Inhibition of IDO activity with 1MT treatment following influenza infection enhances the production of IL-6, TNF-α, IL-1β, and IFN-β through modulation of macrophages.
Keywords: 1MT, alveolar macrophages, cytokines, TLR
Abstract
Influenza virus is recognized by PRRs, which are critical in the early response to virus infection and induction of proinflammatory cytokines. IDO is increased in the lung of mice immediately following influenza infection, and the presence of IDO has been shown to mediate immune suppression through depletion of trp and reduction in IL-6 production. To determine the role of IDO activity in the early immune response to influenza infection, IDO activity was inhibited using the synthetic analog, 1MT. The results show that IDO inhibition enhanced proinflammatory cytokine gene and protein expression at 24 and 48 h postinfection, respectively, compared with control-treated mice and affected PRR expression. The enhanced proinflammatory response in the presence of 1MT was attributed to macrophages in the airways, as Raw264.7 and primary AMs showed enhanced production of IFN-β, IL-1β, IL-6, and TNF-α in the presence of 1MT. These findings provide important knowledge for the role of IDO during initial host response to influenza infection.
Introduction
Influenza virus belongs to the family Orthomyxoviridae and seasonally causes significant morbidity and mortality worldwide. Influenza virus primarily infects and replicates in airway epithelium, which responds to infection with a robust innate immune response, in part, driven by PRR detection of pattern-associated molecular patterns [1]. Influenza virus is recognized primarily by TLR3, TLR7, and retinoic acid-inducible gene-I, which detect dsRNA, ssRNA, and 5′ triphosphate on ssRNA, respectively [2, 3]. Stimulation of PRRs expressed by epithelial cells, AMs, and DCs induces the expression of proinflammatory cytokines (IL-6, TNF-α, IL-1β), chemokines (MCP-1, RANTES, MIP-1α/β), and type I and III IFNs [4, 5]. The expression of these molecules induces an acute-phase inflammatory response, enhanced recruitment, and activation of immune cells and induces an antiviral state, resulting in viral clearance and immunity [4, 6].
IDO is the first and rate-limiting step in the kyn pathway, where it catabolizes trp into kyn [7]. IDO-mediated depletion of trp and resulting metabolites induces an immunosuppressive environment, in part, through T cell anergy and immune cell death [8]. IDO can be induced in a variety of cell types, including DCs [9], macrophages [10], and respiratory epithelial cells [11]. These cell types are important for controlling virus replication and facilitating the adaptive immunity response [12–15]. Furthermore, IDO activity can be blocked using the pharmacological competitive inhibitor, 1MT [16]. Previous work has shown that in the absence of IDO activity during influenza infection, there is an enhanced Th1-type immune response and robust influenza-specific CD8+ T cell response to influenza virus infection [17]. As the induction and robustness of the adaptive immune response is largely reliant on the innate response, IDO modulation of innate mediators affecting the T cell response, e.g., cytokines and chemokines, can affect or alter adaptive immunity to influenza infection or live-attenuated vaccine.
To understand better the innate features affected by IDO activity, the effects of IDO activity were evaluated at early time-points post influenza infection through 1MT treatment. The results show that IDO inhibition during influenza virus infection modifies PRR expression in the lungs of mice and boosts the proinflammatory cytokine response—in particular, the expression of IL-1β, IFN-β, IL-6, and TNF-α. Raw264.7 macrophage cells and primary murine AMs showed increased cytokine production in the presence of 1MT following influenza infection. These findings show a role of AMs in modulation of the immune response to influenza through IDO inhibition.
MATERIALS AND METHODS
Mice, cell culture, virus, and infection of Raw264.7 cells
Six- to 8-week-old female C57BL/6 mice were received from the Charles River National Cancer Institute program (Raleigh, NC, USA). RAW264.7 cells were maintained in DMEM with 5% FBS. X31 (H3N2) was propagated and titered as described previously [17, 18]. RAW264.7 cells were pretreated with 1MT (750μM; Sigma Aldrich, St. Louis, MO, USA) or molecular-grade water (con) for 24 h and then infected with X31 for 1 h in MEM with 1 μg/ml L-(tosylamido-2-phenyl) ethyl chloromethyl ketone-treated trypsin and 1:100 L-glutamine. Following infection, the cells were rinsed three times with PBS, and fresh infection media were added back. RNA and supernatant were collected at indicated time-points.
