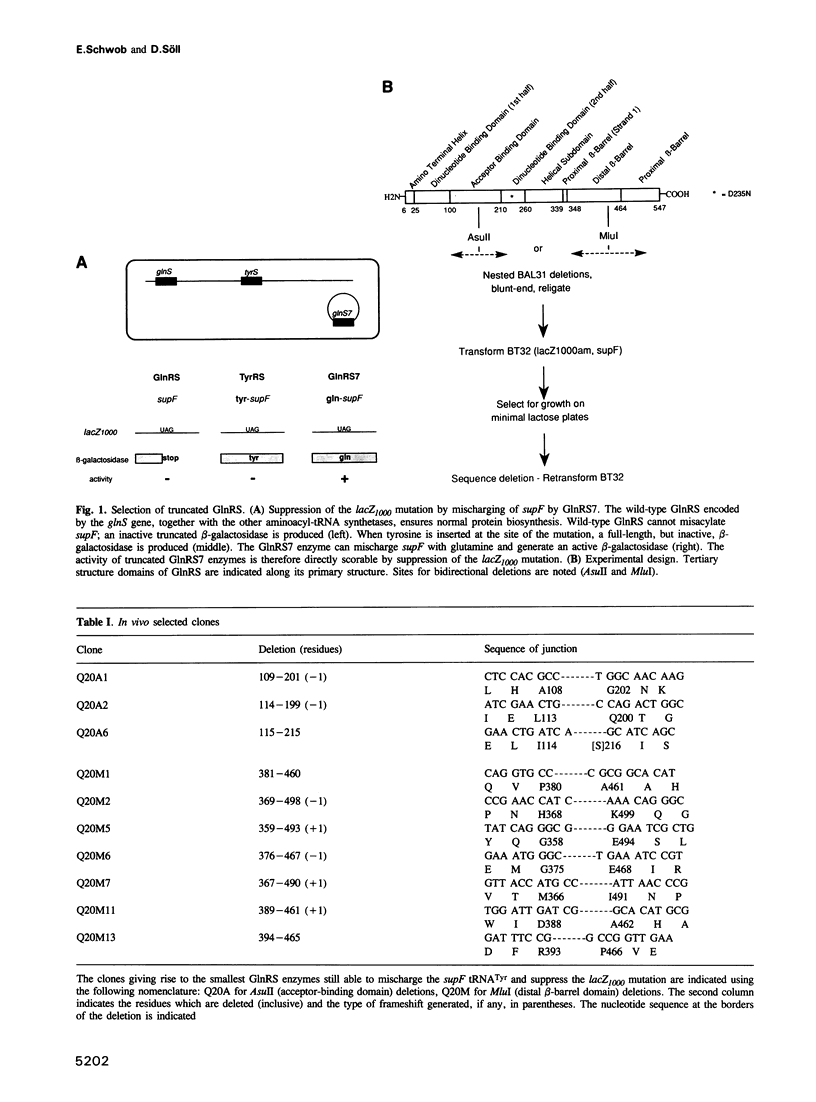
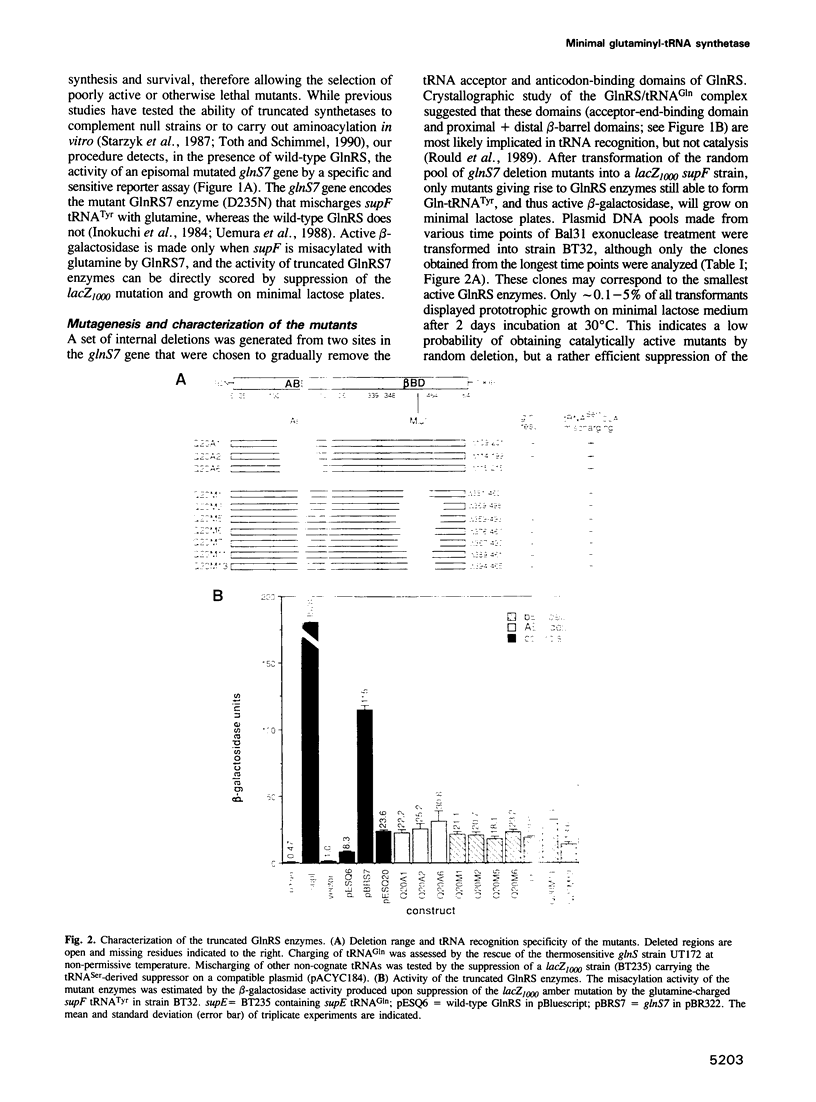
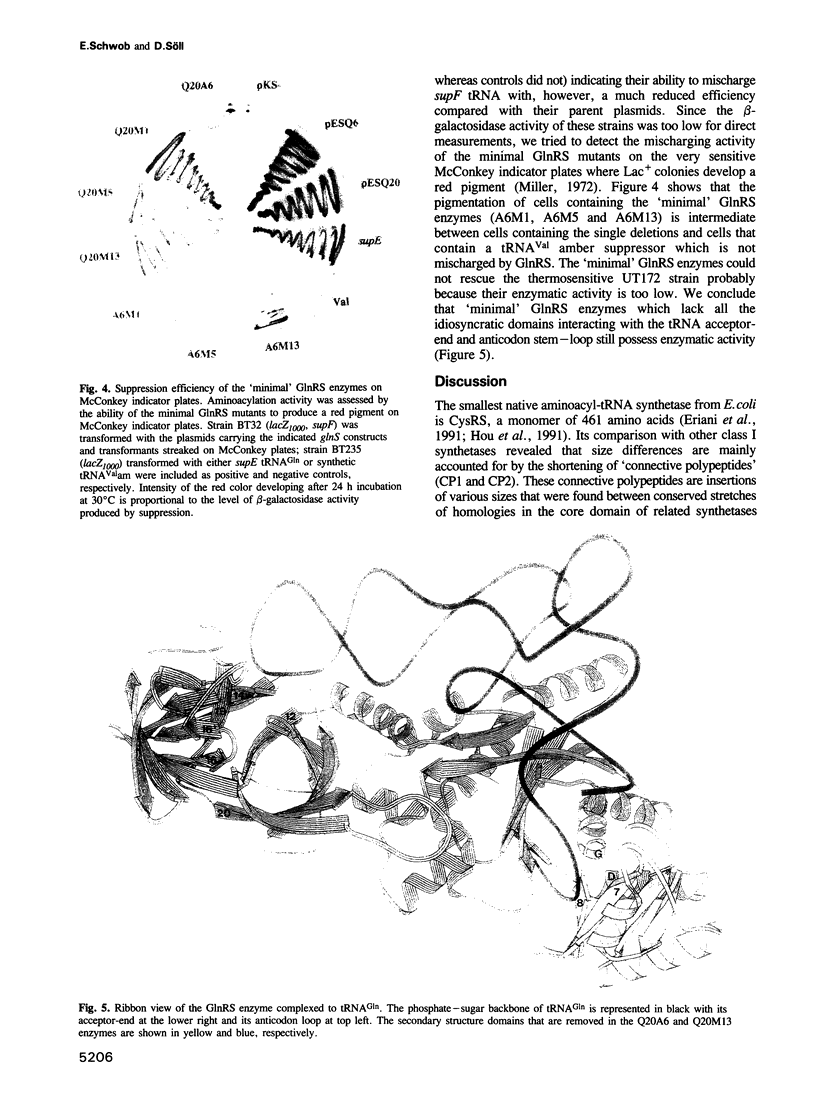
Abstract
The evolution of the aminoacyl-tRNA synthetases is intriguing in light of their elaborate relationship with tRNAs and their significance in the decoding process. Based on sequence motifs and structure determination, these enzymes have been assigned to two classes. The crystal structure of Escherichia coli glutaminyl-tRNA synthetase (GlnRS), a class I enzyme, complexed to tRNA(Gln) and ATP has been described. It is shown here that a 'minimal' GlnRS, i.e. a GlnRS from which domains interacting with the acceptor-end and the anticodon of the tRNA have been deleted, has enzymatic activity and can charge a tRNA(Tyr)-derived amber suppressor (supF) with glutamine. The catalytic core of GlnRS, which is structurally conserved in other class I synthetases, is therefore sufficient for the aminoacylation of tRNA substrates. Some of these truncated enzymes have lost their ability to discriminate against non-cognate tRNAs, implying a more specific role of the acceptor-end-binding domain in the recognition of tRNAs. Our results indicate that the catalytic and substrate recognition properties are carried by distinct domains of GlnRS, and support the notion that class I aminoacyl-tRNA synthetases evolved from a common ancestor, jointly with tRNAs and the genetic code, by the addition of non-catalytic domains conferring new recognition specificities.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Lambowitz A. M. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell. 1987 Jul 31;50(3):331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Thompson S., Gesteland R. F. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: programmed reading frame shifts and hops. Annu Rev Genet. 1991;25:201–228. doi: 10.1146/annurev.ge.25.120191.001221. [DOI] [PubMed] [Google Scholar]
