Only about 10% of the Caenorhabditis elegans operons are conserved in Pristionchus pacificus. Those that are conserved are enriched in germline-expressed genes and genes highly expressed during recovery from dauer arrest in both species.
Keywords: operon, trans-splicing, genome evolution, gene order
Abstract
Discovery of trans-splicing in multiple metazoan lineages led to the identification of operon-like gene organization in diverse organisms, including trypanosomes, tunicates, and nematodes, but the functional significance of such operons is not completely understood. To see whether the content or organization of operons serves similar roles across species, we experimentally defined operons in the nematode model Pristionchus pacificus. We performed affinity capture experiments on mRNA pools to specifically enrich for transcripts that are trans-spliced to either the SL1- or SL2-spliced leader, using spliced leader–specific probes. We obtained distinct trans-splicing patterns from the analysis of three mRNA pools (total mRNA, SL1 and SL2 fraction) by RNA-seq. This information was combined with a genome-wide analysis of gene orientation and spacing. We could confirm 2219 operons by RNA-seq data out of 6709 candidate operons, which were predicted by sequence information alone. Our gene order comparison of the Caenorhabditis elegans and P. pacificus genomes shows major changes in operon organization in the two species. Notably, only 128 out of 1288 operons in C. elegans are conserved in P. pacificus. However, analysis of gene-expression profiles identified conserved functions such as an enrichment of germline-expressed genes and higher expression levels of operonic genes during recovery from dauer arrest in both species. These results provide support for the model that a necessity for increased transcriptional efficiency in the context of certain developmental processes could be a selective constraint for operon evolution in metazoans. Our method is generally applicable to other metazoans to see if similar functional constraints regulate gene organization into operons.
INTRODUCTION
The process of trans-splicing accomplishes the intermolecular ligation of two RNA molecules that are transcribed by two separate loci in the genome. The phenomenon of trans-splicing was originally discovered in unicellular trypanosomes and has subsequently been also observed in many other metazoans such as cnidarians, planarians, platyhelminthes, and nematodes (Lasda and Blumenthal 2011). In this process, typically, a short exon-like noncoding RNA derived from one genetic locus gets spliced upstream of pre-mRNA molecules arising from other genetic loci. This short noncoding RNA is called a spliced leader (SL) and varies in length and sequence in different species, ranging from 16–51 nucleotides (nt). The requirement of trans-splicing in such diverse lineages is not completely understood, but existing models propose their roles in various processes such as resolution of individual cistrons from poly-cistronic transcripts (Blumenthal 1995; Blaxter and Liu 1996; Clayton 2002), stabilization of mRNA by the Tri-Methyl-Guanosine (TMG) cap at the 5′ end of the spliced leader (Zwierzynski and Buck 1991), and increased translational efficiency (Maroney et al. 1995; Lall et al. 2004). Trypanosomes have essentially poly-cistronic transcription, and hence all genes are trans-spliced to a spliced leader. In contrast, only a subset of the transcribed genes receives a spliced leader in nematodes like Caenorhabditis elegans. The spliced leader in nematodes is derived from a 100-nt primary SL RNA found in snRNPs that contributes a 22-nt spliced leader to the acceptor mRNA molecules (Blumenthal 2012). Further, the presence of spliced leaders of a single class, the so-called SL1-spliced leaders, is most likely the ancestral state within nematodes, while more derived conditions in C. elegans, Pristionchus pacificus, or Brugia malayi include a second class of SL2 and its variants, the SL2-like splice-leaders (Guiliano and Blaxter 2006).
An additional feature of trans-splicing in nematodes is its correlation with operon-like gene organization, where a poly-cistronic transcript is transcribed from a cluster of closely spaced genes in the same orientation, through a common promoter that exists upstream of the most 5′ genes in the cluster (Zorio et al. 1994). The adaptive advantage of such an operonic organization in C. elegans is not entirely understood, although existing models propose expression coregulation and clustering of functionally related genes (Blumenthal and Gleason 2003). Recently, it has also been shown that genes in operons are enriched for functions related to germline development (Reinke and Cutter 2009), as well as for genes involved in growth and those essential for recovery from arrested life-stages such as dauers and L1 starvation arrest (Zaslaver et al. 2011). In general, operons facilitate better utilization of limited transcriptional resources, especially under recovery from developmental arrest (Zaslaver et al. 2011). Comparing the genome organization and trans-splicing landscape in related species could provide further clues to functions conserved in last common ancestors. Analysis of trans-splicing in Caenorhabditis briggsae, a rhabditid nematode closely related to C. elegans, shows extensive conservation of operons, with earlier estimates of 96% common operons (Stein et al. 2003; Qian and Zhang 2008) to more recent updates suggesting up to 82% of completely or partially conserved operons (Uyar et al. 2012). However, given that a large fraction of genome is syntenic between the two species (Stein et al. 2003), it is not completely clear whether the observed conservation of operons reflects a functional constraint or insufficient divergence between very closely related species (Guiliano and Blaxter 2006; Qian and Zhang 2008).
The diplogastrid nematode P. pacificus is estimated to have shared a last common ancestor with C. elegans ∼200–400 Myr ago (Dieterich et al. 2008). More recent findings suggest that this divergence time estimate might be in fact 70% lower (Cutter 2008; Erwin et al. 2011). P. pacificus has been developed as a model system for studies of evolution, ecology, and comparative analysis with C. elegans (Hong and Sommer 2006; Sommer and McGaughran 2013). Such analyses have uncovered evolutionary diversifications in various processes such as vulva formation (Tian et al. 2008; Wang and Sommer 2011; Kienle and Sommer 2013), gonad development (Rudel et al. 2005, 2008), dauer development (Ogawa et al. 2009, 2011; Sinha et al. 2012a), innate immune response (Rae et al. 2010; Sinha et al. 2012b), and synaptic connectivity (Bumbarger et al. 2013), during the evolutionary time separating the two species. A genome-wide characterization of the trans-splicing landscape and annotation of operonic genes in related nematode models such as P. pacificus is therefore an important first step in identifying conserved roles for operons in these two species.
Whole-genome analysis of the set of all trans-spliced and operonic genes in C. elegans has provided a systematic catalog of annotated operons (Blumenthal et al. 2002; Allen et al. 2011), which can be used for global comparisons with other species. The availability of whole-genome sequence of P. pacificus (Dieterich et al. 2008) and further refinements of genome assembly and gene models via proteo-genomic approaches (Borchert et al. 2010) now facilitate such comparisons. Here, we present the results from the analysis of genome-wide patterns of trans-splicing and operon-like gene organization in P. pacificus and its comparison with the corresponding scenario in C. elegans. Previous comparative studies of operons in C. elegans and P. pacificus have indicated various levels of conservation and synteny between the operons of C. elegans and P. pacificus (Lee and Sommer 2003; Guiliano and Blaxter 2006; Zaslaver et al. 2011) yet were based on limited examples and experimental data.
Here, we take a comprehensive, genome-wide perspective based on experimental data from transcriptome studies in P. pacificus. Specifically, we sequenced the purified SL1 and SL2 trans-spliced subfractions of the P. pacificus transcriptome, obtained by SL-specific pull-downs from the entire poly-A transcriptome, using biotinylated probes complementary to SL1- or SL2-spliced leaders, respectively. We combined the experimentally determined trans-splicing patterns with computational analysis of intergenic distances in the P. pacificus genome to validate 2219 operons out of 6709 potential candidates in the P. pacificus genome and then generated a whole-genome synteny map between C. elegans and P. pacificus to identify all potentially conserved operons. We found very small conservation at the level of individual operons. However, by integrating data from recent transcriptome studies in P. pacificus, we observed that germline-regulated genes are highly enriched in P. pacificus operons, and the genes in operons have higher expression levels during the exit from dauer stage in P. pacificus. These findings are similar to previous reports in C. elegans (Reinke and Cutter 2009; Zaslaver et al. 2011), suggesting a conserved evolutionarily functional constraint on genome evolution and operon organization.