Evaluating TLR-associated genes using a PRR PCR array
Mice were treated 3 days before infection with 1MT (2 mg/ml) or vehicle (con) in water, as described previously [17]. On Day 0, mice were anesthetized by i.p. administration (0.2 mL) of Avertin (2,2,2-tribromoethanol; Sigma Aldrich) and subsequently, intranasally infected with 103 PFU of X31 in PBS given in a 50-μl vol. Lungs were harvested and homogenized in Trizol for RNA extraction 24 hpi, or BAL was collected 48 hpi. cDNA was prepared using the RT2 First Strand cDNA kit (Qiagen, Valencia, CA, USA) with 1 μg RNA for each sample. Samples were run on a RT2 Profiler PCR Array mouse TLR signaling pathway (PAMM-018A; Qiagen) following the manufacturer's protocol on the Mx3005P or Mx3000P real-time machines. All Ct values were determined using a manual baseline and equivalent threshold values. Data were analyzed using the software provided with HPRT as the housekeeping gene. Mice receiving 1MT-X31 were compared to mice treated with con and infected with X31.
qRT-PCR for peli1 and IDO1 gene expression
RNA was isolated using the RNeasy mini kit (Qiagen). RNA was treated with DNase I recombinant (Roche, Indianapolis, IN, USA). cDNA was synthesized using Verso cDNA kits (Thermo Scientific, Lafayette, CO, USA) with equivalent concentrations of RNA. qPCR was used to detect peli1 (Applied Biosystems, Foster City, CA, USA) and IDO1 (primers available upon request). All samples were normalized to a housekeeping gene, HPRT (Applied Biosystems) and compared with control-treated, uninfected controls at respective time-points. mRNA expression was determined using the 2−ΔΔCt method.
Isolation and phenotyping of murine AMs
Mice were treated with 1MT or con water and infected as described above. BAL was collected individually from mock or X31-infected mice at 24 hpi. Isolated cells were plastic-adhered for 3 h. Adherent cells were collected and counted using a hemocytometer with trypan blue exclusion. The same number of cells was used for each group for culture or phenotyping. Cells collected from 1MT-treated mice were maintained in the presence of 1MT (750 μM) during the 3-h adherence and for the 48-h culture. Control cells received an equal volume of molecular-grade water. Supernatants were collected 48 h postculture. Single-cell suspensions from BAL were plated at the same number stained as described previously [17]. Cells were surface-stained for CD45, CD11c, and Siglec-F (BD Pharmingen, San Diego, CA, USA) and intracellularly stained for CD68 (BD Pharmingen). All samples were run on an LSRII flow cytometer and analyzed using FlowJo. Isotype control antibodies were used to set gates for analysis.
Quantification of proinflammatory cytokines
IL-6, IL-1β, and TNF-α concentrations were determined using Ready-Set-Go ELISA kits (eBioscience, San Diego, CA, USA). Concentration of IFN-β was determined using the VeriKine IFN-β ELISA kit (PBL Interferon Source, Piscataway, NJ, USA).
Statistical analysis
Statistics were performed using GraphPad Prism, version 5.04. Significance was assigned using a Student's t-test or ANOVA with a Bonferroni post hoc test, as listed in the figure legends.
