Abstract
To better understand the transcriptional regulation of high molecular weight glutenin subunit (HMW-GS) expression, we isolated four Glu-1Bx promoters from six wheat cultivars exhibiting diverse protein expression levels. The activities of the diverse Glu-1Bx promoters were tested and compared with β-glucuronidase (GUS) reporter fusions. Although all the full-length Glu-1Bx promoters showed endosperm-specific activities, the strongest GUS activity was observed with the 1Bx7OE promoter in both transient expression assays and stable transgenic rice lines. A 43 bp insertion in the 1Bx7OE promoter, which is absent in the 1Bx7 promoter, led to enhanced expression. Analysis of promoter deletion constructs confirmed that a 185 bp MITE (miniature inverted-repeat transposable element) in the 1Bx14 promoter had a weak positive effect on Glu-1Bx expression, and a 54 bp deletion in the 1Bx13 promoter reduced endosperm-specific activity. To investigate the effect of the 43 bp insertion in the 1Bx7OE promoter, a functional marker was developed to screen 505 Chinese varieties and 160 European varieties, and only 1Bx7-type varieties harboring the 43 bp insertion in their promoters showed similar overexpression patterns. Hence, the 1Bx7OE promoter should be important tool in crop genetic engineering as well as in molecular assisted breeding.
Introduction
Hexaploid wheat (Triticum aestivum L.) is one of the most important human food sources. Its complex genetic background leads to great diversity in nutritional and processing qualities among cultivars. High molecular weight glutenin subunits (HMW-GSs) are the main grain storage proteins in the endosperms of wheat and related species [1], [2]. Although HWM-GSs grain storage proteins account for only about 12% of the total protein [3], they play a key role in wheat gluten as the skeletal network that to a large extent determines its structure and formation [4]. The compositions and quantities of allelic variation in HMW-GS genes substantially affect the taste and appearance of dough products, such as Chinese noodles and European bread [5]. Therefore, improvement of flour quality based on superior HWM-GS alleles is necessary to meet changing consumer demands.
Both qualitative and quantitative effects of HMW-GS subunits are important for flour quality [6], [7]. In the process of breeding, high dough strength is used as a predictor of good-quality bread wheat; and overexpression of Glu-1Bx7 by way of allele 1Bx7OE makes an important contribution to high dough strength in some cultivars [8], [9]. Expression of HMW-GS is regulated by three major factors, which are at the genomic level (gene duplication), transcriptional level and translational level [10]–[13]. Transcriptional regulation driven by Glu-1 5′-upstream flanking regions might provide strategies for improving grain quality in wheat breeding programs [14]. A number of crucial cis-acting elements from HMW-GS promoters of various wheat cultivars have been investigated and characterized, as these could affect tissue specificity or expression activity, including conservative endosperm-specific motifs, such as the GCN4 motif [15], the prolamin box [16], AACA/TA motif [17], RY repeat motif [18], and Skn-1 [19], [20], each of which is capable of exerting temporal expression [21], [22]. However, the basis of transcriptional regulation of divergence caused by large insertion and deletion (InDel) alterations in HMW-GS promoter regions is still not clear. As reported earlier, a tandem 54 bp duplication, known as the “cereal-box” located at −400 bp in the 1Bx promoter may enhance endosperm-specific expression [1], suggesting this duplicated region might be a key region for control of gene expression [23], [24]. There is a 185 bp MITE insertion in the promoters of 1Bx14 and 1Bx20, but functional verification indicated that this insertion had little effect on gene expression [25]–[27]. A 43 bp insertion found at −1000 bp in the 1Bx7OE promoter was significantly associated with the overexpression phenotype. It was speculated that the overexpression was brought about by gene duplication mediated by the insertion of a retroelement [13], and there was no further study concerning the 43 bp InDel effect on protein expression. Therefore, more experimental data are needed to clarify the effect of InDels in HMW-GS promoters.
Highly active endosperm-specific promoters serve as an important genetic resource for high-quality and high-yield wheat breeding. Use of seed storage protein gene promoters is an attractive strategy for obtaining target gene products exclusively from crop kernels. A number of seed-specific promoters from barley, rice, maize and other species have been investigated functionally [15], [21], [28]. Transgenic crops with favorable gene stacking require different tissue-specific promoters from various cereals, as this is helpful to reduce homology-based transcriptional gene silencing [29], [30]. HMW-GS promoters from wheat, although containing endosperm-specific motifs, may not be spatially controlled in the same way as in their original genetic backgrounds due to subtle differences in respective regulation systems [31]. Hence, further research of key motifs from tissue-specific promoters would boost applicability in genetic engineering.
Among hexaploid wheat HMW-GSs, Glu-1Bx often shows the highest level of expression [32]. We therefore set out to analyse 1Bx promoter sequence characteristics to uncover the transcriptional regulation mechanism. Based on diverse protein expression levels in six wheat cultivars, we isolated four Glu-1Bx promoters in approximately 2.2 kb of length and further validated their functions. By comparison with these upstream sequences, several large InDels such as a 43 bp InDel, a 54 bp duplication and a 185 bp MITE resulted in major divergences among the four promoters, including the 1Bx7 promoter (Pro-1Bx7), 1Bx7OE promoter (Pro-1Bx7OE), 1Bx13 promoter (Pro-1Bx13) and 1Bx14 promoter (Pro-1Bx14). The promoter sequence variation was shown to be an important factor causing differential expression in transient expression systems and in transgenic rice plant assays. Notably, Pro-1Bx7OE is a highly active endosperm-specific promoter that can be made available for crop improvement by transgenic methods. Moreover, we developed a new specific molecular marker in terms of the 43 bp insertion residing in the 1Bx7OE promoter, with which we screened 505 Chinese and 160 European cultivars [33]. We found that this functional marker is significantly associated with 1Bx7 overexpression. Our results further showed that transcriptional regulation might be responsible for 1Bx expression diversity to a larger extent than initially expected.
Materials and Methods
Plant materials
Hexaploid wheat (Triticum aestivum L.) cultivars (cv.) Yanzhan 1, Atlas 66, Jimai 20, Xiaoyan 54, Yunmai 33 and Chinese Spring were grown in the field. Endosperm of Xiaoyan 54 was prepared for transient expression assays, and rice (Oryza sativa L. ssp japonica) cv. Kita-ake was used to produce stable transformants. Materials used for molecular marker screening included 505 Chinese and 160 European cultivars [33].