RESULTS
Different organization of spliced leader genes in the P. pacificus genome
The C. elegans genome contains a cluster of about 110 SL1 loci on a 1-kb tandem repeat that also harbors a cluster of similar number of 5s rRNA genes (Krause and Hirsh 1987), and about 18 to 20 loci for SL2-like variants (Blumenthal 2012). We identified all the spliced leader genes in P. pacificus genome by using the RNA structure and sequence alignment tool “Infernal” (Nawrocki et al. 2009) to search against the covariance models of SL1 and SL2 RNA families (accessions RF00198 and RF00199, respectively) obtained from Rfam database version 11.0 (Burge et al. 2013). We found 187 loci for SL1 genes and 16 loci for SL2-like variants in P. pacificus genome (Supplemental Table S1). As a positive control, we also used the Infernal program with same parameters to scan the C. elegans genome for spliced leaders and were able to recover all known SL variants as well as a novel SL2 gene, indicating 100% sensitivity and specificity of our prediction method (Materials and Methods) (Supplemental Table S2A). In P. pacificus, 176 out of all the 187 SL1 loci produce an identically processed SL1-spliced leader that has previously been reported as the almost exclusively used variant Pp_SL1a (Guiliano and Blaxter 2006), while the remaining 11 loci produce six other SL1 variants (Supplemental Table S2B). Of the 16 SL2 type loci that we discovered in P. pacificus, 11 produce an identically processed SL2-spliced leader as previously reported to be the most widely used SL2 variant in P. pacificus (Guiliano and Blaxter 2006), while the remaining five SL2 loci produce three other variants (Supplemental Table S2C).
Interestingly, unlike C. elegans, we do not observe a spatial clustering of either the SL1 genes or the 5s rRNA genes in P. pacificus, or an overlap or proximity between the positions of SL1 and 5s rRNA genes (Supplemental Table S1). Thus, apart from their primary sequence differences (Lee and Sommer 2003; Guiliano and Blaxter 2006), the number and organization of SL genes in P. pacificus are also different from that in C. elegans.
SL-specific pull-downs on entire poly-A RNA pool yield mRNA samples enriched for respective splice variants
For a comprehensive view of the trans-splicing landscape, we sequenced mRNA pools enriched for SL1-spliced and SL2-spliced transcripts, using a magnetic-bead–based pull-down approach (schematic in Fig. 1; for details, see Materials and Methods). The biotinylated probe used for enrichment of SL1 transcripts corresponds to the variant Pp-SL1a, which is reported to be observed almost exclusively on SL1-spliced clones in P. pacificus (Guiliano and Blaxter 2006). Similarly, biotinylated probe used for the enrichment of SL2-spliced transcripts corresponds to the variant Pp-SL2b, reported to be most frequently used relative to all other SL2 variants (Guiliano and Blaxter 2006). In our pull-down experiments, we were successful in obtaining about a 1000-fold enrichment for both SL1- and SL2-spliced transcripts, as verified by qPCR experiments using the gene Ppa-rpl-43 known to be trans-spliced to both SL1 and SL2 (Supplemental Fig. S1). To further verify that each biotinylated SL1- or SL2-specific probe is capturing only its corresponding SL splice variant, we synthesized pure RNA transcripts with either SL1 or SL2 at their 5′ end via in vitro transcription (IVT) reactions and then subjected these samples to one round of pull-downs using the same protocol as for the actual sample (schematic in Supplemental Fig. S2). We observed high specificity in these reactions such that each SL-complementary probe was able to pull down only its corresponding SL splice variant, while no RNA could be detected in a pull-down with the probe corresponding to the other SL (Supplemental Fig. S2A,B). Additionally, we tested the specificity of these pull-downs on a mixed pool of equal amounts of synthetic SL1 and spliced SL2 RNA. qPCR analysis of RNA obtained after each SL-specific pull-down exhibits high relative enrichments, typically 100-fold or more (sevenfold or more on a log2 scale), indicating minimal carry-over from nonspecific binding (Supplemental Fig. S2C). In all these experiments, the primers used for either the SL1 or SL2 sequences were also highly specific, as verified in qPCR experiments on pure pools of SL1- and SL2-spliced cDNA obtained from cloned fragments (see Materials and Methods) (Supplemental Fig. S3).
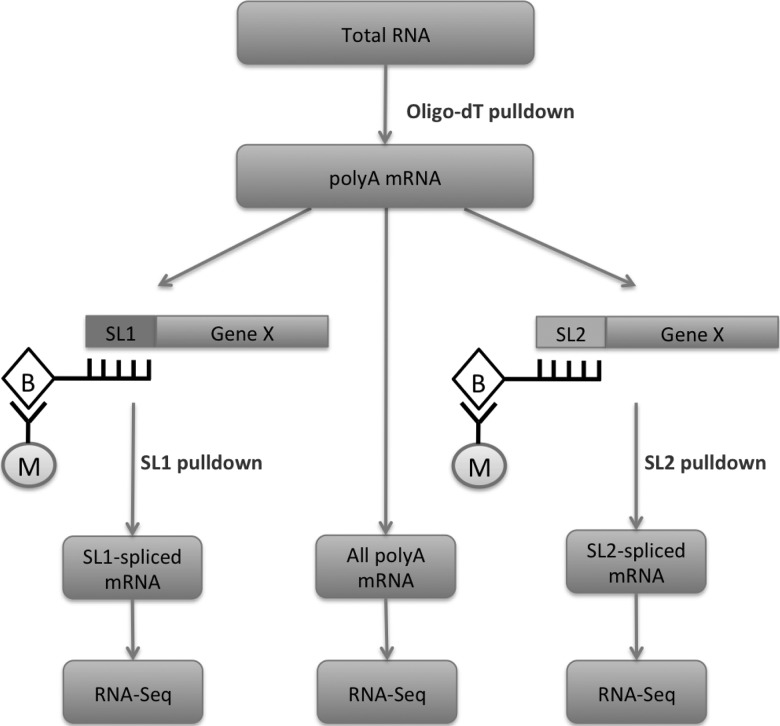
FIGURE 1.
Schematic of experiments for obtaining mRNA pools enriched in SL1- or SL2-spliced variants of all expressed genes. Three pools of poly-adenylated RNA were isolated from total RNA: total polyA mRNA, a SL1-spliced mRNA fraction, and a SL2-spliced mRNA fraction. Briefly, biotinylated oligo-dT probes were used to purify the entire polyA-mRNA from total RNA. The mRNA was split into three aliquots, where one of the aliquot was directly used for RNA-seq, while the SL1-spliced or SL2-spliced mRNA was isolated from the remaining two aliquots, using biotinylated oligo probes with sequences complementary to the respective splice leaders. Streptavidin-coated magnetic beads were used to pull down the biotinylated probes.
About 90% of the expressed genes receive a spliced leader in P. pacificus
The sequencing of each of the three fractions generated around 44 million (Mio) reads, which could be aligned back to the P. pacificus genome/gene-models and consisted of 29 Mio (64% of total) perfectly paired reads and 11 Mio (25%) singletons. Nonzero read-counts (measured in FPKM [fragments per kilobase per million mapped reads]) were observed for 23,693 genes in the entire polyA-fraction, while 21,234 genes in the SL1 fraction and 21,093 genes in the SL2 fraction had nonzero read-counts (Supplemental Table S3). Nine thousand nine hundred genes were detected simultaneously across all the three samples (FPKM > 0), indicating that ∼90% of all detected genes received both spliced leaders in varying proportions. On using a higher detection threshold of FPKM ≥ 1 or FPKM ≥ 2, we still observe trans-splicing of both spliced leaders to 90% of the detected genes. The fraction of genes that receive a spliced leader is estimated to be ∼70%–80% in C. elegans (Allen et al. 2011; Blumenthal 2012). The higher extent of trans-splicing in P. pacificus is most likely due to the higher sequence coverage used in our experiments that could detect most of the trans-splicing events, a possibility consistent with previous observations in C. elegans where higher read-coverage is associated with higher fraction of trans-spliced genes (e.g., Fig. 1; Allen et al. 2011).
