Background: Surface CD157 modulates leukocyte and ovarian cancer cell adhesion and migration through the interaction with an unknown ligand.
Results: CD157 binds heparin-binding domains of numerous extracellular matrix proteins with high affinity.
Conclusion: The interaction of CD157 with extracellular matrix proteins is instrumental in the regulation of cell adhesion.
Significance: These findings provide valuable insights into the biological mechanism responsible for the nonenzymatic functions of CD157.
Keywords: Cell Adhesion, Cell Surface Receptor, Extracellular Matrix, Fibronectin, Membrane Protein, BST-1, CD157, Heparin-binding Domain
Abstract
CD157/BST-1 behaves both as an ectoenzyme and signaling receptor and is an important regulator of leukocyte trafficking and ovarian cancer progression. However, the molecular interactions underpinning the role of CD157 in these processes remain obscure. The biological functions of CD157 and its partnership with members of the integrin family prompted us to assume the existence of a direct interaction between CD157 and an unknown component of the extracellular matrix. Using solid-phase binding assays and surface plasmon resonance analysis, we demonstrated that CD157 binds fibronectin with high affinity within its heparin-binding domains 1 and 2. Furthermore, we found that CD157 binds to other extracellular matrix proteins containing heparin-binding domains. Finally, we proved that the CD157-fibronectin interaction occurs with living cells, where it elicits CD157-mediated cell responses. Indeed, knockdown of CD157 in Met-5A mesothelial cells changed their morphology and cytoskeleton organization and attenuated the activation of intracellular signaling pathways triggered by fibronectin. This led to impaired cell spreading and adhesion to selected extracellular matrix proteins. Collectively, these findings indicate a central role of CD157 in cell-extracellular matrix interactions and make CD157 an attractive therapeutic target in inflammation and cancer.
Introduction
Ectoenzymes are a large and heterogeneous class of cell surface-expressed enzymes (1). Many are expressed in leukocytes and endothelial cells, where they modulate each step of leukocyte trafficking (2). In recent years, we have focused our attention on CD157/BST-1, a glycosylphosphatidylinositol-anchored glycoprotein belonging to the ADP-ribosyl cyclase gene family (3). CD157 is expressed in myeloid, endothelial, and mesothelial cells and in epithelial ovarian cancer cells (4, 5). Since its discovery more than 20 years ago, human CD157 (then known as Mo5) (6) has been implicated in the control of leukocyte migration (7). Subsequently, leukocyte CD157 was found to control adhesion to extracellular matrix (ECM)4 proteins, directional migration, and diapedesis (8, 9) by establishing structural and functional interactions with specific members of the integrin family of adhesion receptors (10, 11). Evidence obtained by in vitro experiments and from correlative clinical studies in ovarian cancer patients suggested an important functional link between high expression of CD157 and the malignant phenotype of tumor cells, including their increased motility and invasion of surrounding tissues (12). It became clear that CD157 could induce a variety of cellular responses both in leukocytes and in ovarian cancer cells, including changes in morphology, spreading, adhesion, motility, and transmigration. These activities of CD157 are apparently unrelated to its enzymatic functions (13–15). So far, the efforts to unravel the nonenzymatic functions of CD157 have been hampered by the lack of a known nonsubstrate ligand, whose identification has proved elusive. This limitation was partially overcome by the use of specific monoclonal antibodies, a strategy commonly adopted to mimic the effects of putative ligands. This approach consistently pointed to an important role of CD157 in cell adhesion and migration, but it failed to identify which interactions were instrumental for its receptor activities in normal and pathological conditions. The crucial role of CD157 in cell adhesion, migration and invasion, and its functional partnership with members of the integrin family in myeloid cells (11) fostered the hypothesis that the ligand of CD157 might be found in the ECM.
The ECM is a complex network of proteins and polysaccharides that is secreted, assembled, and modeled by cells. The ECM constitutes the complex structural scaffold surrounding and supporting cells in all tissues and organs, enabling microenvironmental sensing. The dynamic regulation of cell-ECM interactions is essential for the successful outcome of physiological processes, including embryonic development, morphogenesis and tissue homoeostasis (16). Conversely, aberrant cell-ECM interactions underlie many diseases; for example, the pathogenesis of inflammatory diseases relies on aberrant cell aggregation and/or migration, whereas altered adhesion is a defining characteristic of malignancy (17).
ECM proteins are large and complex and are highly conserved in animal taxa. By virtue of their ordered domain organization, ECM proteins orchestrate the juxtaposition of different receptors, generating multimolecular complexes in the plasma membrane (18). In mammalian cells, the interaction between cells and ECM is coordinated primarily by the members of two gene families, integrins and syndecans (19). These mediate cell-ECM adhesion and regulate intracellular signaling pathways that drive cell proliferation, differentiation, migration, and survival. Integrins and syndecans are flanked by a wide variety of cell surface proteins that bind to specific domains within selected ECM proteins.
In this study, using solid-phase binding assays and surface plasmon resonance (SPR) biosensor, we assessed the possible interactions between CD157 and fibronectin, a prototypic ECM protein that plays a central role in cell adhesion, migration, and differentiation (20).
EXPERIMENTAL PROCEDURES
Proteins and Antibodies
Human plasma fibronectin (FN) and its proteolytic fragments (30 and 45 kDa, corresponding to the heparin (HBD1) and gelatin- and collagen (GBD)-binding domains, respectively), laminin-1 (LM), collagen type I (Coll I), vitronectin (Vn), heparin, human plasma fibrinogen (FB), fibrin, hyaluronic acid, and BSA were purchased from Sigma. The 120- and 40-kDa proteolytic fragments from FN, e.g. the cell-binding domain (CBD) and the heparin-binding domain 2 (HBD2), were from Chemicon (Millipore, Milan, Italy). Recombinant His-tagged human soluble CD157 (rh-sCD157, 4736-AC-050) and PDGF receptor α (rh-PDGFRα, 322-PR-050), both produced in NS0-derived mouse myeloma cells, were purchased from R&D Systems (Milan, Italy). Bacterial recombinant His-tagged human CD157 (b-rhCD157, MBS2010989) was from MyBiosources (Aurogene, Rome, Italy). Anti-CD157 (SY/11B5, IgG1), anti-β1 integrin/CD29 (Moon-4, IgG1), monoclonal antibodies (mAb), and irrelevant murine monoclonal IgG (mIgG) (produced in-house) were affinity-purified on protein G (Sigma). RF3 (anti-CD157, IgG2a) mAb was purchased from MBL International (Milan, Italy); Mo5 (anti-CD157, IgG2a) mAb was kindly provided by R. Todd III (University of Michigan Health System, Ann Arbor, MI). Anti-αv integrin/CD51 (NKI-M9) blocking mAb, HRP-conjugated donkey anti-sheep, goat anti-rabbit, and goat anti-mouse polyclonal antibodies, and HRP-conjugated anti-β-actin antibody were purchased from Santa Cruz Biotechnology (Milan, Italy). Anti-human CD157 affinity-purified polyclonal IgG produced in sheep were from R&D Systems. Affinity-purified FITC-labeled F(ab′)2 fraction from goat antibodies to rabbit IgG was from Jackson ImmunoResearch (LiStarfish, Milan, Italy). TRITC-labeled phalloidin used to detect F-actin was obtained from Sigma.
Cell Line and Transduction
Met-5A nonmalignant pleural mesothelial cell line was purchased from American Type Culture Collection (ATCC, Manassas, VA) and maintained in RPMI 1640 medium supplemented with 10% FCS. CD157 expression was silenced by lentiviral delivery of short hairpin RNA (shRNA) targeting BST-1 mRNA (target sequences 5′-GAGTCAGACTGCTTGTATA-3′ (shCD157) and scramble sequence 5′-TTCTCCGAACGTGTCACGTT-3′) as described previously (15, 21). Transduced cells were selected in 2 μg/ml puromycin (Santa Cruz Biotechnology) for 3 days.
Solid-phase Binding Assay
Solid-phase binding assays were performed in 96-well microtiter plates (Nunc MaxiSorp) by ELISA. Wells were coated overnight at 4 °C with 50 μl/well of 10 μg/ml of the indicated matrix proteins or fibronectin fragments and blocked with 0.1% BSA and 0.05% Tween 20 for 2 h at 37 °C. Following two washes with PBS containing 0.1% Tween 20, rh-sCD157 was added at the indicated concentrations and incubated for 2 h at 37 °C. Bound proteins were detected by specific sheep anti-CD157 polyclonal antibody (1 h at 37 °C), followed by HRP-conjugated anti-sheep IgG polyclonal antibody (1 h at 37 °C). The peroxidase substrate 3,3′,5,5′-tetramethylbenzidine (Sigma) was added to the wells for 10 min; the reaction was stopped with H2SO4 (0.5 m), and the absorbance was measured at 450 nm using a microplate reader.
In competition experiments, rh-sCD157 (100 ng/ml) was preincubated for 20 min at room temperature with 10 μg/ml of the indicated anti-CD157 mAb or isotype-matched IgG and then was added to the wells. In binding inhibition assays, rh-sCD157 (100 ng/ml) was preincubated with fibrin, heparin, or hyaluronic acid at the indicated concentrations.
