Background: AgI/II homolog interaction with GP340 is crucial for bacterial attachment to tooth surface.
Results: Tandem SRCR domains efficiently adhere/aggregate bacteria. Calcium-induced conformational switch and O-linked carbohydrates of SRCRs are necessary for the interaction with AgI/II homologs.
Conclusion: High affinity interactions are dictated by calcium and carbohydrates.
Significance: Oral streptococci adhere to specific calcium-induced conformation of immobilized SRCRs and to its carbohydrates.
Keywords: Bacterial Adhesion; Bacterial Pathogenesis; Calcium; Cell Surface Protein; Cell Surface Receptor; Glycosylation; Innate Immunity; Surface Plasmon Resonance (SPR); Bacterial Adherence, Antigen I/II, Streptococcus mutans, Streptococcus gordonii, Oral Streptococci
Abstract
Oral streptococci adhere to tooth-immobilized glycoprotein 340 (GP340) via the surface protein antigen I/II (AgI/II) and its homologs as the first step in pathogenesis. Studying this interaction using recombinant proteins, we observed that calcium increases the conformational stability of the scavenger-rich cysteine repeat (SRCRs) domains of GP340. Our results also show that AgI/II adheres specifically with nanomolar affinity to the calcium-induced SRCR conformation in an immobilized state and not in solution. This interaction is significantly dependent on the O-linked carbohydrates present on the SRCRs. This study also establishes that a single SRCR domain of GP340 contains the two surfaces to which the apical and C-terminal regions of AgI/II noncompetitively adhere. Compared with the single SRCR domain, the three tandem SRCR domains displayed a collective/cooperative increase in their bacterial adherence and aggregation. The previously described SRCRP2 peptide that was shown to aggregate several oral streptococci displayed limited aggregation and also nonspecific adherence compared to SRCR domains. Finally, we show distinct species-specific adherence/aggregation between Streptococcus mutans AgI/II and Streptococcus gordonii SspB in their interaction with the SRCRs. This study concludes that identification of the metal ion and carbohydrate adherence motifs on both SRCRs and AgI/II homologs could lead to the development of anti-adhesive inhibitors that could deter the adherence of pathogenic oral streptococci and thereby prevent the onset of infections.
Introduction
The attachment of bacteria to human tissue and other surfaces within the oral cavity is thought to be an essential first step in pathogenesis, and microbes utilize surface proteins (pili and fimbrae) to effectively adhere to a variety of molecules and surfaces. The oral cavity is home to a number of microbes, including oral streptococci, where the mutans streptococci (Streptococcus mutans and Streptococcus sobrinus) are the known etiological agents in dental caries, and the viridians streptococci (Streptococcus gordonii and Streptococcus sanguis) are considered to be the commensal flora. Among the surface proteins on oral streptococci, antigen I/II (AgI/II)2 homologs (also known as P1, PAc, SpaP, and SR) in S. mutans (1–3), SspA and SspB in S. gordonii (4–7), and Pas in Streptococcus intermedius, etc. (8, 9) are the most well studied. These AgI/II homologs adhere to tooth-immobilized salivary agglutinin (SAG) secreted by the parotid gland (10, 11). Typically, AgI/II homologs carry a signal sequence at the N terminus, followed by the alanine-rich (A), variable (V), and proline-rich (P) regions, followed by the C-terminal region and the membrane spanning domain that anchors to the bacterial cell wall (Fig. 1). In earlier studies, we identified two SAG adherence regions on AgI/II (12, 13), one at the apex of the molecule (A3VP1) and the other at the C terminus (C123), specifically the C1 and C2 domains that adopt the DEv-IgG fold, a variant of the classical IgG-fold (14). More importantly we determined that these two regions adhere to SAG in a non-competitive manner, indicating the presence of two different surfaces on SAG, pointing toward bacterial heterogeneity (multivalency) in adherence (13). So far, these interactions were studied with extracted and purified SAG (some groups address purified SAG as GP340) (15, 16) from single or multiple donors and in some cases with saliva itself (17, 18). The presence of allelic variability among donors and their associated post-translational modifications, an established standard has been lacking to quantify these interactions. This study now establishes the first benchmark using recombinantly expressed subdomains of SAG that are involved in microbial adherence.
FIGURE 1.
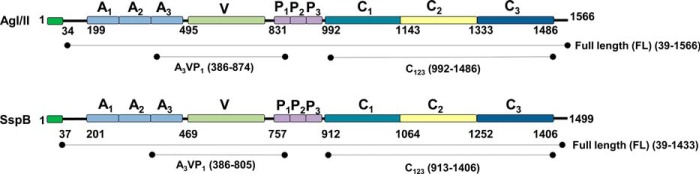
Schematic representation of primary sequence of S. mutans UA159 AgI/II and S. gordonii DL1 SspB, including the extents of the recombinant fragments shown here.
SAG is a large glycoprotein complex that contains glycoprotein 340 (GP340), sIgA, and an unknown 80-kDa protein (10, 19). Among these, the major component GP340 is known to aggregate several species of bacteria, including mutans, viridans streptococci, and Helicobacter pylori (18, 20), and is thereby considered an innate immune response factor. GP340 is a 340-kDa protein that contains 14 SRCR domains, two C1r/C1s Uegf Bmp1 (CUB) domains, and one zona pellucida (ZP) domain (Fig. 2). The 13 SRCR domains are present in tandem at the N terminus, followed by an intriguingly nested 14th SRCR domain between two CUB domains, with a ZP domain at the C terminus. The SRCR domains are interspersed with regions termed SID, an acronym for the SRCR interspersed domains. Except between the 4th and the 5th SRCR domain, all other tandem repeats contain the SID domain (11). These SRCR domains belong to an ancient class of proteins and are present in protozoan parasites like Cryptosporidium, Toxoplasma, Plasmodium, and in the algae Chlamydomonas (21, 22). They also appear in the entire animal kingdom, beginning with sponges, and are highly conserved, where a single SRCR domain usually contains 100–110 amino acids (23). GP340 SRCR domains were recently shown to aid in transcytosis of HIV into vaginal epithelial cells (24, 25). This highlights the role of the GP340 SRCR domains in infection, where it serves as a portal of entry into the host for both bacteria and viruses that result in various human diseases (26–29).
FIGURE 2.
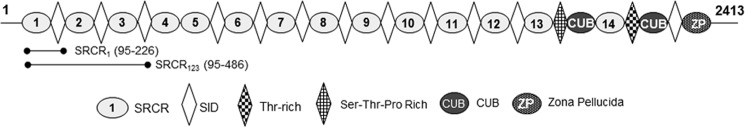
Schematic representation of primary sequence of GP340's 13 tandem SRCR domains, the CUB, and ZP are shown here. The figure also includes the extents of the recombinantly expressed iSRCR1 and iSRCR123 (33).
In a systematic study conducted with various oral streptococci, Loimaranta et al. (30) classified bacterial recognition properties of GP340 into three different groups as follows: group I strains were both aggregated by and adhered to GP340; group II preferentially adhered, and group III preferentially aggregated. Using a peptide-based approach, Bikker et al. (17, 31) identified a consensus peptide SRCRP2 (QGRVEVLYRGSWGTVC) derived from the 14 SRCR domains of GP340, which aggregated several species of bacteria, and also inhibited the adherence of AgI/II to SAG (32). In a subsequent study using alanine scanning, the most important residues involved in aggregation were deduced to reside within the “VEVLXXXXW” motif (31). In these studies, the SID domains that are predicted to host the glycosylation sites were classified into two different groups, namely SID20 and SID22 based on sequence homology, and neither one displayed aggregation nor adherence to bacteria (17, 31).
In this study, we have reported the adherence characteristics and the multicomponent adherence mechanisms adopted by S. mutans AgI/II and S. gordonii SspB with the recombinantly expressed SRCR domains. We also have presented comparative results on the adherence and aggregation properties of the SRCRP2 peptide and recombinantly expressed SRCRs. The results emanating from this study would foster the development of inhibitors to AgI/II homologs on oral streptococci.
EXPERIMENTAL PROCEDURES
Expression and Purification of Proteins
iSRCR1 and iSRCR123 were expressed and purified as described recently by Purushotham et al. (33), and AgI/II fragments used in this study were prepared as described previously (12, 13). SspB constructs Full length (FLSspB) (39-1433), A3VP1SspB (386-805), and C123SspB (913-1406), were cloned into the pET23d vector (Novagen) using primers listed in supplemental Table S1, restriction enzymes NcoI, NotI, BamHI, and XhoI, and the template plasmid containing the SspB gene (Fig. 1). Similar to methods described earlier for S. mutans AgI/II (12, 13), the SspB fragments were purified over three columns, HisPrep nickel affinity, Mono Q, and Superdex 200 10/300 GL gel filtration. The purified fragments FLSspB (154.8 kDa), A3VP1SspB (47.7 kDa), and C123SspB (56.2 kDa) were qualitatively assessed to be >95% pure from SDS-polyacrylamide gels (supplemental Fig. S1).