Extent of trans-splicing to a gene does not depend on its level of gene expression
For each of the detected genes, we calculated the %SL1 and %SL2 as the ratio of FPKM values of SL1 vs. poly-A samples and the SL2 vs. poly-A samples, respectively. A plot of %SL1 or %SL2 vs. absolute polyA FPKMs shows that the extent of trans-splicing to a gene is independent of its expression level (Supplemental Fig. S4). Although most of the genes receive both the spliced leaders SL1 and SL2, the relative proportions of the two splicing variants for any particular gene (measured as log2(SL1_fpkm/SL2_fpkm)) vary from −6 to 5 and show a bimodal distribution (Supplemental Fig. S5). For any given mRNA, the extent of trans-splicing to the two spliced leaders (measured as log2(SL_fpkm/poly_fpkm)) is not strongly correlated (Pearson correlation = 0.14) (Supplemental Fig. S6), suggesting a gene-specific regulation of nature and extent of trans-splicing.
The P. pacificus genome has 6709 putative operons, with various trans-splicing patterns
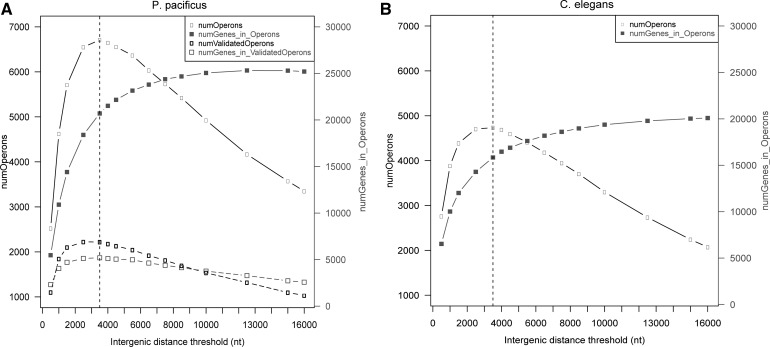
A key feature of operon-like gene organization in C. elegans is the small intergenic distance between adjacent genes (median intergenic distance = 156 nt, 80% of all annotated operons have intergenic distances <500 nt, based on WS239) (Zorio et al. 1994; Huang et al. 2007) that is typically transcribed as a poly-cistronic transcript from a common single promoter found upstream of the most 5′ gene in the operon, which is then resolved into individual cistrons via trans-splicing (Blumenthal 2012 Wormbase). Sometimes, downstream genes within the so called “hybrid operons” might additionally be transcribed from an internal promoter, which promotes an expression pattern different to the poly-cistronic counterpart originating from the operon (Huang et al. 2007). Thus a common strategy to computationally predict potential operons is to search for closely spaced gene clusters with an intergenic distance threshold of maximum 500 nt (Zaslaver et al. 2011). More recently, a small number of noncanonical operons have been discovered in C. elegans. Herein, poly-cistronic transcripts are derived from genes that can be as far as ∼10 kb apart, which is a substantial deviation from the canonical ∼500-nt intergenic distance (Morton and Blumenthal 2011). Thus, in order to predict potential operons in P. pacificus genome, we first systematically analyzed the variation in number of operons and variation in the number of genes within these operons as a function of intergenic distance threshold (Fig. 2A). Remarkably, the number of predicted operons, as well as the number of genes in operons, increases with intergenic distance, up to a threshold of 3500 nt. Beyond this threshold, we found that the number of potential operons decreased sharply. This threshold of 3500 nt is close to the median value of intergenic distances of 3330 nt between all collinear gene pairs in P. pacificus genome.
FIGURE 2.
Effect of different intergenic distance thresholds on the number of predicted operons and the number of genes within operons in P. pacificus (A) and C. elegans (B) suggests an optimal threshold at 3500 nt for predicting operons. The number of predicted operons (left y-axes, black curves with open boxes) in both species initially increases with the maximum intergenic distance allowed between consecutive genes within operons but starts to decrease beyond a 3500-nt cut-off (dotted vertical lines). The total number of genes predicted to be within these operons (right y-axes, gray curves with filled squares) increases with increasing intergenic distance threshold and saturates at ∼13,000 nt when almost all genes become part of predicted operons. In P. pacificus, the number of operons that could be validated using our RNA-seq data (dotted black curve, open black rectangles in A) also show the same trend as predicted operons, with a maximum at 3500 nt. Interestingly, the total number of genes included within these validated operons (dotted gray curve, open gray squares in A) also decreases beyond the 3500-nt threshold, which was thus chosen as the optimal threshold for computational prediction of operonic gene clusters.
We found the same trend for C. elegans, where the number of predicted operons again had a peak at about 3500 nt but decreased beyond the 3500-nt threshold (Fig. 2B), which is close to the median intergenic distance of 2996 nt between all collinear gene pairs in C. elegans genome (WS239). Also, the 4727 C. elegans operons that were predicted at a 3500-nt distance threshold included 1354 out of the 1376 annotated operons in C. elegans (Supplemental Table S4). We therefore decided to use this 3500-nt threshold as a cut-off for intergenic distance in our search of potential operons in P. pacificus. Our computational analysis identified about 6709 operon-like gene clusters containing 19,024 genes in total (average number of genes per operon = 2.83; minimum two genes per operon, and maximum 14 genes per operon) (see Supplemental Table S5). The median intergenic distance within the set of all these predicted operons in P. pacificus was observed to be 1149 nt, while 4204 operons (∼63% of the total 6709 operons) have genes with intergenic distances <500 nt (Supplemental Fig. S7).
Analysis of trans-splicing patterns of predicted operon validates 2219 operons in P. pacificus
We used our mRNA-seq data on SL1- and SL2-spliced fraction of the P. pacificus transcriptome to identify genome-wide patterns of polyA-normalized trans-splicing across all genes (see below). We integrated this information with our list of predicted operons to arrive at a final set of 2209 validated operons in P. pacificus.
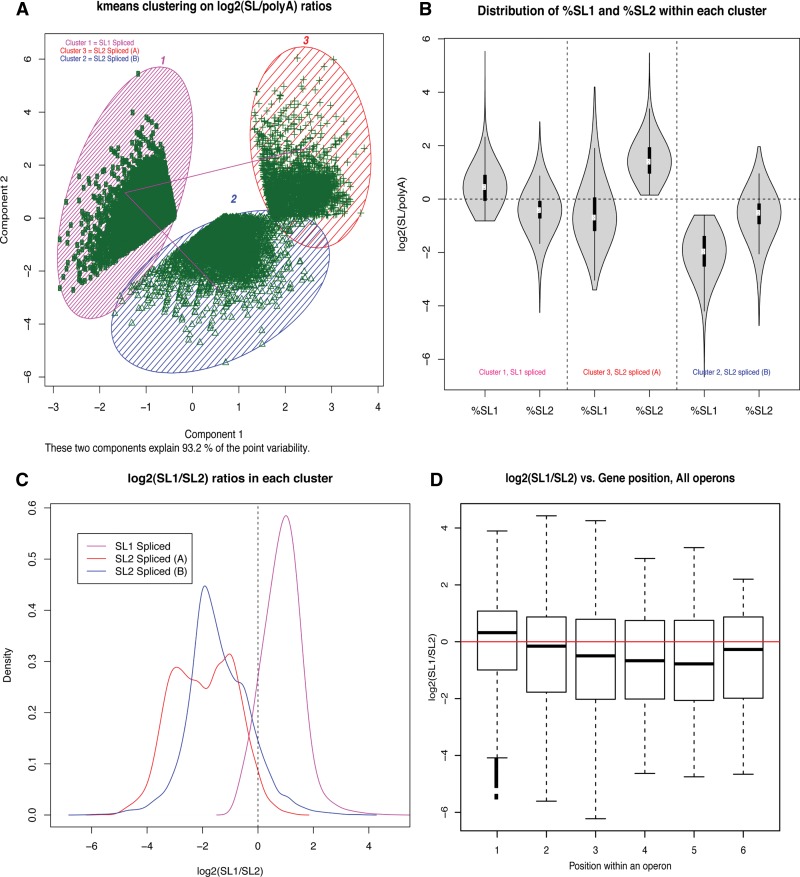
As a first step, we carried out an unbiased k-means clustering on the normalized SL1-spliced and SL2-spliced fractions (log(SL1/polyA) and log(SL2/polyA)) for each gene with FPKMs > 0. This led to the discovery of three distinct clusters (Fig. 3A) with different trans-splicing patterns (Fig. 3B) and with different relative levels of SL1 vs. SL2 trans-splicing (log (SL1/SL2)) (Fig. 3C). Based on these clustering results, we can classify the genes in Cluster 1 as primarily “SL1 spliced” (log(SL1/SL2) > 0) (Fig. 3C), while Clusters 2 and 3 are comprised of genes that are predominantly “SL2 spliced” (log(SL1/SL2) < 0) (Fig. 3C). In general, we observed that the first gene of an operon tends to have higher SL1 reads than the SL2 reads, while the trend is opposite for more downstream genes (Fig. 3D).
