Abstract
We have analyzed base pairing interactions between the U5 snRNA and 5' exon sequences during pre-mRNA splicing in a mammalian in vivo system. We constructed synthetic U5 genes with mutations that alter four bases (C3, U4, U5 and U6) within the invariant 9 nt U5 sequence GCCUUUUAC; transient transfection of HeLa cells with these U5 sequences cloned into a U1 expression vector yielded high levels of the mutant snRNAs. To test their function, we cotransfected a rabbit beta-globin gene containing one of two mutations (G1-->A or T2-->A) in the essential GT dinucleotide at the 5' end of the second intron. Certain U5 loop mutants activated novel 5' splice sites only in mutant rabbit beta-globin transcripts. One novel site surprisingly resides in the first exon; its use is invariably coupled to utilization of a particular cryptic 5' splice site in the second exon. All of the newly activated cryptic 5' splice sites exhibit complementarity with the mutant U5 loop in the exon 1-5 nt upstream of the cryptic site, extending previous results in yeast. However, the register of the potential pairing is not identical at the various novel cryptic 5' splice sites, indicating that the interaction between the U5 loop and the 5' exon may be more flexible than previously believed.
Full text
PDF

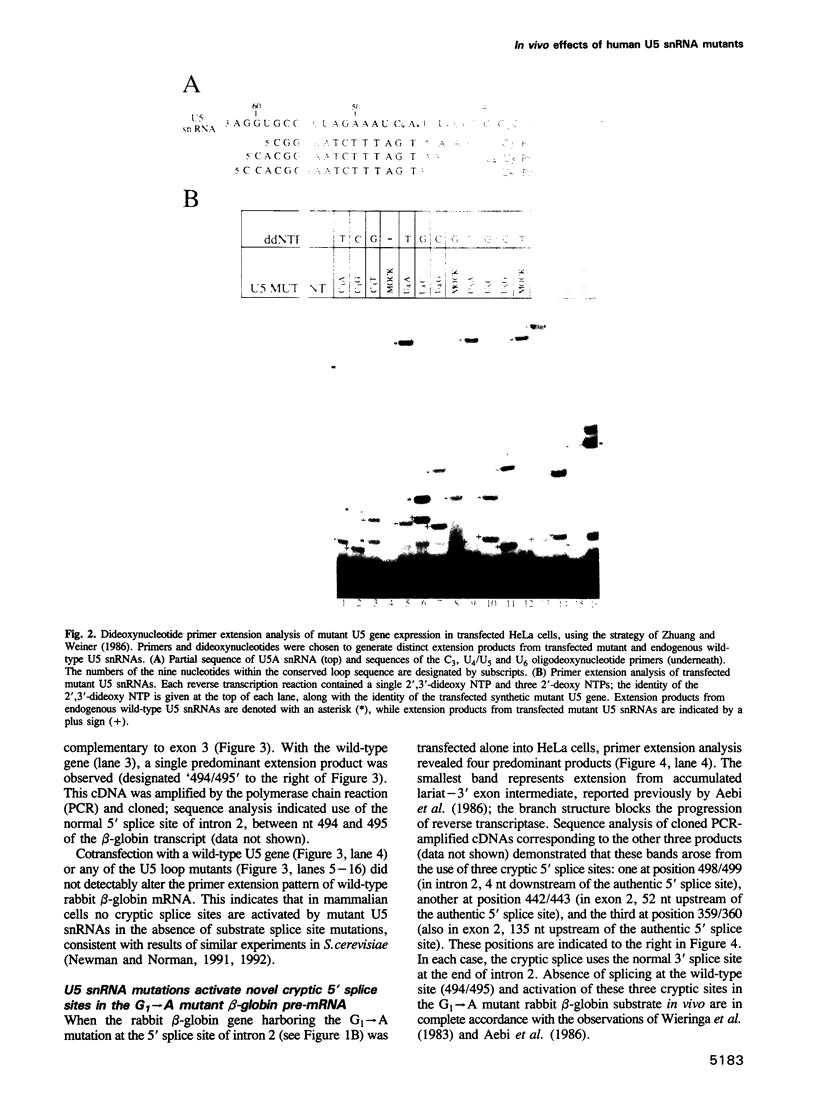


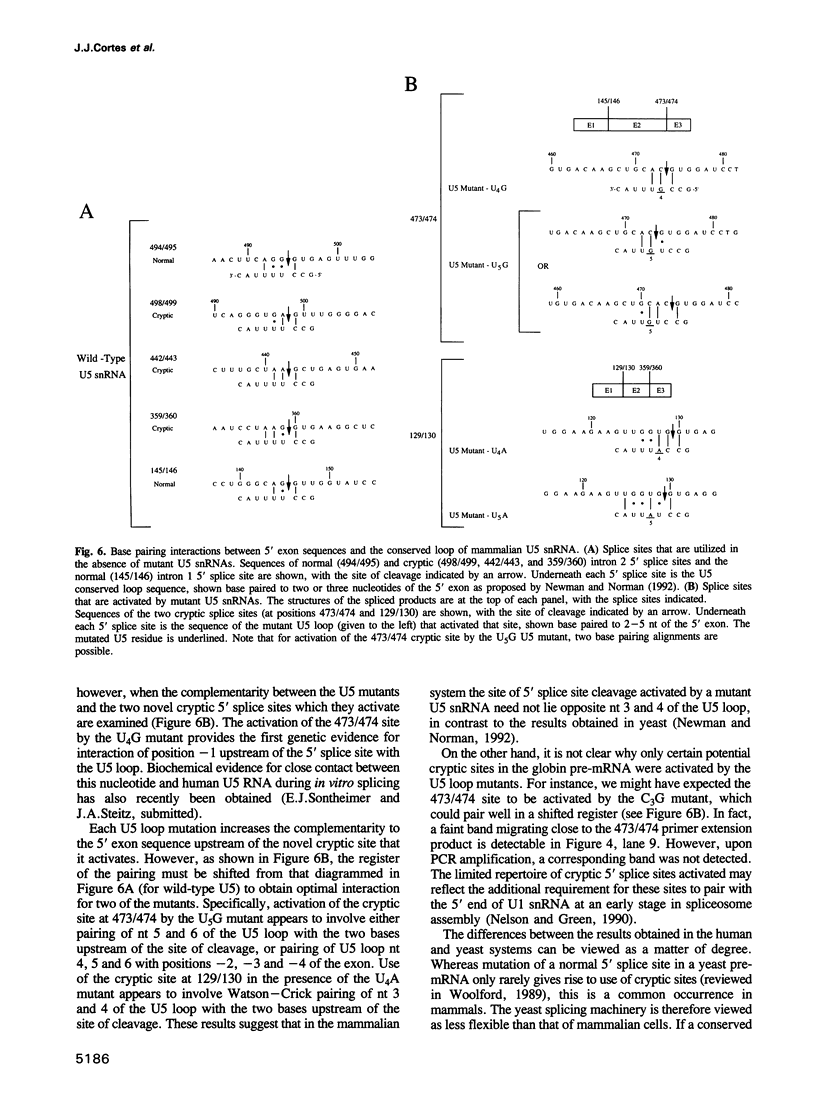



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ach R. A., Weiner A. M. The highly conserved U small nuclear RNA 3'-end formation signal is quite tolerant to mutation. Mol Cell Biol. 1987 Jun;7(6):2070–2079. doi: 10.1128/mcb.7.6.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Goldsmith M. E., Humphries R. K., Ley T., Cline A., Kantor J. A., Nienhuis A. W. "Silent" nucleotide substitution in a beta+-thalassemia globin gene activates splice site in coding sequence RNA. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2318–2322. doi: 10.1073/pnas.80.8.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Hoffman B. E., Grabowski P. J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3' splice site by a network of interactions spanning the exon. Genes Dev. 1992 Dec;6(12B):2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Jacquesson-Breuleux N. Splice site selection and role of the lariat in a group II intron. J Mol Biol. 1991 Jun 5;219(3):415–428. doi: 10.1016/0022-2836(91)90183-7. [DOI] [PubMed] [Google Scholar]