- Breton R., Watson D., Yaguchi M., Lapointe J. Glutamyl-tRNA synthetases of Bacillus subtilis 168T and of Bacillus stearothermophilus. Cloning and sequencing of the gltX genes and comparison with other aminoacyl-tRNA synthetases. J Biol Chem. 1990 Oct 25;265(30):18248–18255. [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Schimmel P. Structural relationships and the classification of aminoacyl-tRNA synthetases. J Biol Chem. 1991 Sep 15;266(26):16965–16968. [PubMed] [Google Scholar]
- Cassio D., Waller J. P. Modification of methionyl-tRNA synthetase by proteolytic cleavage and properties of the trypsin-modified enzyme. Eur J Biochem. 1971 May 28;20(2):283–300. doi: 10.1111/j.1432-1033.1971.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Englisch-Peters S., Conley J., Plumbridge J., Leptak C., Söll D., Rogers M. J. Mutant enzymes and tRNAs as probes of the glutaminyl-tRNA synthetase: tRNA(Gln) interaction. Biochimie. 1991 Dec;73(12):1501–1508. doi: 10.1016/0300-9084(91)90184-3. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Dirheimer G., Gangloff J. Cysteinyl-tRNA synthetase: determination of the last E. coli aminoacyl-tRNA synthetase primary structure. Nucleic Acids Res. 1991 Jan 25;19(2):265–269. doi: 10.1093/nar/19.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Shi J. P., Schimmel P. Overlapping nucleotide determinants for specific aminoacylation of RNA microhelices. Science. 1992 Feb 28;255(5048):1121–1125. doi: 10.1126/science.1546312. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Shiba K., Mottes C., Schimmel P. Sequence determination and modeling of structural motifs for the smallest monomeric aminoacyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):976–980. doi: 10.1073/pnas.88.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi H., Hoben P., Yamao F., Ozeki H., Söll D. Transfer RNA mischarging mediated by a mutant Escherichia coli glutaminyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5076–5080. doi: 10.1073/pnas.81.16.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M., Rogers M. J., Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Jasin M., Regan L., Schimmel P. Modular arrangement of functional domains along the sequence of an aminoacyl tRNA synthetase. Nature. 1983 Dec 1;306(5942):441–447. doi: 10.1038/306441a0. [DOI] [PubMed] [Google Scholar]