SDS-PAGE and quantification of HMW-GSs
Protein fractions were extracted from single wheat kernels using a previously reported HMW-GS extraction protocol [34]. Identical amounts of protein extracted from seeds of different varieties were separated by SDS-PAGE and visualized by Coomassie Blue staining as described by Zhang et al. [35]. Densitometric analyses of 1Bx subunits were carried out by Quantity One software (Bio-rad, USA). The value of the optical density multiplication area was used to quantify HMW-GS expression.
Promoter isolation and cis-element prediction
DNA extraction was performed as previously described [36]. Using a pair of specific primer sets, 1Bx2258F/R (Table S1), four full-length 1Bx promoters, ∼2.3 kb in size, from six wheat varieties were isolated, gel purified and sequenced. Putative regulatory elements within the 1Bx promoter were predicted using the Plant Cis-acting Regulatory DNA elements (PLACE) database [37] combined with a previous report [38].
Construction of promoter-GUS chimeric genes and subsequent transformation
Several full-length and truncated 1Bx promoters were obtained by PCR amplification with primers introducing DNA restriction enzyme sites for convenient subcloning (Table S1), and cloned into a modified vector PAHC25 [39] for transient expression, and then subcloned into binary vector pCAMBIA1391z, containing the reporter gene GUS under the control of different 1Bx promoters. In transient expression experiments, immature embryos harvested at 12–14 days post anthesis (DPA) were used for bombardment as described by Ortiz et al. [40]. Different 1Bx promoter-GUS constructs were tested in transient expression assays as described previously [41]. For stable transformation, the binary vector constructs were first introduced into Agrobacterium tumefaciens strain EHA105, and then rice transformation was carried out as described by Cho et al. [42]. Transgenic rice plants were selected on medium containing 50 mg L−1 of hygromycin, and positive lines were grown in the field for further analysis.
PCR and Southern blot analyses of transformed rice plants
Genomic DNA was isolated from leaf tissues of transformed rice plants as previously described [36]. PCR analysis for molecular identification of transgenic rice plants was performed using a set of specific primers for the 1Bx promoter and GUS gene (listed in Table S1).
For Southern blot analysis, genomic DNA (10 µg) from different transgenic lines were digested with BamHI or HindIII (New England Biolabs, USA). Digested DNA was separated by electrophoresis in 0.8% (w/v) agarose gels, and then transferred to Hybond-N+ membranes (Amersham Biosciences, USA) and hybridized with a GUS gene fragment labeled with [α-32P]dCTP as described previously [36].
Histochemical GUS assay
Wheat endosperms undergoing transient expression and different tissues of T3 transgenic rice were used for histochemical GUS assays. GUS staining was performed as described by Kosugi et al. [43]. Images of stained samples were captured using an MZ16 High-tech Stereomicroscope (Leica, Germany). Stained GUS spots were counted, and for statistical comparison, data of each sample was expressed as the mean number of blue spots per endosperm.
Quantification of expression of the GUS gene under control of 1Bx promoters in transgenic rice plants
Total RNA was extracted from kernels of 3 independent positive T3 transgenic rice lines at 10–18 DPA. Transcriptional levels of GUS in all stable transgenic lines were quantified by quantitative real-time PCR (qRT-PCR) with a 7300 Real-time PCR system (Applied Biosystems, USA) using Power SYBR Green PCR Master Mix (Applied Biosystems, USA). Details of primer pairs used for qRT-PCR are given in Table S1. The specificity of the primer sets was assured by confirmation that the resulting products appeared as single peaks in real-time melting temperature curves and as single fragments after separation by agarose gel electrophoresis. To confirm adequate amplification PCR efficiency was assessed using a sample dilution series as templates [44]. Amplification plots and predicted threshold cycle values were obtained from three independent biological replicates with SDS software version 2.1 (Applied Biosystems, USA). GUS gene expression levels were presented as fold-changes calculated using the comparative threshold cycle (CT) method as described [45] with rice GAPDH used as the internal control.
Results
Identification of HMW-GSs by SDS-PAGE
HMW-GSs of six wheat cultivars were separated by SDS-PAGE (Figure 1A). Their subunit compositions varied from each other (Table S2). Evidently, expression levels of Glu-1A and Glu-1D are generally lower than Glu-1B. There were four allelic variants of 1Bx among the six cultivars, namely 1Bx7OE, 1Bx14, 1Bx13 and 1Bx7. The protein level of 1Bx7OE in Yunmai 33 was much higher than that of 1Bx in other cultivars, about 2.2-fold that of 1Bx13 and 1Bx7 and 1.8-fold that of 1Bx14 (Figure 1B).
Figure 1. HMW-GSs from six common wheat cultivars.
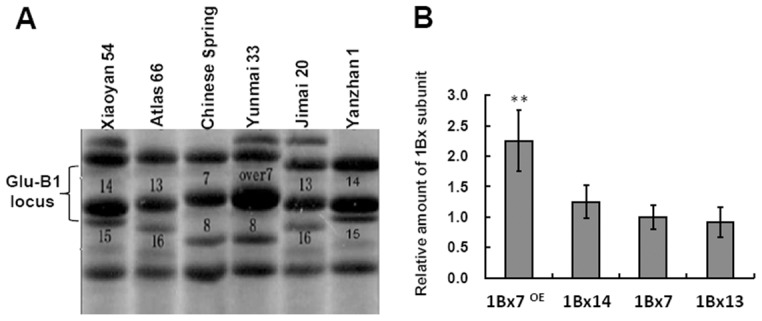
(A) SDS-PAGE profiles of HMW-GSs. 1Bx7 (Chinese Spring), 1Bx13 (Atlas 66 and Jimai 20), 1Bx14 (Xiaoyan 54 and Yanzhan 1) and 1Bx7OE (Yunmai 33). (B) Relative amounts of four 1Bx subunits estimated by densitometric analysis.