Surface Plasmon Resonance
Real time binding analyses of the SRCR domains with AgI/II fragments were carried out using the BIAcore 2000 system. The CM5 chip was labeled with ligands iSRCR1 or iSRCR123 or SAG as described previously (34, 35), using the amine coupling kit (GE Healthcare). The control and experimental surfaces were blocked using 1 m ethanolamine. Various concentrations of analytes (0.125 to 2.5 μm) of S. mutans AgI/II or S. gordonii SspB fragments (supplemental Table S2) were injected over the prepared chip surfaces, and dissociations were measured for 8–10 min at a flow rate of 20 μl/min of binding buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 2.5 mm CaCl2) at 25 °C. Self-adhesion of iSRCR1 or iSRCR123 (2 μm) was also determined in a similar manner as described above. Between experiments, the regeneration of the chip surface was accomplished using solutions as shown in supplemental Table S2. Finally, to determine the effect of calcium, SPR analysis was carried out by dialyzing the analytes and ligands in binding buffer devoid of CaCl2.
On-chip enzymatic deglycosylation of the iSRCR1 and iSRCR123 was carried out to remove N- and O-linked carbohydrates. Briefly, after immobilization of iSRCR1 and iSRCR123 on the CM5 sensor chip, the deglycosylation was carried out by incubating the chip surface with a mixture containing a total reaction volume of 40 μl made up of 4 μl of 10× G7 reaction buffer, 4 μl of 10% Nonidet P-40, 4 μl of neuraminidase (Sigma), 18 μl of water, and 10 μl of O-glycosidase (New England Biolabs) and similarly following the manufacturer's protocols for endoglycosidase H (New England Biolabs). Later, the chip was sealed and incubated overnight at 37 °C. Subsequently, the chip was thoroughly washed with binding buffer (20 mm HEPES, 150 mm NaCl, 2.5 mm CaCl2) to remove the deglycosylating enzymes and other remnants. Similar to the experimental procedure described above, in a separate experiment an off-chip deglycosylation of SRCRs was carried out and later checked to verify deglycosylation using MALDI-TOF MS analysis (data not shown). Binding studies with FLAgI/II and FLSspB and their subfragments were then carried out as described above and regenerated as described in supplemental Table S2. All experiments were carried out in triplicate, and the kinetics of the association (KA) and dissociation (KD) rate constants were deduced using the 1:1 Langmuir kinetic model on the BIA-evaluation software (36).
The utilization of a bivalent adherence model to elucidate the kinetics had inherent difficulties in clearly distinguishing affinities for each region, particularly for FLAgI/II and FLSspB. In addition, the SRCR holding two distinct surfaces compounded the elucidation of individual kinetics, and presently there are no modeling protocols available to determine the individual affinities for such a system; therefore, for simplicity we have utilized a single site 1:1 Langmuir model. The larger χ2 values observed for the full-length AgI/II and SspB are directly attributable to the multiple binding sites.
The concentration (C in micromolars) of analyte (FLAgI/II or FLSSpB at 2 μm) that adhered to the immobilized ligand (iSRCR1 and iSRCR123) within the flow cell was calculated using the formula C = (RU/Mr) × (1/V), where RU is resonance unit (1 RU = 1 pg of bound protein); Mr is molecular weight of analyte, and V is volume of flow cell (1.2 × 10−10 liters).
Competition Adherence Assay
To determine whether AgI/II domains bound to the same site on SRCR domains, competitive binding SPR experiments were conducted in triplicate as described previously (13), where each fragment, 2 μm AgI/II (FL, A3VP1, or C123) or SspB (FL, A3VP1, or C123), was initially passed over the CM5 chip surface immobilized with either iSRCR1, iSRCR123, or SAG for 60 s to saturate available binding sites. The response curve of AgI/II or SspB fragment was first recorded, where the maximal RU (RU1) was considered as the base line for the second injection, and thereafter, the competing fragment was injected, and its response was recorded as RU2. The adherence of the second fragment was then calculated (RU2 − RU1) for all SPR competing assay as reported earlier (13).
ELISA
The binding between commercially synthesized (Think Peptides, Inc.) SRCRP2 peptide (QGRVEVLYRGSWGTVCK-(FAM)) with fluorescein amidite (FAM) at the carboxyl end and AgI/II homologs was analyzed. Briefly, AgI/II homologs (10 μg/well) in carbonate/bicarbonate buffer, pH 9.6, were coated on a black ELISA plate individually, washed with binding buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, and 2.5 mm CaCl2), and blocked with 3% nonfat dry skim milk. Serial dilution of the SRCRP2 peptide (200 μl) ranging from 0.03 to 1 mg/ml in binding buffer were then incubated with all fragments of AgI/II and SspB coated on wells for 3 h at room temperature. The coated wells without fluorescently labeled SRCRP2 peptides were used as controls. Later, the wells were washed with binding buffer, and the data were recorded at an excitation wavelength of 495 nm and emission at 519 nm using Synergy 2- multimode microplate reader and the results were then analyzed (Synergy, Inc.).
Adherence/Inhibition Studies
SRCRP2 peptide at different concentrations (5, 10, 50, 100, and 200 μm) was incubated with 2 μm each of FL, A3VP1, and C123 of AgI/II and SspB at room temperature for 3 h, and then their interaction with immobilized iSRCR1 or iSRCR123 on the CM5 chip was analyzed. Running buffer containing 20 mm HEPES, pH 7.4, 150 mm NaCl, and 2.5 mm CaCl2 at 25 °C with a flow rate of 20 μl/min was used throughout the experiment. The CM5 chip was regenerated with buffer containing 1 m NaCl and 20 mm EDTA, pH 7.4, after each reaction cycle. Direct adherence of 200 μm SRCRP2 peptide alone served as the control, and all calculations on the adherence inhibition were assessed using the BIAevaluation software.
Aggregation Assay
Aggregation assays were performed as described earlier (34) with slight modifications. Briefly, S. mutans UA159 and S. gordonii DL1 cells were grown in TSY broth media (30 g/liter of trypticase soy broth and 0.5 g/liter yeast extract, pH 7.2) overnight at 37 °C in the presence of 5% CO2. The bacteria were centrifuged at 5000 × g and washed with a buffer containing 20 mm HEPES, pH 7.4, 150 mm NaCl and resuspended to an approximate OD700 of 1. The bacterial suspension (900 μl) was mixed with 6 μl of 0.1 m CaCl2 and 100 μl of SAG or iSRCR1 or iSRCR123 (10 μm) or SRCRP2 peptide (400 μg/ml). The aggregation of bacteria was then measured by recording OD700 over 60 min at 5-min intervals, where the buffer alone was used as control. All experiments were carried out at least five times, and the results were analyzed with one-way analysis of variance. Post hoc testing at p < 0.05 was considered statistically significant, and results were presented as the percentage of cells aggregated.
Confocal Microscopy
S. mutans UA159 and S. gordonii DL1 were grown overnight in TSY media on an eight-well LabTek chamber slide system (Sigma). The cells were fixed with 3% paraformaldehyde, washed with binding buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, and 2.5 mm CaCl2), and thereafter iSRCR1 or iSRCR123 (10 μm) was added to the cells and incubated for 60 min. The unbound SRCRs were removed by repeated washing using the binding buffer. Subsequently, Alexa Fluor 488-conjugated anti-His tag antibody (EMD Millipore) (1:50 dilution) that can bind to the His tag on SRCRs was added. After 60 min of incubation, the unbound antibody was washed away thoroughly using binding buffer, and the chamber walls were gently removed. Coverslips were then mounted with 15 μl of Fluoromount-G with DAPI (Southern Biotech Inc.,) to stain bacterial nuclei and were sealed until ready to be imaged. The experiment without SRCRs served as control. All slides were imaged using a Leica SP1 UV confocal laser scanning microscope and a Zeiss LSM 710 confocal laser scanning microscope at the High Resolution Imaging Facility (University of Alabama at Birmingham).