- Koch G. L., Bruton C. J. The subunit structure of methionyl-tRNA synthetase from Escherichia coli. FEBS Lett. 1974 Mar 15;40(1):180–182. doi: 10.1016/0014-5793(74)80922-1. [DOI] [PubMed] [Google Scholar]
- Martinis S. A., Schimmel P. Enzymatic aminoacylation of sequence-specific RNA minihelices and hybrid duplexes with methionine. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):65–69. doi: 10.1073/pnas.89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Moras D. Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem Sci. 1992 Apr;17(4):159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K., Usman N., Scaringe S., Doudna J., Green R., Schimmel P. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991 Aug 16;253(5021):784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- Nagel G. M., Doolittle R. F. Evolution and relatedness in two aminoacyl-tRNA synthetase families. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8121–8125. doi: 10.1073/pnas.88.18.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Rould M. A., Steitz T. A., Risler J. L., Zelwer C., Brunie S. Structural similarities in glutaminyl- and methionyl-tRNA synthetases suggest a common overall orientation of tRNA binding. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2903–2907. doi: 10.1073/pnas.88.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Söll D. Discrimination between glutaminyl-tRNA synthetase and seryl-tRNA synthetase involves nucleotides in the acceptor helix of tRNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6627–6631. doi: 10.1073/pnas.85.18.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sanni A., Walter P., Boulanger Y., Ebel J. P., Fasiolo F. Evolution of aminoacyl-tRNA synthetase quaternary structure and activity: Saccharomyces cerevisiae mitochondrial phenylalanyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8387–8391. doi: 10.1073/pnas.88.19.