RESULTS AND DISCUSSION
1MT inhibition of IDO augments proinflammatory cytokine expression in the lungs of influenza-infected mice
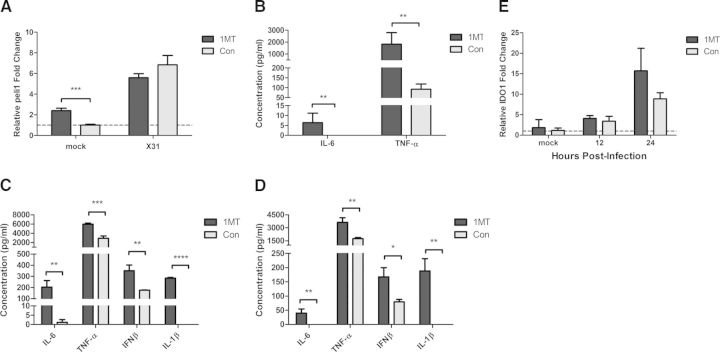
IDO has been shown to attenuate the immune response to infectious diseases, including influenza virus infection, and modulation of IDO activity through 1MT treatment has been shown to inhibit IDO activity [19, 20]. IDO inhibition during influenza infection resulted in an enhanced Th1-type and influenza virus-specific CD8 T cell response [17]. To understand better the innate features that affect the immune response to influenza virus infection, IDO modulation of PRR expression and proinflammatory cytokine expression in the lungs of influenza virus-infected mice was investigated. Mice were orally treated with 1MT, a competitive inhibitor of IDO, or con water for 3 days before intranasal infection with 103 PFU of X31. At 12 and 24 hpi, RNA was isolated from lung homogenate to evaluate the expression of IDO1. There was no increase in IDO1 mRNA expression at 12 hpi; however, there was a significant (P=0.042) increase of IDO1 mRNA expression at 24 hpi compared with 12 hpi (Fig. 1A). As shown previously, treatment with 1MT reduced IDO activity dramatically, as determined by the ratio of kyn:trp (metabolite to substrate) concentrations in the lung homogenate (data not shown). IDO2 mRNA levels were not altered in any of the lung samples, and there was also no difference in IDO1 gene expression between 1MT and con-treated mice (data not shown).
Figure 1. 1MT treatment enhances proinflammatory cytokine expression and modestly increases IDO1 expression in the lungs following influenza infection.
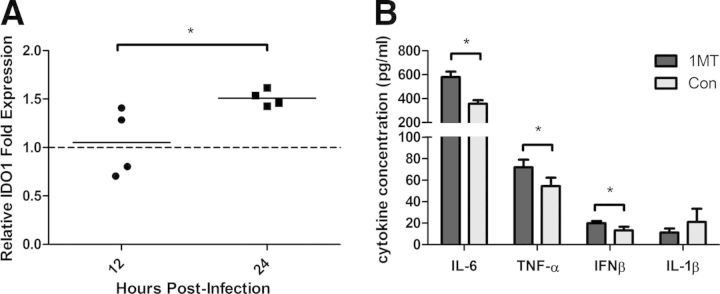
Mice were pretreated with 1MT or con water for 3 days before intranasal infection with 103 PFU of X31. (A) RNA was harvested from lungs and analyzed for IDO1 mRNA expression by qRT-PCR. (B) Concentration of indicated cytokines in the BALF at 48 hpi. Significance was assigned using a Student's t-test, comparing 1MT with con treatment. Graphs represent the mean and sd from a representative experiment that was performed with three mice and was repeated with at least two independent experiments using three to five mice/experiment (*P<0.05).
As IDO1 expression was increased at 24 hpi, the expression of PRR genes from the lungs of 1MT or con-treated mice was determined at 24 hpi using a PCR array. A substantial increase in PRR or cytokine gene expression was indicated if the fold-change from 1MT-treated mice compared with con treatment were >2. Several PRR and proinflammatory cytokine genes were differentially regulated postinfluenza virus infection in 1MT treatment compared with con treatment in mouse lungs (Table 1). Particularly, 1MT treatment enhanced the gene expression of IL-6, IFN-β, and IL-1β, as well as TNF-α-induced pathways, compared with con-treated mice (Table 1). To confirm the increase in gene expression translated to protein for these genes (IL-6, TNF-α, IFN-β, and IL-1β), their concentration was determined in BALF of 1MT or con-treated mice at 48 hpi. 1MT treatment significantly (P<0.05) increased IL-6, TNF-α, and IFN-β levels compared with con treatment (Fig. 1B), a finding consistent with the PCR array data (Table 1). There was no significant difference in the levels of IL-1β expression (Fig. 1B). These results show that 1MT treatment enhances the proinflammatory response in the lung airways following influenza virus infection in mice.
Table 1. Genes Differentially Regulated Post-X31 Infection in 1MT Treatment Compared with con Treatment in Mouse Lungs.
