Abstract
Autoimmune diseases are often intractable because their causes are unknown. Identifying which genes contribute to these diseases may allow us to understand the pathogenesis, but it is difficult to determine which genes contribute to disease. Recently, epigenetic information has been considered to activate/deactivate disease-related genes. Thus, it may also be useful to study epigenetic information that differs between healthy controls and patients with autoimmune disease. Among several types of epigenetic information, promoter methylation is believed to be one of the most important factors. Here, we propose that principal component analysis is useful to identify specific gene promoters that are differently methylated between the normal healthy controls and patients with autoimmune disease. Full Automatic Modeling System (FAMS) was used to predict the three-dimensional structures of selected proteins and successfully inferred relatively confident structures. Several possibilities of the application to the drug discovery based on obtained structures are discussed.
Keywords: Autoimmune disease, drug discovery, FAMS, principal component analysis, promoter methylation.
INTRODUCTION
Autoimmune disease is defined as "a clinical syndrome caused by the activation of T cells or B cells, or both, in the absence of an ongoing infection or other discernible cause" [1]. As this robust definition suggests, the causes of autoimmune diseases are not generally known. However, the genetic background of patients is generally believed to be important [2].
Furthermore, since the concordance rate between monozygotic (MZ) twins is not always high [3], some genetic mechanisms other than simple coincidence between DNA sequences have been sought. Recently, epigenetics was regarded to be a key factor in understanding the regulation of autoimmune diseases [3, 4]. Among these mechanisms, promoter methylation is regarded as the most critical factor [5]. Although studies have suggested that common genes underlie the pathogenesis of multiple autoimmune disorders [2], there have been no reports of common methylation patterns in more than one autoimmune disease.
Recently Javierre et al. [6] investigated white blood cells extracted from the patients of three autoimmune diseases, systemic lupus erythematosus (SLE), rheumatoid arthritis (RA) and dermatomyositis (DM). However, disease-specific promoter methylation was only identified in SLE [6]. Thus, it appears there are no common-methylated promoters between these three autoimmune diseases. In this paper, we reanalyzed Javierre et al. [6]’s methylation data of autoimmune diseases and found disease-specific promoter methylation in RA and DM. We also identified a common methylation pattern in the three autoimmune diseases. The selected genes were evaluated based on their protein structure prediction by FAMS [7, 8]. The selected genes were also compared with previous studies regarding the individual gene. This study investigated the relationship between the selected genes and the proteins commonly expressed in more than one autoimmune disease [9]. The results presented here strongly suggest the validity of our findings. The possibility of using this application for drug discovery based upon FAMS will also be discussed.
MATERIALS AND METHODS
Promoter Methylation Profile and Principal Component Analysis (PCA) Based Selection of Genes
The supplementary file Supp_Data_2.xls, which can be downloaded from the journal website [6], was used for promoter methylation profile. A tab-limited csv file was generated and was loaded to R [10] by read.csv function. All 59 samples were employed for further analysis (Table 1).
Table 1.
Information of samples used. The numbers in parentheses indicate age. M and F in the column identified as gender are male and female, respectively. Samples annotated as SLE, RA and DM are twins with disease. Those identified as Healthy correspond to healthy twins. Control 1 and Control 2 are age and gender matched controls.
| ID | gender | ||||
|---|---|---|---|---|---|
| SLE | |||||
| 34 | F | SLE (34) | Healthy (34) | Control1 (33) | Control2 (36) |
| 20.1 | F | SLE (20) | Healthy (20) | Control1 (23) | Control2 (22) |
| 11 | F | SLE (12) | Healthy (12) | Control1 (10) | — |
| 29 | F | SLE (29) | Healthy (29) | Control1 (24) | Control2 (31) |
| 20.2 | F | SLE (20) | Healthy (20) | Control1 (22) | Control2 (22) |
| RA | |||||
| 4 | F | RA (43) | Healthy (43) | Control1 (41) | Control2 (48) |
| 12 | M | RA (9) | Healthy (9) | Control1 (9) | Control2 (7) |
| 80 | M | RA (12) | Healthy (12) | Control1 (14) | Control2 (12) |
| 85 | F | RA (18) | Healthy (18) | Control1 (21) | Control2 (24) |
| 86 | F | RA (6) | Healthy (6) | Control1 (6) | Control2 (10) |
| DM | |||||
| 16 | M | DM (34) | Healthy (34) | Control1 (33) | Control2 (34) |
| 33 | M | DM (13) | Healthy (13) | Control1 (11) | Control2 (17) |
| 103 | F | DM (3) | Healthy (3) | Control1 (9) | Control2 (9) |
| 127 | F | DM (11) | Healthy (11) | Control1 (8) | Control2 (12) |
| 129 | F | DM (5) | Healthy (5) | Control1 (9) | Control2 (8) |
PCA is a kind of ordination method. In ordination method, objects placed in the high dimensional space is aligned onto the newly generated lower dimensional space. In PCA, projection to the lower dimensional space is performed by simple linear transformation. PCA was applied to promoter methylation profiles. Principal component (PC) scores were attributed to probes that expressed the gene promoters. PCA was also applied to promoter methylation profiles subdivided into sample sets, which corresponded with each disease, SLE, RA and DM.
Probes (promoters of genes) that were outliners along either the second or the third PC scores were selected as being significantly and differently methylated between the normal control and disease groups. The selection of PCs depended upon which axis represented the difference between disease and normal controls. Further details about thresholds and criteria for the selection can be found in Supporting Information.
t Test for Different Methylation of Probes
t test was used to determine whether a diseased or healthy twin had differently methylated promoters for the selected probes. The algorithm used was as follows: N is the number of probes and xijk is the amount of methylation of the ith promoter for the jth individual who belongs to the kth subgroup (k corresponds to disease, either SLE, RA, or DM). Here, j is either 1 (disease twin) or 0 (healthy twin). Then we defined the amount of differential methylation ∆xik between the diseased twin and the healthy twin as
∆xik ≡ xi1k − xi0k .
When we selected probes i' ∈ S’ as being differently methylated and i’' ∈ S’’ as not, P values were determined by the comparison between the probes in S’ and the probes in S’’ by t test.
