Abstract
The interface between HIV-1 gp120 envelope glycoprotein and CD4 receptor contains an unusual interfacial cavity, the “Phe43 cavity”, which miniprotein mimetics of CD4 with non-natural extensions can potentially utilize to enhance their neutralization of HIV-1. Here we report co-crystal structures of HIV-1 gp120 with miniproteins M48U1 and M48U7, which insert cyclohexylmethoxy and 5-hydroxypentylmethoxy extensions, respectively, into the Phe43 cavity. Both inserts displayed flexibility and hydrophobic interactions, but the M48U1 insert showed better shape complementarity with the Phe43 cavity than the M48U7 insert. Subtle alteration in gp120 conformation played a substantial role in optimizing fit. With M48U1, these translated into a YU2-gp120 affinity of 0.015 nM and neutralization of all 180-circulating HIV-1 strains tested, except clade-A/E isolates with non-canonical Phe43 cavities. Ligand chemistry, shape complementary, surface burial, and gp120 conformation act in concert to modulate binding of ligands to the gp120-Phe43 cavity and, when optimized, can effect near pan-neutralization of HIV-1.
The gp120 envelope glycoprotein of the human immunodeficiency virus type 1 (HIV-1) is conformationally flexible and undergoes substantial structural rearrangements during HIV-1 entry into cells (Guttman et al., 2012; Myszka et al., 2000; Wyatt et al., 1998). Several autonomously folded domains namely, the outer domain, the inner domain, the bridging sheet, and the V1/V2 region, constitute gp120, and these domains may adopt different relative spatial arrangements in response to ligand binding or oligomeric interactions within the functional viral spike (Chen et al., 2005; Chen et al., 2009; McLellan et al., 2011; Pancera et al., 2010; Zhou et al., 2007). In the conformation of gp120 induced by engagement of the CD4 receptor, an interfacial cavity forms at the nexus of the inner domain, outer domain, bridging sheet, and the CD4 receptor (Kwong et al., 1998). This interfacial cavity, named the “Phe43” cavity, is capped on one end by the phenyl ring of residue 43CD4, is bordered by conserved gp120 residues, and comprises a volume of ~150 Å3 (for clarity, residue numbers are labeled with macromolecule as subscript).
Natural amino acids cannot fully access the Phe43 cavity, which extends ~8 Å beyond the phenyl ring of residue 43CD4 (Kwong et al., 1998). Synthetic chemistry offers the opportunity to interact fully with this cavity via non-natural, cavity-filling ligands (Curreli et al., 2012; Huang et al., 2005; Lalonde et al., 2011; LaLonde et al., 2012; Stricher et al., 2008). One such synthetic system was developed by transplanting critical elements of the gp120-interactive region of CD4 to a structurally compatible scyllatoxin scaffold (Vita et al., 1999). Structure-guided optimizations of the miniprotein backbone (Martin et al., 2003; Stricher et al., 2008), as well as the side-chain of residue 23miniprotein, which is located in a position analogous to residue 43CD4 (Huang et al., 2005), resulted in M48 and M47, both with nM affinity for gp120 (Stricher et al., 2008). M48 contains a Phe at position 23M48 (Table 1), whereas M47 inserts a biphenylalanine ~5 Å into the Phe43 cavity. The structure of gp120 bound to M48 (Stricher et al., 2008) closely resembled the structure of gp120 bound to CD4, both in overall conformation as well as locally near the Phe43 cavity. In the M47-bound gp120 structure, insertion of a rigid biphenyl appeared to distort gp120 conformation in the region surrounding the insertion (Stricher et al., 2008). This observation spurred the design of CD4-mimetic miniprotein variants with flexible inserts into the Phe43 cavity (Van Herrewege et al., 2008).
Table 1.
CD4 and CD4-mimetic miniproteins: sequences, Phe43 cavity inserts, YU2 gp120 affinity and X-ray crystal structures with HIV-1 gp120
| Sequence | Phe43 cavity insert | KS to YU2 gp120 (nM) | Structural resolution of gp120 complex (Å) | |
|---|---|---|---|---|
| CD4 | 22-..KSIQFHWKNSNQIKILGNQGS F LTK.…-47 |
|
7.68±0.33 (Stricher et al., 2008) | 2.9 (Huang et al., 2004) |
| M48a | 1-TpaNLHFCQLRCKSLGLLGRCAdPT F CACVNH2 |
|
3.87±0.10 (Stricher et al., 2008) | 2.4 (Stricher et al., 2008) |
| M47a, b | 1-TpaNLHFCQLRCKSLGLLGRCAdPTBipCACVNH2 |
|
2.12±0.05 (Stricher et al., 2008) | 2.2 (Stricher et al., 2008) |
| M48U1a, c | 1-TpaNLHFCQLRCKSLGLLGRCAdPT U1CACVNH2 |
|
0.0154±0.0008 (this study) | 1.5 (this study) |
| M48U7a, d | 1-TpaNLHFCQLRCKSLGLLGKCAdPT U7CACVNH2 |
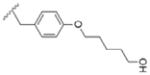
|
0.18±0.03 (this study) | 2.1 (this study) |
Tpa= thiopropionic acid, dP=D-proline,
Bip= biphenylalanine,
U1= cyclohexylmethoxy phenylalanine,
U7= hydroxypentylmethoxy phenylalanine.
One of these variants, M48U1, contained a cyclohexylmethoxy phenylalanine at position 23M48U1 and displayed remarkably potent neutralization of three HIV-1 isolates (Van Herrewege et al., 2008) and a recent study demonstrated its efficacy as a microbicide (Dereuddre-Bosquet et al., 2012). To determine the structural basis for the potent antiviral activity of M48U1, we determined its crystal structure in complex with HIV-1 gp120. For comparison, we also determined the structure of M48U7, which contained a 5-hydroxypentylmethoxy phenylalanine at residue 23M48U7. Structural, biochemical, and virological analyses of M48U1, M48U7, as well as those of previously reported miniproteins, M48 and M47, enabled the structural basis for potent neutralization at the Phe43 cavity of gp120 to be elucidated.
Results
Surface plasmon resonance (SPR) analysis of M48U1 and M48U7
To place the neutralization properties of M48U1 into a biochemical context, we used SPR to determine binding affinity of M48U1 for YU2 gp120 (Figure 1A, Table 1, Table S1). With gp120 immoblized on a CM5 chip, we observed extraordinary affinity of 0.0154±0.0008 nM (Figure 1A, Table 1), and association and dissociation rates of 2.53×106 M−1s−1 and 3.9×10−5 s−1, respectively (Figure 1A, Table S1). To verify affinity, we reversed the experimental format by coupling M48U1 to a CM5 chip via Lys11M48U1. Binding affinity of 0.014±0.005 nM was observed (Figure S1), comparable to the affinity obtained with gp120 immobilized. gp120 affinity for the closely related CD4-mimetic miniprotein, M48U7, was 0.18±0.03 nM (Figure 1B), with an association rate of 2.42×106 M−1s−1 and an off-rate of 4.46×10−4 s−1. For comparison, sCD4 and the CD4-mimetic miniproteins, M48 and M47 have gp120 binding affinities of 7.7 nM, 4.0 nM and 2.1 nM, respectively (Stricher et al., 2008). M48U1, therefore, displayed more than 100-fold tighter binding to YU2 gp120 than sCD4, and previously characterized CD4-mimetic miniproteins, and 10-fold tighter binding compared to M48U7 (Table 1 and Table S1). The remarkable affinity of M48U1 for gp120 was contributed by both its higher rate of association with gp120 as well as slower dissociation rate. While M48U1 and M48U7 both showed tighter affinities to gp120 compared to sCD4, M48 and M47, the discriminating factor between these two miniproteins were their off-rates, with M48U1 dissociating from gp120 about 10-times slower than M48U7.