Surface Plasmon Resonance
The carboxylate cuvettes, N-hydroxysuccinimide, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, and ethanolamine, were obtained from Neosensors (Crewe, UK). Biosensor analyses were performed on an IAsys plus device (Affinity Sensors, Cambridge, UK). The carboxylate cuvette was thermostatted at 25 °C, and impurities were removed by washing with PBS-T (0.05% Tween 20). Prior to activation, the carboxylate surface was washed with detergent-free PBS to ensure the establishment of a base line for the sensor trace at the set temperature and to avoid the “mask” effect of carboxyl groups due to the presence of the detergent. Surface carbonyl groups were activated with 7 min of incubation with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide (freshly prepared by mixing v/v 100 mm N-hydroxysuccinimide and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide solutions) (35). Each protein to be immobilized was dissolved in 10 mm CH3COONa (300 μg/ml), at the proper pH, which was chosen on the basis of the isoelectric point (e.g. 7.7 for FN, 9.7 for HBD1, 5.5–6 for HBD2, and 4.9 for CBD). In detail, we used pH 5 for full-length FN, pH 6 for HBD1 and HBD2, and pH 4.5 for CBD. Finally, each protein was anchored to the carboxylic surface via nucleophilic attack of their N terminus to the activated carbonyl groups. After 20 min of incubation, noncoupled ligand was removed by PBS washes. Free carboxyl groups were inactivated, and any electrostatically bound material was removed by 1 m ethanolamine. The surface was finally re-equilibrated with PBS. These conditions were found to best suit an optimal immobilization of the proteins without affecting their functional properties. For each surface, a normally distributed population of the immobilized protein in the monolayer was obtained by repeating spot-sampling at different sites (n = 10) on the biosensor surface (data not shown). rh-sCD157 and b-rhCD157 were independently added at the indicated concentrations, and binding kinetics were followed up to equilibrium. Dissociation steps and surface regeneration were performed by addition of PBS. Binding was measured at 5-s intervals. Binding data were analyzed by using the Fast Fit software (Fison Applied Sensor Technology). In reverse binding experiments, rh-sCD157 was dissolved (300 μg/ml) in 10 mm CH3COONa, pH 5, and incubated for 20 min on a 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide-activated carboxylate surface to a final surface density of 1.4 ng mm−2. Next, interaction experiments were performed by addition of either soluble HBD1, HBD2, or FN, as described above.
SDS-PAGE and Western Blot Analysis
rh-sCD157 and b-rhCD157 (1 μg) were run on 10% SDS-PAGE under reducing and nonreducing conditions and stained with Coomassie Brilliant Blue. For Western blot analysis, total cell lysates were obtained by incubation of cells in RIPA lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm EDTA, 0.1% SDS) supplemented with 50 μg/ml aprotinin and leupeptin. Protein concentration was determined using the Bradford assay (Bio-Rad). Protein extracts (30 μg) were separated by 10% SDS-PAGE under nonreducing conditions and transferred onto a PVDF membrane (Millipore, Milan, Italy). The membrane was blocked with 5% milk and probed with the SY/11B5 anti-CD157 mAb, followed by HRP-conjugated anti-mouse IgG. The immunoreactive bands were visualized using the ECL detection system (Cyanagen, Bologna, Italy). Images were captured with a ChemiDocTM XRS+ system, and densitometry analysis was performed with ImageLabTM software (Bio-Rad).
Immunofluorescence and Confocal Microscopy
For FACS analysis, cells (2 × 105/sample) were incubated for 30 min at 4 °C with 5 μg/ml of the indicated antibody, washed, and incubated for 30 min at 4 °C with F(ab′)2 GαMIg-FITC. Fluorescence was determined using a FACSCalibur flow cytometer and analyzed with CellQuest software (BD Biosciences). Background mAb binding was estimated by means of an isotype-matched negative control mAb. Ten thousand events were considered for each analysis.
For confocal microscopy analysis, glass coverslips were coated with FN (10 μg/ml) and blocked with 1% BSA. Cells were preincubated with cycloheximide (20 μg/ml, Sigma) for 2 h in serum-free medium, detached with EDTA, and plated on FN-coated coverslips (1 × 105/well) for 4 h in the presence of cycloheximide to prevent synthesis of endogenous matrix. Cells were washed, fixed with 2% paraformaldehyde (PFA), permeabilized with 0.2% Triton X-100, and stained with TRITC-labeled phalloidin. Samples were analyzed using an Olympus FV300 laser scanning confocal microscope (Olympus, Milan, Italy) and imaged using a ×60 oil immersion objective (1.4 NA).
RNA Extraction and RT-PCR
Total RNA (2 μg) was extracted from 70 to 80% confluent cultures using TRI Reagent (Sigma) and reverse-transcribed with the Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo(dT) primers. cDNA was amplified using KAPA2G Fast HotStart DNA polymerase (Kapa Biosystems, Cambridge, MA). Each cycle consisted of denaturation at 95 °C for 10 s, annealing for 10 s, and extension at 72 °C for 1 s. The primers used were as follows: CD157, 5′-CCAAAGTTCCCCGATGGCGGCC-3′ (forward) and 5′-GGTTAGAGCAACACAGTTTCC-3′ (reverse); β-actin, 5′-CCAAGGCCAAACCGCGAGAAGATGAC-3′ (forward) and 5′-AGGGTACATGGTGGTGCCGCCAGAC-3′ (reverse). PCR products were analyzed by electrophoresis on 1% agarose gel stained with ethidium bromide (Sigma).
Cell Adhesion Assays
Met-5A cells (3 × 104 in serum-free medium) were plated onto 96-well plates coated with the indicated ECM protein or FN fragment (10 μg/ml) and blocked with 2% BSA. Cells were incubated for 30 min (or 2 h on FN domains) at 37 °C, and then nonadherent cells were removed; adherent cells were fixed with 4% PFA and stained with 0.5% crystal violet. Where indicated, cells were pretreated for 20 min with 10 μg/ml anti-CD157, anti-CD29, or anti-CD51 mAbs or an isotype-matched irrelevant IgG. In binding competition studies, ECM-coated wells were saturated for 1 h at 37 °C with rh-sCD157 (100 ng/ml). Adherent cells were assessed by light microscopy, and five randomly selected fields (×4) for each well were photographed and counted. Each condition was carried out in triplicate, and experiments were repeated at least three times.
Phosphorylation Assays
Met-5A cells were serum-starved overnight, detached with 2 mm EDTA in PBS, suspended in serum-free medium (1 × 106/ml), and allowed to recover for 30 min at 37 °C. Then 1 ml of cell suspension was plated onto wells coated with FN (5 μg/ml) and blocked with heat-denatured BSA 1% in PBS. After 1 h of adhesion, cells were placed on ice, washed with ice-cold RPMI 1640 medium supplemented with 1 mm pervanadate to stop the reaction, and lysed in ice-cold RIPA buffer. Total lysates (20 μg/lane) were run on 10% SDS-PAGE under reducing conditions and transferred onto PVDF membranes. After blocking, membranes were probed with anti-phospho-c-SRC (Tyr-530), anti-phospho-FAK (Tyr-397) or anti-phospho-Akt (Ser-493) (Santa Cruz Biotechnology), stripped, and re-probed with anti-c-SRC, anti-FAK, anti-Akt1 mAbs (Santa Cruz Biotechnology), followed by incubation with the appropriate HRP-conjugated antibodies. Specific bands were visualized by ECL and captured with a ChemiDoc analyzer, and densitometry analysis was performed with ImageJ software (National Institutes of Health, Bethesda).
Statistical Analysis
Individual experiments were performed at least in triplicate, and unless otherwise stated, the data presented are the means ± S.E. of 3–4 independent experiments. Comparisons between two groups were carried out using an unpaired Student's t test for normal distributed variables. Comparisons between three or more groups were performed with one-way ANOVA analysis with Dunnett's or Bonferroni post-test. Statistical analysis was performed using GraphPad Prism 5 software (San Diego). For all analyses, differences were considered significant at p < 0.05 (*, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control).
RESULTS
CD157 Binds to Fibronectin
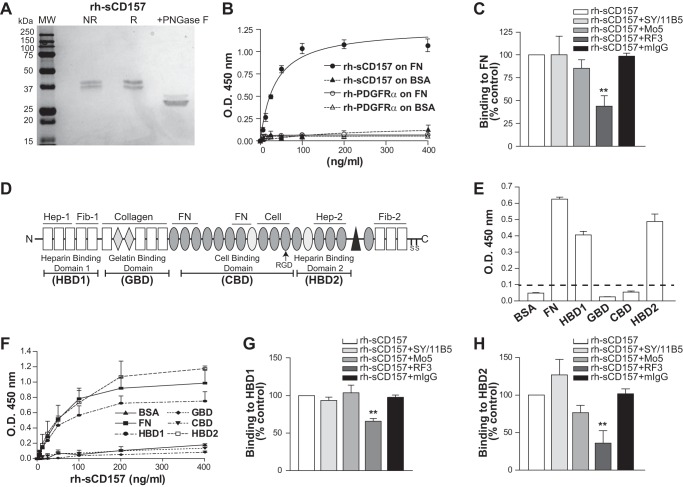
We developed a solid-phase immunoenzymatic assay based on the binding of rh-sCD157 produced in mammalian cells (Fig. 1A) to microtiter plates coated with selected human ECM proteins or BSA as control. We first focused on fibronectin, a ubiquitous component of the ECM that plays a crucial role in mediating cell attachment, because of previous evidence underscoring a functional and structural partnership between CD157 and β1 (and β2) integrin, which are elective receptors for fibronectin (11). Purified rh-sCD157 displayed concentration-dependent binding to FN. His-tagged rh-PDGFRα, used as a specificity control protein, did not bind to FN (Fig. 1B). To further corroborate the specificity of the observed binding, the assay was repeated in the presence of a panel of anti-CD157 mAbs. Incubation of rh-sCD157 with the RF3 mAb significantly reduced binding to FN, whereas incubation with SY/11B5 and Mo5 mAbs did not (Fig. 1C). These findings argue in favor of a specific interaction between purified CD157 and FN proteins.