Glycoprotein Staining and GC-MS Analysis of SRCRs Carbohydrates
The iSRCR1 and iSRCR123 proteins were electrophoretically separated on a 12.5% SDS-polyacrylamide gel and stained by glycoprotein staining kit (Pierce), where horseradish peroxidase (HRP) and soybean trypsin inhibitor were used as positive and negative controls, respectively. The glycosyl composition analysis of purified iSRCR1 and iSRCR123 was done by the preparation and gas chromatography-mass spectrometry (GC-MS) of trimethylsilyl methyl glycosides as described previously (37).
Circular Dichroism
Spectroscopic studies were carried out on an Olis DSM 100 circular dichroism spectrophotometer with a 0.2-mm path length quartz cell. Control recombinant iSRCR1 or iSRCR123 at a concentration of 1 mg/ml in a buffer containing 20 mm HEPES, pH 7.4, 150 mm NaCl at 22 °C was scanned between 200 and 260 nm, and the spectra was recorded (10 times). Similarly, the conformational changes of SRCRs on addition of different concentrations of calcium (1, 2.5, 10, and 100 mm) in binding buffer were analyzed. Using CONTIN/LL algorithm implemented in CDPRO (38), the protein secondary structures were analyzed.
Differential Scanning Calorimetry
The thermostability of SRCRs in the presence of calcium ions was analyzed using MicroCal MC-II differential scanning calorimeter (GE Healthcare) as described earlier (39). Briefly, iSRCR1 or iSRCR123 at a concentration of 1 mg/ml was mixed and incubated with different concentrations of CaCl2 ranging from 0 to 100 mm to final volume of 400 μl of buffer containing 20 mm HEPES and 150 mm NaCl. Buffer without SRCRs served as control. Data were recorded with calorimetric scanning rates that ranged from 30 to 90 °C/h at 30 p.s.i. pressure. The data collected were analyzed for the unfolding temperature (Tm) and the calorimetric (ΔHcal) and van't Hoff (ΔHv) unfolding enthalpies using the Origin 7.0383 software package (MicroCal).
Analytical Ultracentrifugation
iSRCR1 or iSRCR123 (0.5 mg/ml) in a buffer containing 20 mm Tris, pH 8.0, 150 mm NaCl, and 1 mm EDTA were subjected to sedimentation velocity experiments on a Beckman Optima XL-A as described previously (12). Briefly, the samples were centrifuged to 45,000 rpm with the temperature maintained at 20 °C and absorbance at 280 nm across the cell recorded every 5 min. Using Sednterp, buffer density values of 1.0052 g/ml, protein partial specific volumes of 0.720 and 0.714 g/ml, and hydration values of 0.365 and 0.370 g/g for iSRCR1 and iSRCR123, respectively, were calculated (40, 41).
RESULTS
Adherence Assays and Quantitation
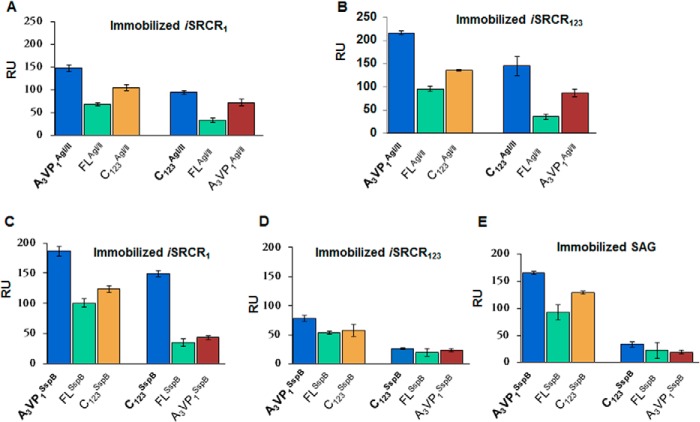
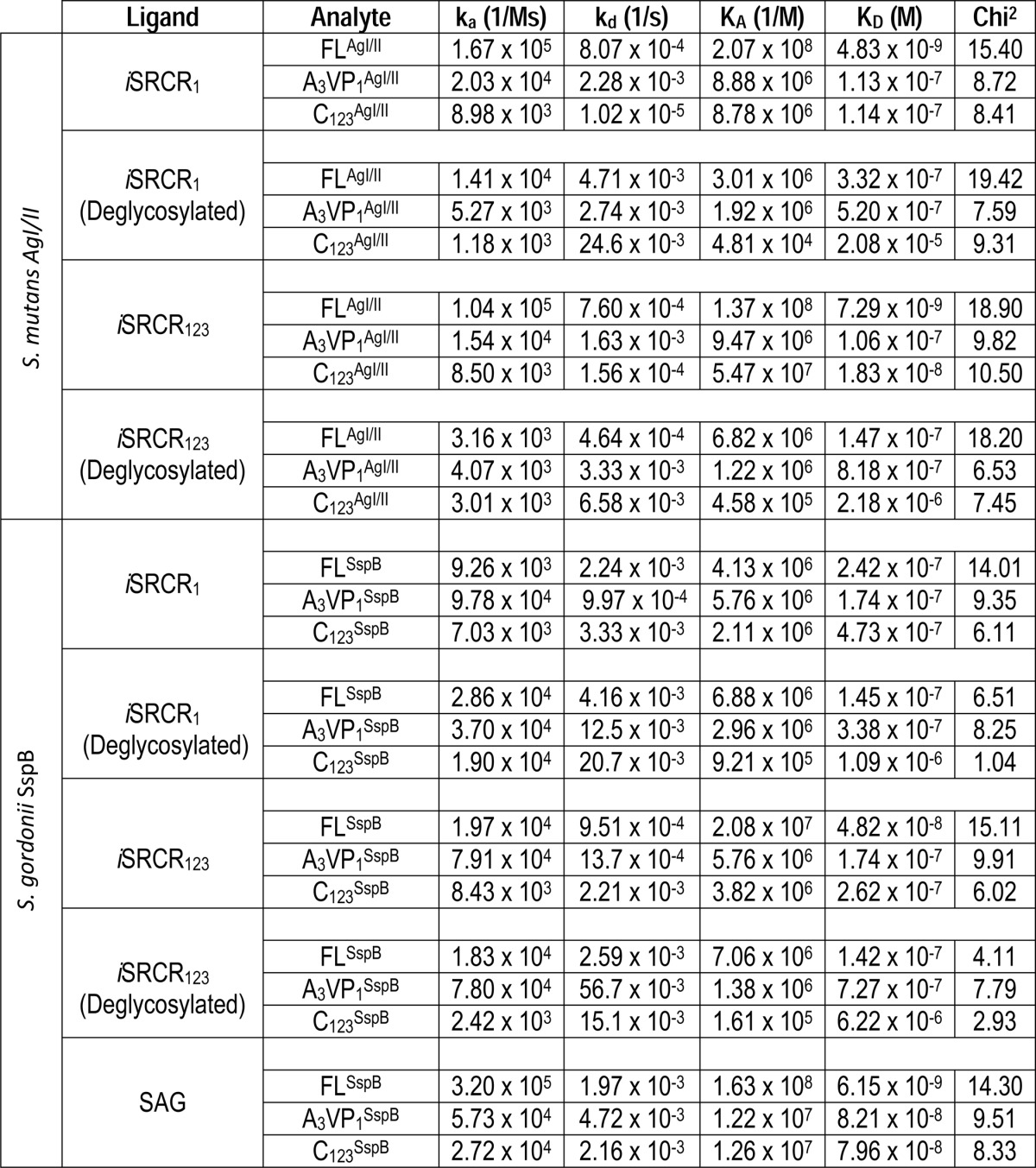
The adherence affinities between immobilized iSRCRs and the analytes FL, A3VP1, and C123 of AgI/II and SspB are summarized in Table 1 and supplemental Fig. S2, A–C. All fragments of AgI/II and SspB interact with nanomolar affinity to immobilized SRCRs. Although FLAgI/II and FLSspB displayed similar affinities with iSRCR1 and iSRCR123, the quantity of protein that adhered to iSRCR123 was higher (16% for FLAgI/II and 43% for FLSspB) than that of iSRCR1 (supplemental Fig. S3).
TABLE 1.
Surface plasmon resonance studies
Competitive Binding Experiments
FLAgI/II was able to inhibit the binding of A3VP1AgI/II and C123AgI/II by 46 and 36% respectively, whereas FLSspB inhibited A3VP1SspB and C123SspB by 54 and 23% with immobilized iSRCR1 (Fig. 3). In all other cases, A3VP1 or C123 of AgI/II and SspB did not significantly inhibit the adherence of each other. These results validate that a single SRCR domain contains the two distinct surfaces that specifically bind A3VP1 as well as C123 fragments of the AgI/II homologs.