Protein Structure Prediction and Protein Complex Inference Based on FAMS
Selected genes’ amino acid sequences were downloaded from SWISS Prot. Then their three-dimensional protein structures were inferred by FAMS [7, 8].
Whether a pair of model proteins used for structure prediction could form protein complexes or not was determined as follows. First, Protein Data Bank (PDB) files that contained at least one model protein as a member of a protein complex were downloaded. Then, the model proteins that were included in the common PDB files were investigated. Next, inter-atomic distances between pairs of model proteins that belonged to the same PDB file were computed. If there was at least one pair of atoms whose distances were less than 3.5 Å, the pair of model proteins was listed as a candidate to form a protein complex.
“in silico” Ligand Screening
First, amino acid sequences registered in PDB with more than 95 % sequence similarity with a reference protein were listed. Among those PDB structures, those with more than 30 heavy atoms and without any atoms within Van der Waals radius were selected. Then we analyzed PDB files that had at least one ligand binding. After removing ligands from these selected PDB files, chooseLD [11] was computed with alternative ligand candidates. Then, FingerPrint Alignment scores (FPAscores) were used to measure ligand candidates.
Validation of “in silico” Ligand Screening with ChEMBL Database
To validate the results of “in silico” ligand screening, we used an assay where ligand binding to the reference protein was investigated in ChEMBL [12]. Then Ki values of ligands were obtained from ChEMBL [12]. ChEMBL is an Open Data database containing binding, functional and “Absorption, Distribution, Metabolism, Excretion, Toxicity” (ADMET) information for a large number of drug-like bioactive compounds. Ki is the inhibition constant and represents the ability to reduce the function of a protein. The correlation coefficients between Kis and FPAScore were used as a measure to represent the accuracy of “in silico” ligand screening.
RESULTS
PCA Application for Whole Samples
First, PCA was applied to all 59 samples (Fig. S1 (1.6MB, pdf) ). It produced a one-dimensional structure and a barb-like structure was observed that extended to the right end of the one-dimensional structure towards the middle-bottom. To understand what this barb-like structure means, we drew the contribution of each sample to PCs (Fig. S2 (1.6MB, pdf) ). In contrast to the first PC, which had almost constant contributions from each sample (Fig. S2 (1.6MB, pdf) (a (1.6MB, pdf) )), the second PC had contributions with opposite signs, dependent upon the gender of each sample (Fig. S2 (1.6MB, pdf) (b (1.6MB, pdf) )). Thus, it is probable that the barb-like structure expresses differences between genders. This is likely, since genes located on the X chromosome are concentrated in this barb-like structure (red dots in Fig. S1 (1.6MB, pdf) (a (1.6MB, pdf) )). Once genes located on the X chromosome are removed, the barb-like structure disappears (Fig. S1 (1.6MB, pdf) (b (1.6MB, pdf) )).
PCA Application to SLE Samples
To select genes differently expressed between normal controls and SLE patients, we applied PCA to SLE samples (IDs 34, 20.1, 11, 29 and 20.2 in Table 1). Figure S3 (1.6MB, pdf) (b (1.6MB, pdf) ) shows two-dimensional embeddings of probes spanning the second and third PC scores. Since the first PC did not represent a difference among probes but had an almost constant value for all probes, we hereafter excluded the first PC scores from further analysis. Although it is not as clear, a bump-like structure expands from the origin toward the negative direction of the second PC scores. To understand the biological meaning of this bump-like structure, we drew the contribution of each sample to the second PC (Fig. S4 (1.6MB, pdf) ). SLE samples were subdivided into five subgroups, each of which consisted of twins and age/gender-matched controls. Disease twins (○) always had the largest second PC scores within each subgroup (Fig. S4 (1.6MB, pdf) (a (1.6MB, pdf) )). Thus, the second PC scores were expected to express any differences in gene promoter methylation between diseased twins and controls.
To understand the differences, we selected 58 probes located in the outer region along the second PC (red dots in Fig. S3 (1.6MB, pdf) ), and applied the t test to determine whether methylation was distinct between probes in the outer regions and others (Fig. S5 (1.6MB, pdf) ). For all five subgroups, the selected probes (red) were significantly demethylated in diseases from other probes (black). This supports the general belief that genes that cause autoimmune diseases should be overexpressed, because methylation is believed to suppress gene expression. Thus, PCA-based probe selection correctly selected the differently expressed probes.
PCA Application to RA Samples
We repeated the same procedure for 20 RA samples (IDs 4, 12, 80, 85 and 86 in Table 1). The results are shown in (Figs. S6 (1.6MB, pdf) , S7 (1.6MB, pdf) and S8 (1.6MB, pdf) ). Although it is less obvious than for the SLE samples, the 53 selected probes form a bump-like structure along the second PC score axis (Fig. S6 (1.6MB, pdf) (b (1.6MB, pdf) )). These probes were significantly demethylated in the diseased twins (○) compared with the healthy twins (△) as expected (Fig. S8 (1.6MB, pdf) ), excluding the subgroup with ID: 85. The striking difference between the SLE samples and the RA samples is that the RA diseased twins did not have distinct second PC scores compared with the age/gender matched controls (+ and ×, Fig. S7 (1.6MB, pdf) ), while the SLE diseased twins did. This suggests that RA diseased twins do not have genes that are methylated differently from normal controls and that the difference between RA diseased twins and the group consisting of healthy twins and normal controls was not significant. This may be the reason why Javierre et al. [6] did not find any probes expressed differently or significantly between controls and RA patients. Again, the PCA-based probe selection successfully selected differently expressed probes.