| Genes | Protein | 24 hpi |
|---|---|---|
| csf3 | CSF-3 | 2.11 |
| il6 | IL-6 | 2.50 |
| il1b | IL-1β | 2.61 |
| ifnb1 | IFN-β 1 | 3.43 |
| ptgs2 | Cyclooxygenase 2 | 2.3 |
| peli1 | peli1 | 7.19 |
| tnfaip3 | TNF-α-induced protein 3 | 13.83 |
| cd80 | CD80 antigen | 2.29 |
| Ita | Lymphotoxin A | 4.76 |
| traf6 | TRAF6 | 2.17 |
| tlr6 | TLR6 | 3.40 |
| myd88 | MyD88 | 2.04 |
Enhanced proinflammatory cytokine expression is mediated through macrophages
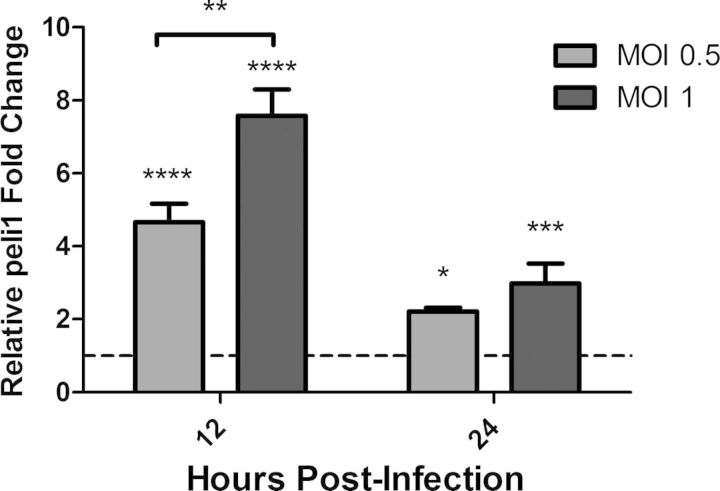
As 1MT treatment during influenza infection increased the proinflammatory cytokine response in the lungs and in BALF, the cell types that could be attributed to these responses were investigated. The immune cell types present in the lungs of naive mice consist primarily of AMs with a low percentage of pulmonary DCs and lymphocytes [21]. Influenza virus primarily replicates in alveolar epithelial cells, stimulating an antiviral response with minimal replication in AMs, but infection of the epithelial or macrophage cell type induces a robust proinflammatory response [13, 22, 23]. Thus, mouse lung epithelial cells (MLE-15) and a murine macrophage cell line (Raw264.7) were evaluated for their cytokine responses following influenza virus infection. Studies have shown that the E3 ubiquitin ligase peli1 is an important adaptor molecule in TLR3 signaling [24], as well as mediating interaction between IL-1R-associated kinase 4 and TRAF6 following IL-1β stimulation, which induces production of cytokines, including TNF-α and IFN-β [25]. Furthermore, peli1 has been shown to be a critical effector molecule during rhinovirus infection of epithelial cells [26]. In this study, peli1 was increased in 1MT- compared with con-treated mice in the lung PRR screen (Table 1) and was associated with the other genes identified with the array (Supplemental Fig. 1). Therefore, peli1 expression was evaluated as a marker for indicating an enhanced cytokine response. MLE-15 or Raw264.7 cells were infected with X31 at varying MOIs, and peli1 expression was assessed at 12 and 24 hpi. There was no increase in peli1 expression in the MLE-15 cells following influenza virus infection (data not shown), suggesting that epithelial cells are not likely key cell types mediating the enhanced proinflammatory response. However, there was a significant (P<0.0001) increase in peli1 expression at 12 and 24 hpi in Raw264.7 cells, peaking at 12 hpi, a feature that was dependent on the MOIs (Fig. 2).
Figure 2. Increased peli1 expression is mediated through macrophages.