FIGURE 1.
rh-sCD157 binds fibronectin. A, SDS-PAGE of human recombinant sCD157 produced in murine myeloma NS0-derived cells. Purified rh-sCD157 run under nonreducing and reducing conditions was detected as 38–40-kDa bands. rh-sCD157 was denatured by boiling 10 min with 0.2% (w/v) SDS in sodium phosphate buffer, pH 7.5, and incubated for 3 h at 37 °C with 1 unit of peptide N-glycosidase F (PNGase F) (Sigma), according to the manufacturers' protocol. The reaction was stopped by heating at 100 °C for 5 min, and samples were suspended in Laemmli Sample Buffer and run on SDS-PAGE. The molecular weight of rh-sCD157 after peptide N-glycosidase F treatment was consistent with the predicted molecular weight of 31 kDa based on the deduced amino acid sequence. MW, molecular weight markers (Precision Plus Protein All Blue standards, Bio-Rad); NR, nonreducing conditions; R, reducing conditions. B, concentration-dependent binding of rh-sCD157 to immobilized FN analyzed by solid-phase binding assay. Microtiter plates coated with 10 μg/ml FN or BSA were incubated with increasing concentrations of rh-sCD157 or rh-PDGFRα (6.25–400 ng/ml), used as control. Matrix-bound rh-sCD157 or rh-PDGFRα was assessed by anti-CD157 or anti-PDGFRα polyclonal antibody, followed by HRP-labeled anti-rabbit IgG. The mean ± S.D. of one representative experiment performed in triplicate is shown (n = 3). The best fits for FN (solid line) and BSA (dashed line) are shown. O.D. = optical density. C, inhibition of rh-sCD157 binding to FN by anti-CD157 mAbs. The indicated mAb or murine monoclonal IgGs (10 μg/ml) were incubated with 100 ng/ml of rh-sCD157 for 20 min at room temperature, and rh-sCD157 binding was analyzed. Results are expressed as relative ratio to the absorbance of rh-sCD157 without inhibitors (100%, white histogram). The mean ± S.E. of three independent experiments is shown. **, p < 0.01, ANOVA with Dunnett's multiple comparison test. D, schematic representation of the modular structure of one chain of the dimeric human fibronectin. Rectangles represent type I; diamonds represent type II, and ovals represent type III motifs. These structural motifs include 12 FN type I repeats (FN1), 2 FN type II repeats (FN2), 15 constitutively expressed and 2 alternatively spliced FN type III (FN3) repeats, and a nonhomologous variable (V) region. Sets of adjacent motifs form binding domains for a number of proteins. The heparin- (HBD1 and HBD2), gelatin- (GBD), and cell (CBD)-binding domains generated by proteolytic cleavage and used in the solid-phase binding assays are underlined. E, binding of rh-sCD157 to FN domains measured by solid-phase binding assays. rh-sCD157 (100 ng/ml) was added onto microtiter plates coated with 10 μg/ml FN or its domains. The mean ± S.D. of one representative experiment is shown (n = 3). Dashed line indicates significant binding over baseline (2-fold binding to BSA). F, concentration-dependent binding of rh-sCD157 to immobilized FN domains. Microtiter plates coated with GBD, HBD1, HBD2, and CBD of FN or BSA (10 μg/ml) were incubated with increasing concentrations of rh-sCD157 (6.25–400 ng/ml). The mean ± S.E. of three experiments performed in duplicate is shown. G and H, inhibition of rh-sCD157 binding to HBD1 and HBD2 by anti-CD157 mAbs. After incubation of the indicated mAbs and control IgG (10 μg/ml) with rh-sCD157 (100 ng/ml) for 20 min at room temperature, rh-sCD157 binding to HBD1 and HBD2 was determined. Results are expressed as relative ratio to the absorbance of rh-sCD157 in the absence of mAb (100%, white histogram). The mean ± S.E. of three independent experiments is shown. **, p < 0.01, ANOVA with Dunnett's multiple comparison test.
CD157 Binds to FN Heparin-binding Domains
Fibronectin is composed of three different types of structural motifs, namely types I, II, and III motifs. Although first identified in fibronectin (22), these motifs have since been found in other ECM proteins. Sets of adjacent motifs form binding domains for a variety of proteins and carbohydrates, including other FN molecules, other ECM components, heparin, fibrin, growth factors, and a dozen members of the integrin receptor family (23, 24). To identify the FN region(s) involved in the binding of rh-sCD157, microtiter plates were coated with FN fragments obtained by proteolytic cleavage of the full-length protein as follows: the N-terminal 30-kDa HBD1; the 45-kDa GBD; the 120-kDa CBD containing the integrin-binding motif; and the C-terminal 40-kDa HBD2 containing the high affinity heparin-binding site (Fig. 1D). The solid-phase binding assays showed that rh-sCD157 (100 ng/ml) was able to bind both HBD1 and HBD2. Conversely, no measurable binding to GBD and CBD fragments was observed (Fig. 2, E and F). As with full-length FN, rh-sCD157 binding to both HBD1 and HBD2 was concentration-dependent (Fig. 1F). This binding was significantly inhibited by the RF3 mAb, whereas Mo5 and SY/11B5 mAbs were unable to significantly interfere with CD157-HBD1 or CD157-HBD2 interactions (Fig. 1, G and H). Collectively, these data indicate that rh-sCD157 has two binding sites in FN, located within its N-terminal (HBD1) and C-terminal (HBD2) heparin-binding domains, respectively. The CD157-HBD interactions likely involve the CD157 epitope recognized by the RF3 mAb.
FIGURE 2.
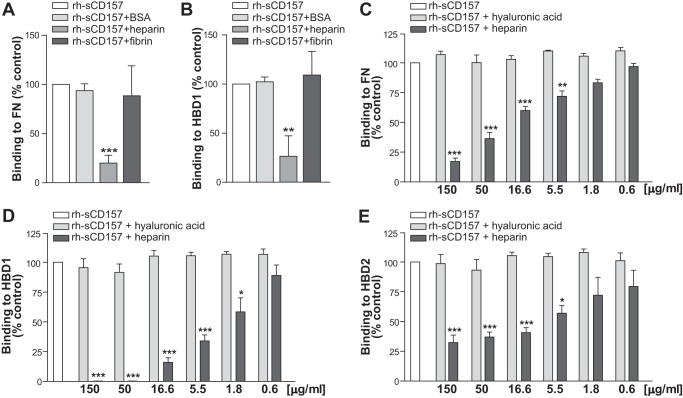
Heparin inhibits binding of rh-sCD157 to fibronectin. A and B, competition experiments were carried out using rh-sCD157 (100 ng/ml) in solid-phase binding assays to FN or HBD1, respectively, in the presence of 10 μg/ml heparin, fibrin, or BSA. C–E, effect of heparin on rh-sCD157 (100 ng/ml) binding to FN (C), HBD1 (D), and HBD2 (E) was assessed in the presence of increasing concentrations (0.6–150 μg/ml) of heparin (dark gray histograms) or hyaluronic acid (light gray histograms). Results are expressed as a relative ratio to the absorbance of rh-sCD157 without competitors (100%, white histogram). Mean ± S.E. of three independent experiments is shown. Asterisks indicate significant differences of rh-sCD157 binding in the presence of each concentration of competitor compared with untreated control (***, p < 0.001; **, p < 0.01; *,p < 0.05, ANOVA with Dunnett's multiple comparison test).
CD157 Binding to FN Is Prevented by Heparin
The N-terminal HBD1 consists of five type I (FN1) homologous repeats that form two distinct regions, the N-terminal FN1(1–3) region containing the binding site for heparin and the FN1(4–5) region that includes the binding site for fibrin (25) (Fig. 1D). To map more precisely the binding site of rh-sCD157 within HBD1, competition experiments were carried out using solid-phase binding assays in the presence of excess amounts of either heparin or fibrin. Binding of rh-sCD157 to insolubilized full-length FN and HBD1 was prevented by addition of heparin but not of fibrin, indicating that the binding site for CD157 is within the FN1(1–3) region, close to or overlapping with that of heparin (Fig. 2, A and B). Next, we assessed the ability of heparin to interfere with the interaction between rh-sCD157 and FN, HBD1, or HBD2 by performing binding assays in the presence of increasing concentrations of heparin (range, 0.6 to 150 μg/ml). The presence of heparin inhibited the binding of rh-sCD157 to full-length FN (Fig. 2C) and to both its heparin-binding domains (HBD1 and HBD2) in a dose-dependent manner (Fig. 2, D and E). Notably, 1.8 μg/ml heparin significantly inhibited and 50 μg/ml heparin completely prevented rh-sCD157 binding to HBD1. At least 5.5 μg/ml heparin were required to significantly inhibit the binding of rh-sCD157 to HBD2. These results suggest that there could be a second CD157-binding site in HBD2 that is not blocked by heparin or that the binding affinity between CD157 and HBD2 is very high (likely higher than that between CD157 and HBD1). Hyaluronic acid, another glycosaminoglycan abundantly expressed in the ECM, did not perturb the binding of rh-sCD157 to FN, HBD1, or HBD2 (Fig. 2, C-E), indicating that the inhibitory effect of heparin is specific.
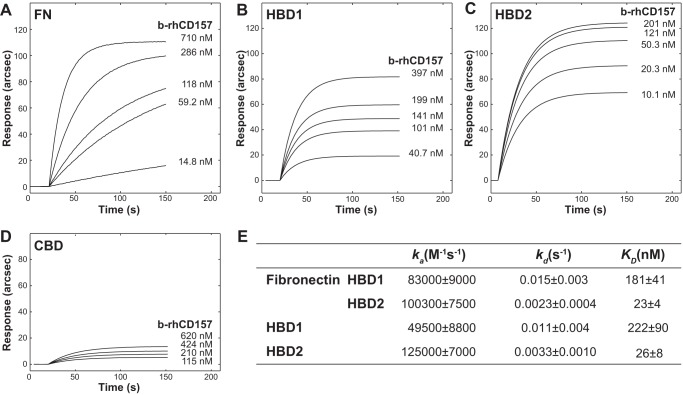
Analysis of CD157-Fibronectin Interaction by Surface Plasmon Resonance
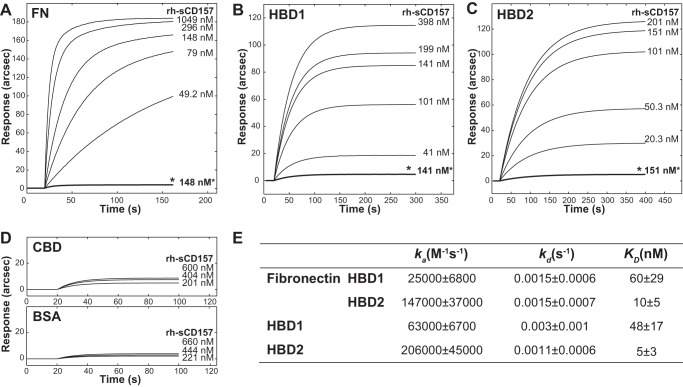
The interactions between rh-sCD157 and FN were further characterized by SPR analysis allowing real time monitoring of binding kinetics. Sensing surfaces containing either full-length FN or its proteolytic fragments were obtained as described under “Experimental Procedures.” From the readout in the range 800–1400 arcsec, we calculated surface densities of ∼2.2 and 1.2 ng mm−2 for full-length FN and for single domains, respectively; these conditions globally resulted in the coupling of a “Langmuir” partial monolayer of capturing macromolecules. These protein immobilization levels (∼3/5 of the monolayer) were adequate to minimize possible steric hindrance or shielding effects, which could affect the analysis. For each sensing surface, binding experiments were carried out at increasing concentrations of rh-sCD157 (range, 20.3 nm to 1.049 μm, corresponding to 710 ng/ml to 52.5 μg/ml), as indicated.