FIGURE 3.
Competition experiments with FLAgI/II, A3VP1AgI/II, and C123AgI/II were conducted with immobilized iSRCR1 (A) and immobilized iSRCR123 (B) to determine the multiple adherence sites of the SRCRs. Although the direct adherence of the fragment is shown in bold (such as A3VP1AgI/II), the observed adherence inhibition in the presence of competing fragments (FLAgI/II and C123AgI/II) are plotted in subsequent bar graphs. Similarly, the competition of FLSspB, A3VP1SspB, and C123SspB with immobilized iSRCR1, iSRCR123, and SAG are shown in C–E. All experiments were carried out in triplicate, and the error bars represent standard deviations.
Similar inhibition was observed with immobilized iSRCR123 domains where FLAgI/II inhibited the binding of A3VP1AgI/II and C123AgI/II by 44 and 25%. However, FLSspB had limited inhibitory effects with immobilized iSRCR123, where A3VP1 and C123 displayed 68 and 76% inhibition respectively. This shows that AgI/II and SspB are different in their adherence characteristics, although they are highly homologous (57% identity and 70% homology).
The results from the competitive adherence experiments conclusively provide evidence for multiple adherence sites within a single SRCR domain and have thus narrowed down the region of adherence to the SRCR domains of GP340. This concurs with our earlier observation of multiple sites on SAG (12).
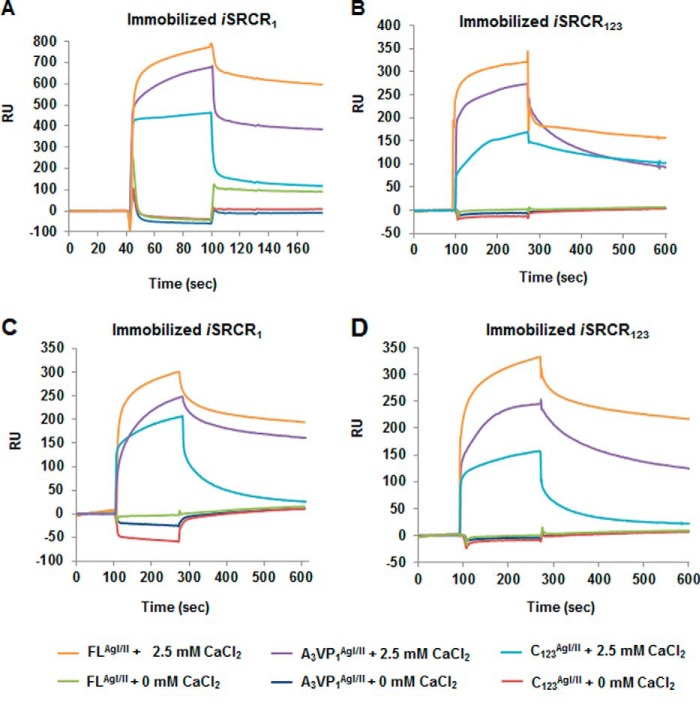
Role of Calcium (Calcium-mediated Adherence/Stability)
In the absence of calcium, there was no adherence between AgI/II homologs and SRCRs (Fig. 4, A–D). In the presence of calcium, CD spectra had recorded a significant reduction in α-helices that was correspondingly compensated with an increase in β-sheet content (Table 2), whereas no such changes were observed with AgI/II or SspB (data not shown). Upon calcium addition, the stability (thermal unfolding) of iSRCR1 increased in a dose-dependent manner (Table 3), and at 100 mm CaCl2, the SRCR domain unfolded at an uncharacteristically surprising 90 °C. Although the thermal unfolding curves of iSRCR1 were simple and easy to interpret, iSRCR123 was more complex to interpret as it involved multiple domains (data not shown).
FIGURE 4.
Illustrated here are the calcium-mediated adherences of (2 μm) AgI/II of S. mutans with iSRCR1 (A) and iSRCR123 (B) on a CM5 sensor chip. The adherence of AgI/II fragments was observed only in the presence of calcium. Devoid of calcium, AgI/II did not adhere to SRCRs. A similar result was observed with SspB of S. gordonii (C and D).
TABLE 2.
CD analysis of SRCRs at various calcium concentrations
| CaCl2 | Helix (%) |
β-Sheet (%) |
Turn (%) |
Random coil (%) |
||||
|---|---|---|---|---|---|---|---|---|
| iSRCR1 | iSRCR123 | iSRCR1 | iSRCR123 | iSRCR1 | iSRCR123 | iSRCR1 | iSRCR123 | |
| 0 mm | 15.9 | 14.8 | 28.3 | 22.9 | 20.4 | 18.2 | 35.4 | 44.2 |
| 1 mm | 6.3 | 5.9 | 34.2 | 34.4 | 24.0 | 20.3 | 35.5 | 37.3 |
| 2.5 mm | 6.6 | 6.8 | 37.4 | 34.2 | 24.2 | 20.4 | 31.9 | 38.6 |
| 10 mm | 6.8 | 5.4 | 35.9 | 34.9 | 24.3 | 19.5 | 33.0 | 40.3 |
| 100 mm | 7.2 | 4.0 | 39.3 | 36.2 | 22.2 | 18.8 | 31.3 | 41.1 |
TABLE 3.
Differential scanning calorimetric analysis of SRCRs at various calcium concentrations
| Samples | Calorimetric enthalpy (ΔHcal) | Van't Hoff enthalpy (ΔHV) | Tm |
|---|---|---|---|
| Cal/mol | Cal/mol | °C | |
| iSRCR1 | 5.703 × 104 ± 216 | 6.016 × 104 ± 281 | 56.5 |
| iSRCR1 + 2.5 mm CaCl2 | 7.828 × 104 ± 280 | 8.455 × 104 ± 374 | 78.1 |
| iSRCR1 + 10 mm CaCl2 | 6.708 × 104 ± 412 | 11.17 × 104 ± 854 | 86.4 |
| iSRCR1 + 100 mm CaCl2 | 9.736 × 104 ± 1.55 × 103 | 11.11 × 104 ± 2.24 ×103 | 92.8 |
The homologous structures of SRCR domains from both group A (with six cysteines) and group B (with eight cysteines) have been determined (42, 43); however, to date there are no crystal structures of the SRCR domains of GP340. Both iSRCR1 and iSRCR123 domains possess similar secondary structures compared with the solved crystal structures PDB2JA4, PDB1BY2, and PDB1P57 (23, 42, 44), thus indicating a possible adoption of similar SRCR folds.
Effect of Carbohydrates on the Adherence of AgI/II
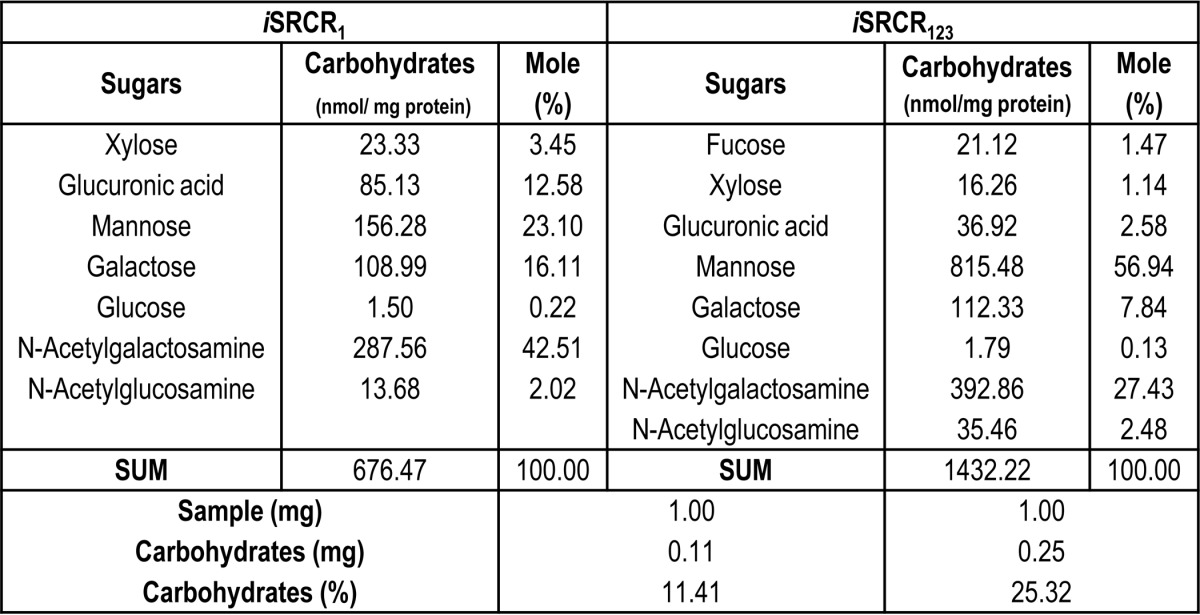
The presence of glycosylation on iSRCR1 and iSRCR123 was initially confirmed using glycoprotein staining (supplemental Fig. S4). Further glycan profile analysis of both iSRCR1 and iSRCR123 indicated that they are predominantly O-glycosylated with Galβ1–3-GalNac and mannose carbohydrates (Table 4). Deglycosylation of iSRCR1 and iSRCR123 by O-glycosidases did not affect the adherence characteristics of A3VP1AgI/II but decreased the adherence of the C123AgI/II by 2 orders of magnitude (Table 1), implicating the involvement of domains closer to the cell surface in carbohydrate binding. Similar results were observed with C123SspB. Although O-glycosidases had profound effects on the adherence kinetics, endoglycosidase H (N-glycosidase) did not have any measurable effect (data not shown).