Figure 1. Surface-plasmon resonance (SPR) analysis of the binding of CD4-mimetic miniproteins, M48U1 and M48U7, to HIV-1 YU2 gp120.
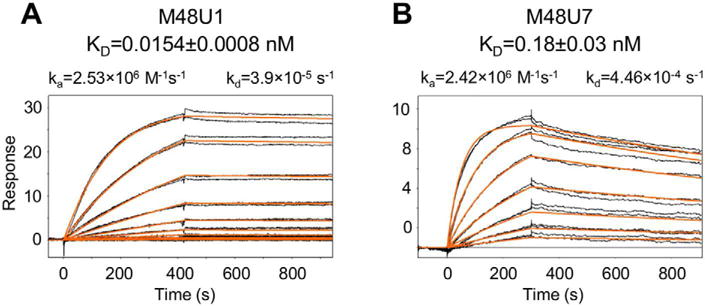
A very slow dissociation rate contributes to the extraordinary affinity of M48U1 for gp120. M48U7 binds with a similar on-rate but dissociates from gp120 about 10-times faster than M48U1. SPR profiles of the binding of (A) M48U1 and (B) M48U7 to full-length HIV-1 gp120 from the clade B YU2 strain. The black lines indicate independent injections of the CD4-mimetic miniproteins with concentrations sampled at 2-fold dilution. Concentrations from 3 nM to 0.006 nM and from 8 nM to 0.06 nM were sampled for M48U1 and M48U7, respectively. Each concentration was sampled in duplicate. The red lines show the global fit of the data to a Langmuir 1:1 binding model. SPR profile of HIV-1 YU2 gp120 binding to M48U1 immobilized on a CM5 chip is shown in Figure S1.
Crystal structures of gp120 bound to CD4-mimetic miniproteins
To gain structural information on the gp120 interactions of M48U1 and M48U7, we crystallized the miniproteins in complex with a variable loops-truncated version of YU2 gp120 (YU2coree) (Kwon et al., 2012). Complexes crystallized as plates in two related space groups, P212121 and C2221 (Table 2). For M48U1, the C2221 crystal form contained one complex in the asymmetric unit and diffracted to 1.49 Å, and the P212121 crystal form contained 2 molecules in the asymmetric unit and diffracted to 1.8 Å. M48U7 crystallized in the C2221 crystal form and diffracted to 2.1 Å. Unless otherwise noted, the C2221 crystal forms of each complex were used for structural comparisons.
Table 2.
Crystallographic data collection and refinement statistics
| PDB accession code | 4JZW | 4JZZ | 4K0A |
|---|---|---|---|
| Complex | |||
| CD4-mimetic miniprotein gp120 | M48U1 YU2 gp120 coree | M48U1 YU2 gp120 coree | M48U7 YU2 gp120 coree |
| Data collection | |||
| Space group | P212121 | C2221 | C2221 |
| Cell constants | |||
| a, b, c (Å) | 63.65, 78.01, 163.25 | 65.09, 164.72, 78.03 | 65.52, 164.66, 78.00 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 1 | 1 | 1 |
| Resolution (Å) | 50.0–1.8 (1.86–1.8) | 50.0–1.49 (1.52–1.49) | 50.0–2.10 (2.14–2.10) |
| Rsyma | 9.2 (43.8) | 11.7 (58.5) | 18.0(60.7) |
| I / σI | 22.9 (2.2) | 20.5 (2.1) | 8.7(1.3) |
| Completeness (%) | 83.7 (74.7) | 99.3 (93) | 90.6(61.1) |
| Redundancy | 4.1 (2.9) | 7.6 (3.5) | 5.4(2.3) |
| Refinement | |||
| Resolution (Å) | 24.66–1.78 (1.81–1.78) | 35.3–1.49 (1.51–1.49) | 36.41–2.13 (2.22–2.13) |
| Unique reflections | 64634 (1672) | 67986(2445) | 21349(1464) |
| Rwork / Rfree (%)b | 0.178/0.227 | 0.173/0.198 | 0.180/0.230 |
| No. atoms | |||
| Protein | 5396 | 2962 | 2882 |
| Sugar (NAG) | 238 | 126 | 126 |
| M48U1/U7 | 441 | 223 | 197 |
| U1/U7 | 38 | 19 | 24 |
| Ligand/ion | 36 | 42 | 0 |
| Water | 934 | 407 | 91 |
| B-factors (Å2) | |||
| Overall | 34.39 | 29.93 | 49.82 |
| Protein | 30.91 | 27.38 | 48.41 |
| Sugars (NAG) | 58.15 | 50.92 | 87.51 |
| M48U1/M48U7 | 42.29 | 41.76 | 72.3 |
| U1/U7 | 25.1 | 25.56 | 37.6 |
| Ligand/ion | 51.27 | 40.06 | – |
| Water | 44.1 | 40.76 | 43.84 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | 0.006 | 0.009 |
| Bond angles (°) | 1.086 | 1.062 | 1.163 |
| Chiral volume (Å2) | 0.146 | 0.072 | 0.076 |
| Estimated coordinate error (maximum likelihood e.s.u.) (Å) | 0.45 | 0.15 | 0.29 |
| Ramachandran | |||
| Favored regions (%) | 98 | 98 | 97 |
| Additional allowed regions (%) | 2 | 2 | 2.8 |
| Disallowed regions (%) | 0 | 0 | 0.2 |
Values in parentheses are for highest-resolution shell.
Rsym = Σ|I − <I>|/Σ<I>, where I is the observed intensity, and <I> is the average intensity of multiple observations of symmetry-related reflections.
R = Σhkl||Fobs| − |Fcalc||/Σhkl|Fobs|.
Rfree is calculated from 5% of the reflections excluded from refinement.
Structures were solved by molecular replacement using gp120-M48 coordinates from PDB ID: 2I60 (Stricher et al., 2008) as search model. Unoccupied density was visible in the Phe43 cavity, into which either U1 or U7 inserts were built. Multiple cycles of simulated annealing, TLS, ADP, positional refinement and automated water picking were employed and alternated with manual model building to obtain final Rfree/Rcryst values of 22.7%/17.8% and 19.8%/ 17.3% for gp120-M48U1 structures in space groups P212121 and C2221, respectively, and 23.0%/18.%, for the gp120-M48U7 structure (Table 2).
M48U1 and M48U7 bound at the gp120 CD4-binding site, at the intersection of three gp120 domains (Figures 2A and 2B). The observed gp120 conformations in the M48U1 and M48U7 bound states were similar to each other with an overall Cα-rmsd of 0.31 Å, and within the range of variation observed for CD4 and other CD4-mimetic miniproteins (Huang et al., 2005; Stricher et al., 2008).
Figure 2. Structures of M48U1 and M48U7 bound to HIV-1 gp120 show ligand inserts penetrating and filling the gp120 Phe43 cavity.