Raw264.7 cells were infected with X31 at indicated MOIs. Expression of peli1 was determined by qRT-PCR. Significance was assigned using a one-way ANOVA for each time-point. Graphs represent the mean and sd from a representative experiment that was performed in triplicate and repeated with three independent experiments (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
As peli1 was up-regulated in Raw264.7 cells, the effect of 1MT treatment on the proinflammatory cytokine response was determined (Fig. 3). Raw264.7 cells were pretreated for 24 h with 1MT or con, followed by X31 infection (MOI=1). RNA was collected at 12 hpi to assess peli1 gene expression. There was no difference in peli1 expression with 1MT treatment compared with con-treated cells following influenza infection (Fig. 3A). Interestingly, there was a significant increase in the expression of peli1 in Raw264.7 cells that were treated with 1MT and mock-infected (Fig. 3A). To determine if increased levels of peli1 correlated with an enhanced proinflammatory cytokine response, Raw 264.7 were pretreated with 1MT or con for 24 h and subsequently infected (MOI=1) with X31 or mock-infected. Cell supernatants were collected 12 and 24 hpi, and cytokine concentrations were evaluated using ELISA. Consistent with the level of peli1 expression, there was a significant increase in IL-6 and TNF-α levels following 1MT treatment compared with con treatment in mock-infected cells (Fig. 3B). There was no detectable level of IL-1β or IFN-β in mock-infected supernatants (data not shown). At 12 hpi, 1MT treatment was associated with increased concentrations of IL-6, TNF-α, IFN-β, and IL-1β compared with control-treated cells (Fig. 3C). The increase in IL-6, TNF-α, IFN-β, and IL-1β in 1MT- compared with con-treated Raw264.7 cells was also observed at 24 hpi, although the overall levels of cytokine expression were lower (Fig. 3D). At 12 and 24 hpi, there was no detectable level of IL-1β in control-treated cells, whereas the addition of 1MT induced high levels of IL-1β from influenza virus-infected cells (Fig. 3C and D). These results suggest that 1MT treatment modulates the activation of the inflammasome in macrophages. Inflammasome activation and subsequent IL-1 secretion are important for enhancing the adaptive immune response, including antigen-specific CD8+ and CD4+ T cell effector function [27, 28]. The enhancement of the adaptive immune response in the absence of IDO activity through 1MT treatment during influenza infection [17] may be related to this increase in IL-1β secretion, although the increased IL-1β production was not observed 48 hpi in BALF (Fig. 1B). Whereas there was no difference in peli1 gene expression in the presence of 1MT after infection, there was a substantial difference in the expression of cytokines, suggesting that the lack of change in peli1 expression may be related to the kinetics of its induction and expression of IDO1.
Figure 3. 1MT enhances proinflammatory cytokine expression.
Raw264.7 cells were pretreated with 1MT or con for 24 h and then infected with X31 (MOI=1) or mock-infected. (A) Expression of peli1 was assessed by qRT-PCR at 12 hpi. (B–D) Cytokine levels were determined using an ELISA for (B) mock-infected cells, (C) 12 hpi, or (D) 24 hpi. Significance was assigned using a Student's t-test, comparing 1MT with con treatment. (E) IDO1 expression was evaluated using qRT-PCR. Graphs represent the mean and sd from a representative experiment that was performed in triplicate and repeated with three independent experiments (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
To relate the effects of 1MT treatment to blockade of IDO, qRT-PCR was used to evaluate IDO1 mRNA expression. There was minimal IDO1 mRNA detected in mock-infected cells, but levels gradually increased through the course of infection (Fig. 3E). There was no difference in IDO1 expression between 1MT and con-treated cells at either time-point. The paucity of IDO1 expression in mock-infected cells suggests that the increase in cytokine expression could be triggered by an IDO1-independent mechanism, i.e., a 1MT-nonspecific effect. However, the IDO1-independent cytokine increase was only seen for IL-6 and TNF-α expression, which suggests that the increase in peli1 expression with 1MT treatment impacts the induction of IL-6 and TNF-α. Macrophages are known to secrete high levels of TNF-α [13] and IL-1β [29] following influenza virus infection, and these results indicate that 1MT modulation of the proinflammatory cytokine response is likely linked to the macrophage population.
AMs are responsible for enhanced TNF-α and IL-6 expression
Although the Raw264.7 cells showed increased cytokine expression with 1MT treatment, it was important to confirm these results using primary mouse AMs. Mice were treated with 1MT or con water for 3 days before X31 or mock infection. Twenty-four hours postinfection, AM macrophages were harvested from the BAL of individual mice by plastic adherence. The adherent cells were phenotyped for AM markers, CD45+CD68hiCD11c+ Siglec-F+, as described previously, to confirm the population and purity [30, 31]. Representative dot plots show the gating scheme for AM phenotyping (Supplemental Fig. 2A). Greater than 93% of the adherent cells collected from each group (1MT or con treatment and X31- or mock-infected) were AMs (Supplemental Fig. 2B). Furthermore, there was no difference in the number of AMs between 1MT and con treatment following plastic adherence, with or without X31 infection (Supplemental Fig. 2C). Interestingly, AMs harvested from mice treated with 1MT and infected with X31 had significantly increased levels of IL-6 and TNF-α in the supernatant compared with con-treated, X31-infected mice following ex vivo culture for 48 h (Fig. 4A and B). There were no detectable levels of IFN-β or IL-1β present with either treatment group (data not shown). This may relate to only IL-6 and TNF-α increasing with 1MT treatment in the mock-infected Raw264.7 cells with no effect on IFN-β or IL-1β. Alternatively, the time-point used to evaluated cytokine levels following ex vivo culture of the AMs may have resulted in degradation of IL-1β and IFN-β in the absence of stimuli, whereas IL-6 and TNF-α were maintained without stimuli.