FIGURE 3.
Distinct patterns of trans-splicing observed in P. pacificus transcriptome. (A) A k-means clustering on the fraction of SL1 and SL2 splicing for each gene reveals three distinct clusters. (B) A violin plot for the %SL1 and %SL2 for all genes within each of the three clusters shows that genes in Cluster 1 get higher SL1 than SL2, while the opposite is true for genes in Clusters 2 and 3. (C) The same trend is more clearly visible in density plots of relative SL levels (log2(SL1/SL2) for all genes within each of the three clusters. (D) Boxplots for relative trans-splicing levels for all the genes within all predicted operons indicate that the first gene in an operon tends to receive higher levels of SL1 than SL2, although outliers can be seen for all gene positions. Only gene positions 1 to 6 are shown in this plot, but the trend remains the same down to gene position 14, which is the maximum number of genes in an operon.
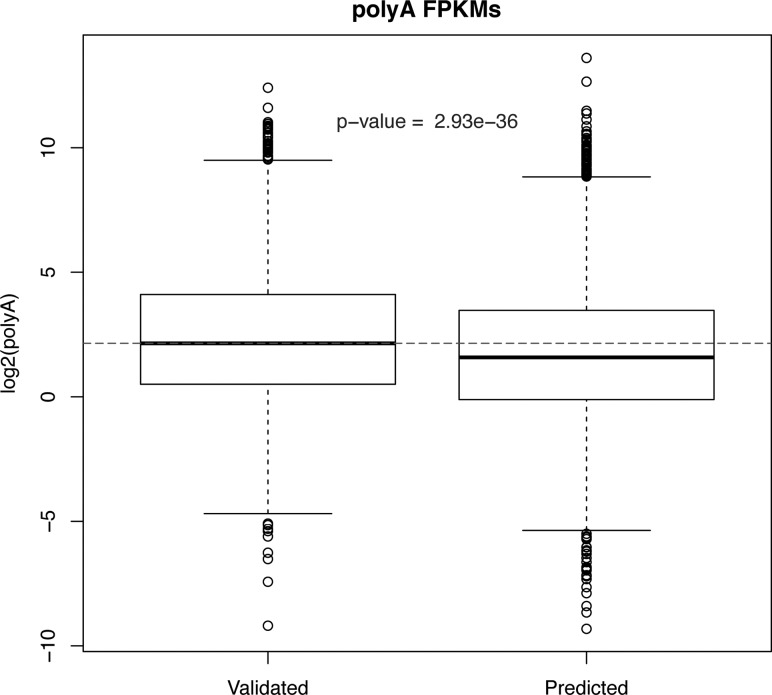
With this information at hand, we looked at gene cluster assignments within each of the previously predicted operons and could identify three different classes of operons: “Type 1,” “Type 2,” and “Type 3” operons, respectively. In “Type 1” operons, the first gene belongs to Cluster 1 and hence receives higher levels of SL1, while the second and more downstream genes belong to either Cluster2 or Cluster 3 and hence receive higher levels of SL2 trans-splicing (n = 925 operons) (Supplemental Table S5). In “Type 2” operons, all genes in an operon are predominantly trans-spliced to SL2-spliced leader (n = 778 operons) (Supplemental Table S5), while in “Type 3” operons, the first gene was not observed to be spliced to either the SL1- or SL2-spliced leaders and the downstream genes predominantly received the SL2-spliced leader (n = 516 operons) (Supplemental Table S5). The remaining operons (n = 4490) (see Predicted Operons in Supplemental Table S5) had a mixture of trans-splicing patterns on genes at different positions (Supplemental Fig. S8). We further observed that the genes in these “predicted” operons had, on average, lower levels of expression than the genes belonging to the Type 1, 2, and 3 operons (Fig. 4). It is thus likely that these are bona fide operons, but their trans-splicing patterns could not be clearly discerned due to the low expression of their constituent genes. In summary, we could validate ∼33% of predicted operons in P. pacificus by using our RNA-Seq data combined with bioinformatic prediction of operon-like clusters using an intergenic threshold of 3500 nt.
FIGURE 4.
Genes within predicted operons are expressed at lower levels compared with the genes within operons validated to be of Types 1, 2, or 3.
Further support in favor of using the 3500-nt threshold came from the observation that when we used the same strategy to identify Type 1, 2, or 3 classes of operons in P. pacificus at various intergenic distance thresholds, not only the number of validated operons but also the number of genes included within these validated operons decreased beyond the 3500-nt threshold (Fig. 2A, dotted lines), suggesting a potential underlying constraint on the maximum intergenic distance typically allowed between consecutive genes within an operon.
Only a small number of operons are conserved between P. pacificus and C. elegans
To identify the operons that are conserved between P. pacificus and C. elegans, we first computed regions of conserved synteny over all genes in a pairwise genome comparison using CYNTENATOR software (Rödelsperger and Dieterich 2010). We then filtered these results to include only the genes included in operons of at least one species and then looked for partial or complete overlaps between C. elegans and P. pacificus operons. Any two operons with at least two genes in common were counted as a conserved syntenic operon between the two nematode species. Surprisingly, we found very little overlap between the operons across the two species. Specifically, only 37 operons are syntenic between the 1376 annotated operons in C. elegans and the 2219 validated operons in P. pacificus (Supplemental Table S6), where only seven operons were fully conserved across the two species, while the remaining 30 operons were only partially conserved. The validated operons in P. pacificus, which are conserved in C. elegans are either “Type 1 operons” (n = 21) or “Type 2 operons” (n = 16) only (Supplemental Table S6). Even when comparing all the 4731 predicted C. elegans operons at the 3500-nt threshold against the 6709 P. pacificus operons, the total number of operons conserved across the two species is just 128, of which only 22 are fully conserved (Supplemental Table S6). In this comparison of all predicted operons across the two species, the P. pacificus operons, which are conserved in C. elegans, are predominantly of the type “predicted operons” (n = 76), followed by Type 1 operons (n = 28), Type 2 operons (n = 20), and Type 3 operons (n = 4) (Supplemental Table S6).
Operon genes are overrepresented within syntenic genes
In our genome-wide analysis of synteny across the two species, we found a total of 623 C. elegans genes that are part of conserved syntenic blocks across the genomes of C. elegans and P. pacificus. We observed that more than one-third of these genes (n = 222) are also part of known C. elegans operons, a highly significant enrichment (P-value from Fisher's exact test ∼10−31). In other words, even though only a few genes are present within the syntenic blocks in C. elegans, they are highly likely to be a part of an operon. This suggests that operons are very highly enriched within syntenic blocks and perhaps constitute an important constraint in maintaining synteny.
Genes in operons conserved across P. pacificus and C. elegans are enriched for detoxification-related genes and nuclear hormone receptor activity
Next, we analyzed if the set of the 109 genes included in these 37 conserved operons was enriched for any particular functions. The results from an enrichment analysis for the PFAM domains and the gene families together identify significant overrepresentation of genes encoding gap junction proteins innexins (inx gene family, Pfam domain Innexin) and UDP-glucuronosyl transferases (ugt gene family, Pfam domain UDPGT) that are potentially involved in detoxification pathways (Supplemental Table S7). A similar analysis on the larger set of 389 genes from the 128 operons conserved between all predicted operons across the two species again identified significant overrepresentation of genes and domains involved in detoxification pathways (glutathione S-transferase gene family gst-, UDP-glucuronosyl transferases encoded by the ugt gene family, cytochrome P450 enzymes encoded by the cyp gene family, and P-glycoprotein–related ABC transporters of the pgp gene family; corresponding Pfam domains “GST_N,” “GST_C,” “UDPGT,” and “p450”) (Supplemental Table S7), as well as nuclear hormone receptor activity (nhr gene family, Pfam domains “Hormone_recep” and “zf-C4”) (Supplemental Table S7). The genes of gst, ugt, and cyp families work together during phase I and phase II metabolism of lipophilic substances such as xenobiotic toxins (Jakoby and Ziegler 1990; Oesch and Arand 1999), and many of the nuclear hormone receptors serve as xenobiotic sensors that can regulate expression of cytochrome P450 enzymes and UDP-glucuronosyl transferases (Zhou et al. 2005; Wallace and Redinbo 2013). It is therefore remarkable to see their enrichment within the set of conserved operons and suggests that their concerted coregulation might be one of the adaptive functions of operonic organization and thus retained across the evolutionary time separating P. pacificus and C. elegans.