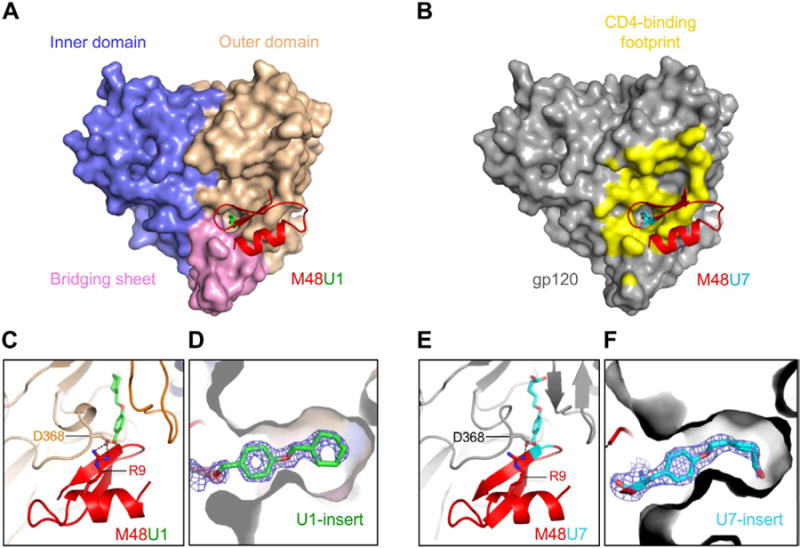
(A) M48U1 (red and green) binds to a conserved gp120 cavity at the intersection of the outer domain (wheat), inner domain (slate) and bridging sheet (pink). (B) M48U7 (red and cyan) binds to the same site on gp120 as M48U1. gp120 is shown as grey surface with the CD4 footprint on gp120 colored yellow. (C) Zoomed-in view of the M48U1 binding site showing hydrogen bonding (black dotted lines) of M48U1 Arg 9 with the conserved Asp 368 of gp120. Figures (A) and (C) are related by an approximately 90° rotation about a horizontal axis. (D) gp120 cross-section showing the U1 side chain (green sticks with the oxygen atom colored red) penetrating the Phe43 cavity. Figures (A) and (D) are related by an approximately 90° rotation about a vertical axis. (E) Zoomed-in view of the M48U7 binding site showing the salt bridge (black dotted) of M48U7 Arg 9 with gp120 Asp 368. Figures (B) and (E) are related by an approximately 90° rotation about a horizontal axis. (F) gp120 cross-section showing U7 side chain (cyan sticks with the oxygen atom colored red) penetrating the Phe43 cavity. The blue mesh surrounding is the 2Fo-Fc map contoured at 2 sigma for M48U1 and 1.6 sigma for M48U7. Figures (B) and (F) are related by an approximately 90° rotation about a horizontal axis.
CD4-mimetic features in M48U1 and M48U7 co-crystal structures
As a first step towards understanding the structure-function relationship of the CD4-mimetic miniproteins, we compared the structural mimicry of their gp120 recognition with that of CD4. In specific, we compared gp120-M48U1 and gp120-M48U7 structures to CD4-bound gp120 structures, as well as to gp120 bound to CD4-binding site antibodies like VRC01 that bind the CD4-bound conformation of gp120 and others such as b12, b13 and F105, which have been shown to induce non-CD4-bound conformations of gp120 (Figure S2). Overall, the conformation of gp120 core bound to M48U1 or M48U7 appeared similar to the CD4-bound conformation of gp120 (Figures 2A and 2B) with main chain rmsds of 0.73 and 0.82, respectively, with CD4-bound gp120 (PDB ID: 1RZK), and differed from other structurally characterized non CD4-bound gp120 conformations (b12-bound, b13-bound, F105-bound, unliganded SIV conformation) (Chen et al., 2005; Chen et al., 2009; Zhou et al., 2007).). The exposed edge of the C-terminal β-strand of M48U1 and M48U7 hydrogen bond to the β15-strand of gp120 in an antiparallel manner similar to the CDR2-like loop of CD4 (Figures 2C and 2E). Both miniprotein use Arg9miniprotein to engage the highly conserved Asp368gp120 in a salt bridge interaction reminiscent of the interaction CD4 makes via its Arg59CD4 (Kwong et al., 1998). A critical interaction at the gp120-CD4 interface is made by the Phe43CD4 side chain, which caps the mouth of the Phe43 cavity. This phenyl interaction is preserved with each of the CD4-mimetic miniproteins (Table 1 and Figures 2C to 2F) (Huang et al., 2005; Stricher et al., 2008) via residue 23miniprotein, which is a Phe in M48 and a Phe-derivative in the other miniproteins. Thus, like the CD4-mimetic miniproteins previously described (Huang et al., 2005; Stricher et al., 2008), M48U1 and M48U7 mimic critical structural elements of CD4 recognition of gp120 including an intermolecular antiparallel β-strands interaction with the β15-strand of gp120, salt bridge with Asp368gp120, and insertion of a phenyl cap at the mouth of the gp120 Phe43 cavity.
Interactions of M48U1 and M48U7 within the Phe43 pocket
The only chemical difference in the miniproteins are their Phe43-cavity inserts (Table 1). Analysis of the miniprotein interface with gp120 showed that interactions outside the Phe43 cavity were overall similar in the different miniprotein complexes (Table S2), with variation between the different miniproteins equivalent to the variation between different complexes of the same miniprotein. This led us to reason that the remarkable gp120-binding and neutralization properties of M48U1, and its differences with the other miniproteins, were likely related to their Phe43 cavity interactions. We, therefore, focused our attention on the Phe43 cavity and sought to understand the details of its interaction with residue 23miniprotein that penetrates the cavity.
With a phenyl capping the mouth of the Phe43 cavity, the cyclohexylmethoxy (U1) in M48U1 and the 5-hydroxypentylmethoxy (U7) in M48U7 extended 6.5 Å and 6.2 Å, respectively, into the Phe43 cavity (Figures 2D and 2F). The Phe43 cavity is predominantly non-polar with islands of polarity contributed by (a) polar atoms from residues lining the cavity walls (main chain O atoms of Ser375gp120, Val255gp120, Phe376gp120 and Asn425gp120; main chain N atoms of Ser256gp120, Phe376gp120 and Asn377gp120, side chain O atoms of Ser375gp120 and Tyr384gp120 (b) a conserved solvent channel about 6.2 Å from the cavity mouth, and (c) an opening at the interior apex of the cavity capped by a water molecule (W1) (Figure 3).
Figure 3. Interactions of M48U1 and M48U7 with the gp120 Phe43 cavity.
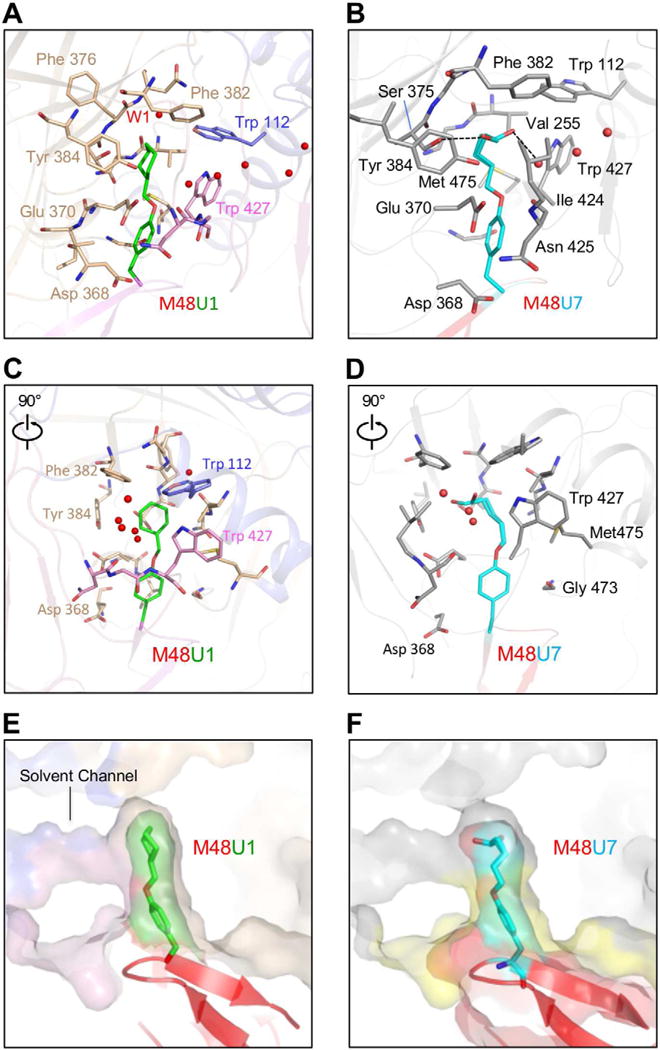
Flexible inserts use shape and chemical complementarity to optimize fit to the Phe43 cavity. (A) M48U1 (red and green) binds to a conserved gp120 cavity at the intersection of the outer domain (wheat), inner domain (slate) and bridging sheet (pink). Red spheres are water molecules. (B) M48U7 (red and cyan) binds to the same site on gp120 as M48U1. gp120 is shown as grey surface. The black dotted lines show hydrogen bonds made by the terminal hydroxyl group of residue 23M48U7 side-chain in the Phe43 cavity. (C) and (D) are 90° rotated views of (A) and (B), respectively. (E) Zoomed-in view of U1 moiety inserting into the Phe43 cavity. (F) Zoomed-in view of U7 moiety inserting into the Phe43 cavity. CD4 binding footprint is colored yellow. In the multilayer transparent surface representation shown in (E) and (F), the outer surface is gp120 and the inner surface is the ligand. Interactions of M48U1 and M48U7 outside the Phe43 cavity are shown in Table S2.
