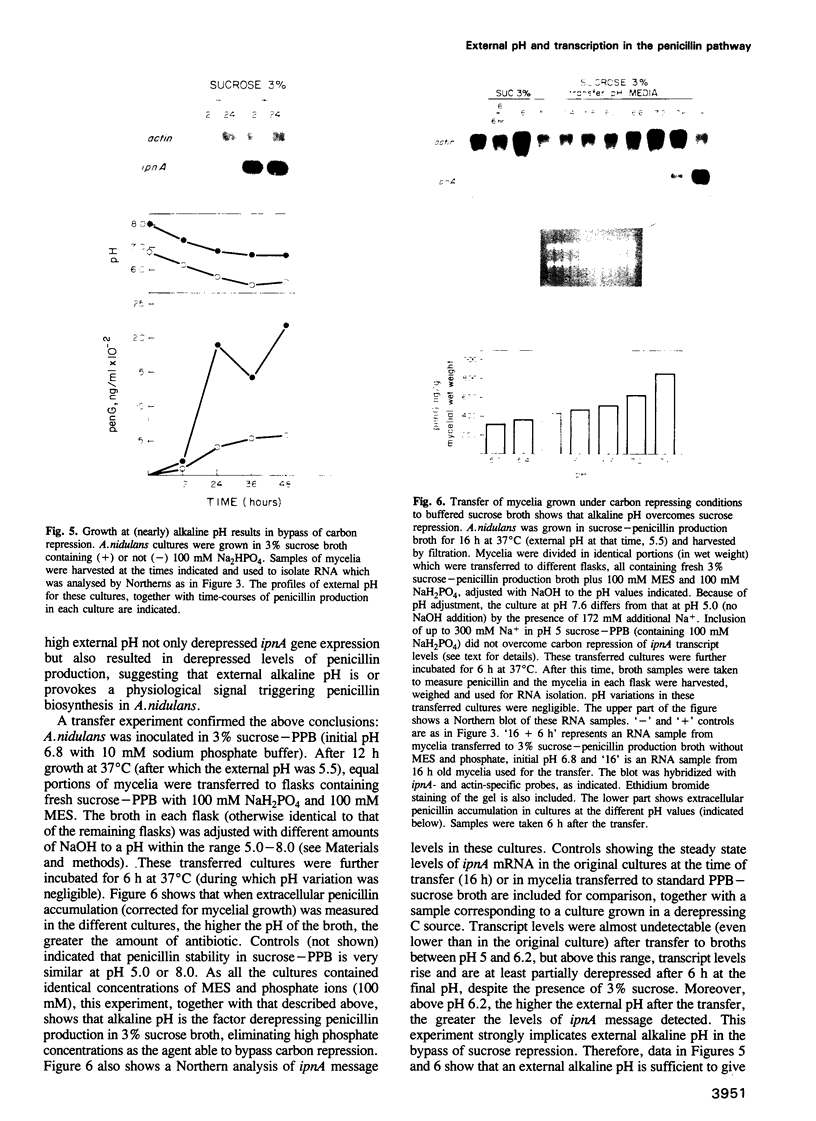
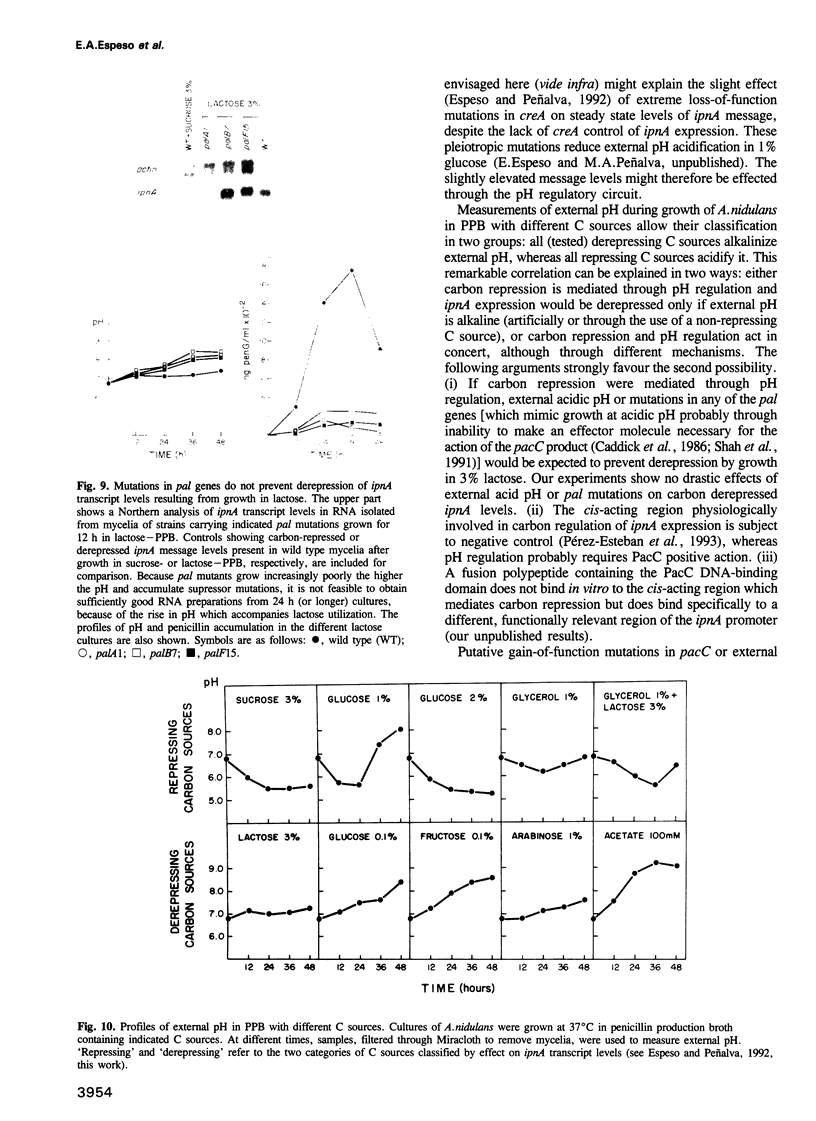
Transcription of the ipnA gene encoding isopenicillin N synthetase, an enzyme of secondary metabolism, is under the control of the pH regulatory system in the fungus Aspergillus nidulans. External alkaline pH or mutations in pacC, the wide domain regulatory gene which mediates pH regulation, override carbon regulation of ipnA transcript levels, resulting in elevation of the levels of this message in sucrose broth. Strains carrying these mutations, which mimic growth at alkaline pH, produce higher levels of penicillins when grown in sucrose broth compared with the wild type strain grown under carbon derepressing conditions. ipnA transcription is regulated by carbon (C) source, but extreme mutations in creA (the regulatory gene mediating carbon catabolite repression) only slightly increase repressed transcript levels. Precise deletion of the only in vitro CreA binding site present in a region of the ipnA promoter involved in carbon regulation has no effect on ipnA expression. The levels of ipnA transcript in broths with acetate or glycerol as principal C sources are inconsistent with direct or indirect creA-mediated transcriptional control of the gene. We conclude that a second, creA-independent mechanism of carbon repression controls expression of this gene. All derepressing C sources tested result in alkalinization of the growth media. In contrast, all repressing C sources result in external acidification. Neither acidic external pH nor pal mutations, mimicking the effects of growth at acid pH, prevent carbon derepression, providing strong support for independent