Operons in both P. pacificus and C. elegans show enrichment for germline-expressed genes
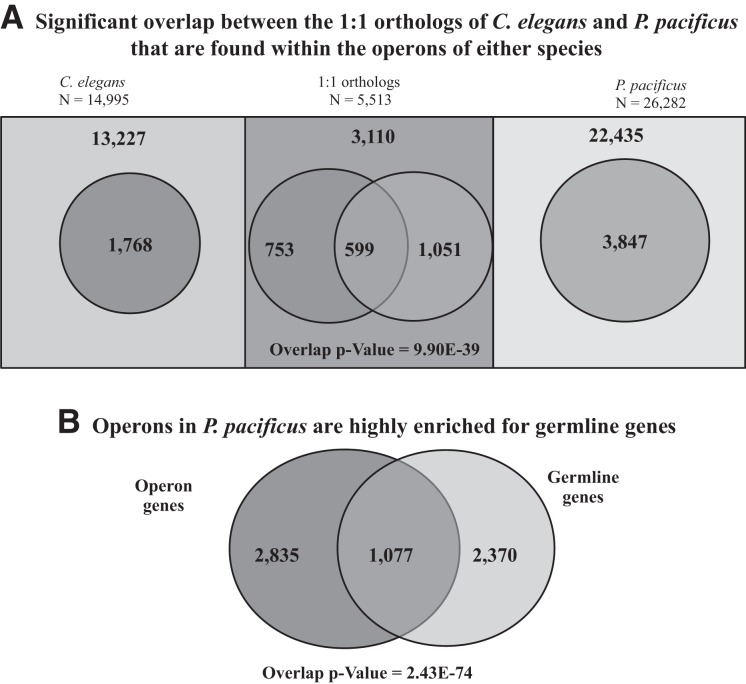
Given that the gene order is poorly conserved between the operons of P. pacificus and C. elegans, we looked for broader levels of conservation by comparing the set of all operonic genes in both species against each other to check if they contain more 1:1 orthologs in common than expected by chance alone. A total of 5513 unambiguous pairs of 1:1 orthologs could be identified between P. pacificus and C. elegans (genome release ws239) using the InParanoid tool (Ostlund et al. 2010). Of these 5513 ortholog gene pairs, 1352 genes are members of C. elegans operons, while 1650 genes are a member of the Type 1, Type 2, or Type 3 operons in P. pacificus. We observed 599 1:1 orthologs to be common between the operons in the two species, a small but highly significant overlap (Fisher's test P-value = 9.90 × 10−39) (Fig. 5A). This significant overlap indicates that gene order within operons is not a crucial constraint in evolution of operon function.
FIGURE 5.
Conservation of gene content and function between operons of C. elegans and P. pacificus. (A) Significant overlap between the 1:1 orthologs of C. elegans and P. pacificus that are found within the operons of either species (Fisher's test P-value = 9.90 × 10−39). The rectangular boxes represent the set of genes within C. elegans and P. pacificus, and their overlap indicates the 1:1 orthologs (n = 5513). The oval regions represent the corresponding subset of genes that are part of operons in respective species. (B) Operons of Type 1, Type 2, and Type 3 in P. pacificus are highly enriched for germline genes (Fisher's test P-value = 2.43 × 10−74).
We further analyzed whether the operonic genes in the two species are enriched for genes with similar biological functions. Operons in C. elegans are enriched for genes involved in germline development (Reinke and Cutter 2009). We assessed the overlap between genes included in P. pacificus operons and genes reported to be enriched in germline tissue in P. pacificus by integrating data from a previous study of P. pacificus germline enriched genes (Rae et al. 2012). We found that similar to C. elegans, P. pacificus operons also show an overrepresentation of germline expressed genes (Fig. 5B). The germline function is also significantly enriched in the subset of genes that are part of the 37 validated operons conserved across the two species (Supplemental Fig. S9). These results suggest that at least one of the functional constraints on operon evolution is inclusion of genes expressed in the germline.
Dauer-exit–induced genes exhibit higher up-regulation if they are part of an operon in both P. pacificus and C. elegans
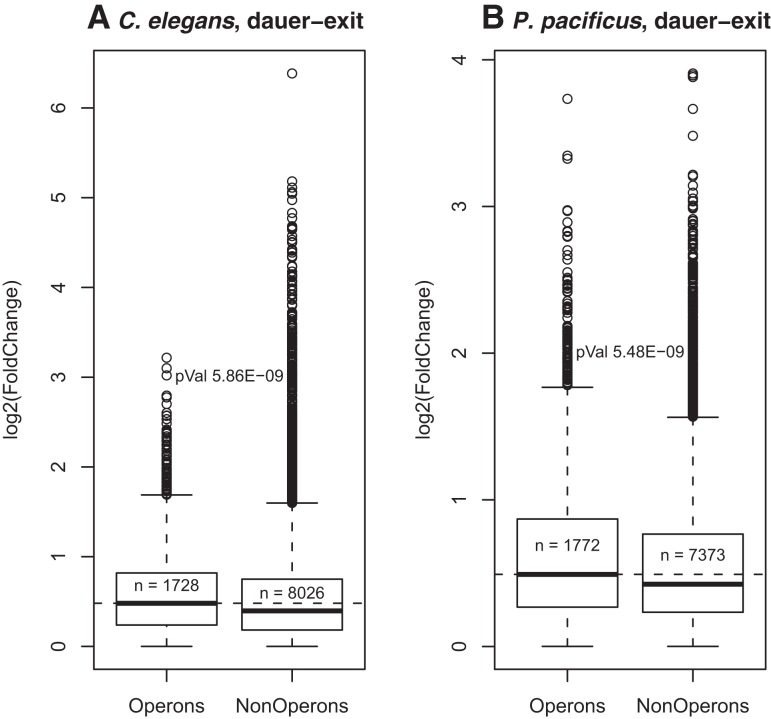
A proposed adaptive advantage for operon-like gene organization is optimization of transcriptional resources during recovery from growth-arrested stages such at the dauer stage in nematode, and the genes contained within operons were found to be expressed at higher levels than the genes not inside the operons during the dauer-exit time course in C. elegans (Zaslaver et al. 2011). We thus analyzed the dauer-exit gene expression profiles of P. pacificus and C. elegans from our previous study (Sinha et al. 2012a). In agreement with previous reports (Zaslaver et al. 2011), we also see a higher expression level of operonic genes compared with nonoperonic genes during dauer-exit in C. elegans (Supplemental Fig. S10). We find this phenomenon to be conserved in P. pacificus as well (Supplemental Fig. S10). Moreover, exit from the dauer stage is accompanied by a global increase in transcriptional activity (Dalley and Golomb 1992). Up-regulated genes inside operons (n = 1728) show a higher response (i.e., gene expression fold-change) than up-regulated genes outside operons (n = 8026) for C. elegans (P-value = 5.86 × 10−9) (see Fig. 6A). This is also true for dauer-exit gene profiles in P. pacificus (P-value = 5.48 × 10−9) (Fig. 6B). The similarity of this trend is even more remarkable given the fact that the dauer-related genes are quite different between C. elegans and P. pacificus (Sinha et al. 2012a). This pattern is not seen with the set of predicted operons (see Supplemental Fig. S11).
FIGURE 6.
Higher fold-change induction of genes inside the annotated/validated operons vs. genes outside of operons, upon dauer-exit in C. elegans (A) and P. pacificus (B).
