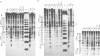Abstract
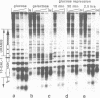
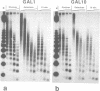
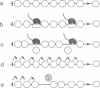
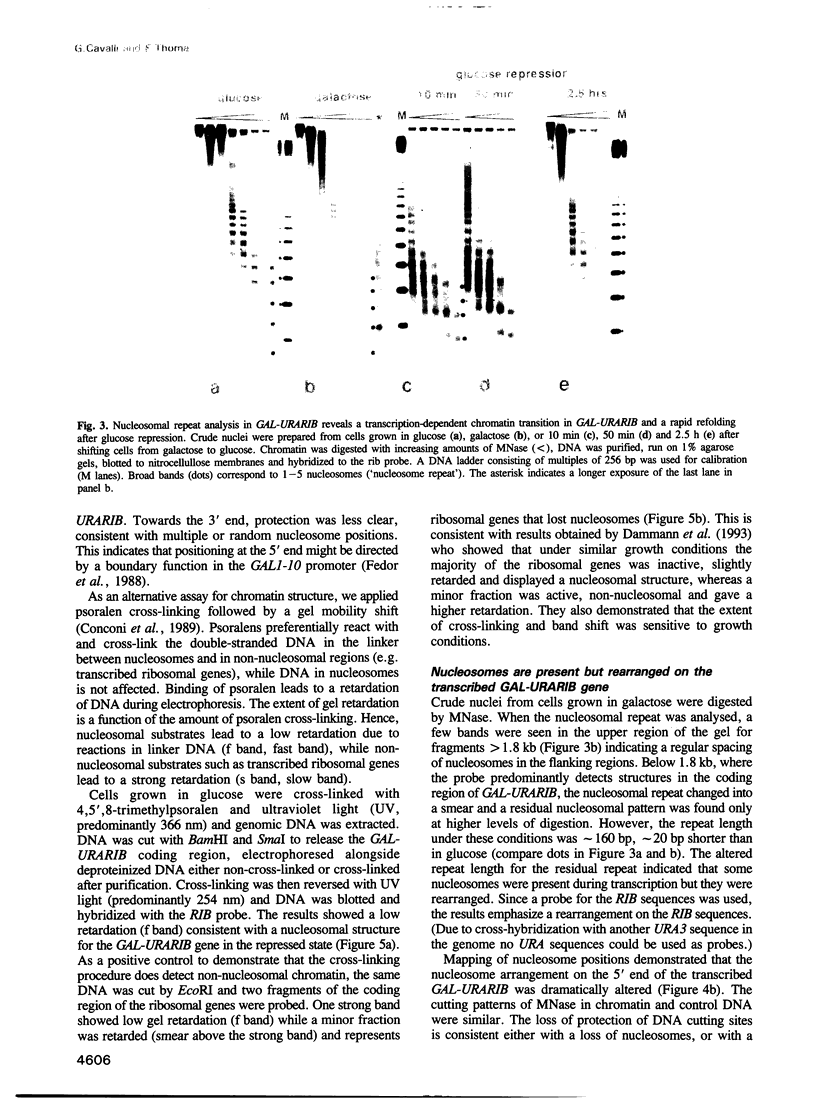
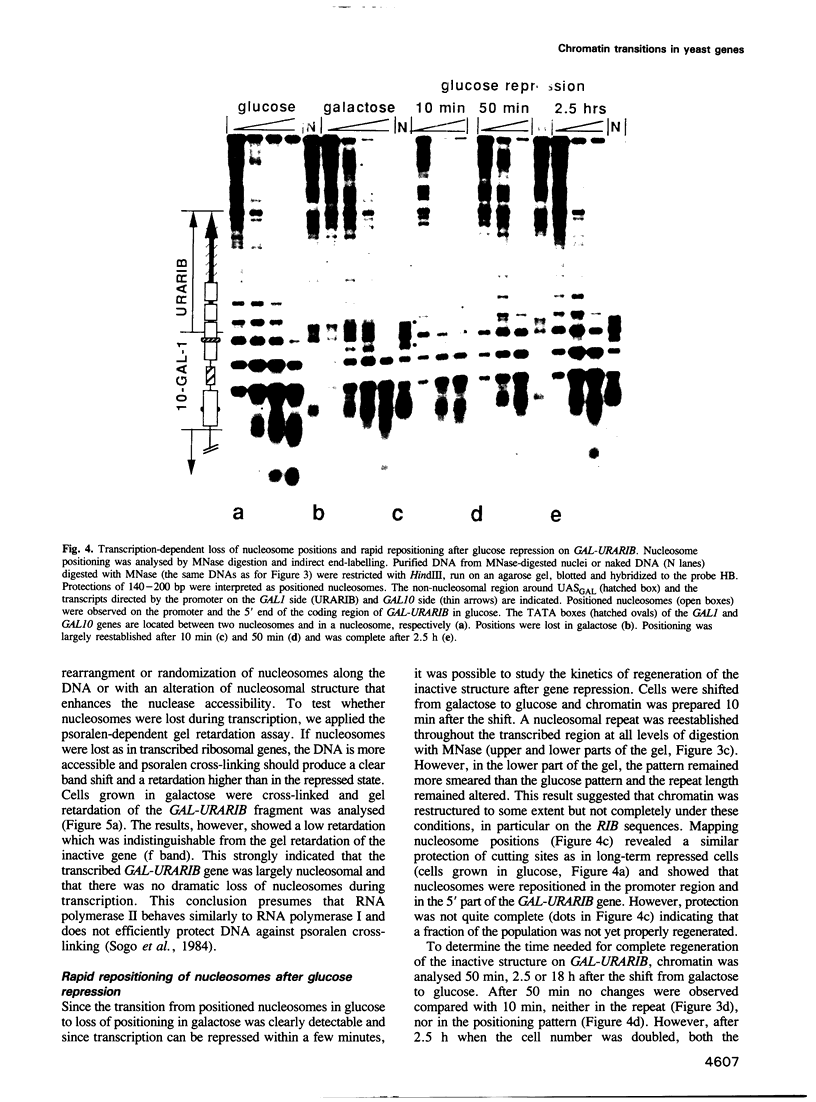
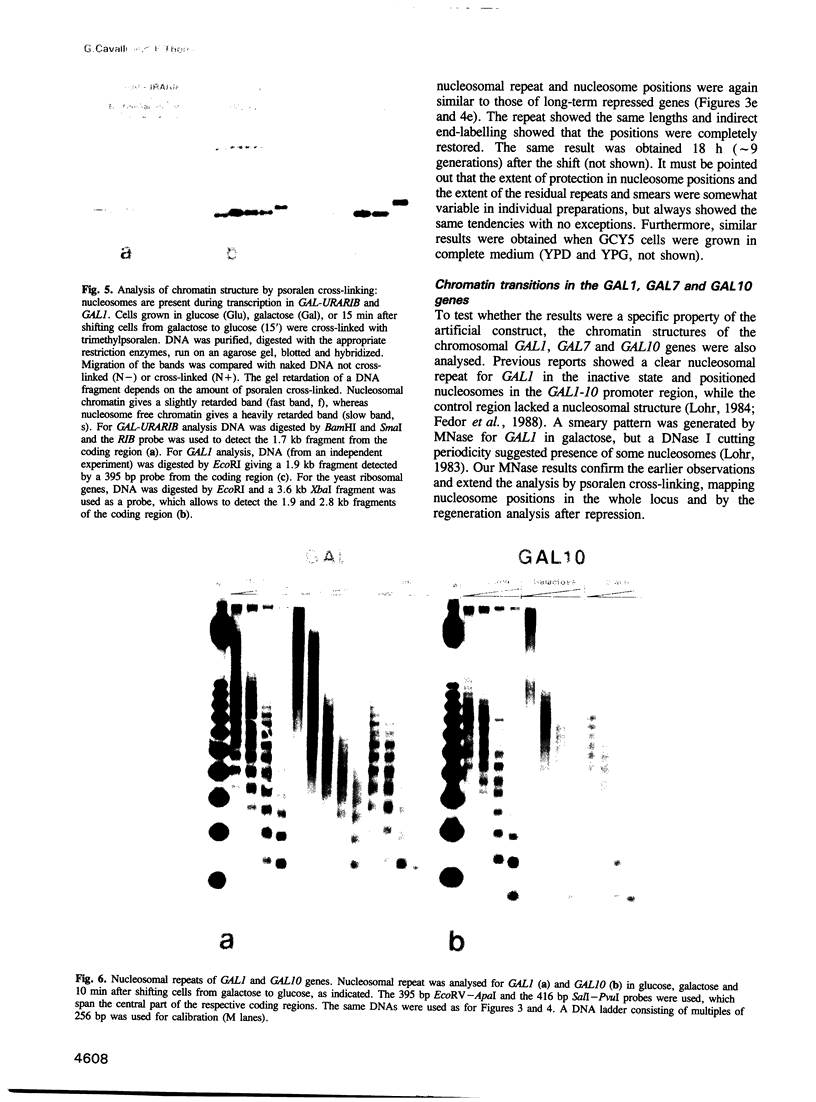
To study the fate of nucleosomes during transcription, a yeast gene 'GAL-URARIB' was constructed which is tightly regulated by the GAL1 promoter and shows in its inactive state a series of positioned nucleosomes that are sensitive for monitoring structural changes by micrococcal nuclease. Upon transcriptional activation, nucleosome positions were lost, but a residual nucleosomal repeat with an altered repeat length and no changes in psoralen accessibility measured by a band shift assay indicated that nucleosomes were present but rearranged on the transcribed gene. When chromatin was prepared 10 or 50 min after glucose repression, nucleosomes were repositioned in a large fraction of the population by a rapid process which most likely did not depend on histone synthesis or DNA replication. However, complete regeneration of the inactive structure and repeat length was observed after one cell generation (2.5 h) suggesting that in this step some missing histones were replaced. The results are consistent with a local dissociation of nucleosomes at the site of the polymerase followed by a rapid reassembly into nucleosomes behind it. The data are further supported by analysis of the chromosomal GAL1, GAL7 and GAL10 genes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
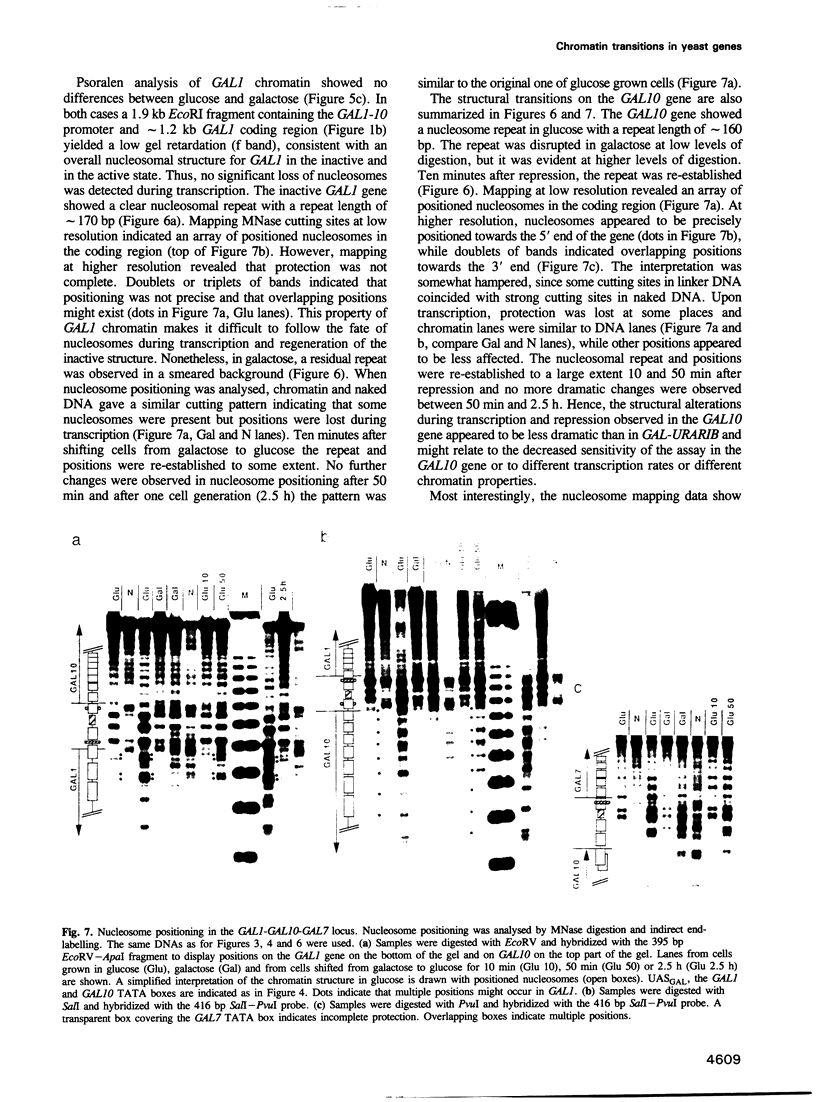