U1 is predominantly non-polar and makes hydrophobic interactions with residues lining the Phe43 cavity walls. The only polar region on U1 is its ether oxygen, which is oriented towards the gp120 solvent channel. The observed distance between the ether oxygen and the nearest water of the solvent channel is 3.6 Å in all three asymmetric units, slightly beyond standard hydrogen bonding distance. The cyclohexyl group fills the inner recess of the Phe43 cavity interacting with Phe382gp120, Trp112gp120, and Val255gp120 that form a hydrophobic cluster at the cavity ceiling (Figures 3A to 3D).
U7 is an alkyl alcohol with greater conformational freedom compared to U1. U7 accommodates its terminal hydroxyl group in a predominantly hydrophobic pocket by directing the hydroxyl way from hydrophobic regions and orienting it towards polar regions of the Phe43 cavity. While the alkyl chain interacts with the hydrophobic walls of the Phe43 cavity, the hydroxyl moiety appears in two distinct conformations, one hydrogen bonding with the Ser375gp120 hydroxyl group and the other with the gp120 solvent channel (Figure 3B).
M48U1 and M48U7 are two examples of how Phe43 cavity penetrating ligands can utilize different chemistries and conformations to interact with this cavity. In summary, the U1 moiety in M48U1 makes 11 hydrophobic interactions with gp120, and the U7 moiety in M48U7 makes 8 hydrophobic interactions and 1 hydrogen bond interaction in each of its two conformations, showing an overall qualitative similarity between the number of contacts that each insert makes with the Phe43 cavity.
Residual flexibility of U1 and U7 in their bound states
While X-ray crystal structures principally offer static snapshots of macromolecular states, information on relative flexibility, disorder, and movement can sometimes be gleaned from features presented by electron density maps. Residue 23M48U7 phenyl ring shows a well-defined electron density (Figure 2F), whereas its alkyl chain shows two distinct conformations, with the terminal hydroxyl in each conformation oriented towards a polar region in the Phe43 cavity.
The electron density of gp120-bound U1 also revealed residual flexibility (Figure 2D). The U1-cyclohexane ring showed asymmetric electron density, with some regions showing well-defined electron density and other regions with poorly defined electron density, the latter suggestive of multiple conformations, unconstrained by interactions with the Phe43 cavity. The methoxy cyclohexyl moiety in the the C2221 crystal form was more disordered than the complexes in the P212121 crystal form, and was modeled as two alternate conformations. For clarity, the predominant conformation found in both molecules in the asymmetric unit of the P212121 crystal form as well as the C2221 crystal form is shown in figures. In contrast, the U1 phenyl ring shows symmetric and continuous electron density suggestive of a defined structure, constrained by its interactions with gp120. This is similar to the well-defined electron densities observed for Phe43CD4 in CD4-bound gp120 structures, for Phe23M48 in the M48-bound gp120 structure, and for the less buried phenyl ring in Bip23 in M47-bound gp120 structure (Kwong et al., 1998; Stricher et al., 2008).
Residual flexibility of U1 in its bound state may play a role in the remarkable neutralization properties of M48U1, by allowing the insert to adapt to changes in the Phe43 cavity conformation as a result of strain-to-strain variation (Shrivastava and LaLonde, 2011). Similar beneficial effect of inhibitor flexibility has been shown for HIV-1 proteases where flexible inhibitors better resist active site variations as well as suffer less entropic penalty compared to rigid inhibitors (Vega et al., 2004).
M48U1 and M48U7 present two examples of ligands utilizing their flexibility to maximize interactions and avoid potential clashes in protein binding sites. While U1 and U7 moieties are both flexible, and both retain mobility in their bound states, M48U1 and M48U7 differ substantially in their binding affinities and neutralization potencies. This suggested that not only is ligand flexibility important, but the precise shape or geometry of the ligand may also contribute to its binding affinity to the Phe43 cavity.
Shape complementarity and surface burial of the CD4 mimetic miniproteins at the gp120 Phe43 cavity
To estimate the goodness-of-fit of each miniprotein insert to the Phe43 cavity, we calculated the shape complementarity (SC) (Lawrence and Colman, 1993) of the insert to the binding pocket, and its buried surface area (BSA) (Krissinel and Henrick, 2007) (Table 3). SC measures the geometric surface complementarity of interfaces and depends both on the relative shape of the surfaces and the extent to which the interaction brings individual elements of the opposing surfaces into proximity. We calculated SC for Phe43 cavity inserts from CD4 and the CD4-mimetic miniproteins M48, M47, M48U1 and M48U7 (Table 3). Phe43CD4, a natural Phe43 cavity ligand, showed highest SC statistic indicating best shape complementarity. SC of Phe23M48 matched closely with that of Phe43CD4, consistent with published reports that show M48 to be a close structural mimic of CD4 (Stricher et al., 2008). Of the other three CD4-mimetic miniproteins (Table 1), M48U1 had the highest SC (Table 3) of 0.762, followed by M47, and M48U7 showed the lowest SC of 0.703. These results suggest that non-natural excursions into the Phe43 cavity resulted in reduced overall shape complementarity compared to the fit of the natural phenyl, and that of these non-natural excursions, U1 showed best shape complementarity (Figures 3E and 3F, Table 3).
Table 3.
Structural properties and biological activity of CD4 and CD4-mimetic miniproteins
| Shape Complementarity (in Phe43 cavity) (SC) | Buried Surface Area (in Phe43 cavity) (BSA) (Å2) | [SC]×[BSA] | Mean Difference Distancesa versus unliganded HIV-1 gp120 Phe43 cavity | Neutralization Potency in μg/ml (Breadth in %)
|
||
|---|---|---|---|---|---|---|
| 24-isolate panel | 180-isolate panel | |||||
| CD4 | 0.823 | 196 | 161 | 0.29 | 3.7 (79.0) | 3.4 (79.2) |
| M48 | 0.822 | 196 | 161 | 0.28 | 5.4 (62.5) | 2.1(70.2) |
| M47 | 0.735 | 310 | 228 | 0.33 | 5.0 (70.8) | 2.3 (76.2) |
| M48U1 | 0.762 | 353 | 269 | 0.21 | 0.39 (83.0) | 0.13 (90.4) |
| M48U7 | 0.703 | 352 | 247 | 0.20 | 5.2 (63.0) | N.D. |
Mean difference distances shown in figures (4) and (5)
Extent of surface burial is another determinant of intermolecular interaction, with a larger shared interfacial area between two molecules suggesting more interfacial contacts and better binding. We calculated buried surface areas for all the Phe43 cavity inserts (Table 1). As expected CD4 and M48 showed the least surface burial (Table 3) since they have the smallest Phe43 cavity insert. Phe43CD4 and Phe23M48 bind the Phe43 cavity mouth, leaving the rest of the cavity unoccupied. M48U1 and M48U7 showed highest surface burial, followed by M47.