First, the interaction between rh-sCD157 and full-length fibronectin was analyzed, and biphasic association time courses were observed (Fig. 3A); this behavior was ascribed to the ability of rh-sCD157 to bind two different sites on FN with comparably high affinity. Measured dissociation equilibrium constants obtained for this interaction were KD, 1 = 60 ± 29 nm and KD, 2 = 10 ± 5 nm (where KD indicates the equilibrium dissociation constant), which could not be associated with any precise binding domain at this stage, although it was plausible that the two binding sites were HBD1 and HBD2. To substantiate this assumption, binding experiments were performed after saturating surface-bound FN with 1 mm heparin; this presaturation step prevented any significant binding of rh-sCD157 (Fig. 3A, *). To further corroborate our evidence, CD157 binding experiments were replicated independently on surface-bound HBD1 and HBD2. In both cases, monophasic time course sensorgrams were observed (Fig. 3, B and C), with KD, HBD1 and KD, HBD2 being 48 ± 17 and 5 ± 3 nm, respectively (Fig. 3E). Presaturation of both HBDs with heparin hindered any binding of rh-sCD157 (Fig. 3, B, *, and C, *), ruling out the existence of a second CD157-binding site in HBD2 not blocked by heparin. It is noteworthy that rh-sCD157 was shown to target HBDs both in full-length and fragmented FN with comparable affinity, confirming that no other FN regions were significantly involved in the binding events. Finally, the CBD fragment was immobilized and used as a negative control for binding; negligible responses were obtained upon addition of rh-sCD157 (0.2–0.6 μm range), as inferred from the affinity of the complex, which was more than 2000-fold lower with respect to HBD-CD157 complexes and comparable with that of BSA-CD157 complexes (Fig. 3D).
FIGURE 3.
Analysis of binding of rh-sCD157 to immobilized fibronectin and its proteolytic fragments by surface plasmon resonance. IAsys binding curves for the interaction of rh-sCD157 with full-length FN, HBD1, and HBD2 immobilized to the surface of a carboxymethyl dextran sensor. Increasing concentrations of rh-sCD157 were added to immobilized FN (A), HBD1 (B), or HBD2 (C), and their association was monitored over time. Binding of rh-sCD157 to FN, HBD1, and HBD2 was blocked by presaturating each protein with 1 mm heparin (*). D, sensorgrams obtained by probing immobilized CBD (top panel) or BSA (bottom panel) with rh-sCD157 at the indicated molarities. E, table of binding kinetics values (ka, kd, and KD reported as the means ± S.E.) determined from the experimental curve fits.
Next, we determined the kinetic parameters of rh-sCD157 binding to FN. The calculation of association (ka) and dissociation (kd) rate constants further dissected the mechanistic properties of their macromolecular recognition process. The interaction between rh-sCD157 and FN was characterized by preferential association kinetics to HBD2; indeed, the association rate at the HBD2 site was 6-fold higher compared with the association at HBD1. In contrast, the dissociation kinetics were comparable. Therefore, the difference in terms of KD (FN-HBD1 = 60 ± 29 nm versus FN-HBD2 = 10 ± 5 nm) could be uniquely attributed to the association phase. This behavior was confirmed with isolated HBDs, with major differences existing only in ka values (Fig. 3E). It is important to note that differences in association kinetics were observed between HBD1 in the context of full-length FN and isolated HBD1. These results revealed that neighboring domains in full-length FN hindered recognition of rh-sCD157, an effect that was otherwise hidden in the equilibrium studies.
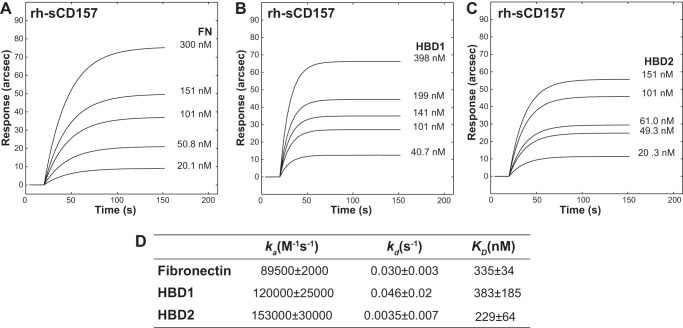
To demonstrate that the FN-CD157 interaction was not influenced by the conformation of FN and HBDs after immobilization, reverse binding experiments were performed on surface-bound rh-sCD157 (Fig. 4, A–C). Kinetic analysis revealed that ka values were fully comparable with those of the original binding experiments, whereas kd values were more than 10-fold higher (Fig. 4D). Based on this evidence, we can reasonably infer that the binding interface of immobilized rh-sCD157 was available for interaction with soluble FN and HBDs and that the destabilization of the complexes mainly relied upon steric hindrance (likely due to the proximity to the surface). This hindering effect was more evident with FN (Fig. 4).
FIGURE 4.
Reverse binding analysis of FN and its proteolytic fragments to immobilized rh-sCD157 by surface plasmon resonance. IAsys-binding curves for the interaction of increasing concentrations of full-length FN (A), HBD1 (B), or HBD2 (C) to rh-sCD157 immobilized to the surface of a carboxymethyl dextran sensor. Their association was monitored over time. D, table of binding kinetics values (ka, kd, and KD reported as the means ± S.E.) determined from the experimental curve fits. A single significant binding event was observable with FN.
Analysis of the Contribution of CD157 Glycosylation to Fibronectin Binding
CD157 is a glycoprotein with four N-glycosylation sites (26). To assess the implications of the glycosidic chains in the CD157-FN interaction, SPR analysis was performed using increasing concentrations of recombinant His-tagged CD157 produced in Escherichia coli (Fig. 5; range 10.1 nm to 0.71 μm, corresponding to 300 ng/ml to 20 μg/ml). These analyses revealed that nonglycosylated b-rhCD157 maintained the ability to selectively bind to HBDs of FN (Fig. 6, A–C) but not to CBD (Fig. 6D). A slight reduction in binding affinity was observed compared with glycosylated rh-sCD157; in particular, the KD was ∼3- and ∼2-fold lower for HDB1 and for HBD2 in full-length FN and ∼5-fold lower for isolated HBDs (Fig. 6E).
FIGURE 5.
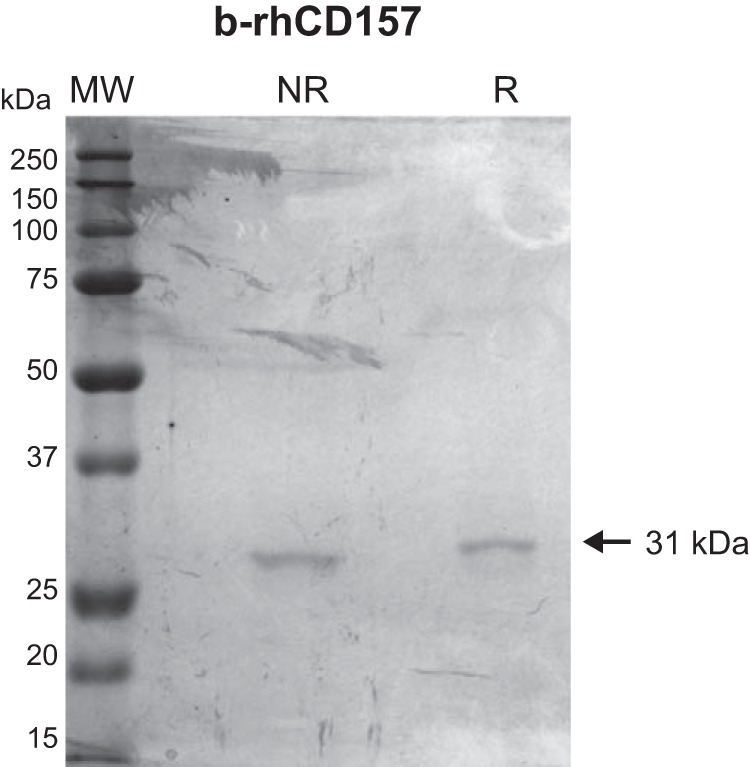
SDS-PAGE of human recombinant sCD157 produced in E. coli. Purified b-rhCD157 produced in E. coli (b-rhCD157, with two N-terminal tags, His tag, and T7 tag) run on a 10% (w/v) polyacrylamide gel and stained with Coomassie Brilliant Blue (CBB) was detected as a single 31-kDa band. MW, molecular weight markers; NR, nonreducing conditions; R, reducing conditions.
FIGURE 6.
Analysis of binding of bacterial rhCD157 to immobilized fibronectin and its proteolytic fragments by surface plasmon resonance. IAsys-binding curves for the interaction of nonglycosylated b-rhCD157 with full-length FN, HBD1, and HBD2 immobilized to the surface of a carboxymethyl dextran sensor. Increasing concentrations of b-rhCD157 were added to immobilized FN (A), HBD1 (B), or HBD2 (C), and their association was monitored over time. D, sensorgrams obtained by probing immobilized CBD with b-rhCD157 at the indicated molarities. E, table of binding kinetic values (ka, kd, and KD reported as the means ± S.E.) determined from the experimental curve fits.