TABLE 4.
Glycan profile analysis of iSRCR1 and iSRCR123
SRCRP2 (Bikker Peptide)
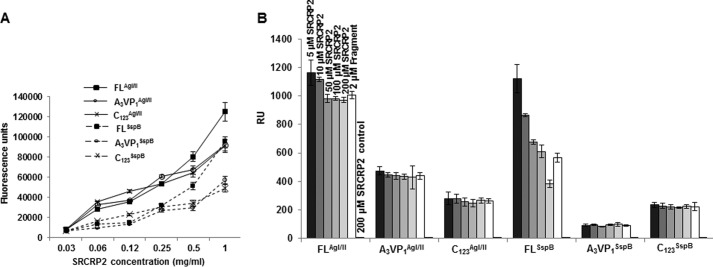
Initial ELISAs (Fig. 5A) demonstrated the adherence of the SRCRP2 peptide to AgI/II, SspB, and their subfragments. When incubated at low concentration (5 μm) with FLAgI/II and A3VP1AgI/II, the SRCRP2 peptide improved the adherence by 8 and 13% respectively to iSRCR1 (data not shown) and 8 and 16% respectively to iSRCR123, whereas C123AgI/II had no changes in adherence (Fig. 5B). Only FLSspB improved the adherence by 97% with iSRCR123 at lower concentration (5 μm of SRCRP2 peptide), whereas A3VP1SspB and C123SspB did not alter the adherence characteristics to either iSRCR1 (data not shown) or iSRCR123 (3 and 5%), respectively (Fig. 5B). Also, the SRCRP2 alone (control) did not show any binding with SRCRs. These results indicated that SRCRP2 peptide does not inhibit the adherence of AgI/II and SspB to iSRCR1 and iSRCR123 and that the adherence site was different from that of the aggregation sites present on AgI/II and SspB.
FIGURE 5.
A, these curves illustrate the direct adherence of SRCRP2 peptide to FLAgI/II A3VP1AgI/II, and C123AgI/II of S. mutans (bold lines) as well as FLSspB, A3VP1SspB, and C123SspB of S. gordonii (dotted lines). Fluorescein-tagged SRCRP2 (FAM) was serially diluted (0.03–1 mg/ml), and its interaction with immobilized AgI/II and SspB and their subfragments was measured at OD519. The results illustrated that the SRCRP2 peptide adheres well with AgI/II and SspB. B, adherence/inhibition of FLAgI/II and FLSspB and subfragments (2 μm) in the presence of SRCRP2 peptide at various concentrations (0.005 to 0.200 mm) with immobilized iSRCR123. In control experiments, SRCRP2 peptide alone does not display any measurable interaction with iSRCR123. More importantly SRCRP2 does not inhibit the adherence of AgI/II and SspB (and their subfragments) to the SRCRs. Surprisingly, SRCRP2 increased the adhesiveness of FLSspB confounding us, as it could be interpreted as nonspecific adherence/aggregation.
SRCR Self-adhesion
Interaction of SRCRs with each other was tested using SPR. The iSRCR1 and iSRCR123 strongly interacted with each other. Analytes iSRCR1 (KD = 1.13 × 10−10 m) and iSRCR123 (KD = 5.68 × 10−9 m) demonstrated high affinity with immobilized iSRCR1. Similarly, analytes iSRCR1 (KD = 1.2 × 10−9 m) and iSRCR123 (KD = 6.72 × 10−9 m) interacted with immobilized iSRCR123 with nanomolar affinities (Fig. 6, A–D). These results showed that the SRCRs have self-adhesion property as well.
FIGURE 6.
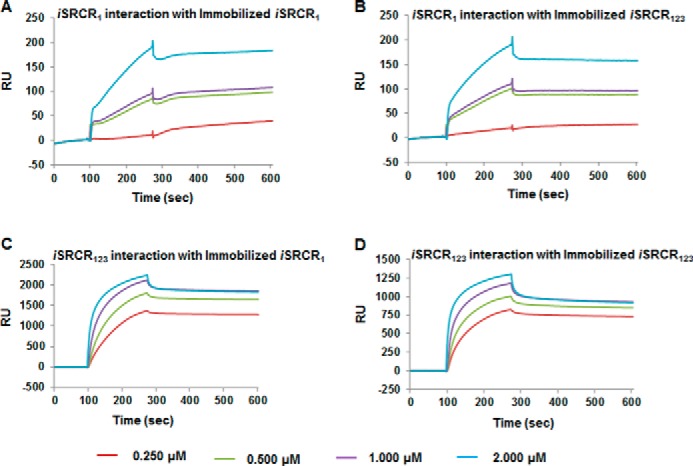
Self-interactions of SRCR domains at various concentrations (0.250–2 μm) with immobilized iSRCR1 (A and C) and iSRCR123 (B and D) on CM5 sensor chip were analyzed using BIA-Evaluation software. The analytes iSRCR1 and iSRCR123 demonstrated nanomolar affinity interaction with SRCRs, clearly indicating the self-adhesion property among SRCRs.
Aggregation Assays
In the presence of iSRCR123, 69% of S. mutans and 48% of S. gordonii aggregated, whereas iSRCR1 aggregated 17% of S. mutans and 12% of S. gordonii (Fig. 7, A and B). The positive control SAG aggregated S. mutans by 74% and S. gordonii by 72%. Earlier studies with the consensus peptide, SRCRP2 derived from SRCR domains, aggregated a variety of bacteria (17, 31); however, in this study compared with iSRCR123, the SRCRP2 peptide displayed very limited aggregation with S. mutans (13%) and S. gordonii (11%) similar to that of a single SRCR domain. Even at higher concentrations, the SRCRP2 peptide did not aggregate S. mutans and S. gordonii (data not shown).
FIGURE 7.
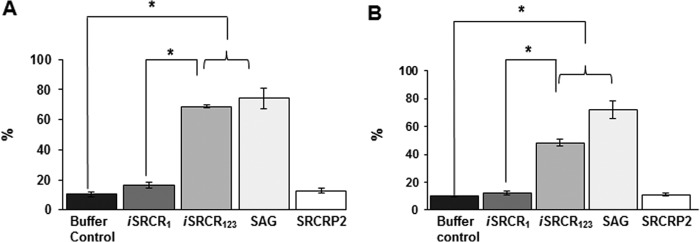
Aggregation of S. mutans UA159 (A) and S. gordonii DL1 (B) cells in the presence of iSRCRs, SAG, and SRCRP2 peptide was analyzed. The results are plotted as percentage of aggregation measured at OD700 at 5-min intervals for 1 h. Bacterial cells in buffer alone were used as control. Differences in aggregation detected between groups were analyzed using one-way analysis of variance, where *, p < 0.05 was considered significant, and error bars represent the standard deviation.
Confocal Microscopy
The Z view of the confocal microscopy pictures show the adherence of SRCRs to the upper surface (surface proteins) of immobilized S. mutans UA159 and S. gordonii DL1 cells (Fig. 8, A and B). From the X-Y panel view, it is noticeable that iSRCR1 adhered poorly compared with iSRCR123, underscoring that multiple SRCR domains have better adherence capability compared with that of a single SRCR domain.
FIGURE 8.