Shape complementarity and surface burial are two independent parameters, with shape complementarity being a measure of local fit of the two binding partners and surface burial a measure of their total interfacial area. Since both parameters contribute to overall fit, we empirically derived a combined fit parameter by multiplying SC with the buried surface area for each pair of interactions (Table 3). This combined fit parameter showed that M48U1 had the best overall fit to the Phe43 cavity, followed by M48U7, M47 and M48 in decreasing order of overall fit.
Structural changes in gp120 related to ligand insertion into the Phe43 cavity
We next sought to understand Phe43 cavity conformational changes in response to ligand binding. While overall the conformation of gp120 coree bound to M48U1 or M48U7 resembled the CD4-bound conformation (Figures 2A and 2B and Figure S2), local conformational changes in the vicinity of the bound ligand may affect ligand fit and influence binding affinity. To understand the effect of ligand insertion on the conformation of the Phe43 cavity we analyzed gp120 conformational changes in the vicinity of the U1 or U7 insertion.
For precise definition of the cavity, we used the algorithm fpocket (Le Guilloux et al., 2009). The highest scoring pocket was a 505 Å3 space made up of two intersecting regions- the Phe43 cavity and a conserved gp120 solvent channel (Figures 2 and 3, Figure S3). This bifurcated cavity is made up of residues from the bridging sheet, the α1 and α3 (commonly known as the CD4 binding loop) helices and loop B. A conserved solvent channel starts from an opening in the Phe43 cavity, about 6.2 Å from the cavity mouth, and connects the gp120 CD4 binding site to the coreceptor binding site (Figures 3A and 3D and Figure S3).
U1 and U7 bind to the Phe43 cavity, extending into the gp120 hydrophobic core, thereby filling a region of topological mismatch in gp120’s hydrophobic interior (Figures 2 and 3). To study local effects of ligand binding, we first calculated Phe43 cavity volume in each of the complexes in absence of ligand (Figure S4). The calculated volumes were overall similar between the miniprotein complexes. We then analyzed ligand-induced changes in the Phe43 cavity at the residue level by constructing distance sorted difference distance matrices (Huang et al., 2005). This structural analysis technique was earlier used to study gp120 conformational changes on binding to M48 and M47 (Huang et al., 2005; Stricher et al., 2008). Difference distance matrices allow superposition independent and residue specific structural comparison, and overall summations are obtained by calculating the rms of matrix elements.
A distance-sorted double sieve was used to derive the subset of atoms for difference distance matrix calculations. First, atoms lining the cavity were selected using the program fpocket. Next, atoms from this subset that were ≤ 6Å from M48U1 were selected. Finally, these atoms were sorted by their distance from the 23M48U1 Cα atom. The Ser375 side chain hydroxyl and Ile424 CG1 was found at different positions in various CD4- and mimetic-bound structures as a result of stochastic rotameric variation, and were therefore disregarded for the analysis. Figure 4 tabulates the set of atoms used for this analysis.
Figure 4. Distance-sorted difference distance matrices reveal conformational changes in the gp120 Phe43 cavity as a result of M48U1 and M48U7 binding.
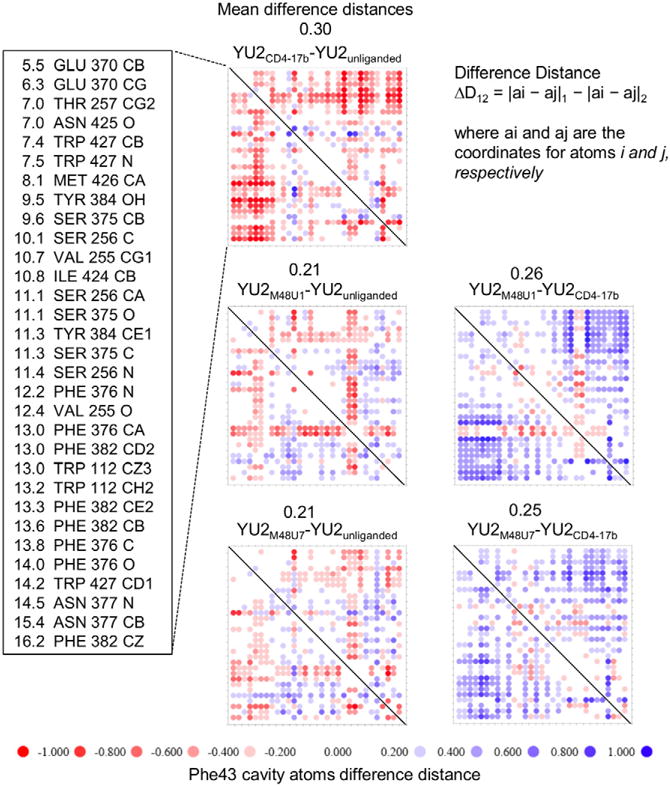
gp120 Phe43 cavity bound to M48U1 or M48U7 shows greater resemblance to the Phe43 cavity in unliganded gp120 than the Phe43 cavity in gp120 bound to CD4 and CD4-induced monoclonal antibody 17b. Distance-sorted difference distance matrices of gp120 were constructed using coordinates of atoms lining the gp120 Phe 43 cavity selected with fpocket. Atoms within 6 Å of U1 were sorted in increasing order of distance from the Phe43 Cα atom of CD4 and are shown along with the corresponding gp120 residue in the far-left panel. The columns in this panel, from left to right, denote distances in Å from Cα of CD4 Phe43, residue names, residue numbers, and atom names. Difference distance matrices composed of these gp120 residues were calculated for unliganded YU2 gp120 and for various gp120 complexes with CD4, M48U1, and M48U7. Each i,j matrix element shows the distance between atom i and atom j in the first specified structure minus the distance between the same atoms in the second specified structure. The difference distance matrices was quantified by mean difference distances, and is shown for each comparison. Physical and chemical properties of the gp120 Phe43 cavity properties are summarized in Figure S3.
We first analyzed differences in the Phe43 cavity between unliganded YU2 gp120 (PDB ID: 3TGQ) and YU2 gp120 bound to CD4 and 17b, a CD4-induced (CD4i) antibody that stabilizes the CD4-bound conformation (PDB ID:1RZK), by constructing difference distance matrices. We detected CD4-binding induced movement in the gp120 Phe43 cavity (Figure 4 and 5A). The largest conformational perturbations were found, as expected, closest to the region of Phe43CD4 insertion in gp120, with changes progressively decreasing in magnitude at increasing distances from the region of Phe43CD4 insertion. It is pertinent to note that the net relative change on CD4 binding suggests overall contraction of the Phe43 pocket, especially surrounding the region of Phe43CD4 insertion. This qualitatively represents a transition from a relaxed state of the gp120 domains to a state constrained by Phe43CD4 insertion at the mouth of the Phe43 cavity, with the gp120 domains brought closer by their interaction with Phe43CD4.
Figure 5. Ligand induced motion in the gp120 Phe43 cavity.