- Adams B. G. Induction of galactokinase in Saccharomyces cerevisiae: kinetics of induction and glucose effects. J Bacteriol. 1972 Aug;111(2):308–315. doi: 10.1128/jb.111.2.308-315.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
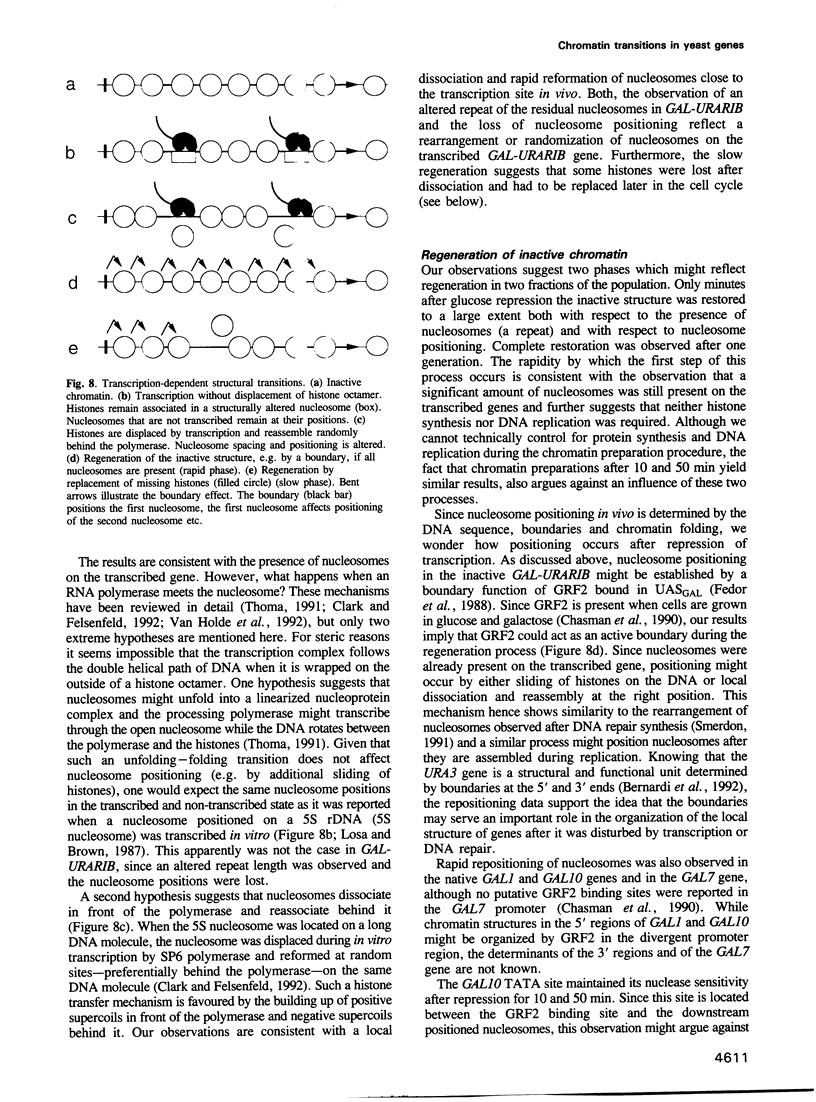
- Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M. L., Lacroute F., Botstein D. Evidence for transcriptional regulation of orotidine-5'-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi F., Koller T., Thoma F. The ade6 gene of the fission yeast Schizosaccharomyces pombe has the same chromatin structure in the chromosome and in plasmids. Yeast. 1991 Aug-Sep;7(6):547–558. doi: 10.1002/yea.320070603. [DOI] [PubMed] [Google Scholar]
- Bernardi F., Zatchej M., Thoma F. Species specific protein--DNA interactions may determine the chromatin units of genes in S.cerevisiae and in S.pombe. EMBO J. 1992 Mar;11(3):1177–1185. doi: 10.1002/j.1460-2075.1992.tb05158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Chasman D. I., Lue N. F., Buchman A. R., LaPointe J. W., Lorch Y., Kornberg R. D. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev. 1990 Apr;4(4):503–514. doi: 10.1101/gad.4.4.503. [DOI] [PubMed] [Google Scholar]
- Chen T. A., Sterner R., Cozzolino A., Allfrey V. G. Reversible and irreversible changes in nucleosome structure along the c-fos and c-myc oncogenes following inhibition of transcription. J Mol Biol. 1990 Apr 5;212(3):481–493. doi: 10.1016/0022-2836(90)90327-I. [DOI] [PubMed] [Google Scholar]
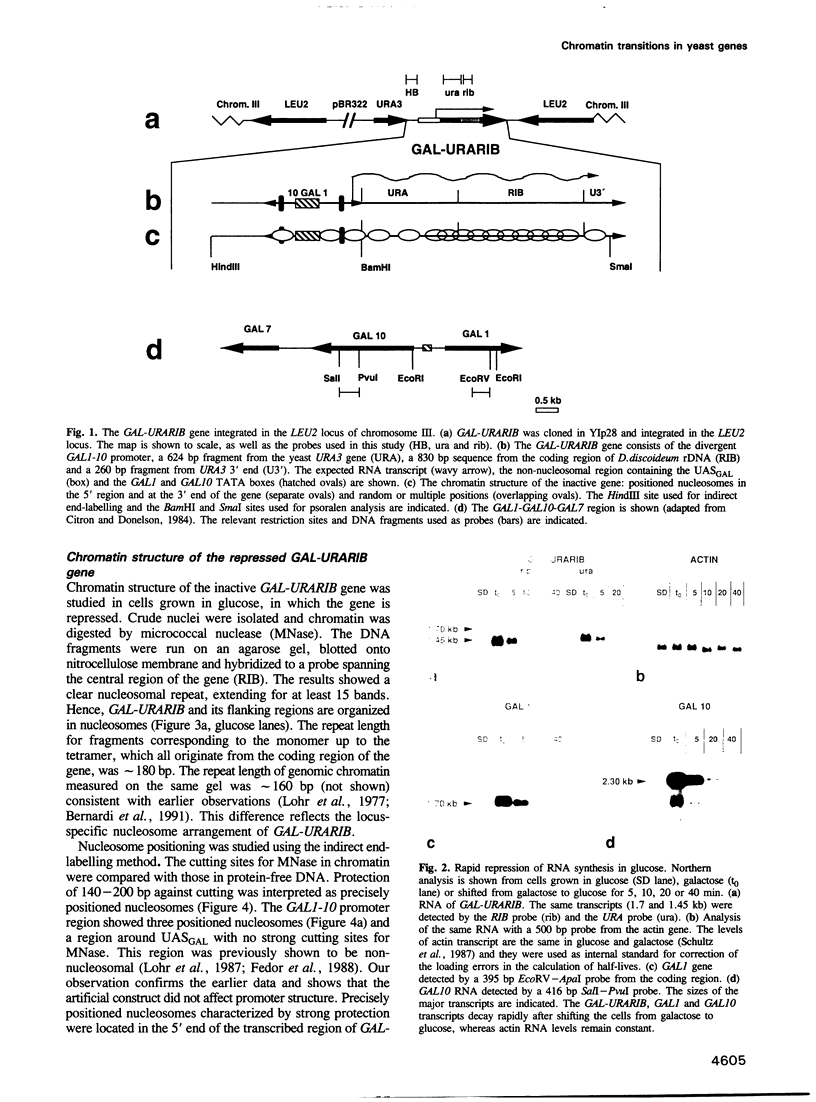