regulatory mechanisms, one mediating carbon regulation (via thus far unidentified genes) and another mediating pH regulation (through the pacC-encoded transcriptional regulator). External pH measurements suggest that these two independent forms of regulation normally act in concert. We propose that external alkalinity represents a physiological signal which triggers penicillin biosynthesis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arst H. N., Jr, Cove D. J. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973 Nov 2;126(2):111–141. doi: 10.1007/BF00330988. [DOI] [PubMed] [Google Scholar]
- Arst H. N., Jr, MacDonald D. W. A gene cluster in Aspergillus nidulans with an internally located cis-acting regulatory region. Nature. 1975 Mar 6;254(5495):26–31. doi: 10.1038/254026a0. [DOI] [PubMed] [Google Scholar]
- Arst H. N., Jr, Tollervey D., Dowzer C. E., Kelly J. M. An inversion truncating the creA gene of Aspergillus nidulans results in carbon catabolite derepression. Mol Microbiol. 1990 May;4(5):851–854. doi: 10.1111/j.1365-2958.1990.tb00656.x. [DOI] [PubMed] [Google Scholar]
- Bailey C., Arst H. N., Jr Carbon catabolite repression in Aspergillos nidulans. Eur J Biochem. 1975 Feb 21;51(2):573–577. doi: 10.1111/j.1432-1033.1975.tb03958.x. [DOI] [PubMed] [Google Scholar]
- Caddick M. X., Brownlee A. G., Arst H. N., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986 May;203(2):346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- Cove D. J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966 Jan 11;113(1):51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- Dowzer C. E., Kelly J. M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991 