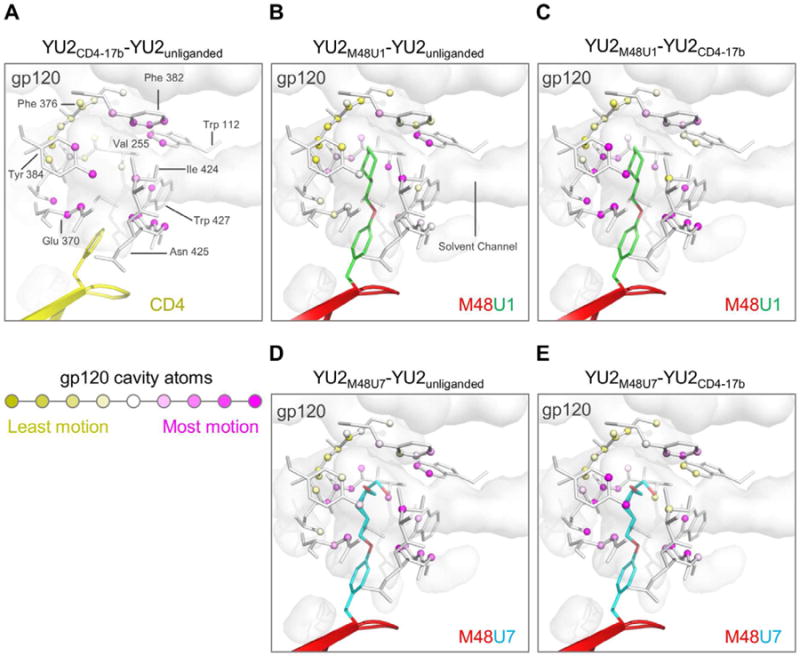
A cross-section of gp120 is shown as a transparent grey surface. Phe43 cavity atoms are shown as spheres, and the residues connecting them as sticks. Distance-sorted difference distance matrices of gp120 Phe43 cavity, bound to different ligands shown in Figure 4, were used to calculate net motion at each cavity atom. The cavity atoms were colored with a yellow-white-magenta gradient, where yellow represents least motion and magenta represents highest differences between (A) gp120 bound to CD4 (yellow) and 17b IgG vs unliganded gp120, (B) gp120 bound to M48U1 vs unliganded gp120, (C) gp120 bound to M48U1vs gp120 bound to CD4 and 17b IgG, (D) gp120 bound to M48U7 (red and cyan) vs unliganded gp120, and (E) gp120 bound to M48U1vs gp120 bound to CD4 and 17b IgG. Analysis of Phe43 cavity volume shown in Figure S4.
We compared M48U1-bound and M48U7-bound gp120 to unliganded (Kwon et al., 2012) and CD4-bound gp120 (Kwong et al., 1998) (Figures 4 and 5B–E), and found that the Phe43 cavity, when bound to these CD4-mimetics, showed greater resemblance to the Phe43 cavity in unliganded gp120 (gp120 coree) than that of CD4-bound gp120. This finding is consistent with the propensity of the Phe43 cavity to be filled as inferred from crystal structures that show small molecules from crystallization solutions occupying the Phe43 cavity (Figure S4), and suggests that cavity filling leads to stabilization of the Phe43 cavity. Residue 23M48U1/M48U7 insertions fill the Phe43 cavity, thereby stabilizing its relaxed or ground state by counteracting the partial “Phe43 cavity collapse” induced by CD4-binding. We also observed localized movement in the Phe43 cavity in response to U1 and U7 insertion. These changes were most dominant at the deeper end of the Phe43 cavity, and in the case of U1, involved residues Phe382gp120, Trp112gp120 and Val255gp120 that form a hydrophobic/aromatic cluster near the ceiling of the cavity, and Ile424gp120 (Figures 3A, 3C, 4 and 5).
We expanded this analysis to CD4-mimetic miniproteins M48 and M47 (Stricher et al., 2008) (Figure 6). We showed that the M48-bound Phe43 cavity resembled the CD4-bound cavity more than the unliganded gp120 cavity. Binding of CD4-mimetic miniprotein M47 distorted the Phe43 cavity more than any of the other CD4-mimetic miniproteins. These findings are consistent with published studies that show M48 is the closest mimic of CD4 (Stricher et al., 2008), and that M47-bound gp120 deviates from the CD4-bound structure around the ligand binding site. We also found that the Phe43 cavity in its M47-bound or M48-bound states are more similar to each other than to the Phe43 cavity bound to either M48U1 or M48U7, and the differences are highest in magnitude at the deeper regions of the Phe43 cavity (≥ 13 Å from Cα of 23M48U1), where U1 and U7 inserts make additional contacts unexplored by both M48 and M47 (Figure 6 and S5).
Figure 6. Comparison of Phe43 cavity conformation between different CD4-mimetic miniproteins.
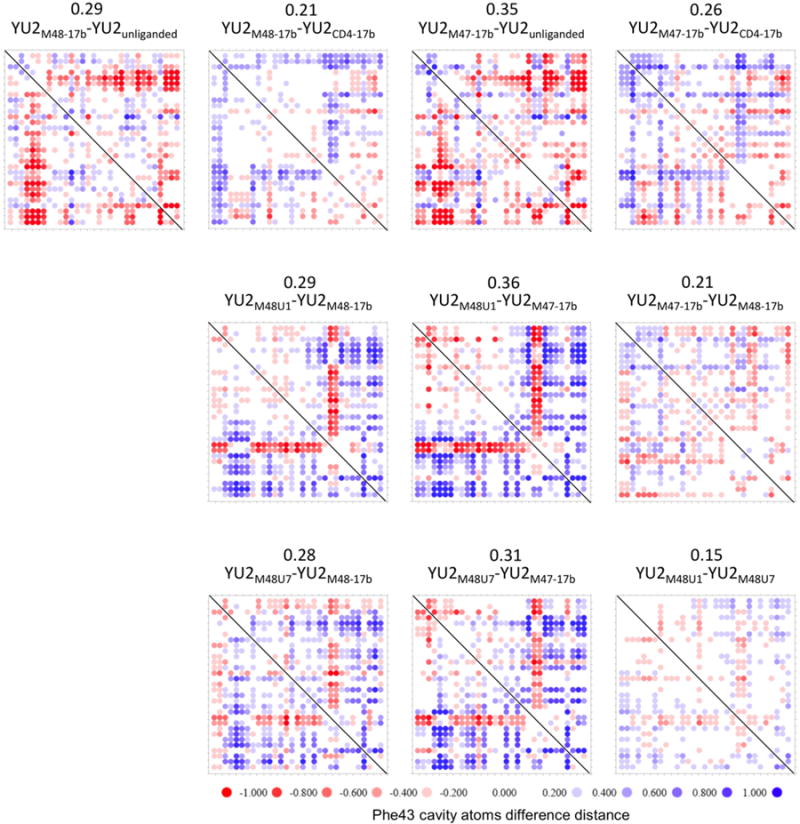
Distance-sorted difference distance matrices of gp120 were constructed using the same set of atoms as in Figure 4. Difference distance matrices were calculated to compare Phe43 cavity conformations of gp120 bound to CD4-mimetic miniproteins M48, M47, M48U1 or M48U7, relative to each other. Each i,j matrix element shows the distance between atom i and atom j in the first specified structure minus the distance between the same atoms in the second specified structure. The difference distance matrices was quantified by overall mean of the difference distances, and is shown for each comparison. Structural changes in the Phe43 cavity conformation upon binding to different cavity filling ligands is shown in Figure S5.
Taken together, these results show that by inserting flexible moieties into the Phe43 cavity, M48U1 and M48U7 induce a Phe43 cavity conformation in core gp120 that is very close to its ground state conformation. This state is similar but distinct from the conformation the cavity assumes when gp120 binds the CD4 receptor. Further, these analyses show that the Phe43 cavity is malleable, and can adapt its conformation to fit an incoming ligand. Despite this malleability, the different affinities of various CD4-mimetic miniproteins suggests that recognition of the ground state appears to be substantially more favorable energetically than of slightly altered conformations.
HIV-1 neutralization by M48U1 and M48U7
To understand the effect of M48U1 binding to the HIV-1 trimeric spike, we assessed its ability to neutralize HIV-1, and compared it to HIV-1 neutralization by sCD4, M48, M47 and M48U7 (Figure 7, Figure S6, Table 3 and Table S3). We tested a panel of 24 pseudoviruses (Figure 7A and Table 3), selected from different clades, for their susceptibility to be neutralized by either M48U1 or M48U7. M48U1 neutralized 20 of the 24 isolates (83%) with geometric IC50 mean of 0.39 μg/mL. In comparison, M48U7 neutralized only 67% of the 24 isolates tested with a geometric IC50 mean of 5.18 μg/mL. M48U1 was also shown to be a better neutralizer than all other currently known CD4-mimetic miniproteins of this class (Van Herrewege et al., 2008).