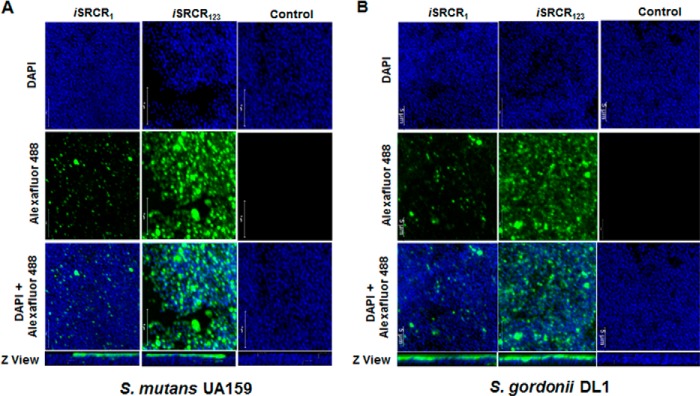
Interaction between S. mutans UA159 (A) and S. gordonii DL1 (B) cells with iSRCR1 and iSRCR123 was analyzed using confocal microscopic images. The images display S. mutans and S. gordonii cells stained with DAPI (blue) and anti-His tag Alexa Fluor 488 antibody (green). The observed green fluorescence depicts the binding of iSRCR1 and iSRCR123 to S. mutans and S. gordonii, whereas the control S. mutans and S. gordonii were counterstained by DAPI alone. Also, it is evident from the images that iSRCR123 adhered more profoundly than iSRCR1.
Analytical Ultracentrifugation
We sought to answer the question of the spatial organization of the SRCR domains, particularly whether they might be elongated similarly to AgI/II through ultracentrifugation experiments. From their observed frictional ratios (iSRCR1 = 1.59 and iSRCR123 = 1.80), resultant prolate ellipsoid ratios (iSRCR1 = 7.18 and iSRCR123 = 10.36), and calculated dimensions (iSRCR1 = 12.60 × 1.75 nm and iSRCR123 = 22.08 × 2.13 nm), it is evident that both iSRCR1 and iSRCR123 will have extended structures (Table 5). However, these are not extended as linear rigid structures but exist in a flexible nonlinear conformation forming curvy tertiary structures.
TABLE 5.
Analytical ultracentrifugation analysis of the SRCRs
r.m.s.d. means root mean square deviation.
| Construct | Theoretical mass | Fit mass | Fit r.m.s.d. | S20 | f/f0 | Stokes radius | a/b (oblate) | a/b (prolate) | Oblate ellipsoid | Prolate ellipsoid |
|---|---|---|---|---|---|---|---|---|---|---|
| Da | Da | OD | S | nm | nm × nm | nm × nm | ||||
| iSRCR1 | 14906 | 17031 | 0.0054 | 1.508 | 1.59 | 2.71 | 7.94 | 7.18 | 6.73 × 0.84 | 12.60 × 1.75 |
| iSRCR123 | 42549 | 51357 | 0.0062 | 2.792 | 1.80 | 4.43 | 11.67 | 10.36 | 10.54 × 0.90 | 22.08 × 2.13 |
DISCUSSION
Oral streptococci primarily attach to tooth-immobilized GP340 via AgI/II homologs and subsequently colonize and infect the host (19, 45). For the past 3 decades, this interaction has been studied using GP340 extracted from the saliva of either single or multiple donors who have inherent allelic variability (30, 46, 47). For the first time in this study using recombinantly expressed SRCR domains of GP340 (Drosophila expression system), we established a benchmark and elucidated the intricate components involved in this bacterial adhesion.
Nanomolar affinity interactions between the SRCRs and AgI/II homologs (Table 1 and supplemental Fig. S2, A–C) were deduced from SPR data. The adherence kinetics of the C123SspB (present near cell the wall) had distinctive sensorgrams, where they did not remain bound to the immobilized SAG or SRCR domains (supplemental Fig. S2C). Overall, these results imply that SAG-binding protein AgI/II of pathogenic S. mutans contains a locking mechanism to remain bound, whereas the C-terminal region of the commensal S. gordonii SspB does not. In isothermal titration calorimetry experiments (supplemental Fig. S5), the affinity between the AgI/II and SRCR in solution was in the micromolar range and indicated that the nature of the interaction is very different in solution compared with an immobilized state.
This study also demonstrates that the adherence surfaces for A3VP1 and C123 of AgI/II are contained within a single SRCR domain, and it is therefore the minimal adherence region (Fig. 3, A–E).
The concentration of calcium within the oral cavity has been estimated to be between 1.2 and 2.8 mmol/liter, and using similar concentrations, we discovered that calcium induces secondary structural changes (Table 2) and increases the thermal stability of the SRCRs (Table 3). Because the oral cavity is subject to environmental changes, including pH and temperature changes (hot and cold food and beverages), perhaps the observed thermal stability could be a direct consequence of evolution. It would be interesting to see whether the SRCR domains from sea urchin, which has 57% sequence identity with human GP340 SRCR domain, possess these thermal properties that would directly link it to evolution of the SRCR domains within the human oral cavity. In SPR experiments (supplemental Fig. S6), calcium induced a large change in RUs while interacting with the immobilized SRCRs. Such phenomena have been observed in other proteins that endure calcium-induced structural and conformational changes (48). This opens up the possibility that the SRCRs undergo a distinct conformational change in the immobilized state, to which the bacterial AgI/II homologs adhere with nanomolar affinity, whereas in a solution state they adhere with micromolar affinity (supplemental Fig. S5). Physiologically, this could represent an evolution of streptococci in the oral cavity, where GP340 (being an innate immunity molecule) would aggregate the microbes and clear them into the gut, and to survive, bacteria have developed specific higher affinity to the tooth-immobilized conformation of GP340.
The positive glycostaining (supplemental Fig. S4) and the glycan profile analysis of recombinant SRCRs (Table 4) indicated that they are predominantly O-glycosylated. Deglycosylation of SRCRs with O-glycanase reduced the adherence affinity (Table 1), and therefore, for the first time we have now quantitatively determined that the high affinity interactions observed between AgI/II homologs and SRCRs/SAG is directly attributed to the carbohydrate adherence.
Compared with iSRCR1 and iSRCR123, the SRCRP2 peptide (Fig. 5B) showed limited aggregation with AgI/II homologs. In addition, SRCRP2 did not offer significant inhibition. There are two possibilities, one is that the aggregation site is different from that of the adhesion in AgI/II homologs, or this could be interpreted as nonspecific adherence by the peptide. We lean toward nonspecific adherence as SRCRP2 increased the adhesiveness of only FLSspB. These results now demonstrate that the peptide does not aggregate well nor does it inhibit the SRCR/GP340-binding motif/site on AgI/II homologs.
Confocal microscopic images (Fig. 8, A and B) and aggregation assays (Fig. 7, A and B) show that iSRCRs bind to S. mutans and S. gordonii cells, where iSRCR123 attaches extensively compared with iSRCR1. Detailed SPR analysis based on protein adherence to chip surface indicated higher amounts of FLAgI/II and FLSspB adhered to immobilized iSRCR123 than iSRCR1 (supplemental Fig. S3). Based on these observations, we conclude that longer tandem SRCR domains would more efficiently agglutinate various bacteria. With GP340 being an innate immunity molecule, the number of tandem repeats it takes to efficiently agglutinate bacteria could have been evolutionarily determined, and it is interesting to note that in humans GP340 contains 14 SRCR domains, of which 13 are tandem repeats, whereas in other vertebrates the number of tandem repeats are comparatively lower (26, 27).
One surprising result that came out of these studies is that the SRCR domains self-associate with nanomolar affinities, thus indicating that this association is highly specific, as nonspecific interactions traditionally appear to fall within the micromolar range (Fig. 6, A–D). GP340 is known to exist as a higher order complex, and these aggregates could be as large as 5000 kDa (19, 49). Thus far, the aggregation property of GP340 has been attributed to the ZP domain; as in other mammalian proteins, the ZP domain was shown to be involved in self-aggregation (50). Furthermore, the tertiary architecture analysis of tandem SRCR domains indicate that they may not strictly form a linear elongated structure (Table 5) but could form a curvy centipede-like extended structure, similar to that observed in electron microscopy images of GP340 (28). Combined together, these results open up several possible models for bacterial aggregation/adherence, wherein one potential model could simulate the bacterial proteins to be sandwiched between two SRCR domains (GP340s) (Fig. 9).
FIGURE 9.