Comparative analysis of upstream sequences of Glu-1Bx
Four types of 5' flanking sequences of 1Bx alleles were isolated with the specific primer pair, 1Bx2258F/R (Table S1 and Figure S1). They were 2,294, 2,253, 2,185 and 2,433 bp in length for Pro-1Bx7OE, Pro-1Bx7, Pro-1Bx13, and Pro-1Bx14, respectively. The four 5' proximal flanking regions contained five common motifs, including DOF recognition sites, bZIP recognition sites, MYB recognition sites, VP1 recognition sites and basal promoter elements (Figure 2 and Table S3), which are conserved in promoters of genes that encode most seed storage proteins [38]. In addition to several single-base substitutions or small deletions, the presence of sequence insertions or deletions (InDels) constituted the main differences among the four entire promoter regions (Figure S2). By comparison with the 1Bx7 promoter, the 1Bx13 promoter has a 54 bp deletion at −400 upstream from the start codon (54 bp duplication position), the 1Bx14 promoter contains a 185 bp MITE insertion at −874, consistent with previous reports [26], [46], and the 1Bx7OE promoter possesses a 43 bp insertion at −1047, which is always associated with the overexpression phenotype [12].
Figure 2. Schematic structure of four 1Bx promoters.
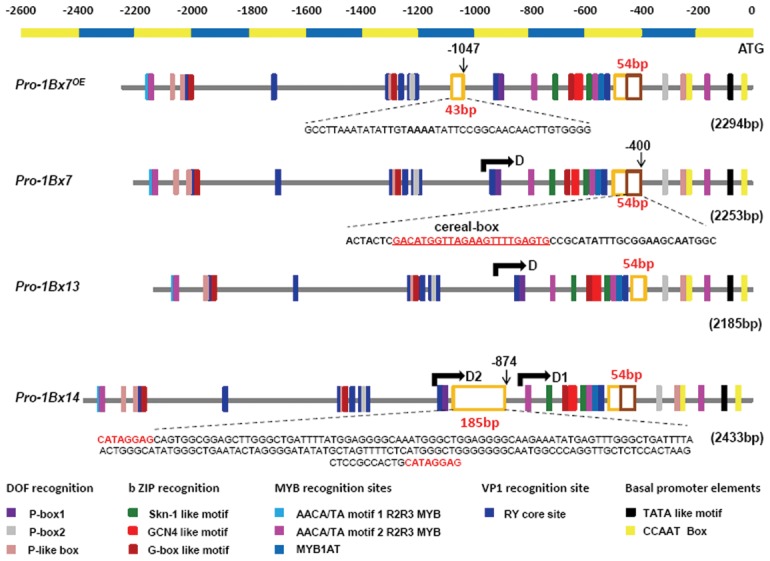
Regulatory elements are indicated by colored rectangles. InDels are labeled with hollow boxes and core sequences are listed in detail under the sketch map. The red underlined sequence shows the “cereal box” in the 54 bp duplication. The 8 bp bases in red at both ends indicate the target site duplication (TSD) of the MITE. Positions of upstream primers used for obtaining truncated promoters are indicated with black arrows.
The transient expression results for different 1Bx promoters
We compared the expression efficiencies of the full-length promoters of 1Bx7, 1Bx13 and 1Bx7OE by means of transient expression assays in wheat endosperms (Figure S3). and GUS driven by the 1Bx7OE promoter exhibited much higher activity than when driven by the 1Bx7 and 1Bx13 promoters (Figure 3A). Since the 43 bp InDel represents the difference between the 1Bx7 and 1Bx7OE promoter sequences, we speculated that the 43 bp insertion enhanced the endosperm-specific expression. In addition, the 1Bx13 promoter activity was lower than that of the 1Bx7 promoter, which lacks the 54 bp duplication present in the 1Bx13 promoter. We further investigated the effect of the 185 bp MITE on 1Bx14 expression. Two truncated 1Bx14 promoters were fused to GUS, and transient expression results showed that GUS expression driven by a 1,192 bp Pro-1Bx14-D2 was slightly higher than that driven by the 873 bp Pro-1Bx14-D1 (Figure 3B), confirming that the MITE might positively but weakly affected transcription of 1Bx14 [26].
Figure 3. Schematic representation of constructs used for transient expression assays in wheat endosperms and GUS activities driven by full-length and truncated 1Bx promoters.
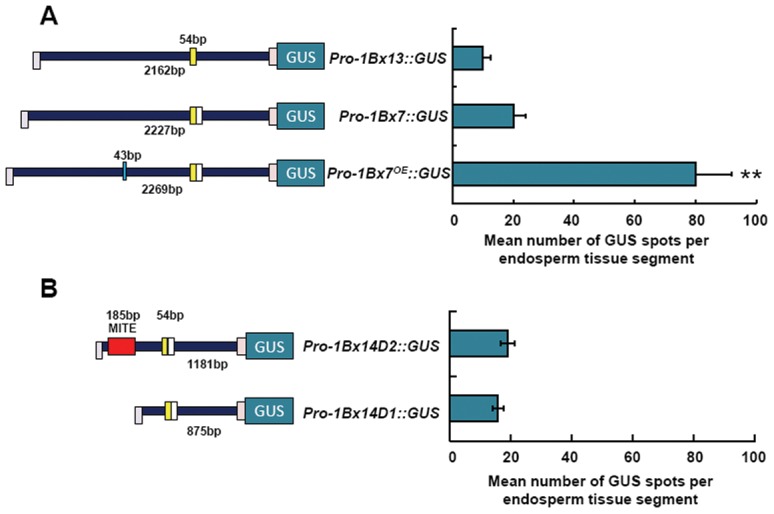
(A) Effects of full-length 1Bx7, 1Bx13 and 1Bx7OE promoters on transient expression. (B) Effect of the 185 bp MITE from the 1Bx14 promoter on transient expression. **P<0.01 (student’s t-test) indicates significant difference from others.
Histochemical and quantitative assays in transgenic rice
Chimeras were constructed using different 1Bx promoters fused to GUS and then transformed into rice. Through GUS staining assays in the T3 generation of stable transgenic plants, we detected GUS activity driven by the three full-length 1Bx promoters only in the seeds and not in stems or leaves collected at 15 DPA. These results indicated that the promoters are endosperm-specific (Figure 4A). In contrast, GUS staining was observed in all tissues of transgenic rice carrying the Ubiquitin promoter-GUS construct (Figure 4A). Therefore, the full-length 1Bx promoters contained necessary cis-elements that specify endosperm-specific regulation in both wheat and rice. Consistent with the transient expression results, the full-length 1Bx7 OE promoter with the 43 bp insertion exhibited much higher GUS activity than either the full-length 1Bx7 or 1Bx13 promoters (Figure 4A). Southern blot analysis confirmed that the transgenic rice lines had single copies of the GUS gene (Figure S4); therefore the comparative results of promoter activities were convincing.