Figure 7. HIV-1 neutralization by CD4-mimetic peptides M48U1 and M48U7.
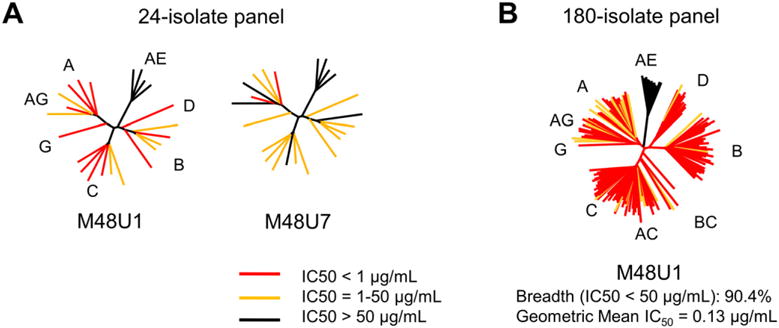
Neutralization profiles of (A) M48U1 and M48U7 depicted with 24-isolate dendrograms and (B) M48U1 depicted with 180-isolate dendrogram representative of circulating HIV-1 Tier 1 and Tier 2 viruses. Neutralization IC50 values are tabulated in Table S3. Neutralization dendrograms of sCD4, M48 and M47 are shown in Figure S5. The molecular weights of M48U1 and M48U7 are 3048 Da and 3038 Da, respectively.
For a more comprehensive estimate of its neutralization efficacy, M48U1 was tested in an expanded panel of 180 HIV-1 isolates representative of all circulating Tier 1 and Tier 2 viruses. M48U1 neutralized all isolates tested, except viruses from Clade A/E, with 90% overall breadth and geometric IC50 mean of 0.13 μg/mL (Figure 6B and Table 3). In comparison, CD4 neutralizes a similar HIV-1 panel with 79% breadth (Table 3, Figure S6) and the CD4-binding site broadly neutralizing antibody VRC01 neutralizes HIV-1 with 91% breadth (Wu et al., 2010).
M48U1 neutralized all viruses tested other than isolates belonging to Clade A/E, a clade prevalent in Central Africa and Southeast Asia. Additionally, none of the clade A/E viruses tested in this study were neutralized by M48U7. Analysis of gp120 sequences showed that all the Clade A/E viruses tested had His at position 375 (Kwon et al., 2012); all the other HIV-1 clades tested contain the canonical Ser at position 375. Clade A/E CA10.3, harboring a Ser375, was shown to be efficiently neutralized by M48U1 (Van Herrewege et al., 2008). As inferred from the structure of HIV-1 Clade A/E 93Th057 gp120 (PDB ID: 3TGT) (Kwon et al., 2012), His375 partially fills the Phe43 pocket, thereby hindering access of the U1 and U7 moieties, allowing the virus to resist a cavity penetrating ligand yet remaining entry competent by retaining binding to CD4, which only accesses the mouth of the Phe43 pocket. Additionally, S375R/N mutations were found to be involved in resistance towards M48U1 (Grupping et al., 2012) consistent with the substrate binding envelope hypothesis for drug design from Schiffer and colleagues (Prabu-Jeyabalan et al., 2002) that postulates that inhibitors that venture out of the envelope defined by natural susbtrates are more susceptible to developing viral resistance under selective pressure of the inhibitor (King et al., 2004). Viral resistance to ligands that access the inner recesses of the Phe43 pocket when a bulky amino acid is placed at position 375 of gp120 has been reported earlier for the NBD class of compounds (Kwon et al., 2012).
Discussion
In this study we have compared the gp120 Phe43 cavity conformation in unliganded and CD4-bound gp120 with its conformation when bound to two flexible ligands of differing chemistries that insert into this cavity. Although the ligands varied in their respective ability to neutralize HIV-1, the conformation of the Phe43 cavity in both cases resembled the unliganded conformation of core gp120 more than the CD4-bound conformation. Analysis of the affinities of the various CD4-mimetic miniproteins as well as of their recognized or induced Phe43-cavity conformations suggest that deviations of the Phe43 cavity from the presumed ground state requires substantial energy. Only by optimizing a combination of chemistry, Phe43-cavity filling, and complementary shape can the potential of the Phe43 cavity be realized – in this case, by near-pan neutralization of HIV-1 and by a gp120 affinity for M48U1 of 0.015 nM.
CD4 makes a functionally critical interaction with HIV-1 envelope, yet soluble CD4 only neutralizes HIV-1 moderately. This is believed to be related to “conformational masking”, the requirement of conformational change of the viral spike to permit CD4 binding, a requirement that trades CD4 affinity in the monomeric gp120 context for induction of the CD4-bound state in the viral spike (Kwong et al., 2002). Two prior mechanisms have been identified for CD4-binding-site ligands that nevertheless achieve broad and potent HIV-1 neutralization: (1) Avidity – both dodecameric CD4 and tetravalent CD4 versions of CD4 neutralize diverse strains of HIV-1 potently (Arthos et al., 2002; Trkola et al., 1995), presumably by spreading the energy required to change spike conformation over several binding sites; (2) Avoiding conformation change – antibody VRC01 is able to bind without inducing conformational changes in the viral spike, allowing this antibody to neutralize over 90% of circulating HIV-1 isolates (Zhou et al., 2010). Here we focus on a third mechanism: optimization of the interaction of the CD4-binding site ligand with the interfacial Phe43 cavity. In particular, we show that a CD4-mimetic ligand that gains affinity by optimizing contacts in the Phe43 cavity can effect broad and potent HIV-1 neutralization. This observation that high affinity binding can overcome the restrictions imposed by conformational masking may have important implications for neutralization at the CD4-binding site of HIV-1 envelope. It may be possible to combine this third mechanism with the prior two. A multimeric M48U1, for example, might have higher potency than monomeric M48U1. Chemical modifications of a Phe43Cys variant of CD4 have also been explored (Xie et al., 2007). Also, recent studies have shown that engaging the gp120 Phe43 cavity can lead to enhanced neutralization by CD4 binding site antibodies (Diskin et al., 2011). Thus, while engagement of the Phe43 cavity may lead to a conformational change of the HIV-1 envelope, our results demonstrate that optimizing ligand fit in the Phe43 cavity can nonetheless lead to potent, near-pan neutralization of HIV-1. Induction of conformational change further allows the possibility of combining these potent inhibitors with entry inhibitors that target the CD4i site of coreceptor binding on gp120 (Acharya et al., 2011; Lagenaur et al., 2010) or with inhibitors that target transient viral envelope intermediates further downstream in the HIV-1 entry pathway (Chen et al., 2002; Reeves et al., 2005).
In summary, our studies provide a structural explanation for the broad and potent neutralization by CD4-mimetic miniprotein M48U1. They show that achieving optimal fit to the Phe43 cavity can be an effective means to enhance the neutralization capability of a CD4-binding site ligand. These results provide frameworks, both mechanistic and structural, for optimizing the interaction of CD4-binding-site ligands with the Phe43 cavity, a conserved site of HIV-1 vulnerability and proven target for HIV-1 neutralization.
Materials and Methods
Synthesis of CD4-mimetic miniproteins, gp120 expression amd purification
M48U1, M48U7, M48 and M47 were synthesized by solid-phase methods as described (Martin et al., 2003). The concentrations of the peptides in solution were determined by hydrochloric acid hydrolysis followed by amino acid analysis. HIV-1 clade B YU2 gp120 coree (Kwon et al., 2012) was expressed, purified and deglycosylated as previously described (Kwon et al., 2012). Complexes with M48U1 and M48U7 were prepared following methods described in the Supplemental Experimental Procedures.