PCA Application to DM Samples
We repeated the same procedure using 20 DM samples (IDs 16, 33, 103, 127 and 129 in Table 1). The results are shown in (Figs. S9 (1.6MB, pdf) , S10 (1.6MB, pdf) , S11 (1.6MB, pdf) and S12 (1.6MB, pdf) ). Compared with the former two cases using SLE and RA samples, the DM sample was harder to analyze. First, as can be seen in (Fig. S11 (1.6MB, pdf) ), the second PC did not express differences between the control and disease groups but rather between genders. This suggested that the difference between disease and control groups was not even of secondary importance. The difference between disease and control groups was only observed in the third PC (Fig. S11 (1.6MB, pdf) (b (1.6MB, pdf) )). Second, along the third PC score, the bump-like structure was less clear, even compared with the RA samples (Fig. S9 (1.6MB, pdf) (b (1.6MB, pdf) )). This prevented us from selecting a clear threshold for outliers. Thus, we tentatively selected 44 probes (red dots in Fig. S9 (1.6MB, pdf) ) with negative third PC scores. However, close inspection of the distribution of the third PC scores highlighted a slight hump in the distribution of the third PC scores (red regions and a red arrow in Fig. S10 (1.6MB, pdf) ). Thus, we selected probes that belonged to this small hump as outliers, and as the probes that were differently expressed between disease and control groups. Third, the observed differences of the third PC scores between disease and control groups were dependent upon gender (Fig. S11 (1.6MB, pdf) (b (1.6MB, pdf) )). Although female twins (red) had larger third PC scores for DM twins (○) compared with healthy twins (△), male twins (black) had smaller PC scores for DM twins (○) compared with healthy twins (△). This gender-dependence makes it difficult to identify significant differences between disease and control groups, because samples with equal number of males and females can have lower third PC scores. This possibly also reduced the amount of methylation/demethylation differences between control and disease groups. Differential methylation is reversed between males (ID: 16 and 33 in Fig. S12 (1.6MB, pdf) ) and females (ID: 103, 127, and 129 in Fig. S12 (1.6MB, pdf) ). This may explain why Javierre et al. [6] did not find probes expressed differently and significantly between control and DM groups. Thus, the PCA-based method can select promoters that are methylated differently between normal twins and diseased twins from DM samples.
Comparisons with the Previous Feature Selection Methods and Comparisons with other Data Sets
Comparisons with the previous feature selection methods and comparisons with other (independent or validation) data set were not discussed here but were better to be discussed. However, since it is lengthy and is not directly related to the outcome obtained in the present study, we moved these to supporting information. As a result, we confirmed that our methods outperformed previously proposed methods [13, 14] (no previously proposed methods tested provided us commonly selected genes for three diseases), and our methods can work even if it is applied to other (only SLE [15] and RA [16], no DM) samples.
Protein Structures Predicted by FAMS
To understand the functions and structures of the selected genes, we applied FAMS to the genes selected by PCA. In Table 2, we listed the genes selected in the present study. These genes were used as reference proteins for FAMS. Together with the reference proteins, we listed the model proteins that were inferred to have similar structures to each of the selected genes by FAMS.
Table 2.
Selected genes and model proteins used for structure prediction. Bold ID of PDB indicates that the reference protein itself was detected in PDB.
| Reference gene symbol | Model PDB ID |
P-value | gene symbol |
|---|---|---|---|
| AIM2 | 3RN5_A | 4 ×10−84 | INTERFERON-INDUCIBLE PROTEIN AIM2 |
| CARD15 | 3CIY_B | 1 ×10−58 | TOLL-LIKE RECEPTOR 4, VARIABLE LYMPHOCYTE (TLR4) |
| CD82 | 2BG9_A | 0.48 | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN |
| CSF1R | 3B43_A | 5 ×10−58 | TITIN |
| CSF3 | 1GNC_A | 8 ×10−66 | GRANULOCYTE COLONY-STIMULATING FACTOR |
| CSF3R | 3DMK_A | 4 ×10−64 | DOWN SYNDROME CELL ADHESION MOLECULE (DSCAM) |
| DHCR24 | 2Q4W_A | 1 ×10−114 | CYTOKININ DEHYDROGENASE 7 (CKO7) |
| ERCC3 | 2W74_D | 1 ×10−151 | TYPE I RESTRICTION ENZYME ECOR124II R PROTEIN (HSDR) |
| GRB7 | 3HK0_B | 3 ×10−73 | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 10 (GRB10) |
| HGF | 4DUU_A | 1 ×10−179 | PLASMINOGEN |
| HOXB2 | 2D5V_A | 1 ×10−23 | HEPATOCYTE NUCLEAR FACTOR 6 (HNF-6) |
| IFNGR2 | 3T1W_A | 4 ×10−43 | FOUR-DOMAIN FIBRONECTIN FRAGMENT |
| LCN2 | 1X71_A | 4 ×10−52 | NEUTROPHIL GELATINASE-ASSOCIATED LIPOCALIN (NGAL) |
| LMO2 | 2XJY_A | 2 ×10−33 | RHOMBOTIN-2 |
| LTB4R | 2KS9_A | 2 ×10−77 | SUBSTANCE-P RECEPTOR |
| MMP14 | 1SU3_B | 1 ×10−160 | INTERSTITIAL COLLAGENASE (MMP-1) |
| MMP8 | 1SU3_B | 1 ×10−171 | INTERSTITIAL COLLAGENASE (MMP-1) |
| MPL | 3L5H_A | 8 ×10−61 | INTERLEUKIN-6 RECEPTOR SUBUNIT BETA (IL6RB) |
| PADI4 | 2DEW_X | 0.0 | PROTEIN-ARGININE DEIMINASE TYPE IV |
| PECAM1 | 3DMK_A | 1 ×10−104 | DOWN SYNDROME CELL ADHESION MOLECULE (DSCAM) |
| PI3 | 1TWP_A | 2 ×10−19 | WHEY ACIDIC PROTEIN (WAP) |
| RARA | 3DZY_A | 4 ×10−95 | RETINOIC ACID RECEPTOR RXR-ALPHA |
| S100A2 | 2RGI_A | 4 ×10−19 | PROTEIN S100-A2 |
| SEPT9 | 3FTQ_A | 1 ×10−137 | SEPTIN-2 |
| SLC22A18 | 1PW4_A | 1 ×10−108 | GLYCEROL-3-PHOSPHATE TRANSPORTER |
| SPI1 | 1GVJ_B | 1 ×10−21 | C-ETS-1 PROTEIN (ETS1) |
| SPP1 | 1D2T_A | 1 ×10−12 | ACID PHOSPHATASE (ACP) |
| STAT5A | 1Y1U_A | 0.0 | SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION (STAT5A) |
| SYK | 2OZO_A | 1 ×10−170 | TYROSINE-PROTEIN KINASE ZAP-70 |
| TIE1 | 3DMK_A | 2 ×10−88 | DOWN SYNDROME CELL ADHESION MOLECULE (DSCAM) |
| TM7SF3 | 1AR1_A | 6 ×10−88 | CYTOCHROME C OXIDASE |
| TRIP6 | 1B8T_A | 2 ×10−32 | CYSTEINE-RICH PROTEIN 1 (CRP1) |
| VAMP8 | 2KOG_A | 1 ×10−21 | VESICLE-ASSOCIATED MEMBRANE PROTEIN 2 (VAMP2) |
First, FAMS successfully listed the model proteins with small P-values for most of the reference proteins. Fig. S13 (1.6MB, pdf) shows a representative example of the combination of a model protein and a reference protein: the model protein 2OQ0_B and the reference protein AIM2. Regions of alignment included a 192 amino acid sequence (total length = 209 amino acids) of 2OQ0_B and 191 amino acid sequence (total length = 343 amino acids) of AIM2. Sequence similarity between the two alignment regions was 44 %. P-value attributed was 2 × 10-90. 2OQ0_B is annotated as IFI-16. Although AIM2 itself is included in the present PDB, the structure predicted by FAMS using IFI-16 as a model protein was very similar to the true structure, although sequence similarity between AIM2 and IFI-16 is less than 50 %. This demonstrated that structure prediction using FAMS is very accurate. Although this is only one example of a typical relationship between model/reference proteins, it was generally representative of the quality of structural similarities we observed for other proteins. This suggests that the structural homology between models and references is reliable.