Figure 4. GUS staining of various tissues and quantitative analysis in developing endosperm of transgenic rice.
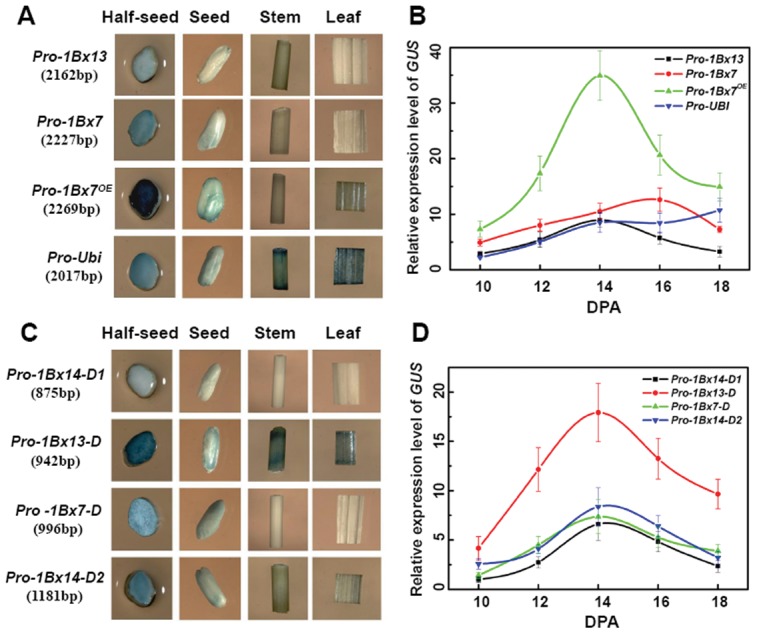
(A) and (C) Histochemical analysis of GUS driven by different promoters in transgenic rice tissues collected at 15 DPA; (B) and (D) Relative expression levels of GUS in seeds from different transgenic lines during 10 to 18 DPA based on qRT-PCR. Rice GAPDH was used as the internal control. Values are shown as means ± s.d (standard deviation) of three independent experiments and three biological replicates. Colored lines at the top right corner represent different transgenic plants.
We also investigated the activities of different truncated 1Bx promoters in transgenic rice. Except for the Pro-1Bx13-D promoter, other truncated promoters, including Pro-1Bx14-D1/D2 and Pro-1Bx7-D, retained endosperm-specific expression activity (Figure 4C). Absence of the 54 bp duplication led to loss of endosperm-specific expression controlled by the Pro-1Bx13-D promoter, thus indicating that the fragment removed from the 1Bx13 promoter (−943∼−2162) contained necessary cis-elements that restricted expression to the endosperm. Like transient expression results, GUS expression directed by Pro-1Bx14-D2 harboring the 185 bp MITE was higher than that directed by Pro-1Bx14-D1.
To confirm the results of GUS staining in seeds of transgenic rice, we applied qRT-PCR to determine expression levels of GUS. GUS expression detected in seeds during 10–18 DPA showed that expression driven by 1Bx promoters increased rapidly from 10 DPA to 14 DPA, and reached a peak level at 14–16 DPA (Figure 4B and 4D). As the control, GUS expression driven by the Ubiquitin promoter maintained a relatively constant level through 10 to 18 DPA (Figure 4B). The results of GUS expression at 14–16 DPA were highly consistent with those of GUS activities based on histochemical staining (Figure 4A and 4C), confirming that the protein expression pattern was similar to the gene expression pattern at the mRNA level.
Phylogenetic analysis of HMW-GS promoters
To address the question of whether these InDels are present in other HMW-GS promoters, we analyzed promoters from 14 different wheat HMW-GS genes [1], [26], [47] and identified the regulatory motifs related to endosperm-specific expression in the promoter regions about 1,200 bp upstream of the initiation codon. The numbers of regulatory motifs obviously differed among the different HMW-GS promoter sequences (Figure 5). In phylogenetic analysis, all Glu-1Bx promoters clustered together in one branch. The 185 bp MITE insertion was present in both the Glu-1Bx14 and Glu-1Bx20 promoters. The 54 bp tandem duplication was absent in non-Glu-1Bx promoters, but present in all Glu-1Bx promoters except the Glu-1Bx13 promoter. Therefore, InDels contributed to the diversity in HMW-GS promoters, which is an important means for evolution of HMW-GS genes. These large fragment InDels can be used as a potential resource for creating new alleles.
Figure 5. Phylogenetic analysis of 14 HMW-GS promoters.
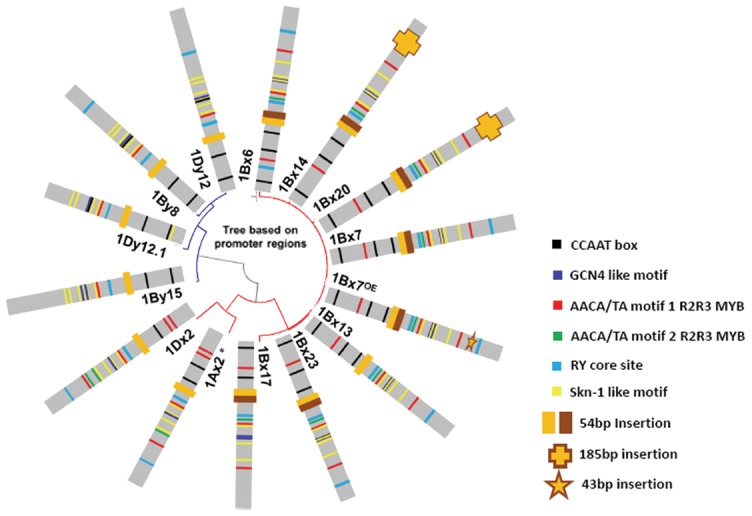
Neighbor-Joining Tree of partial length (-1 ∼ -1200 bp upstream of the start codon) sequences of Glu-1 from Triticum aestivum L. and other wheat-related grass species. This work was done with the MEGA program (Version 5.2). InDels and conserved cis-elements are shown by markers in different colors.