Kinetic analysis indicated that the carbohydrate moieties differentially affect formation and stabilization of the CD157-HBD1 or CD157-HBD2 complexes. Specifically, glycosylation had a modest impact on the recognition process, with a maximal 2-fold increase in kinetic association constants (with the only exception of HBD1 within FN), possibly in virtue of favorable electrostatic interactions across the CD157-HBDs interface (Fig. 6E). Glycosylation did not significantly affect the kinetic dissociation constants of HBD2. The kinetic dissociation constants of HBD1 were up to 10-fold higher in the presence of b-rhCD157 than of rh-sCD157, indicating that the observed lower affinity of nonglycosylated CD157 for HBD1 (both isolated and within FN) is largely due to the significant kinetic destabilization of the complex. These findings suggest that, although not required for CD157 binding, glycosylation contributes to stabilize binding thanks to its dynamic properties and flexibility.
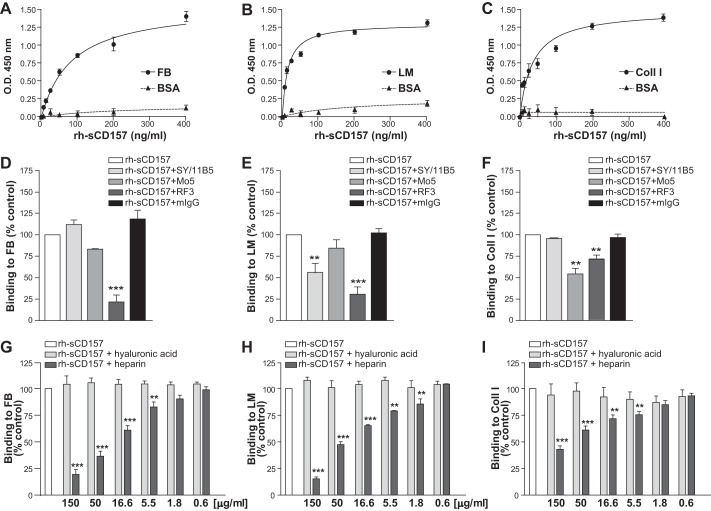
CD157 Binds to Selected ECM Proteins
The ECM is composed of two main classes of macromolecules, fibrous proteins (such as fibronectin, collagen, laminin, among others) and proteoglycans (16), which are proteins covalently attached to one or more glycosaminoglycan chains. Numerous ECM proteins contain heparin-binding domains (27). Therefore, we investigated CD157 binding to heparin-binding domains in ECM proteins other than FN. Solid-phase binding assays were carried out in microtiter plates coated with either FB, Coll I, Vn, or LM, all of which contain at least one HBD. rh-sCD157 bound to immobilized FB, LM, and Coll I but not to Vn or gelatin used as control. No significant binding was observed to polysaccharide components of the ECM (such as heparin and hyaluronic acid) (Fig. 7). Binding of rh-sCD157 to FB, LM, and Coll I was concentration-dependent (Fig. 8, A–C). The specificity of these interactions was strengthened by binding competition experiments performed in the presence of anti-CD157 mAbs. Indeed, addition of the RF3 mAb to the assay consistently inhibited the binding of rh-sCD157 to FB, LM, and Coll I-coated plates. Addition of SY/11B5 or Mo5 mAbs reduced rh-sCD157 binding to LM and Coll I, respectively (Fig. 8, D–F). Moreover, binding of saturating concentrations (100 ng/ml) of rh-sCD157 to FB, LM, and Coll I performed in the presence of increasing concentrations of heparin showed a consistent dose-dependent, heparin-mediated inhibition. No significant interference was detected with hyaluronic acid used as control (Fig. 8, G–I).
FIGURE 7.
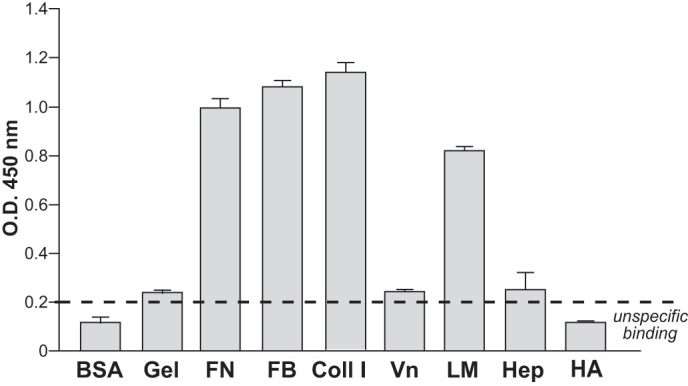
rh-sCD157 binds selected extracellular matrix proteins. Binding of rh-sCD157 (100 ng/ml) to immobilized gelatin, FN, FB, Coll I, Vn, LM, heparin, hyaluronic acid (HA), and BSA (10 μg/ml) was measured by solid-phase binding assays. The mean ± S.D. of one representative experiment performed in triplicate is shown (n = 3). Dashed line indicates significant binding over baseline (2-fold binding to BSA).
FIGURE 8.
Heparin inhibits the binding of rh-sCD157 to fibrinogen, laminin-1, and collagen I. Increasing concentrations of rh-sCD157 (6.25–400 ng/ml) were added to microtiter plates coated with 10 μg/ml FB (A), LM (B), or Coll I (C) (●) or BSA (▴), used as negative control, and incubated for 2 h. The mean ± S.D. of one representative experiment is shown (n = 3). In some cases, the error bars are smaller than the symbols. Lines indicate the nonlinear regression curve fit. D–F, inhibition of rh-sCD157 binding to FB, LM, or Coll I, by anti-CD157 mAbs. The indicated mAb and control murine IgG (10 μg/ml) were incubated with 100 ng/ml rh-sCD157, and then rh-sCD157 binding to matrix was analyzed. Results are expressed as relative ratio to the absorbance of rh-sCD157 in the absence of antibody (100%, white histogram). The influence of heparin on rh-sCD157 binding to FB (G), LM (H), and Coll I (I) was assessed by means of binding competition assays performed in the presence of increasing concentrations (0.6–150 μg/ml) of heparin (dark gray histograms) or hyaluronic acid (light gray histograms), used as control. Results are expressed as relative ratio to the absorbance of rh-sCD157 in the absence of inhibitors (100%, white histogram). The mean ± S.E. of three independent experiments is shown. ***, p < 0.001; **, p < 0.01, ANOVA with Dunnett's multiple comparison test.
These findings indicate that CD157 binds multiple ECM proteins at their heparin-binding sites and rule out the existence of a direct binding between rh-sCD157 and heparin, corroborating the results obtained by SPR. The absence of glycosaminoglycans attached to sCD157 (28) or to the rh-sCD157 used in this study (Fig. 9) allowed us to exclude a contribution of heparan sulfate in mediating the interaction between CD157 and ECM.
FIGURE 9.
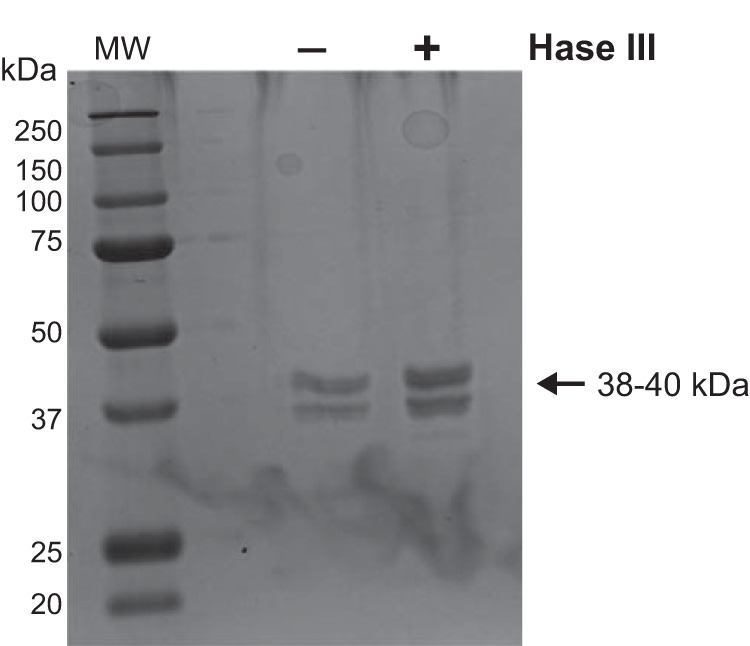
Heparinase treatment of rh-sCD157. Purified rh-sCD157 was treated with heparinase III (Hase III) (2 Sigma units/ml) to remove heparan sulfate (HS) chains potentially decorating the protein. The recombinant protein was run on 10% (w/v) polyacrylamide gel under reducing conditions and stained with Coomassie Brilliant Blue. No shift in molecular weight is appreciable, excluding the presence of heparan sulfate chains. MW, molecular weight markers.
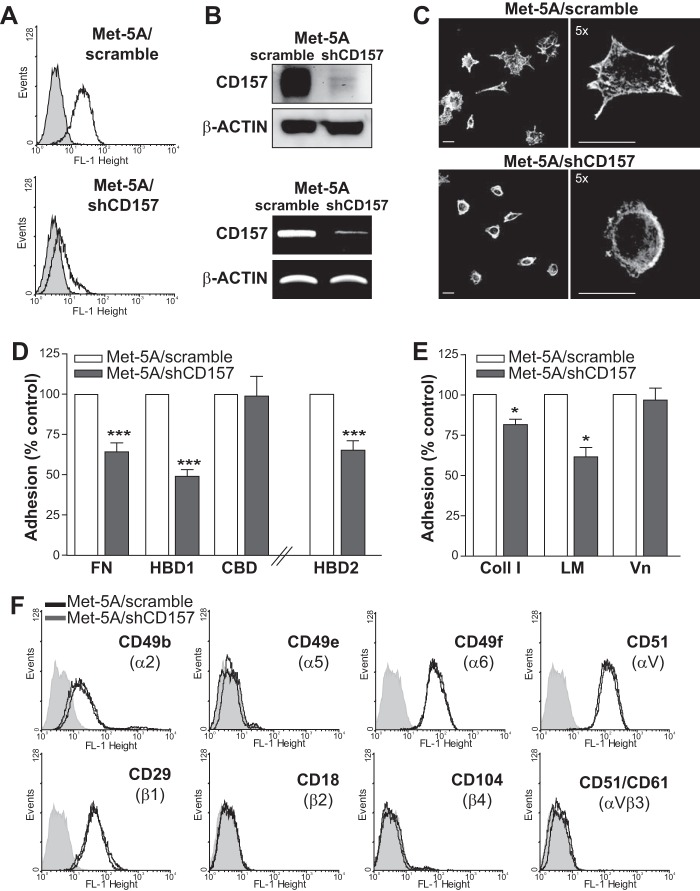
CD157 Regulates Mesothelial Cell Adhesion to Fibronectin
To confirm that the interaction between CD157 and FN observed in vitro using purified proteins was biologically significant in a cellular context, we used Met-5A non-neoplastic human pleural mesothelial cells that express membrane CD157 as an in vitro experimental model. Met-5A cells adhered to both FN and HBD1, although with different efficiencies, and to HBD2 with poor efficiency (Fig. 10A).