We propose that the conserved higher expression and higher expression change of dauer-exit–induced genes, despite the nonconservation of gene expression profiles or operon organization across the two species, are common evolutionary constraints influencing clustering of genes into operons in both the species.
P. pacificus–specific pioneer genes are also incorporated in operons
About one-third of the P. pacificus gene set is composed of lineage-specific genes, the so-called pioneer genes, which do not show any sequence similarity to any of the known proteins outside the genus Pristionchus (Borchert et al. 2010). We found that out of the 6709 predicted operons, a total of 3515 operons in P. pacificus contain at least one pioneer gene, of which 1490 operons contained two to nine pioneer genes (Supplemental Table S5). The fraction of pioneer genes within operons is slightly less than that expected by chance (total 5692 pioneer genes found in operons, 5743 pioneer genes expected by chance; Fisher's exact test P-value = 3.00 × 10−6). Most of these pioneer gene–containing operons are of the type “Predicted Operon” (n = 2663 out of total 3515, ∼76%), but there still exist 852 validated operons of Type 1, Type 2, or Type 3 that contain at least one pioneer gene. Thus 38% of all the 2219 validated operons incorporate at least one pioneer gene. Furthermore, 315 validated operons have more than one pioneer gene (15% of all validated operons), while 212 validated operons are in fact comprised completely of pioneer genes only (10% of all validated operons). These observations together indicate that relatively novel genes are also incorporated into preexisting operons as well as novel operons at a significant rate.
DISCUSSION
Trans-splicing and operon-like gene organization have been extensively studied in C. elegans and related nematodes (Lasda and Blumenthal 2011; Blumenthal 2012). However, despite its widespread occurrence especially within the nematode phyla, the origin and potentially adaptive functions of eukaryotic operons are not completely understood. Operons in C. elegans were initially discovered based on the observation that SL2-spliced genes were situated on genome in the same orientation with small intergenic distances of ∼100–300 nt (Spieth et al. 1993). Further genome-wide studies estimated the existence of about 1068 such operons that contain 15% of all transcribed genes (Blumenthal et al. 2002; Allen et al. 2011). About 80% of all the annotated C. elegans operons exhibit intergenic distance ≤500 nt (median intergenic distance = 156 nt within operons, based on WS239). Thus close gene spacing coupled with high relative levels of SL2 trans-splicing on downstream genes can be considered a reliable method of operon annotation. Here we have used similar criteria to define operons in P. pacificus, using a combination of intergenic distances of collinear genes in the same orientation and the trans-splicing patterns of genes in these clusters obtained from RNA-Seq experiments. We have used a systematic analysis of the effect of an intergenic distance threshold on the number of predicted operons to arrive at an optimum threshold of 3500 nt.
Since not all trans-spliced genes in nematodes such as C. elegans are part of operons, it is not well understood what factors govern the organization of a few selected genes into operons (Qian and Zhang 2008). Although, many of the operons seem to be highly conserved between C. elegans and the related nematode C. briggsae (Stein et al. 2003; Qian and Zhang 2008; Uyar et al. 2012), it is not entirely clear if this is due to adaptive constraints or a result of insufficient divergence between the two species. An evolutionary comparison to more distantly related nematode species is expected to be more informative in identifying the conserved trends and, hence, potentially adaptive functions of these operons. Previous comparative studies analyzing a few candidate operons have found differences in spliced leader sequences as well as some evidence of operon conservation across the nematode phylogeny (Lee and Sommer 2003; Guiliano and Blaxter 2006). We have now extended this comparison to a genome-wide level in P. pacificus and experimentally validated the genome-wide trans-splicing patterns.
The first difference that we find between C. elegans and P. pacificus is the organization of the genes coding for the SL1- and SL2-spliced leaders themselves. Unlike in C. elegans, the SL1 loci do not appear to be clustered on a single chromosome, nor are they in close proximity to the 5S rRNA loci (Supplemental Table S1). We observed ∼90% of all expressed genes to be trans-spliced to either SL1 or SL2 or to a mixture of both, similar to that in C. elegans (Allen et al. 2011). Interestingly, a k-means clustering on RNA-Seq data identifies three clusters of genes based on their splicing patterns (see Fig. 3). By using this information, we were able to annotate about 2219 operons out of a total of 6709 predicted operons. When we searched for operons syntenic between the two species, we found little conservation of the operon gene order across the two species. Specifically, out of the 1376 known operons in C. elegans and the 2219 P. pacificus operons validated in this study, only 37 operons were found to be partially or completely syntenic across the two species. Taking into account all the 4727 potential operons in C. elegans and about 6709 potential operons in P. pacificus, we still found only 128 syntenic operons. The overlap between the set of all orthologous genes that are part of operons in either species was relatively small but significant, indicating some conservation in gene content of operons on a global level (Fig. 5A). In addition, we found many P. pacificus–specific “pioneer” genes are incorporated into its operons. These data indicate that the genome organization has evolved considerably since the split between the two species lineages, changing the order and the content of operons. It has been argued that operon-like genomic organization places a constraint on gene order rearrangements during genome evolution because genes that are downstream in such operons lack a promoter and would not be transcribed if they move out of it (Nimmo and Woollard 2002; Blumenthal and Gleason 2003). However, more recent studies have predicted higher turnover rates for operons within the Caenorhabditis genus, suggesting that new operons might be formed at a higher rate than they are lost (Qian and Zhang 2008; Cutter and Agrawal 2010). Our observation of nonconserved operons across C. elegans and P. pacificus raises further interesting questions about the mechanism for such genome reorganizations and provides additional data to test various models of operon evolution (Qian and Zhang 2008; Cutter and Agrawal 2010) in a related nematode species.
To gain functional insights and infer potentially enriched functions in operonic genes in both P. pacificus and C. elegans, we analyzed gene expression data from other transcriptome studies in both species. This analysis revealed strong conservation between the functions potentially mediated by the operonic genes in the two species, despite the limited conservation of operons at a gene-by-gene level. We found strong enrichment for germline development and proliferation-related genes in the set of operonic genes in both the species (Fig. 5B). Also, during the dauer-exit in both the species, the genes within operons show higher expression levels than the genes that are not inside operons (Fig. 6). These results together suggest that although the identity of individual genes involved in particular biological processes has diverged, the operons in the two species have restructured accordingly such that they still contain genes related to similar functions, which potentially require bursts of transcription and translation. Such functional constraints have already been proposed to govern the organization of genes into operon-like clusters in nematodes and potentially other metazoans (Zaslaver et al. 2011). Our results now provide further evidence in support of this hypothesis.
A more comprehensive understanding of functions of operon-like gene organization in eukaryotes will require further analysis of such operons and require expression and functional studies of operonic genes across different species. The increasing number of sequenced metazoan genomes and EST or transcriptome data sets available for comparative analysis now provides new opportunities for discovering trans-splicing across new species (Lasda and Blumenthal 2011), as well as identification of operon-like clusters based on bioinformatics analysis. In addition, similar to the approach used in our study, mRNA obtained from spliced leader–specific pull-downs can be subjected to RNA-seq to quantitatively characterize the patterns of trans-splicing and annotate potential operons across different species, especially within the nematode phylum, where a wealth of genomic data is now available for comparative analysis (Guiliano and Blaxter 2006; Sommer and Streit 2011). Further analysis of annotated operons in the context of other functional and gene-expression data from different species can thus help elucidate the functional and structural constraints governing the evolution and maintenance of operons in metazoan lineages.
MATERIALS AND METHODS
mRNA collection and pull-down experiments
About 3 mg total RNA was extracted from mixed-stage cultures of P. pacificus reference strain RS2333 and was used for subsequent mRNA pull-downs. In the first step, a polyA fraction of the total RNA was purified by two rounds of pull-downs using biotinylated oligo-dT probes and magnetic beads coated with streptavidin (Promega polyA-attract kit). One aliquot of ∼150 ng of the purified poly-A mRNA was saved for RNAseq experiments. The remaining mRNA was split into two equal portions, and these were subjected to pull-downs using either a SL1-specific or SL2-specific biotinylated probe, respectively. This resulted in two pools of mRNA highly enriched for the SL1-spliced and SL2-spliced transcripts, respectively, which were then used for RNA-Seq experiments. The biotinylated probes used for enrichment of SL1 and SL2 transcripts correspond to the variants Pp-SL1a and Pp_SL2b, which have been reported to be observed most frequently on trans-spliced transcripts in P. pacificus (97% and 62%, respectively) (Guiliano and Blaxter 2006). Based on the high sequence similarity between all SL2 variants in P. pacificus (Supplemental Fig. S12), we expect the SL2 probe to also hybridize and therefore enrich for transcripts with other SL2 variants at their 5′ ends.