Development of a molecular marker for the 43 bp insertion and its distribution in natural populations
Based on the 43 bp insertion sequence in the 1Bx7OE promoter, we developed a new molecular marker that differed from those previously reported [34]. Our marker can precisely identify the insertion in HWM-GS promoters among common wheat varieties. PCR amplification resulted in two kinds of bands that distinguish promoters with the 43 bp insertion (a 476 bp fragment) from those without (a 433 bp fragment) (Figure 6A). Among 505 Chinese and 160 European accessions surveyed, we found 3 Chinese and 11 European varieties with the 43 bp insertion when we used this marker. HMW-GS profiles of accessions containing the 476 bp marker were later obtained by SDS-PAGE (Figure 6B). The presence of particular 1Bx alleles was determined by densitometric analysis; 10 accessions (3 from China and 7 from Europe) exhibited a 1Bx7 overexpression phenotype relative to Chinese Spring used as a control (Figure 6B and Table S4). We also identified some types of 1Bx6 and 1Bx14 with the 43 bp insertion (Figure 6B and Table S4), and confirmed its presence by DNA sequencing. However, these two types of 1Bx did not show overexpression at the protein level. Therefore, the 43 bp insertion in promoters preferentially enhanced 1Bx7 expression although no obvious differences were found among the upstream regions of 1Bx6, 1Bx14 and 1Bx7OE.
Figure 6. Electrophoretic separation of PCR products from 1Bx promoters with or without the 43 bp insertion, and SDS-PAGE profiles of HMW-GS of wheat cultivars with the insertion.
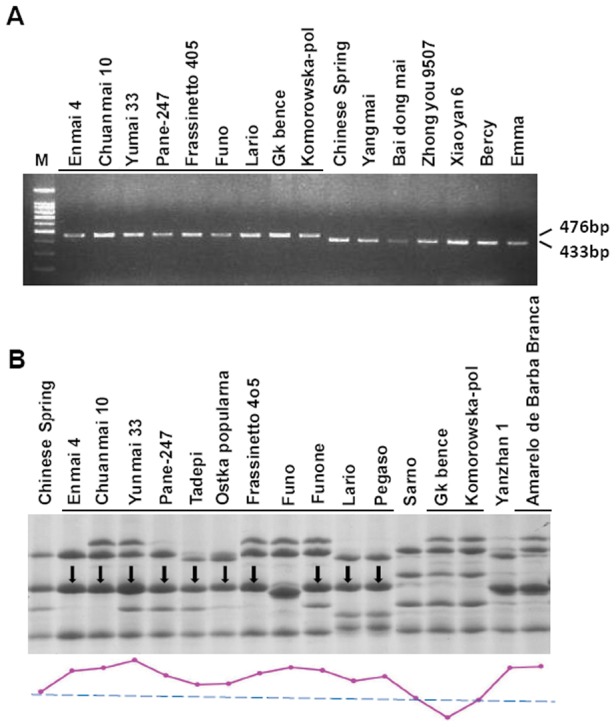
(A) PCR assays for 1Bx promoters on a 2% agarose gel. M: 100 bp DNA Ladder. Underlined accessions possess the 43 bp insertion. (B) SDS-PAGE assay of HMW-GS from different accessions containing the 43 bp insertion. Underlined accessions possess the 43 bp insertion. Down black arrows indicate 1Bx7 with 43 bp insertion in accessions from China and Europe. Chinese Spring, Yanzhan 1 and Samo were used as controls. The purple curve represents the relative amounts from different 1Bx subunits.
Discussion
HMW-GSs represent a set of important seed-storage proteins, and both their composition and quantity significantly affect wheat flour quality [7]. Since gene transcriptional regulation is the dominant means of control in production of proteins [48], we isolated four promoter sequences of Glu-1Bx and investigated their effects on gene expression. Although the reporter gene driven by all the 1Bx promoters exhibited an endosperm-specific expression pattern, the 1Bx7OE promoter from cv. Yunmai 33 produced a markedly stronger activity than other promoters. Previous studies showed that gene duplication was the cause of 1Bx7 overexpression in some wheat cultivars [49]. The connection between the strong activity of the 1Bx7OE promoter and high protein level produced by the 1Bx7OE subunit clearly indicated that transcription regulation is also a factor in 1Bx7 overexpression. We therefore concluded that multiple factors, including gene duplication and transcriptional regulation determine the expression of 1Bx7. This work revealed a complex regulatory network of HMW-GS expression in wheat.
The 43 bp insertion at −1047 bp is closely associated with high expression of 1Bx7OE
Large InDels in promoter regions often result in higher rates of transcriptional divergence [50]. In this study, we identified a 43 bp InDel, a 54 bp duplication and a 185 bp MITE in different Glu-1Bx promoters, and they accounted for the main differences among 1Bx promoter sequences. These InDels affected the expression levels of the genes. The presence of the 43 bp insertion at position −1047 upstream of the start codon was shown to be closely associated with high expression levels of the 1Bx7OE subunit [12], [13]. We verified that the 43 bp insertion can serve as a strong enhancer to improve expression of the gene by comparing the transcriptional activities between full-length 1Bx7 and 1Bx7OE promoters (Figure 3A; Figure 4A and 4B). Since there are no known cis-elements in the 43 bp insertion, this insertion may facilitate evolutionary tuning of gene expression by affecting local chromatin structure and nucleosome positioning [50].
The 185 bp MITE insertion at −874 bp of 1Bx14 might slightly affect transcription
The 185 bp MITE insertion located at −874 bp in the 1Bx14 promoter may be a remnant of an earlier transposition of a large element or of small, highly repeated elements [51]. In the present study, the 1Bx14 promoter with or without the 185 bp MITE, did not produce a significantly different activity in the transient system or in stable transgenic rice assays, suggesting that it might only slightly affect the transcriptional regulation (Figure 3B; Figure 4C and 4D). The 185 bp MITE exists in both hexaploid and tetraploid wheat, and may be linked to the polyploidization event affecting the constitutions and activities of the genomes of grass species [26].