FIGURE 10.
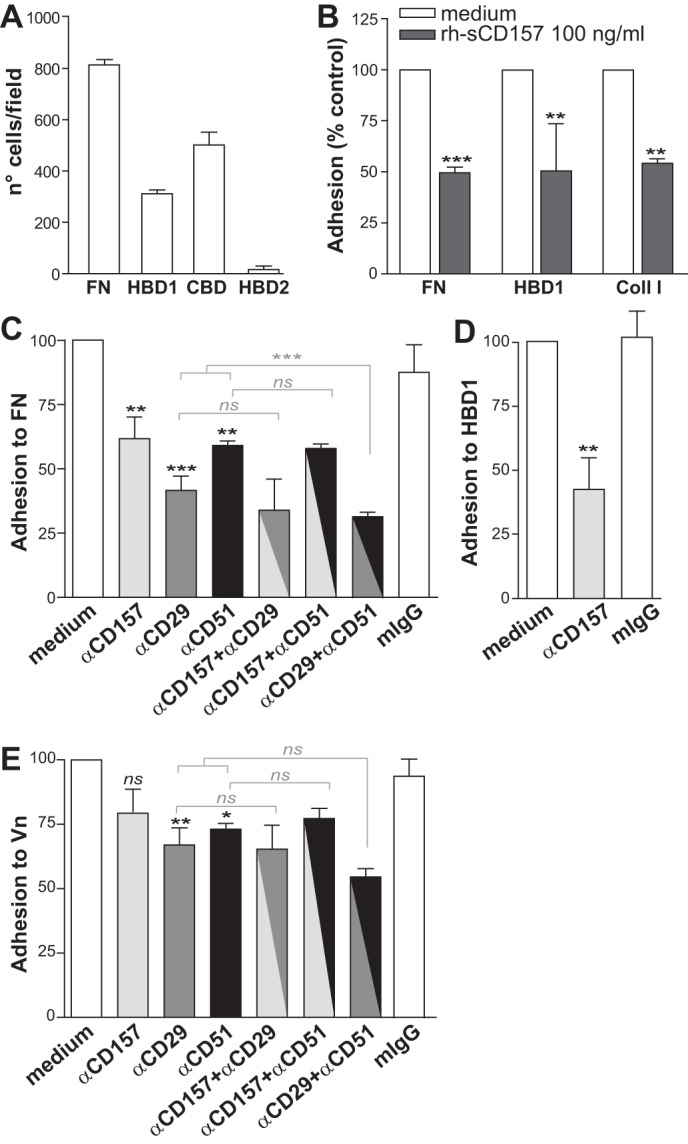
CD157 controls the adhesion of mesothelial cells to extracellular matrix proteins. A, Met-5A cells were plated on full-length fibronectin or on its proteolytic fragments for 2 h. B, ability of membrane CD157 and rh-sCD157 to bind the same sites on FN, HBD1, and Coll I was evaluated by assessing the adhesion efficiency of Met-5A cells to each protein after saturation with rh-sCD157 (100 ng/ml). Results are expressed as percentage of controls (cell adhesion to coated proteins without saturation with rh-sCD157 = 100%, white histogram). ***, p < 0.001; **, p < 0.01, unpaired t test. C–E, ability of membrane CD157 to bind full-length fibronectin, HBD1, and Vn was evaluated in the presence or absence of the RF3 anti-CD157, anti-CD29, or anti-CD51 blocking mAbs (either alone or in combination) or irrelevant murine IgG (10 μg/ml). Results are expressed as percentage of control (number of adherent cells in the absence of antibody = 100%, white histogram). ***, p < 0.001; **, p < 0.01; *, p < 0.05, ns, not significant, ANOVA with Bonferroni's multiple comparison test. Cells were plated for FN, HBD1, Coll, or Vn, and then adherent cells were fixed with 4% PFA, stained with 0.5% crystal violet, and counted by light microscopy in five randomly selected fields (×4) for each well. Results represent the mean values ± S.E. of at least three experiments performed in triplicate.
To investigate whether native membrane-bound CD157 and purified rh-sCD157 recognize the same binding sites, adhesion of Met-5A cells to FN and HBD1 was assessed after saturation of each immobilized protein with rh-sCD157. rh-sCD157 consistently reduced Met-5A cell adhesion to FN and HBD1 and to other ECM proteins containing the binding site for rh-sCD157, such as Coll I (Fig. 10B). This suggested that native membrane-bound CD157 and rh-sCD157 likely share the same binding site(s). The involvement of CD157 in ECM-mediated cell adhesion was confirmed by the ability of RF3 anti-CD157 mAb (but not of isotype-matched mouse IgG) to significantly inhibit Met-5A cell adhesion to FN and HBD1 (Fig. 10, C and D). As expected, cell adhesion to FN was also inhibited by neutralizing antibodies to integrin β1/CD29 and integrin αv/CD51. However, combined block of CD157 and CD29 or CD51 did not further increase inhibition of Met-5A cell adhesion (Fig. 10C). Blocking CD157 did not significantly perturb Met-5A cell adhesion to Vn nor increase the inhibitory effect of neutralizing antibody to β1/CD29 or αv/CD51 integrins (Fig. 10E). To conclusively prove a role of membrane-bound CD157 in the control of cell adhesion to ECM proteins, Met-5A cells were transduced with an shRNA that targeted the CD157 mRNA, resulting in efficient knockdown of CD157 mRNA and protein expression (Met-5A/shCD157), or with a control shRNA (Met-5A/scramble) (Fig. 11, A and B). In cells adhering to fibronectin, loss of CD157 caused changes in F-actin cytoskeleton organization and marked reduction in cell spreading and lamellipodia formation (Fig. 11C). Next, we compared the ability of CD157-positive and CD157-negative Met-5A cells to adhere to FN and to its HDBs and CBD to Coll I, LM, and Vn. Silencing of CD157 resulted in reduced adhesion of Met-5A cells to FN and to HBD1, but it had no appreciable effect on cell adhesion to CBD (mainly mediated through integrin engagement). Adhesion of Met-5A cells to HBD2, although limited (Fig. 10A), was further reduced by the loss of CD157 (Fig. 11D). Impaired adhesion of Met-5A/shCD157 cells compared with control cells was also observed with Coll I and LM but not Vn (Fig. 11E). The reduced adhesive capacity of CD157-negative cells was not attributable to different expression of a set of integrins (Fig. 11F). These findings support the notion that CD157 plays a direct role in cell adhesion to FN, LM, and Coll I but not to Vn.
FIGURE 11.
CD157 silencing reduces spreading and adhesion of mesothelial cells to extracellular matrix proteins. A, FACS analysis of Met-5A cells transduced with a CD157-specific shRNA (bottom panel) or a control shRNA (top panel). Cells were stained with the SY/11B5 anti-CD157 mAb (5 μg/ml) (white peaks) or isotype-matched murine monoclonal IgG (gray peaks) followed by F(ab′)2-GαMIg-FITC. Fluorescence was analyzed using a FACSCalibur flow cytometer and CellQuest software. x axis = fluorescence intensity (FL-1 height), y axis = number of cells (events). B, Western blot analysis (top panel) and RT-PCR (bottom panel) of Met-5A cells transduced with a CD157-specific shRNA. β-Actin was used as loading control. C, CD157 knockdown-induced changes in cell morphology and F-actin cytoskeleton. Met-5A/shCD157 and Met-5A/scrambled cells were plated on FN-coated slides in serum-free medium in the presence of cycloheximide, then fixed with 2% PFA, permeabilized with 0.2% Triton X-100, and stained with TRITC-labeled phalloidin. Samples were analyzed by an Olympus FV300 laser scanning confocal microscope. Cells were imaged using a ×60 oil immersion objective (1.4 NA). Scale bars, 100 μm. Microphotographs were reproduced in black and white. D, comparison of the ability of Met-5A/scramble (white histogram) and Met-5A/shCD157 (gray histogram) cells to adhere to 10 μg/ml of FN, HBD1, and CBD-coated plates or 50 μg/ml HBD2. E, adhesion of Met-5A/scramble and Met-5A/shCD157 to Coll I, LM, or Vn-coated plates. Adherent cells were fixed with 4% PFA, stained with 0.5% crystal violet, and counted by light microscopy in five randomly selected fields (×4) for each well. Results are expressed as percentage of controls (number of adherent Met-5A/scramble cells = 100%, white histogram) and represent the mean ± S.E. of three experiments performed in triplicate. ***, p < 0.001; *, p < 0.05, unpaired t test. F, flow cytometric analysis of selected integrins in Met-5A cells transduced with a CD157-specific shRNA (gray peaks) or a control shRNA (black peaks). Cells were stained with anti-CD49b, CD49e, CD49f, CD51, CD29, CD18, CD104, and CD51/CD61 mAbs (5 μg/ml) or isotype-matched murine monoclonal IgG (shaded peaks) followed by F(ab′)2-GαMIg-FITC. Fluorescence was analyzed using a FACSCalibur flow cytometer and CellQuest software. Ten thousand cells were considered for each analysis. x axis = fluorescence intensity (FL-1 height), and y axis = number of cells (events).