RNA-Seq experiment and analysis of isolated mRNA pools
High-throughput paired-end sequencing of the three mRNA pools (entire polyA transcriptome, SL1-spliced fraction, and SL2-spliced fraction of mRNA) was carried out according to manufacturer-provided standard protocols (2 × 76-nt, Illumina GA IIx). The resulting raw data were processed using the software Flexbar (Dodt et al. 2012) and mapped against the P. pacificus genome with Tophat 1.3.1. Transcript abundance for each predicted gene in the P. pacificus genome was calculated as FPKM read-counts with Cufflinks 1.3.0. All subsequent analysis was carried out on log2-transformed FPKM values, using custom scripts in R and Perl.
qRT-PCR experiments for SL enrichment and fidelity of SL-specific primers
The relative enrichments of SL1 or SL2 trans-spliced transcripts obtained from pull-down experiments were verified by qRT-PCR. Data from previous EST libraries in the laboratory showed the gene Ppa-rpl-43 (Contig15-snapOP.94) was trans-spliced to both SL1 and SL2 and, hence, was used to calculate relative enrichments after pull-down. A 200-bp fragment of this gene was amplified using either the SL1- or SL2-specific primers on cDNA obtained from total RNA as well as SL1- and SL2-enriched pools generated by spliced leader–specific pull-downs. The specificity of SL-specific primers used in these qPCRs was verified by first cloning the SL1- and SL2-splice variants of this fragment and then using pure plasmid DNA from each clone in a qPCR reaction with the opposite SL primer (Supplemental Fig. S1).
In vitro reactions and pull-down experiments on synthetic RNA
To verify the specificity of spliced leader–specific pull-downs used to generate samples for RNA-Seq, we carried out SL1 probe–specific and SL2 probe–specific pull-downs on pools of pure SL-spliced RNA synthesized by IVT reactions (TranscriptAid T7 high-yield transcription kit from Thermo Scientific). For the SL1-spliced template DNA needed in IVT, primer extension was used to add a T7 promoter upstream to the first 500 bp of the SL1-spliced version of Ppa-fib-1 gene (Contig20-snapOP.268). The DNA template for the corresponding SL2-spliced version was obtained as a synthetic gene from Eurofins MWG Operon. After IVT, both SL1- and SL2-specific pull-downs were then carried out on the following three kinds of RNA mixtures: (1) purely SL1-spliced RNA, (2) purely SL2-spliced RNA, and (3) 1:1 mixture of SL1 and SL2-spliced RNA. Using the Qubit RNA Assay Kit and fluorometer (from Life Technologies), no RNA could be detected in either the pull-down of mixture 1 by a SL2-specific probe, or pull-down of mixture 2 by a SL2-specific probe, indicating the high specificity of each probe. The RNA samples obtained from each of the SL1 and SL2 pull-downs on pool 3 were converted to cDNA and then analyzed via qRT-PCR to quantify any nonspecific carry over. The cross-SL contamination was observed to be less than 100-fold (sevenfold on a log2 scale), again indicating high specificity of the probes used in pull-down experiments.
Operon prediction and annotation
By using the “Hybrid1 Assembly” (Borchert et al. 2010) and the latest gene predictions in P. pacificus (GFF file for the gene models used in this study available from www.pristionchus.org/download), all contiguous gene clusters with genes in the same orientation and intergenic distances ≤3500 nt were identified as potential operons in P. pacificus, leading to identification of 6709 putative operons. In the absence of annotated transcription start sites and transcription end sites in P. pacificus, we used the coordinates of the predicted start and stop codon for each gene for this analysis, an approach successfully used in previous studies (Blumenthal et al. 2002; Zaslaver et al. 2011; Uyar et al. 2012). Integrating the splicing patterns of each gene then validated these putative operons, using the RNA-seq data derived from the three mRNA pools as described above. We basically used an unbiased k-means clustering on the relative SL1 and SL2 levels of each gene (log2(SL1/polyA) and log2(SL2/polyA) levels) and could divide the genes into three independent cluster (Fig. 3). The k-means clustering was carried out using the package “cluster” (version 1.14.0) in R. Based on the membership of operonic genes into SL1-spliced or SL2-spliced clusters, 2219 of the 6709 predicted operons could be validated and annotated as a Type 1, Type 2, or Type 3 operon, with distinct splicing patterns as described in the Results section above. We also verified that Type 2 operons are a bona fide class in themselves and not a special case of Type 1 operons where the first SL1-spliced gene might be situated at an intergenic distance greater than the 3500-nt threshold (data not shown). The remaining 4490 operons are labeled as “Predicted_Operons,” for which the splicing patterns could not be reliably discerned from the k-means clustering, most likely due to the low level of expression of constituent genes (Fig. 4).
To analyze the level of synteny between the operons in P. pacificus and C. elegans, we first identified all syntenic regions across the whole genomes of C. elegans (WS239 release) and P. pacificus, using the software CYNTENATOR (Rödelsperger and Dieterich 2010). We then focused on syntenic blocks or regions that contained an operon from at least one species. By using the Inparanoid program (Ostlund et al. 2010), we identified 5513 pairs of 1:1 orthologs between C. elegans and P. pacificus. We then checked for enrichment of these orthologs within the set of operonic genes using a 2 × 2 Fisher's exact test. The operon annotations and chromosomal coordinates for the C. elegans operons were extracted from the gff file corresponding to release WS239 of Wormbase (ftp://ftp.wormbase.org/pub/wormbase/species/c_elegans/gff/).
Using microarray data to identify functions potentially enriched in operonic genes
We used the following microarray-based gene expression data to identify functions enriched within operonic genes: gene expression profiles during dauer-exit in P. pacificus and C. elegans (NCBI GEO accessions GSE30977 and GSE31861) (Sinha et al. 2012a) and germline expressed genes in P. pacificus (Rae et al. 2012). Since the P. pacificus gene models used in this study are derived from the RNA-Seq experiments described in this study and, hence, are different from the ones used in the previous microarray studies, we used the tool cuffcompare (Trapnell et al. 2010) to first map the current gene models onto the older gene set and thereby the microarray data. In the dauer-exit vs. dauer comparisons, log2 of signal intensities from both channels (log2(RG)) was used as a measure for average expression levels, while the fold-change is defined as log2 of ratio of signal intensities (log2(R/G)). Statistical significance of differences in expression levels or fold-changes between operonic and nonoperonic genes was assessed by Wilcoxon test. The germline-enriched genes in P. pacificus were obtained as the list of genes significantly down-regulated in germline-ablated animals compared with germline-intact animals in P. pacificus (Rae et al. 2012). The statistical significance of overlap between operonic genes and germline-enriched genes in P. pacificus was assessed by Fisher's exact test on a 2 × 2 contingency table (gene in operon [yes, no] vs. gene enriched in germline [yes, no]).
DATA DEPOSITION
The RNA-Seq data generated in this study have been deposited to the Short Read Archive under the accession no. SRP039388. All gene expression data from microarray experiments are accessible from Gene Expression Omnibus (GSE30977, GSE31861, and GSE37331). The gene models used in this study are available at www.pristionchus.org/download under the section Hybrid1 Data.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Christian Rödelsperger for bioinformatics support. This study was partially funded by the Federal Ministry for Education and Research (BMBF) and the Senate of Berlin, Berlin, Germany (grant to C.D.).
Author contributions: A.S. collected all RNA samples and carried out all mRNA pull-down experiments and qPCR experiments. C.L. prepared the RNA-seq libraries and performed all sequencing reactions. A.S. and C.D. analyzed the data. A.S., R.J.S., and C.D. wrote the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.041954.113.
