Abstract
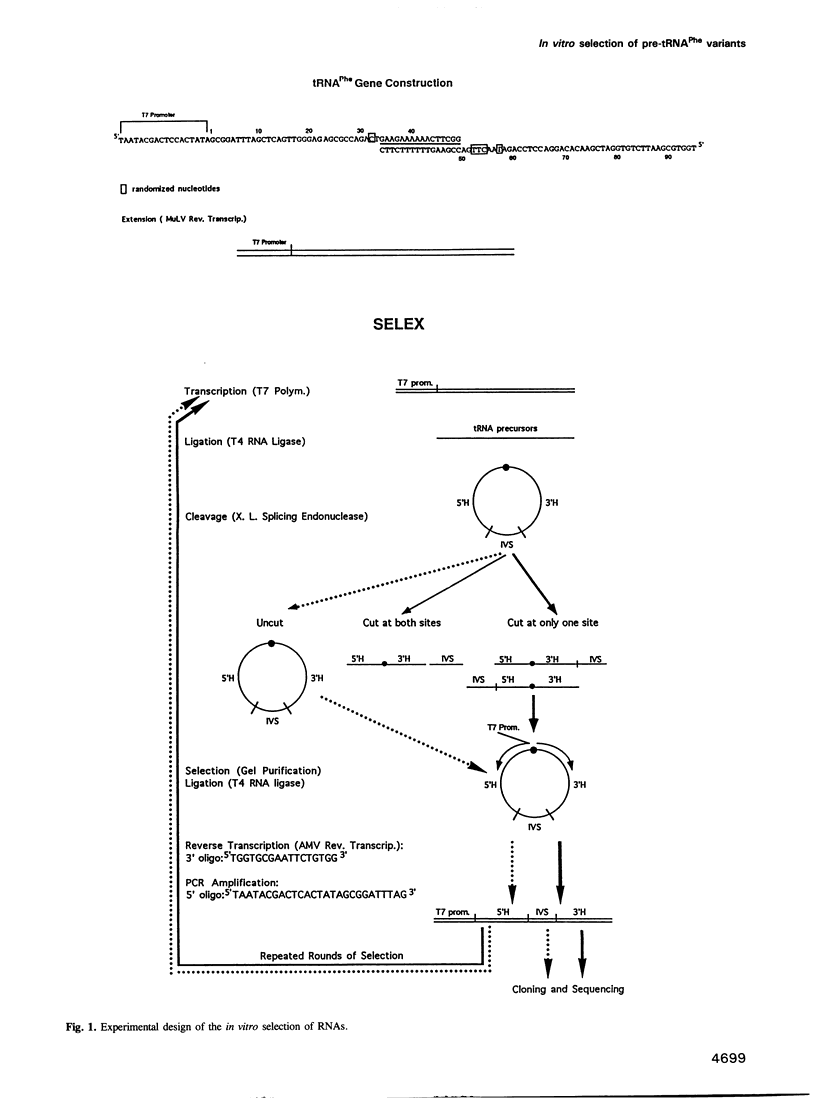
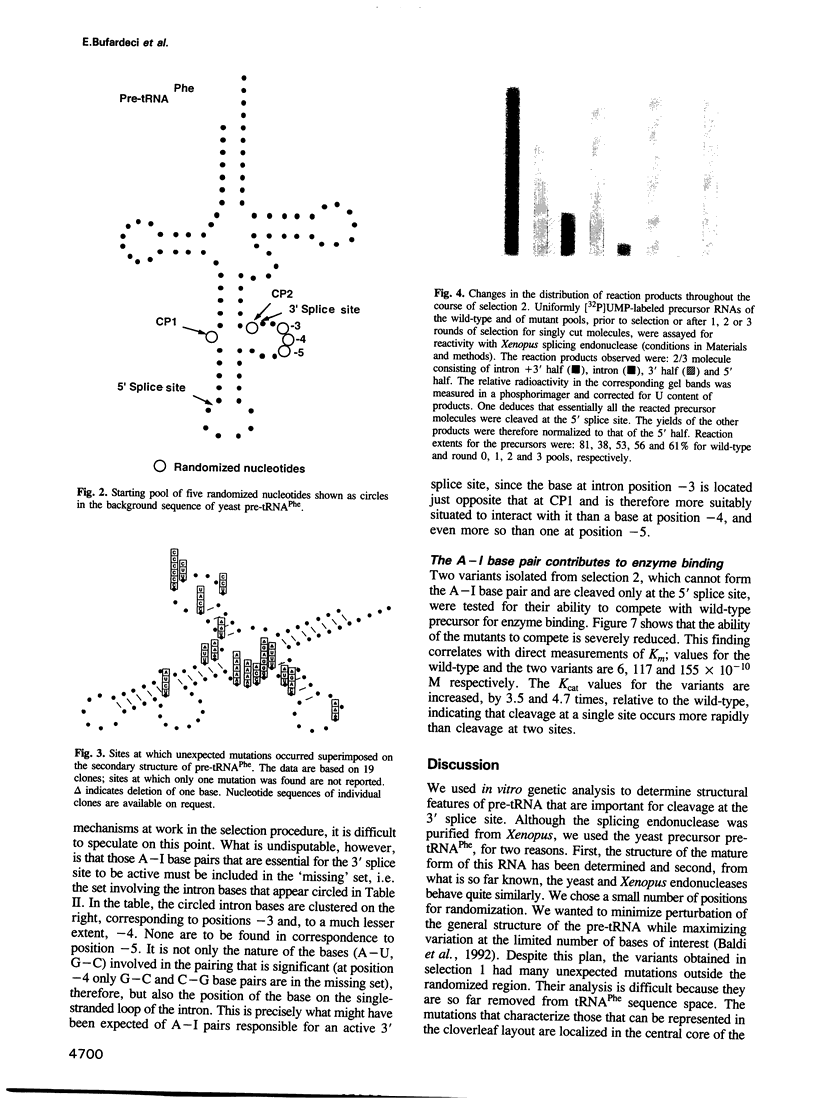
During processing of intron-containing pre-tRNAs, the Xenopus laevis splicing endonuclease binds the precursor and cleaves it at both the 5' and 3' splice sites. In vitro selection was used to determine structural features characteristic of precursor tRNA molecules that are active in this reaction. We performed two types of selection, one for molecules that are not cut, the other for molecules that are cut at only one site. The results shed light on various aspects of the intron excision reaction, including the importance of the three-dimensional structure of the mature domain for recognition and binding of the enzyme, the active role played by the single-stranded region of the intron, and the importance of the cardinal positions which, although not necessarily occupied by the same base in all precursors, nevertheless play a fundamental role in the splicing reaction. A precursor can be cut at the 3' site if a base in the single-stranded loop of the intron is allowed to pair (A-I pair) with the base of the 5' exon situated at the position immediately following the anticodon stem [first cardinal position (CP1)]. The nature of the bases involved in the A-I pair is important, as is the position of the base in the single-stranded loop of the intron. We discuss the role of the cardinal positions in the reaction.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldi M. I., Mattoccia E., Bufardeci E., Fabbri S., Tocchini-Valentini G. P. Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science. 1992 Mar 13;255(5050):1404–1408. doi: 10.1126/science.1542788. [DOI] [PubMed] [Google Scholar]
- Baldi M. I., Mattoccia E., Ciafrè S., Attardi D. G., Tocchini-Valentini G. P. Binding and cleavage of pre-tRNA by the Xenopus splicing endonuclease: two separable steps of the intron excision reaction. Cell. 1986 Dec 26;47(6):965–971. doi: 10.1016/0092-8674(86)90811-1. [DOI] [PubMed] [Google Scholar]