- Jacquier A. Self-splicing group II and nuclear pre-mRNA introns: how similar are they? Trends Biochem Sci. 1990 Sep;15(9):351–354. doi: 10.1016/0968-0004(90)90075-m. [DOI] [PubMed] [Google Scholar]
- Krol A., Gallinaro H., Lazar E., Jacob M., Branlant C. The nuclear 5S RNAs from chicken, rat and man. U5 RNAs are encoded by multiple genes. Nucleic Acids Res. 1981 Feb 25;9(4):769–787. doi: 10.1093/nar/9.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns--a review. Gene. 1989 Oct 15;82(1):5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6253–6257. doi: 10.1073/pnas.87.16.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Antonarakis S. E., Loukopoulos D. Abnormal processing of beta Knossos RNA. Blood. 1984 Jul;64(1):311–313. [PubMed] [Google Scholar]
- Orkin S. H., Kazazian H. H., Jr, Antonarakis S. E., Ostrer H., Goff S. C., Sexton J. P. Abnormal RNA processing due to the exon mutation of beta E-globin gene. Nature. 1982 Dec 23;300(5894):768–769. doi: 10.1038/300768a0. [DOI] [PubMed] [Google Scholar]
- Reich C. I., VanHoy R. W., Porter G. L., Wise J. A. Mutations at the 3' splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell. 1992 Jun 26;69(7):1159–1169. doi: 10.1016/0092-8674(92)90637-r. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Séraphin B. Who's on first? The U1 snRNP-5' splice site interaction and splicing. Trends Biochem Sci. 1991 May;16(5):187–190. doi: 10.1016/0968-0004(91)90073-5. [DOI] [PubMed] [Google Scholar]
- Sawa H., Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5' splice site during the splicing reaction in yeast. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. "Five easy pieces". Science. 1991 Nov 1;254(5032):663–663. doi: 10.1126/science.1948046. [DOI] [PubMed] [Google Scholar]
- Steitz J. A. Splicing takes a holliday. Science. 1992 Aug 14;257(5072):888–889. doi: 10.1126/science.1386941. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Exon mutations uncouple 5' splice site selection from U1 snRNA pairing. Cell. 1990 Nov 2;63(3):619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992 Sep 25;257(5078):1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr Nuclear pre-mRNA splicing in yeast. Yeast. 1989 Nov-Dec;5(6):439–457. doi: 10.1002/yea.320050604. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Sontheimer E. J., Steitz J. A. Site-specific cross-linking of mammalian U5 snRNP to the 5' splice site before the first step of pre-mRNA splicing. Genes Dev. 1992 Dec;6(12B):2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984 May 11;224(4649):574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]