CD157-Fibronectin Interaction Modulates the Transduction of Intracellular Signals
In addition to its activity in promoting cell adhesion and migration (8), CD157 cross-linking by specific mAbs has been shown to elicit a signaling cascade that involves the phosphorylation of FAK (14, 29, 30). Moreover, CD157 is part of the integrin-driven molecular machinery that regulates fundamental functions in neutrophils and monocytes through the activation of a complex signaling network (10, 11). The ability of CD157 to bind FN suggests that this interaction could account for the receptor activity attributed to CD157. Hence, we investigated cell signaling events triggered by FN binding to CD157-positive or CD157-negative Met-5A cells. It is known that integrin binding to FN rapidly activates FAK by phosphorylation of Tyr-397 leading to recruitment of SRC and subsequent activation of the FAK-SRC signaling complex (31). Western blot analysis of cell extracts, obtained after incubating the cells for 1 h on FN, showed reduced FAK phosphorylation at Tyr-397 and an appreciable (although not statistically significant) decrease in phosphorylation of c-SRC in Met-5A/shCD157 cells compared with Met-5A/scramble cells. The absolute amount of FAK or c-SRC proteins was comparable. Moreover, the attenuated phosphorylation of the serine/threonine kinase Akt (a key downstream effector of p110 PI3K) indicated reduced phosphorylation of the downstream PI3K/Akt pathway in Met-5A/shCD157 compared with control cells (Fig. 12, A and B).
FIGURE 12.
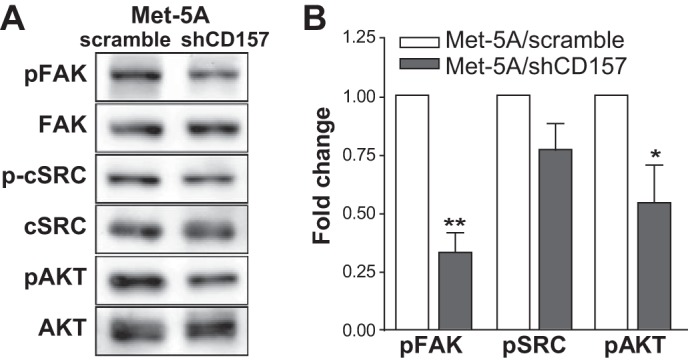
CD157 binding to FN induces FAK and c-SRC activation. A, Met-5A/scramble and Met-5A/shCD157 were starved overnight and plated for 1 h at 37 °C onto FN-coated plates. Adherent cells were lysed, and cleared lysates were subjected to Western blot. Membranes were first probed with anti-phospho-FAK, phospho-c-SRC, or phospho-Akt, stripped, and re-probed with anti-FAK, -c-SRC, or -Akt, followed by incubation with the appropriate HRP-conjugated antibodies. B, phosphorylation of FAK, c-SRC, and Akt in Met-5A/shCD157 compared with Met-5A/scramble was quantified using the software package ImageJ 1.43 (National Institutes of Health). Results are the means ± S.E. of three experiments and are expressed as fold decrease in phosphorylation in Met-5A/shCD157 compared with control cells (Met-5A/scramble, white histograms). **, p < 0.01; *, p < 0.05, unpaired t test.
These results show that engagement of CD157 by fibronectin increases the activation of key adhesion-dependent tyrosine kinases mirroring the effects observed upon CD157 antibody cross-linking (11).
DISCUSSION
Soon after the identification of CD157/BST-1 as a NAD-metabolizing ectoenzyme (EC 3.2.2.5) (32), it became clear that overexpression of CD157 (30) or its clustering by specific monoclonal antibodies (8) induced a variety of cellular responses, including changes in cell adhesion and migration. These effects can be attributed to the ability of CD157 to act as a receptor (29, 33, 34). However, the nature of the physiological interaction(s) implementing these multiple biological activities remained an unresolved issue. The identification of the interaction between CD157 and ECM achieved in this study thus provides valuable insights into the biological mechanism responsible for the nonenzymatic functions of CD157 in physiological conditions and in specific pathological contexts.
Using solid-phase immunoenzymatic and SPR assays, we demonstrated that purified CD157 specifically binds the extracellular protein fibronectin at its N-terminal (HBD1) and C-terminal (HBD2) heparin-binding domains. The equilibrium dissociation constants (KD) in the nanomolar range highlight the fact that the interaction between CD157 and FN at both HBD1 and HBD2 is very strong (far stronger than that of heparin (35)) and that CD157 binds to HBD2 with a KD value ∼6-fold higher than to HBD1. The interaction between CD157 and the HBDs is mediated by the protein core, although the glycosidic chains contribute to stabilize the interaction.
In addition to FN, CD157 bound Coll I, FB, and LM. This binding was consistently and specifically inhibited by heparin and by selected CD157-specific mAbs. However, the differential ability of each mAb to inhibit CD157 binding to discrete ECM proteins observed in this study cannot be considered a general rule and cannot be extrapolated to different experimental contexts. Indeed, antibodies apparently unable to inhibit the interaction between purified CD157 and ECM proteins (for example, the SY/11B5 mAb), effectively interfered with adhesion of CD157-positive normal or neoplastic cells to fibronectin, laminin-1, and collagen I (11, 12), a situation that more faithfully recapitulates the complexity of cell-ECM interaction occurring in vivo.
The lack of CD157 binding to vitronectin indicates that the presence of a heparin-binding domain within an ECM protein is not sufficient to warrant CD157 binding, at least in vitro. Despite some common characteristics, HBDs differ widely in structure, and it is conceivable that the CD157-binding site fits some HBDs but not all. The specific amino acid sequences or spatial arrangement of amino acids within the HBDs critical for interaction with CD157 remain to be defined and deserve further investigation.
Binding competition experiments have enabled us to further narrow the binding sites, highlighting that CD157 interacts with the N-terminal FN1(1–3) motifs comprising the low affinity binding site for heparin, and with the C-terminal type III(13–14) motifs, which accounts for nearly all the heparin binding activity (36, 37). The interaction between CD157 and HBDs of fibronectin that occurs in vitro using purified proteins reflects physiological interactions driving specific biological functions in live cells. Indeed, membrane CD157 expressed by Met-5A mesothelial cells interacts with FN, and anti-CD157 mAbs significantly reduce cell binding to FN and its HBD1, confirming previous observations (11, 12). The ability of rh-sCD157 to significantly reduce the binding of Met-5A cells to FN suggests that native membrane-anchored and recombinant soluble CD157 share the same binding sites. However, we cannot exclude that rh-sCD157 affects cell adhesion by interfering with the binding of other cell surface proteins (such as syndecans) to the heparin-binding domains.
The functional relevance of a direct molecular interaction between CD157 and HBDs has been further substantiated by the observation that loss of CD157 expression in Met-5A cells reduced cell spreading and lamellipodia formation and altered the organization of the matrix-proximal F-actin cytoskeleton. These changes impaired cell adhesion to FN, Coll I, and LM, but not to CBD of FN or to Vn, in Met-5A/shCD157 compared with Met-5A/scramble cells. This result conclusively demonstrates that CD157 contributes to cell adhesion through its interaction with the HBDs located within selected ECM proteins. As expected, CD157 knockdown did not completely prevent cell adhesion to ECM proteins because of the existence of compensatory molecular interactions that dampen the effects of CD157 deficiency. Notably, the opposite effects were observed in ovarian cancer cells in which exogenous expression of CD157 resulted in increased cell spreading and cytoskeletal organization, leading to improved tumor cell motility and dissemination (15).
It has been documented that, despite being a glycosylphosphatidylinositol-anchored protein, CD157 acts as a receptor because it is part of a multimolecular complex ruled by integrins (10, 11). However, how CD157 became part of this molecular complex in physiological and pathological situations was unclear. The results of this study fill this gap, suggesting that fibronectin can drive the formation of this complex. FN (like other ECM molecules), by virtue of its ordered domain organization and its simultaneous interaction with a variety of proteins (including integrins and CD157), can promote the re-organization of multimolecular complexes in the plane of the membrane, thus driving orderly juxtaposition of different receptors that orchestrate and integrate the intracellular signaling network that determine cell fate.
CD157 (like many other glycosylphosphatidylinositol-anchored proteins (38)) is associated with membrane microdomains known as lipid rafts (11); therefore, the concomitant engagement of CD157 and integrin receptors by FN will recruit integrins inside lipid rafts and bring them in close contact with CD157. Lipid rafts are enriched in signal transduction molecules (39); hence, the CD157-FN interaction is instrumental to the delivery of synergistic signals leading to optimal cell adhesion and migration. Consistent with this assumption, knockdown of CD157 in Met-5A cells remarkably reduced the induction of fibronectin-mediated phosphorylation of FAK, SRC, and Akt tyrosine kinases and eventually led to impaired cell adhesion and spreading.
It is tempting to predict that the broad interaction of CD157 with several ECM proteins may be responsible for many of the biological effects exerted by CD157 in different physiological (e.g. leukocyte trafficking) and pathological contexts (e.g. inflammatory diseases and cancer), where the composition of the extracellular matrix dictates the final outcome. For example, in ovarian cancer, the CD157-ECM interaction occurring in the peritoneal cavity rescues cells from anoikis and promotes their migration and metastatic dissemination, ultimately enhancing tumor aggressiveness (15).
Accumulating evidence implicates tumor cell-ECM interaction in many aspects of tumor biology, including tumor growth and metastatic spread (40, 41). Hence, it is reasonable to envision that hindering the interaction between CD157 and fibronectin (or other ECM proteins depending on a specific microenvironment) may represent a promising new avenue for the design of therapeutic strategies to treat inflammation and ovarian cancer.
Acknowledgments
We thank Dr. Elisa Vigna (Istituto per la Ricerca e la Cura del Cancro, Candiolo, Torino, Italy) for providing the PMDLg/pRRE, p-RSV-Rev, and pMd2-VSVG lentiviral packaging plasmids. We are greatly indebted to Enza Ferrero (University of Torino, Italy) for help in stylistic assembly of the article.
This work was supported by Italian Association for Cancer Research Grant IG 11602 (to E. O.), Italian Ministry for University and Scientific Research 60% Projects (to A. F.), and The International Foundation for Research in Experimental Medicine.