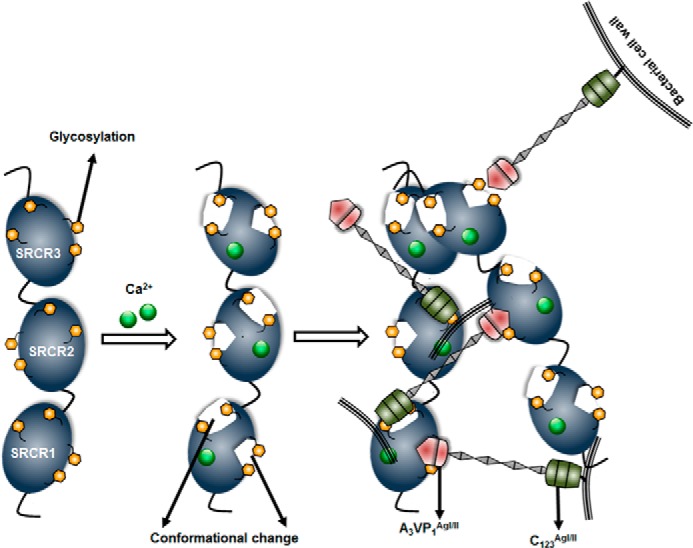
Schematic model displaying the conformational change on the SRCR domains of GP340 in the presence of calcium, and particularly implicating the role of glycosylation in this high affinity adherence to AgI/II homologs. The model also includes the possible self-association of the SRCR domains that could potentially sandwich AgI/II homologs of oral streptococci.
Earlier studies have shown that the SRCR domains of GP340 play a pivotal role in mediating HIV adhesion/clearance through GP120 (16, 51). Although GP340 acted as a clearance mechanism in the oral cavity, the case was very different on the vaginally derived GP340, which is immobilized on the cell surface, where this was shown to mediate transcytosis from apical to basolateral surface in both genital tract epithelial cells in culture and with endocervical tissue (52). Similarly, in SPR experiments, immobilized SRCRs adhere tightly to AgI/II homologs, and in the fluid phase SRCRs aggregate S. mutans and S. gordonii, a double-faceted property, where on the one hand it acts as a portal of entry for microbes while immobilized and on the other hand as a clearance mechanism within the oral cavity in fluid state. This property indicates that SRCRs could possibly adopt different secondary structural conformations in fluid and immobilized states, and this conformation could be induced by calcium ions.
Summarizing our findings, we report that the minimal adherence region is restricted to a single SRCR domain, which carries the two distinct surfaces that adhere to A3VP1 as well as C123 of both AgI/II and SspB. Better adherence and aggregation of bacteria are observed with increasing numbers of SRCR domains. These SRCR domains attain stability in the presence of calcium, and calcium mediates structural changes that are essential for the adherence of AgI/II homologs. Furthermore, glycosylations play a significant role in the adherence to AgI/II and SspB. Interestingly, the SRCRs self-associate and the tandem domains may adopt a curvy centipede-like structure. Although there are similarities in the binding of AgI/II and SspB, there are certainly distinct differences pointing toward species specificity in their adherence.
Overall, these results now point to the fact that focusing on the SRCRs and elucidating the molecular motifs involved in adherence would aid in the development of interventional therapeutics. Such studies could potentially result in the identification and development of small molecule inhibitors or passive immunization therapies that could impede oral streptococcal adherence to tooth surfaces and alleviate the global burden of dental caries.
Supplementary Material
Acknowledgments
We acknowledge the X-ray Core, Comprehensive Cancer Center, and High Resolution Imaging Facilities at the University of Alabama at Birmingham. We are grateful to Dr. Christie Brouillette and Dr. Peter Prevelige for CD, differential scanning calorimetry, and analytical ultracentrifugation instrumentation. We thank Dr. L. Jeannine Brady for SAG and Dr. Poustka for the DMBT1 (GP340) gene template. We also thank Brian Sharon, Manisha Patel, and Shaan Khaled for protein purifications and Zhengrong Yang and Matthew Larson for discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DE017737.

This article contains supplemental Figs. S1–S6 and Tables S1 and S2.
- AgI/II
- antigen I/II
- GP340
- glycoprotein 340
- SRCR
- scavenger receptor cysteine-rich domain
- SAG
- salivary agglutinin
- RU
- resonance unit
- SPR
- surface plasmon resonance
- ZP
- zona pellucida
- FAM
- fluorescein amidite.
REFERENCES
- 1. Lehner T., Russell M. W., Caldwell J. (1980) Immunization with a purified protein from Streptococcus mutans against dental caries in rhesus monkeys. Lancet 1, 995–996 [DOI] [PubMed] [Google Scholar]
- 2. Russell M. W., Childers N. K., Michalik S. M., Smith D. J., Taanman M. A. (2004) A Caries vaccine? The state of the science of immunization against dental caries. Caries Res. 38, 230–235 [DOI] [PubMed] [Google Scholar]
- 3. Robinette R. A., Olli M. W., McArthur W. P., Brady L. J. (2011) A therapeutic anti-Streptococcus mutans monoclonal antibody used in human passive protection trials influences the adaptive immune response. Vaccine 29, 6292–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. El-Sabens A., Demuth D. R., Lamont R. J. (2001) Regulation of Streptococcus gordonii SspB by the SspA gene product. Infect. Immun. 69, 6520–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamont R. J., El-Sabens A., Park Y., Cook G. S., Costantin J. W., Demuth D. R. (2002) Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148, 1627–1636 [DOI] [PubMed] [Google Scholar]
- 6. Demuth D. R., Duan Y., Brooks W., Holmes A. R., McNab R., Jenkinson H. F. (1996) Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20, 403–413 [DOI] [PubMed] [Google Scholar]
- 7. Chung W. O., Demuth D. R., Lamont R. J. (2000) Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect. Immun. 68, 6758–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brady L. J., Maddocks S. E., Larson M. R., Forsgren N., Persson K., Deivanayagam C. C., Jenkinson H. F. (2010) The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 77, 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nobbs A. H., Lamont R. J., Jenkinson H. F. (2009) Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ericson T., Rundegren J. (1983) Characterization of a salivary agglutinin reacting with a serotype c strain of Streptococcus mutans. Eur. J. Biochem. 133, 255–261 [DOI] [PubMed] [Google Scholar]
- 11. Mollenhauer J., Wiemann S., Scheurlen W., Korn B., Hayashi Y., Wilgenbus K. K., von Deimling A., Poustka A. (1997) DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3–26.1, is deleted in malignant brain tumours. Nat. Genet. 17, 32–39 [DOI] [PubMed] [Google Scholar]
- 12. Larson M. R., Rajashankar K. R., Patel M. H., Robinette R. A., Crowley P. J., Michalek S., Brady L. J., Deivanayagam C. (2010) Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of α- and PPII-helices. Proc. Natl. Acad. Sci. U.S.A. 107, 5983–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larson M. R., Rajashankar K. R., Crowley P. J., Kelly C., Mitchell T. J., Brady L. J., Deivanayagam C. (2011) Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J. Biol. Chem. 286, 21657–21666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deivanayagam C. C., Wann E. R., Chen W., Carson M., Rajashankar K. R., Höök M., Narayana S. V. (2002) A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 21, 6660–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leito J. T., Ligtenberg A. J., van Houdt M., van den Berg T. K., Wouters D. (2011) The bacteria binding glycoprotein salivary agglutinin (SAG/gp340) activates complement via the lectin pathway. Mol. Immunol. 49, 185–190 [DOI] [PubMed] [Google Scholar]
- 16. Wu Z., Golub E., Abrams W. R., Malamud D. (2004) gp340 (SAG) binds to the V3 sequence of gp120 important for chemokine receptor interaction. AIDS Res. Hum. Retroviruses 20, 600–607 [DOI] [PubMed] [Google Scholar]
- 17. Bikker F. J., Ligtenberg A. J., Nazmi K., Veerman E. C., van't Hof W., Bolscher J. G., Poustka A., Nieuw Amerongen A. V., Mollenhauer J. (2002) Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J. Biol. Chem. 277, 32109–32115 [DOI] [PubMed] [Google Scholar]
- 18. Prakobphol A., Xu F., Hoang V. M., Larsson T., Bergstrom J., Johansson I., Frängsmyr L., Holmskov U., Leffler H., Nilsson C., Borén T., Wright J. R., Strömberg N., Fisher S. J. (2000) Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J. Biol. Chem. 275, 39860–39866 [DOI] [PubMed] [Google Scholar]