Figure 4. 1MT treatment enhances AM expression of TNF-α and IL-6.
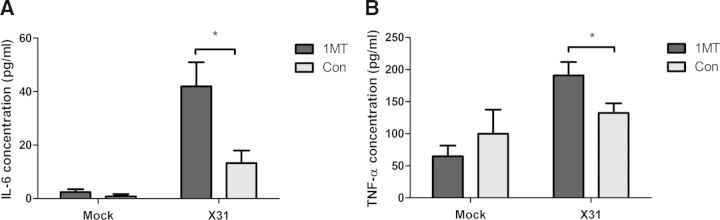
Mice were pretreated with 1MT or con water for 3 days before intranasal infection with 103 PFU of X31 or mock-infected. AMs were isolated from BAL collected 24 hpi. Isolated AMs were cultured for an additional 48 h, and levels of (A) IL-6 or (B) TNF-α were evaluated in the supernatant through ELISA. Significance was assigned using a Student's t-test comparing 1MT with con treatment. Graphs show the mean and sem from two combined independent experiments (n=7–8; *P<0.05).
1MT inhibition of IDO activity was associated with increased IL-6 expression in the BALF (Fig. 1B) and by AMs (Fig. 4A) collected from influenza-infected mice, and IL-6 expression was increased in an influenza-infected macrophage cell line (Fig. 3B–D). IL-6 is prominently expressed in inflammatory environments and functions to increase the recruitment of neutrophils and monocytes [32]. IL-6 is also a key mediator driving the differentiation of CD4+ T cells toward a Th17-type proinflammatory response over a Treg response [33]. IDO activity has been shown to skew the immune response to a Treg phenotype, and inhibition of IDO activity drives a Th17 response through enhanced secretion of IL-6 [34]. Although enhanced IL-6 secretion may be seen as potentially detrimental to the host, studies have shown that increased IL-6 expression primarily affects inducible Treg cell development with little effect on natural Tregs [35]. Furthermore, IL-6 is known to be important for the resolution of influenza infection [36]. Together, these results show that IL-6 and AMs can modulate the Treg phenotype in the lungs.
The results from this study identify a novel mechanism affecting the proinflammatory cytokine response during influenza virus infection, and the findings imply that the use of a pharmacological inhibitor of IDO, i.e., 1MT, could be useful to boost vaccines with poor immune activation, as IDO inhibition would improve cytokine expression. This study connects IDO activity and modulation of the early immune response following influenza virus infection and shows that 1MT is a means to augment the immune response for influenza virus.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the U.S. National Institutes of Health Grant U01 AI083005-01 (to R.A.T.) and the Georgia Research Alliance (to R.A.T.).
We thank Elizabeth O'Connor for her help.
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
- 1MT
- 1-methyl-D, L-tryptophan
- AM
- alveolar macrophage
- BALF
- bronchoalveolar lavage fluid
- con
- control
- Ct
- cycle threshold
- DC
- dendritic cell
- hpi
- hours postinfection
- HPRT
- hypoxanthine guanine phosphoribosyl transferase
- kyn
- kynurenine
- MOI
- multiplicity of infection
- peli1
- Pellino-1
- PRR
- pattern recognition receptor
- qRT-PCR
- quantitative RT-PCR
- TRAF6
- TNFR-associated factor 6
- Treg
- regulatory T cell
- trp
- tryptophan
- X31
- A/HK-X31
AUTHORSHIP
J.M.F. and R.A.T. conceived of and designed experiments and wrote the manuscript. J.M.F., L.K.S, S.P., and S.J. performed experiments. J.M.F., L.K.S., S.M.T., and R.A.T. interpreted results.
DISCLOSURES
The authors have no financial conflicts of interest.