A 54 bp cereal-box motif is necessary for endosperm-specific expression
The tandem 54 bp duplication at position −400 contains the “cereal-box” implicated in seed-specific expression [1]. Our data demonstrated that the 1Bx13 full-length promoter harboring one 54 bp deletion retains endosperm-specific activity, but a 1Bx13 promoter truncated at −942 bp lacks endosperm-specificity accompanied by increased activity (Figure 4C). We speculate that the 54 bp deletion might complement essential cis-elements in the region −940 to −2000 bp of the 1Bx13 promoter to effectively control gene endosperm-specific expression. Without the aid of the cis-elements in the region, only one 54 bp cereal-box motif may not be enough to restrict gene expression to endosperm. Based on phylogenetic analysis of HWM-GS promoters, only the 1Bx13 promoter and non-1Bx promoters contain a 54 bp deletion (Figure 5). This tandem 54 bp duplication must have occurred before hexaploidization because it is also present in tetraploid wheat. Flanking-sequence divergence was also noted from extensive DNA sequencing analysis of a-gliadin genes [52]. The basis of HMW-GS evolution is repeated sequence events that lead to new alleles [53].
A simple PCR marker was developed to target high expression of 1Bx7 and 1Bx7OE
Since previous 43 bp InDel marker covers a region of 1.2∼1.3 kb that also contains other InDels such as the 185 bp MITE and 54 bp duplication [34], a new specific marker based on the 43 bp insertion was developed and used effectively in two independent wheat populations combined with SDS-PAGE electrophoresis analysis to identify 1Bx7 overexpressing cultivars. Interestingly, the 43 bp insertion exists not only in the 1Bx7 promoter but also in other 1Bx promoters such as those of 1Bx14 and 1Bx6 (Figure 6B). Despite harboring the 43 bp insertion in the promoters, the 1Bx14 and 1Bx6 subunits produce no significant increases in protein compared to subunit alleles without the insertion (Figure 6B). The likely reason is that a co-regulatory factor linking the 43 bp insertion to expression efficiency is present in the 1Bx7 alleles or regulation at the translational level might strongly influence the divergence in expression between 1Bx7 and non-1Bx7 subunits.
Putative additive effects of gene duplication and transcriptional regulation on 1Bx7 expression
According to the literature, it is concluded that the 1Bx7 overexpression phenotype is mediated by an LTR retroelement resulting in gene duplication along with the polyploidization event [13]. In the present study, we confirmed that a 43 bp insertion situated in the 1Bx7OE promoter is capable of strengthening transcriptional activity markedly through transient expression and transgenic rice assays. By using molecular markers which can be used to indicate 1Bx7 gene duplication [13], we found that only the cultivar Yunmai 33 has both the 43 bp InDel and two 1Bx7 copies (gene duplication), while other 9 cultivars with the 43 bp InDel have only one 1Bx7 copy (Table S4). Although the 1Bx7 subunit of Yunmai 33 is the most abundant in this study (Figure 6B), other cultivars with the 43 bp InDel demonstrate higher 1Bx7 expression than the control, especially Chinese cultivars Enmai 4 and Chuanmai 10 (Figure 6B). So it can be inferred that both gene duplication and transcriptional regulation can lead to 1Bx7 overexpression, and their effects on 1Bx7 expression can be accumulated.
Endosperm is the storage tissue for starch and protein in cereal crops, which are the major sources of carbohydrates and proteins for humans. Improved yield and quality of crops by genetic modification has huge potential, and some significant achievements have already been accomplished [54]. Because continuous high expression of foreign genes in all tissues may cause detrimental effects in host plants [55], identification and application of strong endosperm-specific promoters will attract interest from breeders and biologists. In the current work, we identified a highly active 1Bx7OE promoter that can enhance endosperm-specific gene expression at the transcriptional level, and it should be useful for wheat quality improvement by means of genetic transformation and molecular assisted breeding.
Supporting Information
PCR amplification of 1Bx promoters by using 1Bx1007-F/R (A) and 1Bx2258-F/R (B) primer pairs. Lane 1–4: Chinese Spring (Pro-1Bx7); Yunmai 33 (Pro-1Bx7OE); Yanzhan 1 (Pro-1Bx14); Atlas 66 (Pro-1Bx13); M is a DNA ladder. The PCR products were separated in 1.5% agarose gels.
(PDF)
Alignment of four 1Bx promoters ( Pro-1Bx7 , Pro-1Bx7OE , Pro-1Bx13 and Pro-1Bx14 ).
(PDF)
Representative transient expression results of GUS driven by Pro-1Bx in wheat endosperms. (A) Pro-1Bx13 ; (B) Pro-1Bx7 .
(PDF)
Southern blot analysis of transgenic rice lines with full-length 1Bx promoters (A) or truncated 1Bx promoters (B). Genomic DNA was digested by BamHI and detected by GUS gene probes.
(PDF)
Primers used in this study.
(PDF)
HMW-GS compositions of six wheat accessions.
(PDF)
Details of 12 known endosperm-specific cis-elements in 1Bx promoters.
(PDF)
Fourteen wheat cultivars harboring the 43 bp insertion in the 1Bx promoter were identified by marker screening.