- Citron B. A., Donelson J. E. Sequence of the Saccharomyces GAL region and its transcription in vivo. J Bacteriol. 1984 Apr;158(1):269–278. doi: 10.1128/jb.158.1.269-278.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Felsenfeld G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell. 1992 Oct 2;71(1):11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- Conconi A., Widmer R. M., Koller T., Sogo J. M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989 Jun 2;57(5):753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J. M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993 May 25;21(10):2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bernardin W., Koller T., Sogo J. M. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J Mol Biol. 1986 Oct 5;191(3):469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Kornberg R. D. Upstream activation sequence-dependent alteration of chromatin structure and transcription activation of the yeast GAL1-GAL10 genes. Mol Cell Biol. 1989 Apr;9(4):1721–1732. doi: 10.1128/mcb.9.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Foe V. E., Wilkinson L. E., Laird C. D. Comparative organization of active transcription units in Oncopeltus fasciatus. Cell. 1976 Sep;9(1):131–146. doi: 10.1016/0092-8674(76)90059-3. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Han S., Udvardy A., Schedl P. Chromatin structure of the 87A7 heat-shock locus during heat induction and recovery from heat shock. Biochim Biophys Acta. 1985 Jun 24;825(2):154–160. doi: 10.1016/0167-4781(85)90099-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R., Sprague G. F., Jr, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci U S A. 1983 May;80(10):3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Garrard W. T. Transcription-induced nucleosome 'splitting': an underlying structure for DNase I sensitive chromatin. EMBO J. 1991 Mar;10(3):607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., Noll M. Chromatin fine structure of active and repressed genes. Nature. 1981 Jan 15;289(5794):198–203. doi: 10.1038/289198a0. [DOI] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Lohr D. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 1984 Nov 26;12(22):8457–8474. doi: 10.1093/nar/12.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. The chromatin structure of an actively expressed, single copy yeast gene. Nucleic Acids Res. 1983 Oct 11;11(19):6755–6773. doi: 10.1093/nar/11.19.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Torchia T., Hopper J. The regulatory protein GAL80 is a determinant of the chromatin structure of the yeast GAL1-10 control region. J Biol Chem. 1987 Nov 15;262(32):15589–15597. [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. On the displacement of histones from DNA by transcription. Cell. 1988 Dec 2;55(5):743–744. doi: 10.1016/0092-8674(88)90128-6. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- Nam H. G., Fried H. M. Effects of progressive depletion of TCM1 or CYH2 mRNA on Saccharomyces cerevisiae ribosomal protein accumulation. Mol Cell Biol. 1986 May;6(5):1535–1544. doi: 10.1128/mcb.6.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T., Hoshikawa Y., Iida Y., Iwabuchi M. Sequence analysis of the transcribed and 5' non-transcribed regions of the ribosomal RNA gene in Dictyostelium discoideum. Nucleic Acids Res. 1984 May 25;12(10):4171–4184. doi: 10.1093/nar/12.