- ECM
- extracellular matrix
- SPR
- surface plasmon resonance
- FN
- fibronectin
- HBD
- heparin-binding domain
- GBD
- gelatin-binding domain
- CBD
- cell-binding domain
- LM
- laminin-1
- Coll I
- collagen type I
- Vn
- vitronectin
- FB
- fibrinogen
- TRITC
- tetramethylrhodamine isothiocyanate
- rh-sCD157
- recombinant His-tagged human soluble CD157
- rh-PDGFRα
- recombinant human PDGF receptor α
- b-rhCD157
- bacterial recombinant His-tagged human CD157
- FAK
- focal adhesion kinase
- PFA
- paraformaldehyde
- ANOVA
- analysis of variance.
REFERENCES
- 1. Goding J. W. (2000) Ecto-enzymes: physiology meets pathology. J. Leukocyte Biol. 67, 285–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salmi M., Jalkanen S. (2012) Ectoenzymes controlling leukocyte traffic. Eur. J. Immunol. 42, 284–292 [DOI] [PubMed] [Google Scholar]
- 3. Malavasi F., Deaglio S., Funaro A., Ferrero E., Horenstein A. L., Ortolan E., Vaisitti T., Aydin S. (2008) Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol. Rev. 88, 841–886 [DOI] [PubMed] [Google Scholar]
- 4. Ortolan E., Vacca P., Capobianco A., Armando E., Crivellin F., Horenstein A., Malavasi F. (2002) CD157, the Janus of CD38 but with a unique personality. Cell Biochem. Funct. 20, 309–322 [DOI] [PubMed] [Google Scholar]
- 5. Quarona V., Zaccarello G., Chillemi A., Brunetti E., Singh V. K., Ferrero E., Funaro A., Horenstein A. L., Malavasi F. (2013) CD38 and CD157: A long journey from activation markers to multifunctional molecules. Cytometry B Clin. Cytom. 84, 207–217 [DOI] [PubMed] [Google Scholar]
- 6. Todd R. F., 3rd, Roach J. A., Arnaout M. A. (1985) The modulated expression of Mo5, a human myelomonocytic plasma membrane antigen. Blood 65, 964–973 [PubMed] [Google Scholar]
- 7. Todd R. F., 3rd (1995) in Leukocyte Typing V (Schlossman S. F., Gilks W., eds) pp. 991–1093, Oxford University Press, New York [Google Scholar]
- 8. Funaro A., Ortolan E., Ferranti B., Gargiulo L., Notaro R., Luzzatto L., Malavasi F. (2004) CD157 is an important mediator of neutrophil adhesion and migration. Blood 104, 4269–4278 [DOI] [PubMed] [Google Scholar]
- 9. Ortolan E., Tibaldi E. V., Ferranti B., Lavagno L., Garbarino G., Notaro R., Luzzatto L., Malavasi F., Funaro A. (2006) CD157 plays a pivotal role in neutrophil transendothelial migration. Blood 108, 4214–4222 [DOI] [PubMed] [Google Scholar]
- 10. Lavagno L., Ferrero E., Ortolan E., Malavasi F., Funaro A. (2007) CD157 is part of a supramolecular complex with CD11b/CD18 on the human neutrophil cell surface. J. Biol. Regul. Homeost. Agents 21, 5–11 [PubMed] [Google Scholar]
- 11. Lo Buono N., Parrotta R., Morone S., Bovino P., Nacci G., Ortolan E., Horenstein A. L., Inzhutova A., Ferrero E., Funaro A. (2011) The CD157-integrin partnership controls transendothelial migration and adhesion of human monocytes. J. Biol. Chem. 286, 18681–18691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ortolan E., Arisio R., Morone S., Bovino P., Lo-Buono N., Nacci G., Parrotta R., Katsaros D., Rapa I., Migliaretti G., Ferrero E., Volante M., Funaro A. (2010) Functional role and prognostic significance of CD157 in ovarian carcinoma. J. Natl. Cancer Inst. 102, 1160–1177 [DOI] [PubMed] [Google Scholar]
- 13. Kaisho T., Ishikawa J., Oritani K., Inazawa J., Tomizawa H., Muraoka O., Ochi T., Hirano T. (1994) BST-1, a surface molecule of bone marrow stromal cell lines that facilitates pre-B-cell growth. Proc. Natl. Acad. Sci. U.S.A. 91, 5325–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hussain A. M., Chang C. F. (1999) Novel kinetics, behaviour and cell-type specificity of CD157-mediated tyrosine kinase signalling. Cell. Signal. 11, 891–897 [DOI] [PubMed] [Google Scholar]
- 15. Morone S., Lo-Buono N., Parrotta R., Giacomino A., Nacci G., Brusco A., Larionov A., Ostano P., Mello-Grand M., Chiorino G., Ortolan E., Funaro A. (2012) Overexpression of CD157 contributes to epithelial ovarian cancer progression by promoting mesenchymal differentiation. PLoS ONE 7, e43649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frantz C., Stewart K. M., Weaver V. M. (2010) The extracellular matrix at a glance. J. Cell Sci. 123, 4195–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nelson C. M., Bissell M. J. (2006) Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 22, 287–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hynes R. O. (2009) The extracellular matrix: not just pretty fibrils. Science 326, 1216–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morgan M. R., Humphries M. J., Bass M. D. (2007) Synergistic control of cell adhesion by integrins and syndecans. Nat. Rev. Mol. Cell Biol. 8, 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berrier A. L., Yamada K. M. (2007) Cell-matrix adhesion. J. Cell Physiol. 213, 565–573 [DOI] [PubMed] [Google Scholar]
- 21. Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D., Naldini L. (1998) A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Potts J. R., Campbell I. D. (1996) Structure and function of fibronectin modules. Matrix Biol. 15, 313–320 [DOI] [PubMed] [Google Scholar]
- 23. Pankov R., Yamada K. M. (2002) Fibronectin at a glance. J. Cell Sci. 115, 3861–3863 [DOI] [PubMed] [Google Scholar]
- 24. Martino M. M., Hubbell J. A. (2010) The 12th–14th type III repeats of fibronectin function as a highly promiscuous growth factor-binding domain. FASEB J. 24, 4711–4721 [DOI] [PubMed] [Google Scholar]
- 25. Potts J. R., Campbell I. D. (1994) Fibronectin structure and assembly. Curr. Opin. Cell Biol. 6, 648–655 [DOI] [PubMed] [Google Scholar]
- 26. Yamamoto-Katayama S., Sato A., Ariyoshi M., Suyama M., Ishihara K., Hirano T., Nakamura H., Morikawa K., Jingami H. (2001) Site-directed removal of N-glycosylation sites in BST-1/CD157: effects on molecular and functional heterogeneity. Biochem. J. 357, 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leahy D. J., Hendrickson W. A., Aukhil I., Erickson H. P. (1992) Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science 258, 987–991 [DOI] [PubMed] [Google Scholar]
- 28. Yamamoto-Katayama S., Ariyoshi M., Ishihara K., Hirano T., Jingami H., Morikawa K. (2002) Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities. J. Mol. Biol. 316, 711–723 [DOI] [PubMed] [Google Scholar]
- 29. Okuyama Y., Ishihara K., Kimura N., Hirata Y., Sato K., Itoh M., Ok L. B., Hirano T. (1996) Human BST-1 expressed on myeloid cells functions as a receptor molecule. Biochem. Biophys. Res. Commun. 228, 838–845 [DOI] [PubMed] [Google Scholar]
- 30. Liang F., Qi R. Z., Chang C. F. (2001) Signalling of GPI-anchored CD157 via focal adhesion kinase in MCA102 fibroblasts. FEBS Lett. 506, 207–210 [DOI] [PubMed] [Google Scholar]
- 31. Schlaepfer D. D., Broome M. A., Hunter T. (1997) Fibronectin-stimulated signaling from a focal adhesion kinase-c-Src complex: involvement of the Grb2, p130cas, and Nck adaptor proteins. Mol. Cell. Biol. 17, 1702–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hirata Y., Kimura N., Sato K., Ohsugi Y., Takasawa S., Okamoto H., Ishikawa J., Kaisho T., Ishihara K., Hirano T. (1994) ADP ribosyl cyclase activity of a novel bone marrow stromal cell surface molecule, BST-1. FEBS Lett. 356, 244–248 [DOI] [PubMed] [Google Scholar]
- 33. Malavasi F., Deaglio S., Ferrero E., Funaro A., Sancho J., Ausiello C. M., Ortolan E., Vaisitti T., Zubiaur M., Fedele G., Aydin S., Tibaldi E. V., Durelli I., Lusso R., Cozno F., Horenstein A. L. (2006) CD38 and CD157 as receptors of the immune system: a bridge between innate and adaptive immunity. Mol. Med. 12, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Funaro A., Ortolan E., Bovino P., Lo Buono N., Nacci G., Parrotta R., Ferrero E., Malavasi F. (2009) Ectoenzymes and innate immunity: the role of human CD157 in leukocyte trafficking. Front. Biosci. 14, 929–943 [DOI] [PubMed] [Google Scholar]
- 35. Ingham K. C., Brew S. A., Atha D. H. (1990) Interaction of heparin with fibronectin and isolated fibronectin domains. Biochem. J. 272, 605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barkalow F. J., Schwarzbauer J. E. (1991) Localization of the major heparin-binding site in fibronectin. J. Biol. Chem. 266, 7812–7818 [PubMed] [Google Scholar]
- 37. Sachchidanand, Lequin O., Staunton D., Mulloy B., Forster M. J., Yoshida K., Campbell I. D. (2002) Mapping the heparin-binding site on the 13–14F3 fragment of fibronectin. J. Biol. Chem. 277, 50629–50635 [DOI] [PubMed] [Google Scholar]
- 38. Mayor S., Riezman H. (2004) Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5, 110–120 [DOI] [PubMed] [Google Scholar]
- 39. Simons K., Toomre D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 40. Barkan D., Green J. E., Chambers A. F. (2010) Extracellular matrix: a gatekeeper in the transition from dormancy to metastatic growth. Eur. J. Cancer 46, 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pirazzoli V., Ferraris G. M., Sidenius N. (2013) Direct evidence of the importance of vitronectin and its interaction with the urokinase receptor in tumor growth. Blood 121, 2316–2323 [DOI] [PubMed] [Google Scholar]