Nov;11(11):5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeso E. A., Peñalva M. A. Carbon catabolite repression can account for the temporal pattern of expression of a penicillin biosynthetic gene in Aspergillus nidulans. Mol Microbiol. 1992 Jun;6(11):1457–1465. doi: 10.1111/j.1365-2958.1992.tb00866.x. [DOI] [PubMed] [Google Scholar]
- Fernández-Cañn J. M., Reglero A., Martínez-Blanco H., Luengo J. M. Uptake of phenylacetic acid by Penicillium chrysogenum Wis 54-1255: a critical regulatory point in benzylpenicillin biosynthesis. J Antibiot (Tokyo) 1989 Sep;42(9):1398–1409. doi: 10.7164/antibiotics.42.1398. [DOI] [PubMed] [Google Scholar]
- Haavik H. I. Studies on the formation of bacitracin by Bacillus licheniformis: effect of glucose. J Gen Microbiol. 1974 Apr;81(2):383–390. doi: 10.1099/00221287-81-2-383. [DOI] [PubMed] [Google Scholar]
- Kulmburg P., Mathieu M., Dowzer C., Kelly J., Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993 Mar;7(6):847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- Kulmburg P., Sequeval D., Lenouvel F., Mathieu M., Felenbok B. Identification of the promoter region involved in the autoregulation of the transcriptional activator ALCR in Aspergillus nidulans. Mol Cell Biol. 1992 May;12(5):1932–1939. doi: 10.1128/mcb.12.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCabe A. P., Riach M. B., Unkles S. E., Kinghorn J. R. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990 Jan;9(1):279–287. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Blanco H., Reglero A., Fernández-Valverde M., Ferrero M. A., Moreno M. A., Peñalva M. A., Luengo J. M. Isolation and characterization of the acetyl-CoA synthetase from Penicillium chrysogenum. Involvement of this enzyme in the biosynthesis of penicillins. J Biol Chem. 1992 Mar 15;267(8):5474–5481. [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