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Classes of aminoacyl-tRNA synthetases and the establishment of the genetic code. Trends Biochem Sci. 1991 Jan;16(1):1–3. doi: 10.1016/0968-0004(91)90002-d. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Shiba K., Schimmel P. Functional assembly of a randomly cleaved protein. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1880–1884. doi: 10.1073/pnas.89.5.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starzyk R. M., Webster T. A., Schimmel P. Evidence for dispensable sequences inserted into a nucleotide fold. Science. 1987 Sep 25;237(4822):1614–1618. doi: 10.1126/science.3306924. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Swanson R., Hoben P., Sumner-Smith M., Uemura H., Watson L., Söll D. Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science. 1988 Dec 16;242(4885):1548–1551. doi: 10.1126/science.3144042. [DOI] [PubMed] [Google Scholar]
- Toth M. J., Schimmel P. Deletions in the large (beta) subunit of a hetero-oligomeric aminoacyl-tRNA synthetase. J Biol Chem. 1990 Jan 15;265(2):1000–1004. [PubMed] [Google Scholar]
- Uemura H., Rogers M. J., Swanson R., Watson L., Söll D. Site-directed mutagenesis to fine-tune enzyme specificity. Protein Eng. 1988 Oct;2(4):293–296. doi: 10.1093/protein/2.4.293. [DOI] [PubMed] [Google Scholar]
- Vidal-Cros A., Bedouelle H. Role of residue Glu152 in the discrimination between transfer RNAs by tyrosyl-tRNA synthetase from Bacillus stearothermophilus. J Mol Biol. 1992 Feb 5;223(3):801–810. doi: 10.1016/0022-2836(92)90991-r. [DOI] [PubMed] [Google Scholar]
- Weygand-Durasević I., Schwob E., Söll D. Acceptor end binding domain interactions ensure correct aminoacylation of transfer RNA. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):2010–2014. doi: 10.1073/pnas.90.5.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F., Inokuchi H., Cheung A., Ozeki H., Söll D. Escherichia coli glutaminyl-tRNA synthetase. I. Isolation and DNA sequence of the glnS gene. J Biol Chem. 1982 Oct 10;257(19):11639–11643. [PubMed] [Google Scholar]