DISCUSSION
First, we will discuss the analysis of our results to determine whether our selection process was correct. Then, the possibilities of the application to drug discovery will be discussed.
Comparison with Previous Studies and between Diseases
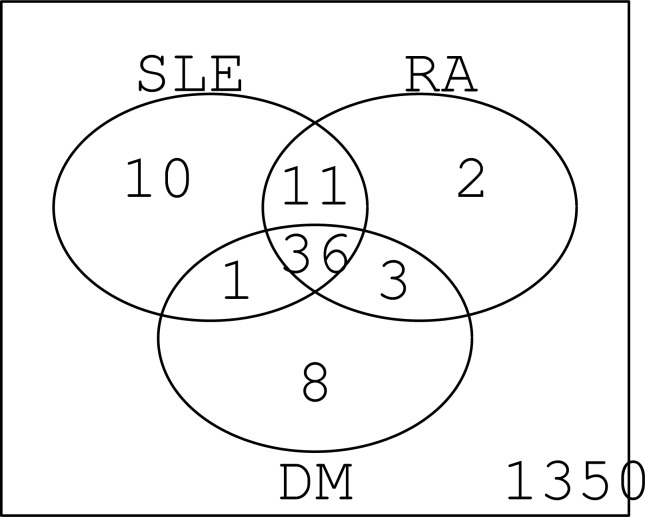
We compared our results with Javierre et al. [6] (Table 3). It is clear that there are substantial overlaps between our studies and Javierre et al. [6]. If one considers that different methods and/or samples were employed, this coincidence is more than remarkable. It may suggest that our analysis was correct. Figure 1 shows the overlap between the distinct diseases. Again, the coincidence is very high. Thus, we can conclude that for the first time we could successfully identify commonly methylated promoters for more than one autoimmune disease.
Table 3.
A comparison between previous studies (*) [6] and the present work. Numbers indicate the number of over-laps between previous studies and the present work. Numbers in parentheses represent the total number of probes that were regarded as being expressed differ-ently between disease and control groups in the pre-sent work
| SLE(*) | SLE | RA | DM |
|---|---|---|---|
| 54 | 51(58) | 48(53) | 37 (44) |
Fig. (1).
Venn diagram of selected probes between diseases.
Biological Relevance of Selected Proteins
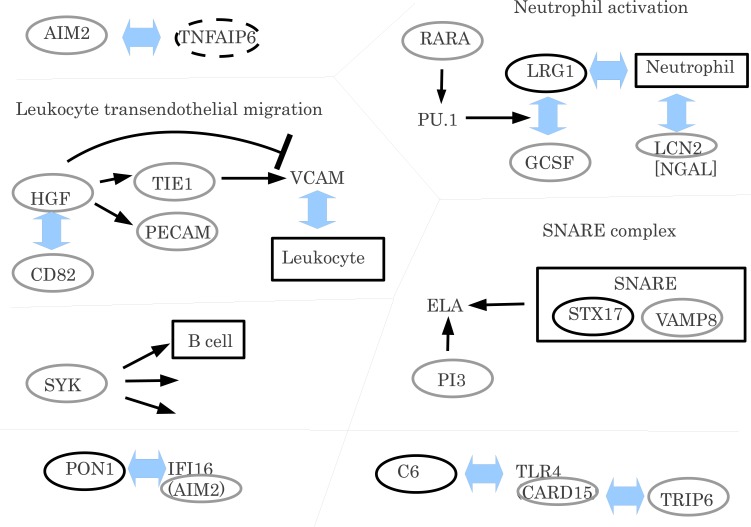
In this subsection, we determine whether the selection of proteins was coincident with what was previously known, which is schematically summarized in (Fig. 2).
Fig. (2).
Suggested relation between proteins selected in the present study and those reported in previous studies. Grey ovals: present study, black ovals: O’Hanlon et al. [9] and a broken oval: O’Hanlon et al. [29]. For more details, see text.
Recently, O’Hanlon et al. [9] investigated plasma proteomic profiles from disease-discordant MZ twins (four pairs discordant for SLE, four pairs discordant for juvenile idiopathic arthritis (juvenile RA) and two pairs discordant for juvenile DM), and found that several proteins were differently expressed between patients and normal controls. Among several proteins found, O’Hanlon et al. [9] identified 11 genes (STX17, MGAM, PON1, C6, SYNE1, PLEKHG5, ZNA2GP, LRG1, PKD1 and APOA2) as important. Many clinical studies suggested that these proteins (genes) are related to the genes selected by the present study. For example, GCSF was reported to upregulate LRG1 [17], while LRG1 was upregulated in patients as shown by O’Hanlon et al. [9]. LRG1 and GCSF were also upregulated in chemia-conditioned mice [18]. Ai et al. [19] also discussed the tight relationship between G-CSF and LRG1, which is located downstream of PU.1 signaling pathways. PU.1 is targeted by RARA [20], one of the promoters we identified as being differently methylated in patients compared with normal controls. (Table 2, see also Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway hsa05200).