REFERENCES
- 1. Sanders C. J., Doherty P. C., Thomas P. G. (2011) Respiratory epithelial cells in innate immunity to influenza virus infection. Cell Tissue Res. 343, 13–21. [DOI] [PubMed] [Google Scholar]
- 2. Le Goffic R., Pothlichet J., Vitour D., Fujita T., Meurs E., Chignard M., Si-Tahar M. (2007) Cutting edge: influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J. Immunol. 178, 3368–3372. [DOI] [PubMed] [Google Scholar]
- 3. Diebold S. S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. (2004) Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531. [DOI] [PubMed] [Google Scholar]
- 4. Julkunen I., Sareneva T., Pirhonen J., Ronni T., Melen K., Matikainen S. (2001) Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev. 12, 171–180. [DOI] [PubMed] [Google Scholar]
- 5. Onoguchi K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., Fujita T. (2007) Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 282, 7576–7581. [DOI] [PubMed] [Google Scholar]
- 6. Conn C. A., McClellan J. L., Maassab H. F., Smitka C. W., Majde J. A., Kluger M. J. (1995) Cytokines and the acute phase response to influenza virus in mice. Am. J. Physiol. 268, R78–R84. [DOI] [PubMed] [Google Scholar]
- 7. Taylor M. W., Feng G. S. (1991) Relationship between interferon-γ, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 5, 2516–2522. [PubMed] [Google Scholar]
- 8. Grohmann U., Fallarino F., Puccetti P. (2003) Tolerance, DCs and tryptophan: much ado about IDO. Trends Immunol. 24, 242–248. [DOI] [PubMed] [Google Scholar]
- 9. Fallarino F., Vacca C., Orabona C., Belladonna M. L., Bianchi R., Marshall B., Keskin D. B., Mellor A. L., Fioretti M. C., Grohmann U., Puccetti P. (2002) Functional expression of indoleamine 2,3-dioxygenase by murine CD8 α(+) dendritic cells. Int. Immunol. 14, 65–68. [DOI] [PubMed] [Google Scholar]
- 10. Yeung A. W., Wu W., Freewan M., Stocker R., King N. J., Thomas S. R. (2012) Flavivirus infection induces indoleamine 2,3-dioxygenase in human monocyte-derived macrophages via tumor necrosis factor and NF-κB. J. Leukoc. Biol. 91, 657–666. [DOI] [PubMed] [Google Scholar]
- 11. Van Wissen M., Snoek M., Smids B., Jansen H. M., Lutter R. (2002) IFN-γ amplifies IL-6 and IL-8 responses by airway epithelial-like cells via indoleamine 2,3-dioxygenase. J. Immunol. 169, 7039–7044. [DOI] [PubMed] [Google Scholar]
- 12. Bender B. S., Small P. A., Jr., (1992) Influenza: pathogenesis and host defense. Semin. Respir. Infect. 7, 38–45. [PubMed] [Google Scholar]
- 13. Seo S. H., Webby R., Webster R. G. (2004) No apoptotic deaths and different levels of inductions of inflammatory cytokines in alveolar macrophages infected with influenza viruses. Virology 329, 270–279. [DOI] [PubMed] [Google Scholar]
- 14. Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267. [DOI] [PubMed] [Google Scholar]
- 15. Wijburg O. L., DiNatale S., Vadolas J., van Rooijen N., Strugnell R. A. (1997) Alveolar macrophages regulate the induction of primary cytotoxic T-lymphocyte responses during influenza virus infection. J. Virol. 71, 9450–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia L., Schweikart K., Tomaszewski J., Page J. G., Noker P. E., Buhrow S. A., Reid J. M., Ames M. M., Munn D. H. (2008) Toxicology and pharmacokinetics of 1-methyl-d-tryptophan: absence of toxicity due to saturating absorption. Food Chem. Toxicol. 46, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fox J. M., Sage L. K., Huang L., Barber J., Klonowski K. D., Mellor A. L., Tompkins S. M., Tripp R. A. (2013) Inhibition of indoleamine 2,3-dioxygenase enhances the T-cell response to influenza virus infection. J. Gen. Virol. 94, 1451–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matrosovich M., Matrosovich T., Garten W., Klenk H. D. (2006) New low-viscosity overlay medium for viral plaque assays. Virol. J. 3, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Donovan M. J., Tripathi V., Favila M. A., Geraci N. S., Lange M. C., Ballhorn W., McDowell M. A. (2012) Indoleamine 2,3-dioxygenase (IDO) induced by Leishmania infection of human dendritic cells. Parasite Immunol. 