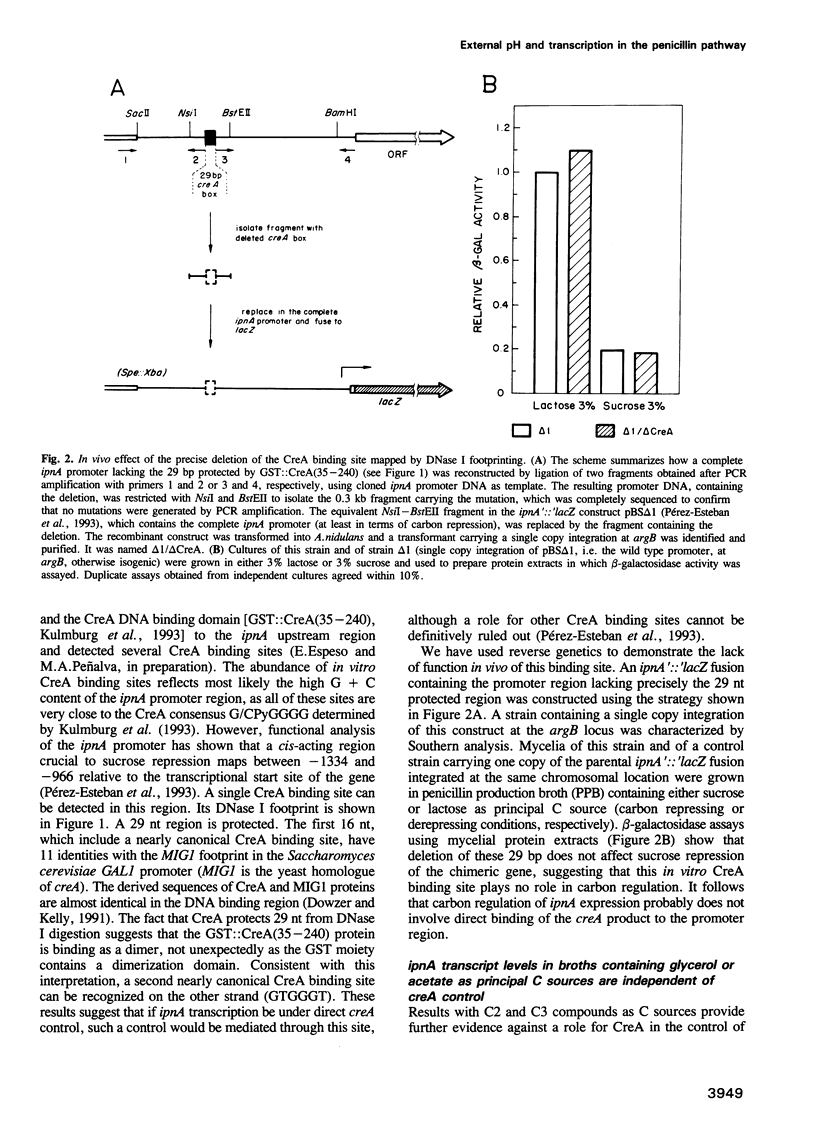
- Pérez-Esteban B., Orejas M., Gómez-Pardo E., Peñalva M. A. Molecular characterization of a fungal secondary metabolism promoter: transcription of the Aspergillus nidulans isopenicillin N synthetase gene is modulated by upstream negative elements. Mol Microbiol. 1993 Aug;9(4):881–895. doi: 10.1111/j.1365-2958.1993.tb01746.x. [DOI] [PubMed] [Google Scholar]
- Rossi A., Arst H. N., Jr Mutants of Aspergillus nidulans able to grow at extremely acidic pH acidify the medium less than wild type when grown at more moderate pH. FEMS Microbiol Lett. 1990 Jan 1;54(1-3):51–53. doi: 10.1016/0378-1097(90)90257-q. [DOI] [PubMed] [Google Scholar]
- Shah A. J., Tilburn J., Adlard M. W., Arst H. N., Jr pH regulation of penicillin production in Aspergillus nidulans. FEMS Microbiol Lett. 1991 Jan 15;61(2-3):209–212. doi: 10.1016/0378-1097(91)90553-m. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sophianopoulou V., Suárez T., Diallinas G., Scazzocchio C. Operator derepressed mutations in the proline utilisation gene cluster of Aspergillus nidulans. Mol Gen Genet. 1993 Jan;236(2-3):209–213. doi: 10.1007/BF00277114. [DOI] [PubMed] [Google Scholar]
- den Hollander J. A., Ugurbil K., Brown T. R., Shulman R. G. Phosphorus-31 nuclear magnetic resonance studies of the effect of oxygen upon glycolysis in yeast. Biochemistry. 1981 Sep 29;20(20):5871–5880. doi: 10.1021/bi00523a034. [DOI] [PubMed] [Google Scholar]