LGR1 is upregulated during neutrophil activation. Neutrophil granulocytes are the most abundant type of white blood cells in mammals and form an essential part of the innate immune system. It is notable that LCN2 (NGAL : neutrophil gelatinase-associated lipocalin) was also selected by our study.
Thus, the selection of CSF1R, CSF3, and CSF3R as critical genes by our research is not accidental but biologically meaningful.
O’Hanlon et al. [9] also emphasized the importance of PON1, a gene that causes IFI16-mediated expression changes in endothelial cells [21]. IFI16 was reported to have a similar structure to AIM2 in our research (see Fig. S13 (1.6MB, pdf) ).
Fischer et al. [22] confirmed that C6 is a toll-like receptor (TLR) 4 agonist. C6 is expressed in autoimmune disease patients and is involved in the key pathway related to autoimmune disease [9], while TLR4 was found to have a similar structure to CARD15 by our study.
Syntaxin 17 (STX17) protein was highlighted by O’Hanlon et al. [9]. Syntaxin forms a SNARE complex with VAMP [23], and VAMP8 was recognized by our analysis. VAMP8 and STX17 signal in the KEGG pathway "SNARE interactions in vesicular transport" (hsa04130). SNARE complexes including VAMP8 were reported to be related to the autoimmune disease, Sjögren’s syndrome [24], and was proposed to be a novel target for the development of new therapies to treat allergic and autoimmune disease [25]. Gordon et al. [26] also studied SNARE complex structures including STX17 and VAMP8 using a newly developed assay. They experimentally confirmed the various functions of SNARE complexes although they could not identify the specific function of either STX17 or VAMP8 for cell secretion processes. Thus, VAMP8 is expected to be a potential drug target for treatment of autoimmune diseases.
Mashima et al. [27] reported that irf2 deficiency caused low serum levels of elastase-1 (ELA1) mediated by SNAREs. PI3 binds to ELA1 [28]. Thus, it is likely that PI3 is involved in mediating the functions of SNARE.
O’Hanlon et al. [29] also investigated gene expression profiles. RNA microarray analyses (Agilent Human 1A(V2) 20K oligo arrays) were used to quantify gene expression in peripheral blood cells from 20 MZ twin pairs discordant for systemic autoimmune diseases. The cohort consisted of six affected probands with SLE, six with RA, eight with idiopathic inflammatory myopathies, and their same-gender unaffected twins. O’Hanlon et al. [29] confirmed that several genes’ expression could be distinguished between normal controls and patients. Genes that O’Hanlon et al. [29] identified as important were TNFAIP6, TNFSF10, MAP2K6, IL1RN, IFI27, ANXA3, CEA- CAM6, DEFA4, EIF2AK2, FCERA1, SYTL2, LGR6, KRTCAP2, LTK, and FYN. Among these genes, TNFAIP6 was observed to be co-expressed with AIM2 during the first 10 weeks of therapy with Pegylated-interferon-alfa2b (PegIntronTM ) and ribavirin (administered by weight) in HCV patients [30].
HGF and TIE1 are receptor tyrosine kinases. The overexpression of HGF results in the up- regulation of TIE1 and PECAM in the 3T3-F442A mouse cell line [31]. PECAM1 functions in the KEGG pathway for leukocyte transendothelial migration (hsa04670). TIE1 upregulates the cell adhesion molecules (CAMs) VCAM-1, E-selectin, and ICAM-1 through a p38-dependent mechanism. VCAM1 mediates adhesion between leukocytes and endothelium prior to leukocyte transendothelial migration. This is important as migrating leukocytes cause RA. However, HGF downregulates VEGF-mediated expression of ICAM-1 and VCAM-1 at the transcriptional level [32]. The association between HGF and CD82 has also been reported [33, 34]. Thus, it is likely that HGF, TIE1, PECAM1 and CD82 work cooperatively and may be potential drug targets for the treatment of autoimmune diseases.
The Nucleotide Oligomerization Domain (NOD)-like receptor (NLR) signaling pathway (hsa04621) includes NOD2 (CARD15) and TRIP6. NLRs are cytoplasmic proteins with a variety of functions for the regulation of inflammatory and apoptotic responses. CARD15 has been identified as a risk factor gene for Crohn’s disease [2].
SYK is a spleen tyrosine kinase [EC:2.7.10.2], which functions in many immune-related KEGG pathways such as natural killer cell mediated cytotoxicity (ko04650), B cell receptor signaling (ko04662), Fc epsilon RI signaling (ko04664), and Fc gamma R-mediated phagocytosis (ko04666).
Natural killer cell mediated cytotoxicity is the directed killing of a target cell by a natural killer cell through the release of granules containing cytotoxic mediators or through the engagement of death receptors.
Taken together, a remarkable number of proteins among those selected by the present study have strong relationships with proteins previously reported to be related to the autoimmune disease- related biological processes.
Biological Significance of Inference by FAMS
In this subsection, we determined whether the biological features attributed to the model proteins selected by FAMS (Table 2) were reasonable. Because of space limitations, we cannot explain all of them singly, therefore we will discuss selected examples.