REFERENCES
- Allen MA, Hillier LW, Waterston RH, Blumenthal T 2011. A global analysis of C. elegans trans-splicing. Genome Res 21: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M, Liu L 1996. Nematode spliced leaders: ubiquity, evolution and utility. Int J Parasitol 26: 1025–1033 [PubMed] [Google Scholar]
- Blumenthal T 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet 11: 132–136 [DOI] [PubMed] [Google Scholar]
- Blumenthal T 2012. Trans-splicing and operons in C. elegans. WormBook Nov 11: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T, Gleason KS 2003. Caenorhabditis elegans operons: form and function. Nat Rev Genet 4: 112–120 [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, et al. 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851–854 [DOI] [PubMed] [Google Scholar]
- Borchert N, Dieterich C, Krug K, Schutz W, Jung S, Nordheim A, Sommer RJ, Macek B 2010. Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res 20: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumbarger DJ, Riebesell M, Rodelsperger C, Sommer RJ 2013. System-wide rewiring underlies behavioral differences in predatory and bacterial-feeding nematodes. Cell 152: 109–119 [DOI] [PubMed] [Google Scholar]
- Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A 2013. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 41: D226–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton CE 2002. Life without transcriptional control? From fly to man and back again. EMBO J 21: 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol 25: 778–786 [DOI] [PubMed] [Google Scholar]
- Cutter AD, Agrawal AF 2010. The evolutionary dynamics of operon distributions in eukaryote genomes. Genetics 185: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalley BK, Golomb M 1992. Gene expression in the Caenorhabditis elegans dauer larva: developmental regulation of Hsp90 and other genes. Dev Biol 151: 80–90 [DOI] [PubMed] [Google Scholar]
- Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, et al. 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet 40: 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M, Roehr J, Ahmed R, Dieterich C 2012. FLEXBAR: flexible barcode and adapter processing for next-generation sequencing platforms. Biology 1: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ 2011. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science 334: 1091–1097 [DOI] [PubMed] [Google Scholar]
- Guiliano DB, Blaxter ML 2006. Operon conservation and the evolution of trans-splicing in the phylum Nematoda. PLoS Genet 2: e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong RL, Sommer RJ 2006. Pristionchus pacificus: a well-rounded nematode. Bioessays 28: 651–659 [DOI] [PubMed] [Google Scholar]
- Huang P, Pleasance ED, Maydan JS, Hunt-Newbury R, O'Neil NJ, Mah A, Baillie DL, Marra MA, Moerman DG, Jones SJ 2007. Identification and analysis of internal promoters in Caenorhabditis elegans operons. Genome Res 17: 1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby WB, Ziegler DM 1990. The enzymes of detoxication. J Biol Chem 265: 20715–20718 [PubMed] [Google Scholar]
- Kienle S, Sommer RJ 2013. Cryptic variation in vulva development by cis-regulatory evolution of a HAIRY-binding site. Nat Commun 4: 1714. [DOI] [PubMed] [Google Scholar]
- Krause M, Hirsh D 1987. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell 49: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Friedman CC, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Davis RE 2004. Contribution of trans-splicing, 5′-leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J Biol Chem 279: 45573–45585 [DOI] [PubMed] [Google Scholar]
- Lasda EL, Blumenthal T 2011. Trans-splicing. Wiley Interdiscip Rev RNA 2: 417–434 [DOI] [PubMed] [Google Scholar]
- Lee K-Z, Sommer RJ 2003. Operon structure and trans-splicing in the nematode Pristionchus pacificus. Mol Biol Evol 20: 2097–2103 [DOI] [PubMed] [Google Scholar]
- Maroney PA, Denker JA, Darzynkiewicz E, Laneve R, Nilsen TW 1995. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA 1: 714–723 [PMC free article] [PubMed] [Google Scholar]
- Morton JJ, Blumenthal T 2011. Identification of transcription start sites of trans-spliced genes: uncovering unusual operon arrangements. RNA 17: 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo R, Woollard A 2002. Widespread organisation of C. elegans genes into operons: fact or function? Bioessays 24: 983–987 [DOI] [PubMed] [Google Scholar]
- Oesch F, Arand M 1999. Xenobiotic metabolism. In Toxicology (ed. Marquardt H, et al. ), pp. 83–109 Academic Press, San Diego [Google Scholar]
- Ogawa A, Streit A, Antebi A, Sommer RJ 2009. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr Biol 19: 67–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A, Bento G, Bartelmes G, Dieterich C, Sommer RJ 2011. Pristionchus pacificus daf-16 is essential for dauer formation but dispensable for mouth form dimorphism. Development 138: 1281–1284 [DOI] [PubMed] [Google Scholar]
- Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL 2010. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res 38: D196–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Zhang J 2008. Evolutionary dynamics of nematode operons: easy come, slow go. Genome Res 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae R, Iatsenko I, Witte H, Sommer RJ 2010. A subset of naturally isolated Bacillus strains show extreme virulence to the free-living nematodes Caenorhabditis elegans and Pristionchus pacificus. Environ Microbiol 12: 3007–3021 [DOI] [PubMed] [Google Scholar]
- Rae R, Sinha A, Sommer RJ 2012. Genome-wide analysis of germline signaling genes regulating longevity and innate immunity in the nematode Pristionchus pacificus. PLoS Pathog 8: e1002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Cutter AD 2009. Germline expression influences operon organization in the Caenorhabditis elegans genome. Genetics 181: 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Dieterich C 2010. CYNTENATOR: progressive gene order alignment of 17 vertebrate genomes. PLoS One 5: e8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel D, Riebesell M, Sommer RJ 2005. Gonadogenesis in Pristionchus pacificus and organ evolution: development, adult morphology and cell-cell interactions in the hermaphrodite gonad. Dev Biol 277: 200–221 [DOI] [PubMed] [Google Scholar]
- Rudel D, Tian H, Sommer RJ 2008. Wnt signaling in Pristionchus pacificus gonadal arm extension and the evolution of organ shape. Proc Natl Acad Sci 105: 10826–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A, Sommer RJ, Dieterich C 2012a. Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans. BMC Genomics 13: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A, Rae R, Iatsenko I, Sommer RJ 2012b. System wide analysis of the evolution of innate immunity in the nematode model species Caenorhabditis elegans and Pristionchus pacificus. PLoS One 7: e44255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ, McGaughran A 2013. The nematode Pristionchus pacificus as a model system for integrative studies in evolutionary biology. Mol Ecol 22: 2380–2393 [DOI] [PubMed] [Google Scholar]
- Sommer RJ, Streit A 2011. Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Annu Rev Genet 45: 1–20 [DOI] [PubMed] [Google Scholar]
- Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T 1993. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell 73: 521–532 [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Schlager B, Xiao H, Sommer RJ 2008. Wnt signaling induces vulva development in the nematode Pristionchus pacificus. Curr Biol 18: 142–146 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyar B, Chu JSC, Vergara IA, Chua SY, Jones MR, Wong T, Baillie DL, Chen N 2012. RNA-seq analysis of the C. briggsae transcriptome. Genome Res 22: 1567–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BD, Redinbo MR 2013. Xenobiotic-sensing nuclear receptors involved in drug metabolism: a structural perspective. Drug Metab Rev 45: 79–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sommer RJ 2011. Antagonism of LIN-17/Frizzled and LIN-18/Ryk in nematode vulva induction reveals evolutionary alterations in core developmental pathways. PLoS Biol 9: e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Baugh LR, Sternberg PW 2011. Metazoan operons accelerate recovery from growth-arrested states. Cell 145: 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhang J, Xie W 2005. Xenobiotic nuclear receptor-mediated regulation of UDP-glucuronosyl-transferases. Curr Drug Metab 6: 289–298 [DOI] [PubMed] [Google Scholar]
- Zorio DA, Cheng NN, Blumenthal T, Spieth J 1994. Operons as a common form of chromosomal organization in C. elegans. Nature 372: 270–272 [DOI] [PubMed] [Google Scholar]
- Zwierzynski TA, Buck GA 1991. RNA-protein complexes mediate in vitro capping of the spliced-leader primary transcript and U-RNAs in Trypanosoma cruzi. Proc Natl Acad Sci 88: 5626–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.