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Herrick D., Peltz S. W., Jacobson A. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:415–423. doi: 10.1016/0076-6879(91)94032-8. [DOI] [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Littau V. C., Allfrey V. G. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983 Oct;34(3):1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Dean A., Simpson R. T. Yeast alpha 2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990 May;10(5):2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A., Fascher K. D., Hörz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992 Nov 27;71(5):853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- Schultz L. D., Hofmann K. J., Mylin L. M., Montgomery D. L., Ellis R. W., Hopper J. E. Regulated overproduction of the GAL4 gene product greatly increases expression from galactose-inducible promoters on multi-copy expression vectors in yeast. Gene. 1987;61(2):123–133. doi: 10.1016/0378-1119(87)90107-7. [DOI] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Effects of DNA sequence and histone-histone interactions on nucleosome placement. J Mol Biol. 1990 Nov 5;216(1):69–84. doi: 10.1016/S0022-2836(05)80061-0. [DOI] [PubMed] [Google Scholar]
- Smerdon M. J. DNA repair and the role of chromatin structure. Curr Opin Cell Biol. 1991 Jun;3(3):422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Ness P. J., Widmer R. M., Parish R. W., Koller T. Psoralen-crosslinking of DNA as a probe for the structure of active nucleolar chromatin. J Mol Biol. 1984 Oct 5;178(4):897–919. doi: 10.1016/0022-2836(84)90318-8. [DOI] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Straka C., Hörz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991 Feb;10(2):361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Thoma F. Nucleosome positioning. Biochim Biophys Acta. 1992 Feb 28;1130(1):1–19. doi: 10.1016/0167-4781(92)90455-9. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]
- Thoma F. Structural changes in nucleosomes during transcription: strip, split or flip? Trends Genet. 1991 Jun;7(6):175–177. doi: 10.1016/0168-9525(91)90429-t. [DOI] [PubMed] [Google Scholar]
- Thoma F., Zatchej M. Chromatin folding modulates nucleosome positioning in yeast minichromosomes. Cell. 1988 Dec 23;55(6):945–953. doi: 10.1016/0092-8674(88)90240-1. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Widmer R. M., Lucchini R., Lezzi M., Meyer B., Sogo J. M., Edström J. E., Koller T. Chromatin structure of a hyperactive secretory protein gene (in Balbiani ring 2) of Chironomus. EMBO J. 1984 Jul;3(7):1635–1641. doi: 10.1002/j.1460-2075.1984.tb02022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Wu T. C., Simpson R. T. Transient alterations of the chromatin structure of sea urchin early histone genes during embryogenesis. Nucleic Acids Res. 1985 Sep 11;13(17):6185–6203. doi: 10.1093/nar/13.17.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K. E., Lohr D. E., Robert C. What happens to nucleosomes during transcription? J Biol Chem. 1992 Feb 15;267(5):2837–2840. [PubMed] [Google Scholar]