34, 464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Larrea E., Riezu-Boj J. I., Gil-Guerrero L., Casares N., Aldabe R., Sarobe P., Civeira M. P., Heeney J. L., Rollier C., Verstrepen B., Wakita T., Borras-Cuesta F., Lasarte J. J., Prieto J. (2007) Upregulation of indoleamine 2,3-dioxygenase in hepatitis C virus infection. J. Virol. 81, 3662–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu X., Wakefield J. K., Liu H., Xiao H., Kralovics R., Prchal J. T., Kappes J. C. (2000) Development of a novel trans-lentiviral vector that affords predictable safety. Mol. Ther. 2, 47–55. [DOI] [PubMed] [Google Scholar]
- 22. Huang L., Li L., Klonowski K. D., Tompkins S. M., Tripp R. A., Mellor A. L. (2013) Induction and role of indoleamine 2,3 dioxygenase in mouse models of influenza a virus infection. PLoS One 8, e66546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hofmann P., Sprenger H., Kaufmann A., Bender A., Hasse C., Nain M., Gemsa D. (1997) Susceptibility of mononuclear phagocytes to influenza A virus infection and possible role in the antiviral response. J. Leukoc. Biol. 61, 408–414. [DOI] [PubMed] [Google Scholar]
- 24. Chang M., Jin W., Sun S. C. (2009) Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 10, 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kappes J. C., Wu X., Wakefield J. K. (2003) Production of trans-lentiviral vector with predictable safety. Methods Mol. Med. 76, 449–465. [DOI] [PubMed] [Google Scholar]
- 26. Manjunath N., Wu H., Subramanya S., Shankar P. (2009) Lentiviral delivery of short hairpin RNAs. Adv. Drug Deliv. Rev. 61, 732–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ben-Sasson S. Z., Hogg A., Hu-Li J., Wingfield P., Chen X., Crank M., Caucheteux S., Ratner-Hurevich M., Berzofsky J. A., Nir-Paz R., Paul W. E. (2013) IL-1 enhances expansion, effector function, tissue localization, and memory response of antigen-specific CD8 T cells. J. Exp. Med. 210, 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ichinohe T., Lee H. K., Ogura Y., Flavell R., Iwasaki A. (2009) Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Silva J. M., Li M. Z., Chang K., Ge W., Golding M. C., Rickles R. J., Siolas D., Hu G., Paddison P. J., Schlabach M. R., Sheth N., Bradshaw J., Burchard J., Kulkarni A., Cavet G., Sachidanandam R., McCombie W. R., Cleary M. A., Elledge S. J., Hannon G. J. (2005) Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 37, 1281–1288. [DOI] [PubMed] [Google Scholar]
- 30. Stevens W. W., Kim T. S., Pujanauski L. M., Hao X., Braciale T. J. (2007) Detection and quantitation of eosinophils in the murine respiratory tract by flow cytometry. J. Immunol. Methods 327, 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaynagetdinov R., Sherrill T. P., Kendall P. L., Segal B. H., Weller K. P., Tighe R. M., Blackwell T. S. (2013) Identification of myeloid cell subsets in murine lungs using flow cytometry. Am. J. Respir. Cell Mol. Biol. 49, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boden D., Pusch O., Silbermann R., Lee F., Tucker L., Ramratnam B. (2004) Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 32, 1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kimura A., Kishimoto T. (2010) IL-6: regulator of Treg/Th17 balance. Eur. J. Immunol. 40, 1830–1835. [DOI] [PubMed] [Google Scholar]
- 34. Baban B., Chandler P. R., Sharma M. D., Pihkala J., Koni P. A., Munn D. H., Mellor A. L. (2009) IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J. Immunol. 183, 2475–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fujimoto M., Nakano M., Terabe F., Kawahata H., Ohkawara T., Han Y., Ripley B., Serada S., Nishikawa T., Kimura A., Nomura S., Kishimoto T., Naka T. (2011) The influence of excessive IL-6 production in vivo on the development and function of Foxp3+ regulatory T cells. J. Immunol. 186, 32–40. [DOI] [PubMed] [Google Scholar]
- 36. Dienz O., Rud J. G., Eaton S. M., Lanthier P. A., Burg E., Drew A., Bunn J., Suratt B. T., Haynes L., Rincon M. (2012) Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol. 5, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.