(PDF)
Acknowledgments
We are grateful to Dr. Jianmin Wan and Xiuping Guo for help with rice transformation, and gratefully acknowledge help with English editing from Prof. Robert A Mclntosh, University of Sydney.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
National Transgenic Research Project (ZX08002-004) and National Natural Science Foundation of China (30671293). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Anderson OD, Greene FC (1989) The characterization and comparative analysis of high-molecular-weight glutenin genes from genomes A and B of a hexaploid bread wheat. Theor Appl Genet 77: 689–700. [DOI] [PubMed] [Google Scholar]
- 2. Reddy P, Apples R (1993) Analysis of a genomic DNA segment carrying the wheat high molecular weight (HMW) glutenin Bx17 subunit and its use as an RFLP marker. Theor Appl Genet 85: 616–624. [DOI] [PubMed] [Google Scholar]
- 3. Halford NG, Forde J, Anderson OD, Greene FC, Shewry PR (1987) The nucleotide and deduced amino acid sequences of an HMW glutenin subunit gene from chromosome 1B of bread wheat (Triticum aestivum L.) and comparison with those of genes from chromosomes 1A and 1D. Theor Appl Genet 75: 117–126. [Google Scholar]
- 4. Shewry PR, Tatham AS (1990) The prolamin storage proteins of cereal seeds: structure and evolution. Biochem J 267: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu JJ, He ZH, Zhao ZD, Pena RJ, Rajaram S (2003) Wheat quality traits and quality parameters of cooked dry white Chinese noodles. Euphytica 131: 147–154. [Google Scholar]
- 6. Barro F, Rooke L, Békés F, Gras P, Tatham AS, et al. (1997) Transformation of wheat with high-molecular-weight subunit genes results in improved functional properties. Nat Biotechnol 15: 1295–1299. [DOI] [PubMed] [Google Scholar]
- 7. Wieser H (2000) Comparative investigations of gluten proteins from different wheat species. I. Qualitative and quantitative composition of gluten protein types. Eur Food Res Technol 211: 262–268. [Google Scholar]
- 8. Gupta RB, Paul JG, Cornish GB, Palmer GA, Bekes F, et al. (1994) Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1, of common wheats. I. Its additive and interaction effects on dough properties. J Cereal Sci 19: 9–17. [Google Scholar]
- 9. Cornish GB, Békés F, Allen HM, Martin DJ (2001) Flour proteins linked to quality traits in an Australian doubled haploid wheat population. Aust J Agr Res 52: 1339–1348. [Google Scholar]
- 10. Harberd NP, Flavell RB, Thompson RD (1987) Identification of a transposon-like insertion in a Glu-1 allele of wheat. Mol Genet Genomics 209: 326–332. [DOI] [PubMed] [Google Scholar]
- 11. Marchylo BA, Lukow OM, Kruger JE (1992) Quantitative variation in high molecular weight glutenin subunit 7 in some Canadian wheats. J Cereal Sci 15: 29–37. [Google Scholar]
- 12. Butow BJ, Gale KR, Ikea J, Juhász A, Bedo Z, et al. (2004) Dissemination of the highly expressed Bx7 glutenin subunit (Glu-B1al allele) in wheat as revealed by novel PCR markers and RP-HPLC. Theor Appl Genet 109: 1525–1535. [DOI] [PubMed] [Google Scholar]
- 13. Ragupathy R, Naeem HA, Reimer E, Lukow OM, Sapirstein HD, et al. (2008) Evolutionary origin of the segmental duplication encompassing the wheat GLU-B1 locus encoding the overexpressed Bx7 (Bx7OE) high molecular weight glutenin subunit. Theor Appl Genet 116: 283–296. [DOI] [PubMed] [Google Scholar]
- 14. Thomas MS, Flavell RB (1990) Identification of an enhancer element for the endosperm-specific expression of high molecular weight glutenin. Plant Cell 2: 1171–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng Z, Kawagoe Y, Xiao S, Li Z, Okita T, et al. (1993) 5′ distal and proximal cis-acting regulator elements are required for developmental control of a rice seed storage protein glutelin gene. Plant J 4: 357–366. [DOI] [PubMed] [Google Scholar]
- 16. Dong G, Ni Z, Yao Y, Nie X, Sun Q (2007) Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol Biol 63: 73–84. [DOI] [PubMed] [Google Scholar]
- 17. Takaiwa F, Yamanouchi U, Yoshihara T, Washida H, Tanabe F, et al. (1996) Characterization of common cis-regulatory elements responsible for the endosperm-specific expression of members of the rice glutelin multigene family. Plant Mol Biol 30: 1207–1221. [DOI] [PubMed] [Google Scholar]
- 18. Fujiwara T, Beachy RN (1994) Tissue-specific and temporal regulation of a β-conglycinin gene: roles of the RY repeat and other cis-acting elements. Plant Mol Biol 24: 261–273. [DOI] [PubMed] [Google Scholar]
- 19. Sha S, Sugiyama Y, Mitsukawa N, Masumura T, Tanaka K (1996) Cloning and sequencing of a rice gene encoding the 13-kDa prolamin polypeptide. Biosci Biotech Bioch 60: 335–337. [DOI] [PubMed] [Google Scholar]
- 20. Wu CY, Suzuki A, Washida H, Takaiwa F (1998) The GCN4 motif in a rice glutelin gene is essential for endosperm-specific gene expression and is activated by Opaque-2 in transgenic rice plants. Plant J 14: 673–683. [DOI] [PubMed] [Google Scholar]
- 21. Müller M, Knudsen S (1993) The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. Plant J 4: 343–355. [DOI] [PubMed] [Google Scholar]
- 22. Onodera Y, Suzuki A, Wu CY, Washida H, Takaiwa F (2001) A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J Biol Chem 276: 14139–14152. [DOI] [PubMed] [Google Scholar]
- 23. Forde J, Malpica JM, Halford NG, Shewry PR, Anderson OD, et al. (1985) The nucleotide sequence of a HMW glutenin subunit gene located on chromosome 1A of wheat (Triticum aestivum L.). Nucleic Acids Res 13: 6817–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halford NG, Ford J, Shewry PR, Kreis M (1989) Functional analysis of the upstream regions of a silent and an expressed member of a family of wheat seed-protein genes in transgenic tobacco. Plant Sci 62: 207–216. [Google Scholar]
- 25. Anderson OD, Larka L, Christoffers MJ, McCue KF, Gustafson JP (2002) Comparison of orthologous and paralogous DNA flanking the wheat high molecular weight glutenin genes: sequence conservation and divergence, transposon distribution, and matrix-attachment regions. Genome 45: 367–380. [DOI] [PubMed] [Google Scholar]
- 26. Li W, Wan Y, Liu Z, Liu K, Li B, et al. (2004) Molecular charaterization of HMW glutenin subunit allele 1Bx14: further insights into the evolution of Glu-B1-1 alleles in wheat and related species. Theor Appl Genet 109: 1093–1104. [DOI] [PubMed] [Google Scholar]