TRIP6 is expected to have a similar structure to CRP1, which is involved in immune responses [35]. TM7SF3 is a cytochrome c oxidase, which was reported to bind to immune gamma-globulins [36]. TIE1, PECAM1 and CSF3R form Down Syndrome Cell Adhesion Molecule (DSCAM), which is an immunoglobulin (Ig)-superfamily receptor in insects [37]. SYK is a tyrosine-protein kinase ZAP-70 and both SYK and ZAP-70 display distinct requirements for Src family kinases in immune response receptor signal transduction [38]. STAT5A is found in PDB, which has a critical role in cytokine responses and normal immune functions [39]. SPI1 is ETS1, and is expressed in SLE and functions in the immune system [40]. S100-A2 is in PDB and is reported to be an anti-body that inhibits the receptor for advanced glycation end products (RAGE) ligands [41]. RARA is structurally similar to RXR-α, which is involved in inflammatory responses [42]. PI3 is a WAP, which is important in innate immunity [43]. PADI4 is in PDB and is important in RA [44]. The structure of MPL structure was inferred to be similar to IL6RB. IL6R is a key mediator of RA [45]. LCN2, also called NGAL, is in PDB. NGAL has been tested as a marker of inflammatory status for the early diagnosis of inflammatory diseases such as DS [46]. HNF-6 is a HOXB2 model proteins and causes immunologically distinct features [47]. AIM2 is structurally similar to IFI16. AIM2 and IFI16 have critical roles in immunology [48]. CARD15 was inferred to be similar to TLR4, which has a role in cell antiviral responses together with TLR3:TICAM1-specific signaling pathways [49]. CD82 is an acetylcholine receptor protein that has functions in the immune system [50]1. CSF1R is assigned as TITAN, and is involved in immune responses [51]. SPP1 is an acid phosphatase and is related to autoimmune prostatitis [52]. LMO2, (rhombotin-2), is related to ZFAT (a zinc-finger gene in autoimmune thyroid disease susceptibility region) an immune-related transcriptional regulator containing 18 C2H2-type zinc-finger domains and one AT-hook) [53]. DHCR24 is a cytokinin dehydrogenase. Cytokines have been referred to as immunomodulating agents. SEPT9 is homologous to septin-2, which is upregulated in cytoskeletal and immune function-related proteome profiles [54]. IFNGR2 is a four-domain fibronectin fragment, which plays a role in autoimmune diseases [55]. CSF3 is in PDB, and is related to immune system functions [56]. GRB7 (GRB10) has roles in the immune system and in cancer [57]. HGF is related to plasminogen, which plays critical roles in autoimmune diseases together with matrix metalloproteinase (MMP) 9 [58]. LTB4R is a substance-P receptor, which mediates immune responses to respiratory syncytial virus infection [59].
This is only a partial list of the immune system-related features that were attributed to each model protein selected by FAMS. Although more examples could be listed, we have omitted them because of space restrictions. Such a coincidence that most of the selected model proteins have some relationship to immune-related molecular functions cannot be accidental. This suggests that the three-dimensional structures inferred by FAMS have the potential for autoimmune disease-related drug discovery.
Possibility of Drug Discovery
To regard the selected proteins as drug targets, it is remarkable to find that all of the three-dimensional structures predicted by FAMS are immune-related. However, it is important to use this information for drug discovery by FAMS. In this subsection, we would like to demonstrate the possibilities of drug discovery for the proteins selected in the present study.
Ligand Binding to “Pocket”
The most popular method of identifying drugs is to find a small molecule to bind a “pocket” of each protein. If FAMS can identify or suggest such a candidate for each gene in Table 2, it will be very useful.
For example, two proteins, MMP8 and MMP14, are shown in Table 2. They coregulate target genes [60]. Both of them are members of the MMP family, which is related to inflammation. For MMP8, using 1XUC_A, which is MMP-13, as a template, FAMS successfully showed that there might be many ligands that bind to MMP8 (Fig. S14 (1.6MB, pdf) ). Similarly, for MMP14, using 1BQO_B, which is MMP-3, as a template, FAMS successfully showed many ligands could also bind MMP14 (Fig. S15 (1.6MB, pdf) ). Although this cannot be described as finding a new drug, it does show the potential for proteins listed in Table 2 that may be novel drug targets.
To determine the possibility of “in silico” ligand screening, we employed MMP8. For MMP8, we identified three PDB structures for model proteins, 1A86_pld, 1I73_pld, and 1MMB_BAT. Table 4 shows the correlation coefficients between FPAscores and Ki values for 15 ligand candidates used in the assay CHEMBL711322. In this calculation, we calculated the mean of the values obtained by more than one model protein that were indicated by three-digit numbers. For example, 100 represents the consideration of only 1A86_pld and the exclusion of 1I73_pld and 1MMB_BAT. Figure S16 (1.6MB, pdf) shows the scatter plots of FPAscores and Ki values. Table S1 (1.6MB, pdf) shows the FPAscores and Kivalues for ligand candidates. Table 4 suggests that the mean over two pairs, i.e., 1A86_pld and 1I73_pld, or 1A86_pld and 1MMB_BAT, can provide a significant correlation independent of the types of correlation coefficients considered.
Table 4.
A comparison between the FPAscores and Kivalues. Three-digit numbers indicated which of the three model PDB files, i.e., 1A86_pld, 1I73_pld,1MMB_BAT, were considered for choosing LD computation. Adjusted P-values were P-values adjusted by Benjamini and Hochberg[63]. Bold numbers indicate adjusted P-values less than 0.05.
| Pearson | Spearman | |||||
|---|---|---|---|---|---|---|
| Combination | corr. | P-value | Adjusted P-value | corr. | P-value | Adjusted P-value |
| 111 | -0.503 | 0.056 | 0.098 | -0.540 | 0.038 | 0.066 |
| 011 | 0.159 | 0.572 | 0.572 | 0.197 | 0.482 | 0.562 |
| 101 | -0.694 | 0.004 | 0.016 | -0.705 | 0.003 | 0.013 |
| 110 | -0.688 | 0.005 | 0.016 | -0.698 | 0.004 | 0.013 |
| 001 | -0.334 | 0.223 | 0.261 | -0.064 | 0.820 | 0.820 |
| 010 | -0.624 | 0.013 | 0.030 | -0.572 | 0.026 | 0.060 |
| 100 | -0.444 | 0.097 | 0.136 | -0.431 | 0.109 | 0.152 |
Based on the results obtained here, we can expect possible “in silico” screenings for drug discovery.