Crystallization, X-ray Data Collection, Structure Determination and Refinement of CD4 mimetic miniproteins in Complex with HIV-1 gp120
Crystallization of miniprotein-gp120 complexes and data collection are described in Supplemental Experimental Procedures. Data integration and scaling, structure solution, refinement, and analysis are described in Supplemental Experimental Procedures.
Assessment of HIV-1 Neutralization
HIV-1 neutralization was measured using single-round-of-infection HIV-1 Env-pseudoviruses and TZM-bl target cells using protocols described in Supplemental Experimental Procedures.
Surface plasmon resonance
Experiments were carried out on a Biacore 3000 or Biacore T200 instrument (GE Healthcare) using protocols described in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
M48U1 displays a gp120 affinity of 0.015 nM and near-pan HIV-1 neutralization.
Remarkable properties of M48U1 result from filling a hydrophobic interfacial cavity.
Optimal cavity fit requires both specific chemistry and complementary shape.
A Phe43-cavity mechanism for enhancing affinity of CD4-binding site ligands.
Acknowledgments
We thank members of the Structural Biology Section and Structural Bioinformatics Core at the NIH Vaccine Research Center for comments on the manuscript, J. Stuckey for assistance with figures. sCD4 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Amino acid analysis for peptide concentration determination was performed at the Amino Acid Analysis and Protein Sequencing facility at the W.M. Keck Foundation Biotechnology Resource Laboratory, Yale University. Support for this work was provided by the Intramural AIDS Targeted Anti-retroviral Program (IATAP) and the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes: Atomic coordinates and structure factors have been deposited in the Protein Data Bank with PDB IDs 4JZW and 4JZZ for gp120-M48U1 in the P212121 and C2221 crystal forms, respectively, and 4K0A for gp120-M48U7.
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- Acharya P, Dogo-Isonagie C, LaLonde JM, Lam SN, Leslie GJ, Louder MK, Frye LL, Debnath AK, Greenwood JR, Luongo TS, et al. Structure-based identification and neutralization mechanism of tyrosine sulfate mimetics that inhibit HIV-1 entry. Acs Chem Biol. 2011;6:1069–1077. doi: 10.1021/cb200068b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthos J, Cicala C, Steenbeke TD, Chun TW, Dela Cruz C, Hanback DB, Khazanie P, Nam D, Schuck P, Selig SM, et al. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J Biol Chem. 2002;277:11456–11464. doi: 10.1074/jbc.M111191200. [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RY, Kilby JM, Saag MS. Enfuvirtide. Expert Opin Investig Drugs. 2002;11:1837–1843. doi: 10.1517/13543784.11.12.1837. [DOI] [PubMed] [Google Scholar]
- Curreli F, Choudhury S, Pyatkin I, Zagorodnikov VP, Bulay AK, Altieri A, Kwon YD, Kwong PD, Debnath AK. Design, synthesis, and antiviral activity of entry inhibitors that target the CD4-binding site of HIV-1. J Med Chem. 2012;55:4764–4775. doi: 10.1021/jm3002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereuddre-Bosquet N, Morellato-Castillo L, Brouwers J, Augustijns P, Bouchemal K, Ponchel G, Ramos OH, Herrera C, Stefanidou M, Shattock R, et al. MiniCD4 Microbicide Prevents HIV Infection of Human Mucosal Explants and Vaginal Transmission of SHIV(162P3) in Cynomolgus Macaques. PLoS Pathog. 2012;8:e1003071. doi: 10.1371/journal.ppat.1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupping K, Selhorst P, Michiels J, Vereecken K, Heyndrickx L, Kessler P, Vanham G, Martin L, Arien KK. MiniCD4 protein resistance mutations affect binding to the HIV-1 gp120 CD4 binding site and decrease entry efficiency. Retrovirology. 2012;9:36. doi: 10.1186/1742-4690-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Kahn M, Garcia NK, Hu SL, Lee KK. Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J Virol. 2012;86:8750–8764. doi: 10.1128/JVI.07224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Stricher F, Martin L, Decker JM, Majeed S, Barthe P, Hendrickson WA, Robinson J, Roumestand C, Sodroski J, et al. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120: Crystal structures, molecular mimicry, and neutralization breadth. Structure. 2005;13:755–768. doi: 10.1016/j.str.2005.03.006. [DOI] [PubMed] [Google Scholar]
- King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagenaur LA, Villarroel VA, Bundoc V, Dey B, Berger EA. sCD4-17b bifunctional protein: extremely broad and potent neutralization of HIV-1 Env pseudotyped viruses from genetically diverse primary isolates. Retrovirology. 2010;7:11. doi: 10.1186/1742-4690-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde JM, Elban MA, Courter JR, Sugawara A, Soeta T, Madani N, Princiotto AM, Kwon YD, Kwong PD, Schon A, et al. Design, synthesis and biological evaluation of small molecule inhibitors of CD4-gp120 binding based on virtual screening. Bioorganic & medicinal chemistry. 2011;19:91–101. doi: 10.1016/j.bmc.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLonde JM, Kwon YD, Jones DM, Sun AW, Courter JR, Soeta T, Kobayashi T, Princiotto AM, Wu X, Schon A, et al. Structure-based design, synthesis, and characterization of dual hotspot small-molecule HIV-1 entry inhibitors. Journal of medicinal chemistry. 2012;55:4382–4396. doi: 10.1021/jm300265j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Le Guilloux V, Schmidtke P, Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, et al. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat Biotechnol. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure. 2002;10:369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- Reeves JD, Lee FH, Miamidian JL, Jabara CB, Juntilla MM, Doms RW. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J Virol. 2005;79:4991–4999. doi: 10.1128/JVI.79.8.4991-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava I, LaLonde JM. Enhanced Dynamics of HIV gp120 Glycoprotein by Small Molecule Binding. Biochemistry-Us. 2011;50:4173–4183. doi: 10.1021/bi2002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricher F, Huang CC, Descours A, Duquesnoy S, Combes O, Decker JM, Do Kwon Y, Lusso P, Shaw GM, Vita C, et al. Combinatorial optimization of a CD4-mimetic miniprotein and cocrystal structures with HIV-1 gp120 envelope glycoprotein. Journal of Molecular Biology. 2008;382:510–524. doi: 10.1016/j.jmb.2008.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF, Burton DR, Ho DD, et al. Cross-Clade Neutralization of Primary Isolates of Human-Immunodeficiency-Virus Type-1 by Human Monoclonal-Antibodies and Tetrameric Cd4-Igg. Journal of Virology. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herrewege Y, Morellato L, Descours A, Aerts L, Michiels J, Heyndrickx L, Martin L, Vanham G. CD4 mimetic miniproteins: potent anti-HIV compounds with promising activity as microbicides. J Antimicrob Chemother. 2008;61:818–826. doi: 10.1093/jac/dkn042. [DOI] [PubMed] [Google Scholar]
- Vega S, Kang LW, Velazquez-Campoy A, Kiso Y, Amzel LM, Freire E. A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease. Proteins. 2004;55:594–602. doi: 10.1002/prot.20069. [DOI] [PubMed] [Google Scholar]
- Vita C, Drakopoulou E, Vizzavona J, Rochette S, Martin L, Menez A, Roumestand C, Yang YS, Ylisastigui L, Benjouad A, et al. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc Natl Acad Sci U S A. 1999;96:13091–13096. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Xie H, Ng D, Savinov SN, Dey B, Kwong PD, Wyatt R, Smith AB, 3rd, Hendrickson WA. Structure-activity relationships in the binding of chemically derivatized CD4 to gp120 from human immunodeficiency virus. J Med Chem. 2007;50:4898–4908. doi: 10.1021/jm070564e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