- 27. Jiang QT, Ma J, Zhao S, Zhao QZ, Lan XJ, et al. (2012) Characterization of HMW-GSs and their gene inaction in tetraploid wheat. Genetica 40: 325–335. [DOI] [PubMed] [Google Scholar]
- 28. Marks MD, Lindell JS, Larkins BA (1985) Quantitative analysis of the accumulation of zein mRNA during maize endosperm development. J Biol Chem 260: 16445–16450. [PubMed] [Google Scholar]
- 29. Butaye KMJ, Cammue BPA, Delaurè SL, De Bolle MFC (2005) Approaches to minimize variation of transgene expression in plants. Mol breeding 16: 79–91. [Google Scholar]
- 30. Qu LQ, Xing YP, Liu WX, Xu XP, Song YR (2008) Expression pattern and activity of six glutelin gene promoters in transgenic rice. J Exp Bot 59: 2417–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Furtado A, Henry RJ, Takaiwa F (2008) Comparison of promoters in transgenic rice. Plant Biotechnol J 6: 679–693. [DOI] [PubMed] [Google Scholar]
- 32. Galili G, Feldman M (1983) Genetic control of endosperm proteins in wheat. Theor appl genet 66: 77–86. [DOI] [PubMed] [Google Scholar]
- 33. Su Z, Hao C, Wang L, Dong Y, Zhang X (2011) Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor Appl Genet 122: 211–223. [DOI] [PubMed] [Google Scholar]
- 34. Radovanovic N, Cloutier S (2003) Gene-assisted selection for high molecular weight glutenin subunits in wheat doubled haploid breeding programs. Mol breeding 12: 51–59. [Google Scholar]
- 35. Zhang X, Huang C, Xu X, Hew CL (2002) Identification and localization of a prawn white spot syndrome virus gene that encodes an envelope protein. J Gen Virol 83: 1069–1074. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- 37. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Juhász A, Makai S, Sebestyén E, Tamás L, Balázs E (2011) Role of conserved non-coding regulatory elements in LMW glutenin gene expression. PloS ONE 6: e29501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218. [DOI] [PubMed] [Google Scholar]
- 40. Ortiz JP, Ravizzini RA, Morata MM, Vallejos RH (1997) A rapid system for studying foreign gene expression in wheat (Triticum aestivum L.). Theor Appl Genet 38: 123–130. [Google Scholar]
- 41. Oñate L, Vicente-Carbajosa J, Lara P, Diaz I, Carbonero P (1999) Barley BLZ2: a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J Biol Chem 274: 9175–9182. [DOI] [PubMed] [Google Scholar]
- 42. Cho HJ, Brotherton JE, Widholm JM (2004) Use of the tobacco feedback-insensitive anthranilate synthase gene ASA2 as a selectable marker for legume hairy root transformation. Plant Cell Res 23: 104–113. [DOI] [PubMed] [Google Scholar]
- 43. Kosugi S, Arai Y, Nakajima K, Ohashi Y (1990) An improved assay for β-glucuronidase (GUS) in transformed cells: methanol almost suppresses a putative endogenous GUS activity. Plant Sci 70: 133–140. [Google Scholar]
- 44.Rasmussen R (2001) Quantification on the Light Cycler instrument. In: Meuer S, Wittwer C, Nakagawara K, eds, Rapid Cycle Real-Time PCR, Methods and Applications. Springer Press, Heidelberg. pp.21–34. [Google Scholar]
- 45. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 46. Yang ZJ, Li GR, Liu C, Feng J, Zhou JP, et al. (2006) Molecular characterization of a HMW glutenin subunit allele providing evidence for silencing of x-type gene on Glu-B1 . Acta Genet Sin 33: 929–936. [DOI] [PubMed] [Google Scholar]
- 47. Yan Y, Zheng J, Xiao Y, Yu J, Hu Y, et al. (2004) Identification and molecular characterization of a novel y-type Glu-Dt1 glutenin gene of Aegilops tauschii . Theor Appl Genet 108: 1349–1358. [DOI] [PubMed] [Google Scholar]
- 48. Shaw LM, McIntyre CL, Gresshoff PM, Xue GP (2009) Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genomics 9: 485–498. [DOI] [PubMed] [Google Scholar]
- 49.Cloutier S, Banks T, Nilmalgoda S (2005) Molecular understanding of wheat evolution at the Glu-B1 locus. In: Proceedings of the international conference on plant genomics and biotechnology: challenges and opportunities, Raipur, India, p 40.
- 50. Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ (2009) Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324: 1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wessler SR (1998) Transposable elements and the evolution of gene expression. Symposia of the Society for Experimental Biology 51: 115–122. [PubMed] [Google Scholar]
- 52.Anderson OD (1997) Applications of molecular biology in understanding and improving wheat quality. In: Steele JL, Chung OK, eds. Proc Int Wheat Quality Conf, Grain Industry Alliance, Manhattan, KS. pp. 205–211.
- 53. SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, et al. (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- 54. Bajaj S, Mohanty A (2005) Recent advances in rice biotechnology towards genetically superior transgenic rice. Plant Biol 3: 275–307. [DOI] [PubMed] [Google Scholar]
- 55. Cheon BY, Kim HJ, Oh KH, Bahn SC, Ahn JH, et al. (2004) Overexpression of human erythropoietin (EPO) affects plant morphologies: retarded vegetative growth in tobacco and male sterility in tobacco and Arabidopsis. Transgenic Res 13: 541–549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR amplification of 1Bx promoters by using 1Bx1007-F/R (A) and 1Bx2258-F/R (B) primer pairs. Lane 1–4: Chinese Spring (Pro-1Bx7); Yunmai 33 (Pro-1Bx7OE); Yanzhan 1 (Pro-1Bx14); Atlas 66 (Pro-1Bx13); M is a DNA ladder. The PCR products were separated in 1.5% agarose gels.
(PDF)
Alignment of four 1Bx promoters ( Pro-1Bx7 , Pro-1Bx7OE , Pro-1Bx13 and Pro-1Bx14 ).
(PDF)
Representative transient expression results of GUS driven by Pro-1Bx in wheat endosperms. (A) Pro-1Bx13 ; (B) Pro-1Bx7 .
(PDF)
Southern blot analysis of transgenic rice lines with full-length 1Bx promoters (A) or truncated 1Bx promoters (B). Genomic DNA was digested by BamHI and detected by GUS gene probes.
(PDF)
Primers used in this study.
(PDF)
HMW-GS compositions of six wheat accessions.
(PDF)
Details of 12 known endosperm-specific cis-elements in 1Bx promoters.
(PDF)
Fourteen wheat cultivars harboring the 43 bp insertion in the 1Bx promoter were identified by marker screening.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.