Inhibition of Protein Complex Formation
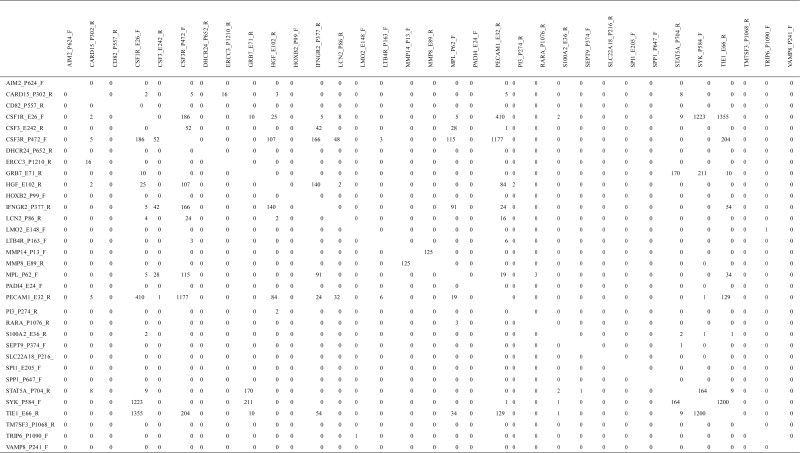
Another new possibility for drug targets is the inhibition of protein complex formation. Many proteins cannot function as a single substance and must form protein complexes with other proteins. Thus, if we inhibit the protein complex formation, we can also inhibit the function of the protein complex. Table 5 lists protein complex candidates inferred by FAMS. Since FAMS uses a representative protein within each cluster that has more than 95 % sequence similarity as a model protein, there are often more than a thousand model proteins, which can bind to other proteins. However, it can be seen that the list includes many reasonable outcomes. For example, there are 52 model protein complexes listed when using both CSF3 and CSF3R as reference proteins. Just by the name, it is obvious that they are possibly a ligand and its receptor. However, there are 186 model proteins between CSF3R and CSF1R. This represents the possibility that each monomer can form protein complexes that can function together, possibly as a receptor. In addition to this, both CSF3R and CSF1R most frequently have non-zero model proteins that bind to other reference proteins. This is reasonable since many proteins can bind as a ligand or can form a receptor. Further analysis of this table will give us fruitful information to find drug targets by inhibiting the formation of protein complexes.
Table 5.
The number of model proteins that can bind to other model proteins
 |
In addition to these known protein complexes, there are many new candidates for protein complex formation. Figure S17 (1.6MB, pdf) shows one such possible candidate. In Table 6, we computed binding energy with FiberDock software [61]. Obtained binding energy was significantly small enough to form protein complex.
Table 6.
Caluculated binding enregy by FiberDock[61] for the binary complex shown in Fig. S17.
| glob | aVdW | rVdW | ACE | inside | aElec | rElec |
| -32.93 | -40.51 | 17.63 | 13.06 | 11.41 | -47.40 | 61.35 |
| laElec | lrElec | HB | piS | catpiS | aliph | BBdeform |
| -7.64 | 11.38 | -0.36 | -13.00 | 0.00 | -5.00 | 0.00 |
In Table 5, there are 410 possible candidate pairs between CSF1R and PECAM1. Among these, there is one pair having 61 atom pairs in contact with each other. This means there is a structure on PDB (2ZJS) that includes monomers whose protein structures are expected to be similar to CSF1R and PECAM1, respectively. 2ZJS is a SecYE translocon, which functions as a protein-conducting channel [62]. Although this protein complex was found in Thermus thermophilus, since this protein is expected to be highly conserved, it is possible that CSF1R and PECAM1 form a protein complex that is secreted across or integrated into membranes and plays a critical role in human autoimmune diseases. Thus, if we can identify a drug that inhibits the formation of a protein complex between CSF1R and PECAM1, it may be used to treat autoimmune diseases.
Although many more protein complex formation candidates were detected, we cannot report all of them here because of space limitations. This will be reported in the future.
Why is PCA Useful to Infer Differential Expression between Control and Disease?
To our knowledge, this is the first report to describe commonly methylated promoters between more than one autoimmune disease and healthy controls. In this subsection, we argue why it is difficult to find commonly methylated promoters between more than one autoimmune disease and healthy controls and why our PCA-based method was successful.
First, to determine whether individual promoters are differently methylated between control and disease groups by the conventional statistical test, more than one replicate is required. However, as shown by Javierre et al. [6], when twins are used, it is impossible to obtain more than one replicate, because they are twins. Twins consist of two genetically identical humans. Thus, if one has the disease and the other does not, there is no way to obtain additional genetically identical individuals. In this case, an additional normal control must be used to obtain biological replicates. However, since additional subjects will have a different genome from the twins, the addition of non-twin control samples weakens the ability of detection of significance.
Another method to apply the statistical test to the set of probes is to divide the probes into two classes: differently expressed probes and others. However, to find suitable classifications with which to divide probes to differently expressed probes and others, multiple trials and errors are required. These trials weaken the sensitivity of the test because of the correction due to the multiple comparisons.
Although these are general problems, there are also some autoimmune-specific problems. For example, there is a strong gender-dependence of promoter methylation related to autoimmune diseases. When PCA is applied to the whole sample, the second PC is related to X chromosome specific methylation (Fig. S1 (1.6MB, pdf) ). While males have only one X chromosome, females have two. Usually, this difference is sustained by methylation of one pair of the female X chromosomes. Thus, one has to exclude genes located on the X chromosome. However, differential promoter methylation is also observed for DM between different genders (see section, “PCA application to DM samples”). This can also weaken the sensitivity of statistical tests.
Usually, there is no way to detect these dependencies in advance. However, PCA correctly detected these and selected differently expressed probes considering these gender specificities. Thus, PCA can separate different dependencies from each other. For example, for DM, the second PC represented gender specific promoter methylation (Fig. S11 (1.6MB, pdf) (a (1.6MB, pdf) )) while the third PC reflected differential methylation between healthy twins and DM twins (Fig. S11 (1.6MB, pdf) (b (1.6MB, pdf) )). It even automatically reflected the reversed methylation dependent upon gender. For RA, the subgroup with ID: 85 did not have differential methylation and it was automatically reflected (Figs. S7 (1.6MB, pdf) (b (1.6MB, pdf) )).
PCA does not assume pre-defined classification but reflects classifications with larger inter- class differences. If there are some classes accompanied with differentially methylation, it is probable that PCA can detect them. This is not guaranteed, but it worked in the present research. This is an explanation for why this study could detect differences by PCA-based promoter methylation selection.