- 19. Oho T., Yu H., Yamashita Y., Koga T. (1998) Binding of salivary glycoprotein-secretory immunoglobulin A complex to the surface protein antigen of Streptococcus mutans. Infect. Immun. 66, 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Demuth D. R., Lammey M. S., Huck M., Lally E. T., Malamud D. (1990) Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb. Pathog. 9, 199–211 [DOI] [PubMed] [Google Scholar]
- 21. Claudianos C., Dessens J. T., Trueman H. E., Arai M., Mendoza J., Butcher G. A., Crompton T., Sinden R. E. (2002) A malaria scavenger receptor-like protein essential for parasite development. Mol. Microbiol. 45, 1473–1484 [DOI] [PubMed] [Google Scholar]
- 22. Abrahamsen M. S., Templeton T. J., Enomoto S., Abrahante J. E., Zhu G., Lancto C. A., Deng M., Liu C., Widmer G., Tzipori S., Buck G. A., Xu P., Bankier A. T., Dear P. H., Konfortov B. A., Spriggs H. F., Iyer L., Anantharaman V., Aravind L., Kapur V. (2004) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441–445 [DOI] [PubMed] [Google Scholar]
- 23. Hohenester E., Sasaki T., Timpl R. (1999) Crystal structure of a scavenger receptor cysteine-rich domain sheds light on an ancient superfamily. Nat. Struct. Biol. 6, 228–232 [DOI] [PubMed] [Google Scholar]
- 24. Stoddard E., Cannon G., Ni H., Karikó K., Capodici J., Malamud D., Weissman D. (2007) gp340 expressed on human genital epithelia binds HIV-1 envelope protein and facilitates viral transmission. J. Immunol. 179, 3126–3132 [DOI] [PubMed] [Google Scholar]
- 25. Cummins J. E., Christensen L., Lennox J. L., Bush T. J., Wu Z., Malamud D., Evans-Strickfaden T., Siddig A., Caliendo A. M., Hart C. E., Dezzutti C. S. (2006) Mucosal innate immune factors in the female genital tract are associated with vaginal HIV-1 shedding independent of plasma viral load. AIDS Res. Hum. Retroviruses 22, 788–795 [DOI] [PubMed] [Google Scholar]
- 26. Ligtenberg A. J., Veerman E. C., Nieuw Amerongen A. V., Mollenhauer J. (2007) Salivary agglutinin/glycoprotein-340/DMBT1: a single molecule with variable composition and with different functions in infection, inflammation and cancer. Biol. Chem. 388, 1275–1289 [DOI] [PubMed] [Google Scholar]
- 27. Ligtenberg A. J., Karlsson N. G., Veerman E. C. (2010) Deleted in malignant brain tumors-1 protein (DMBT1): a pattern recognition receptor with multiple binding sites. Int. J. Mol. Sci. 11, 5212–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Madsen J., Mollenhauer J., Holmskov U. (2010) Review: Gp-340/DMBT1 in mucosal innate immunity. Innate Immun. 16, 160–167 [DOI] [PubMed] [Google Scholar]
- 29. Malamud D., Abrams W. R., Barber C. A., Weissman D., Rehtanz M., Golub E. (2011) Antiviral activities in human saliva. Adv. Dent. Res. 23, 34–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loimaranta V., Jakubovics N. S., Hytönen J., Finne J., Jenkinson H. F., Strömberg N. (2005) Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect. Immun. 73, 2245–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bikker F. J., Ligtenberg A. J., End C., Renner M., Blaich S., Lyer S., Wittig R., van't Hof W., Veerman E. C., Nazmi K., de Blieck-Hogervorst J. M., Kioschis P., Nieuw Amerongen A. V., Poustka A., Mollenhauer J. (2004) Bacteria binding by DMBT1/SAG/gp-340 is confined to the VEVLXXXXW motif in its scavenger receptor cysteine-rich domains. J. Biol. Chem. 279, 47699–47703 [DOI] [PubMed] [Google Scholar]
- 32. Jakubovics N. S., Strömberg N., van Dolleweerd C. J., Kelly C. G., Jenkinson H. F. (2005) Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 55, 1591–1605 [DOI] [PubMed] [Google Scholar]
- 33. Purushotham S., Deivanayagam C. (2013) Cloning, expression and purification of the SRCR domains of glycoprotein 340. Protein Expr. Purif. 90, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brady L. J., Piacentini D. A., Crowley P. J., Oyston P. C., Bleiweis A. S. (1992) Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect. Immun. 60, 1008–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oli M. W., McArthur W. P., Brady L. J. (2006) A whole cell BIAcore assay to evaluate P1-mediated adherence of Streptococcus mutans to human salivary agglutinin and inhibition by specific antibodies. J. Microbiol. Methods 65, 503–511 [DOI] [PubMed] [Google Scholar]
- 36. Schuck P. (1997) Use of surface plasmon resonance to probe the equilibrium and dynamic aspects of interactions between biological macromolecules. Annu. Rev. Biophys. Biomol. Struct. 26, 541–566 [DOI] [PubMed] [Google Scholar]
- 37. Hobb R. I., Tzeng Y. L., Choudhury B. P., Carlson R. W., Stephens D. S. (2010) Requirement of NMB0065 for connecting assembly and export of sialic acid capsular polysaccharides in Neisseria meningitidis. Microbes Infect. 12, 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sreerama N., Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 39. Yang Z. R., Tendian S. W., Carson W. M., Brouillette W. J., Delucas L. J., Brouillette C. G. (2004) Dimethyl sulfoxide at 2.5% (v/v) alters the structural cooperativity and unfolding mechanism of dimeric bacterial NAD+ synthetase. Protein Sci. 13, 830–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lebowitz J., Lewis M. S., Schuck P. (2002) Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodamilans B., Muñoz I. G., Bragado-Nilsson E., Sarrias M. R., Padilla O., Blanco F. J., Lozano F., Montoya G. (2007) Crystal structure of the third extracellular domain of CD5 reveals the fold of a group B scavenger cysteine-rich receptor domain. J. Biol. Chem. 282, 12669–12677 [DOI] [PubMed] [Google Scholar]
- 43. Ojala J. R., Pikkarainen T., Tuuttila A., Sandalova T., Tryggvason K. (2007) Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J. Biol. Chem. 282, 16654–16666 [DOI] [PubMed] [Google Scholar]
- 44. Somoza J. R., Ho J. D., Luong C., Ghate M., Sprengeler P. A., Mortara K., Shrader W. D., Sperandio D., Chan H., McGrath M. E., Katz B. A. (2003) The structure of the extracellular region of human hepsin reveals a serine protease domain and a novel scavenger receptor cysteine-rich (SRCR) domain. Structure 11, 1123–1131 [DOI] [PubMed] [Google Scholar]
- 45. Lamont R. J., Demuth D. R., Davis C. A., Malamud D., Rosan B. (1991) Salivary-agglutinin-mediated adherence of Streptococcus mutans to early plaque bacteria. Infect. Immun. 59, 3446–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Esberg A., Löfgren-Burström A., Ohman U., Strömberg N. (2012) Host and bacterial phenotype variation in adhesion of Streptococcus mutans to matched human hosts. Infect. Immun. 80, 3869–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ligtenberg T. J., Bikker F. J., Groenink J., Tornoe I., Leth-Larsen R., Veerman E. C., Nieuw Amerongen A. V., Holmskov U. (2001) Human salivary agglutinin binds to lung surfactant protein-D and is identical with scavenger receptor protein gp-340. Biochem. J. 359, 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Christopeit T., Gossas T., Danielson U. H. (2009) Characterization of Ca2+ and phosphocholine interactions with C-reactive protein using a surface plasmon resonance biosensor. Anal. Biochem. 391, 39–44 [DOI] [PubMed] [Google Scholar]
- 49. Young A., Rykke M., Smistad G., Rølla G. (1997) On the role of human salivary micelle-like globules in bacterial agglutination. Eur. J. Oral Sci. 105, 485–494 [DOI] [PubMed] [Google Scholar]
- 50. Jovine L., Darie C. C., Litscher E. S., Wassarman P. M. (2005) Zona pellucida domain proteins. Annu. Rev. Biochem. 74, 83–114 [DOI] [PubMed] [Google Scholar]
- 51. Wu Z., Lee S., Abrams W., Weissman D., Malamud D. (2006) The N-terminal SRCR-SID domain of gp-340 interacts with HIV type 1 gp120 sequences and inhibits viral infection. AIDS Res. Hum. Retroviruses 22, 508–515 [DOI] [PubMed] [Google Scholar]
- 52. Stoddard E., Ni H., Cannon G., Zhou C., Kallenbach N., Malamud D., Weissman D. (2009) gp340 promotes transcytosis of human immunodeficiency virus type 1 in genital tract-derived cell lines and primary endocervical tissue. J. Virol. 83, 8596–860319553331 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.