- Bartel D. P., Zapp M. L., Green M. R., Szostak J. W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991 Nov 1;67(3):529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- Behlen L. S., Sampson J. R., DiRenzo A. B., Uhlenbeck O. C. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry. 1990 Mar 13;29(10):2515–2523. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- Gandini-Attardi D., Baldi I. M., Mattoccia E., Tocchini-Valentini G. P. Transfer RNA splicing endonuclease from Xenopus laevis. Methods Enzymol. 1990;181:510–517. doi: 10.1016/0076-6879(90)81148-n. [DOI] [PubMed] [Google Scholar]
- Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem. 1985 Mar 10;260(5):3108–3115. [PubMed] [Google Scholar]
- Mattoccia E., Baldi I. M., Gandini-Attardi D., Ciafrè S., Tocchini-Valentini G. P. Site selection by the tRNA splicing endonuclease of Xenopus laevis. Cell. 1988 Nov 18;55(4):731–738. doi: 10.1016/0092-8674(88)90231-0. [DOI] [PubMed] [Google Scholar]
- Miao F., Abelson J. Yeast tRNA-splicing endonuclease cleaves precursor tRNA in a random pathway. J Biol Chem. 1993 Jan 5;268(1):672–677. [PubMed] [Google Scholar]
- Otsuka A., de Paolis A., Tocchini-Valentini G. P. Ribonuclease "XlaI," an activity from Xenopus laevis oocytes that excises intervening sequences from yeast transfer ribonucleic acid precursors. Mol Cell Biol. 1981 Mar;1(3):269–280. doi: 10.1128/mcb.1.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Gutell R. R., Uhlenbeck O. C. Folding of circularly permuted transfer RNAs. Science. 1991 Nov 29;254(5036):1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- Pan T., Uhlenbeck O. C. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+. Biochemistry. 1992 Apr 28;31(16):3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988 Nov 18;55(4):719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- Swerdlow H., Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem. 1984 Apr 25;259(8):5197–5207. [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Wrede P., Wurst R., Vournakis J., Rich A. Conformational changes of yeast tRNAPhe and E. coli tRNA2Glu as indicated by different nuclease digestion patterns. J Biol Chem. 1979 Oct 10;254(19):9608–9616. [PubMed] [Google Scholar]