CONCLUSIONS
In this study, we applied PCA-based gene selection to promoter methylation measurements to identify promoters of genes differently methylated between healthy twins and autoimmune diseased twins. Significant numbers of gene promoters were commonly demethylated between distinct autoimmune diseases, but differently between healthy twins and diseased twins. The genes whose promoters were commonly methylated had a remarkable relationship to genes that were previously shown as related to autoimmune diseases. Their three-dimensional protein structures were also inferred by FAMS. Most model proteins selected to infer the three-dimensional structures were immune-related proteins. This fact reveals that the inference by FAMS was biologically accurate. The possible applications of the inference by FAMS for drug discovery were discussed. The inference by FAMS may therefore be useful for both ligand discovery and the search for composites that inhibit protein complex formation, which may promote autoimmune disease.
ACKNOWLEDGEMENTS
YT was supported by KAKENHI, 23300357 and 26120528, also by Chuo University Joint Research Grant. SI performed the research. HU, MI, and YT designed and performed the research. YT wrote the paper.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Web site along with the published article.
REFERENCES
- 1.Davidson A, Diamond B. Autoimmune diseases. N. Engl. J. Med. 2001;345:340–350. doi: 10.1056/NEJM200108023450506. [DOI] [PubMed] [Google Scholar]
- 2.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu. Rev. Immunol. 2009;27:363–391. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quintero-Ronderos P, Montoya-Ortiz G. Epigenetics and autoimmune diseases. Autoim- mune Dis. 2012;2012:593720. doi: 10.1155/2012/593720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballestar E. Epigenetic contributions in autoimmune disease. Preface. Adv. Exp. Med. Biol. 2011;711:v–vi. [PubMed] [Google Scholar]
- 5.Richardson B. DNA methylation and autoimmune disease. Clin. Immunol. 2003;109:72–79. doi: 10.1016/s1521-6616(03)00206-7. [DOI] [PubMed] [Google Scholar]
- 6.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreño L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methyla-tion associate with twin discor dance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umeyama H, Iwadate M. Current Protocols in Bioinformatics. John Wiley & Sons Inc. 2002 [Google Scholar]
- 8.Terashi G, Takeda-Shitaka M, Kanou K, Umeyama H. Introduction to Protein Structure Prediction. Wiley. 2010;Chapter 14 pp:299–321. [Google Scholar]
- 9.O’Hanlon TP, Li Z, Gan L, Gourley MF, Rider LG, Miller FW. Plasma pro- teomic profiles from disease discordant monozygotic twins suggest that molecular pathways are shared in multiple systemic autoimmune diseases. Arthritis Res. Ther. 2011;13:R181. doi: 10.1186/ar3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R Core Team R A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna Austria. 2012;ISBN:390005107. [Google Scholar]
- 11.Takaya D, Takeda-Shitaka M, Terashi G, Kanou K, Iwadate M, Umeyama H. Bioin- formatics based Ligand-Docking and in silico screening. Chem Pharm Bull. 2008;56:742–744. doi: 10.1248/cpb.56.742. [DOI] [PubMed] [Google Scholar]
- 12.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Tetko IV, Hall MA, Frank E, Facius A, Mayer KF, Mewes HW. Gene selection from microarray data for cancer classification-a machine learning approach. Comput Biol Chem. 2005;29:37–46. doi: 10.1016/j.compbiolchem.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome- wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythe- matosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, Shchetynsky K, Scheynius A, Kere J, Alfredsson L, Klareskog L, Ekström TJ, Feinberg AP. Epigenome wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–7. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Donnell LC, Druhan LJ, Avalos BR. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein a novel marker of granulocytic differentiation. J Leukoc Biol. 2002;72:478–485. [PubMed] [Google Scholar]
- 18.Gregory AD, Capoccia BJ, Woloszynek JR, Link DC. Systemic levels of G-CSF and interleukin-6 determine the angiogenic potential of bone marrow resident monocytes. J Leukoc Biol. 2010;88:123–131. doi: 10.1189/jlb.0709499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai J, Druhan LJ, Hunter MG, Loveland MJ, Avalos BR. LRG-accelerated differ- entiation defines unique G-CSFR signaling pathways downstream of PU and C/EBPepsilon that modulate neutrophil activation. J. Leukoc. Biol. 2008;83:1277–1285. doi: 10.1189/jlb.1107751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Wang P, Shi J, Zhu X, He M, Jia X, Yang X, Qiu F, Jin W, Qian M, Fang H, Mi J, Yang X, Xiao H, Minden M, Du Y, Chen Z, Zhang J. PML/RARalpha targets promoter regions containing PU consensus and RARE half sites in acute promyelocytic leukemia. Cancer Cell. 2010;17:186–197. doi: 10.1016/j.ccr.2009.12.045. [DOI] [PubMed] [Google Scholar]
- 21.Caposio P, Gugliesi F, Zannetti C, Sponza S, Mondini M, Medico E, Hiscott J, Young HA, Gribaudo G, Gariglio M, Landolfo S. A novel role of the interferon inducible protein IFI16 as inducer of proinflammatory molecules in endothelial cells. J Biol Chem. 2007;282:33515–33529. doi: 10.1074/jbc.M701846200. [DOI] [PubMed] [Google Scholar]
- 22.Fischer H, Ellstrom P, Ekstrom K, Gustafsson L, Gustafsson M, Svanborg C. Ce- ramide as a TLR4 agonist; a putative signalling intermediate between sphingolipid receptors for microbial ligands and TLR4. Cell. Microbiol. 2007;9:1239–1251. doi: 10.1111/j.1462-5822.2006.00867.x. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Tomchick DR, Kovrigin E, Arac D, Machius M, Sudhof TC, Rizo J. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 24.Barrera MJ, Sanchez M, Aguilera S, Alliende C, Bahamondes V, Molina C, Quest AF, Urzua U, Castro I, Gonzalez S, Sung HH, Albornoz A, Hermoso M, Leyton C, Gonzalez MJ. Aberrant localization of fusion receptors involved in regulated exocytosis in salivary glands of Sjögren’s syndrome patients is linked to ectopic mucin secre- tion. J. Autoimmun. 2012;39:83–92. doi: 10.1016/j.jaut.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Woska JR, Gillespie ME. SNARE complex-mediated degranula-tion in mast cells. J. Cell. Mol. Med. 2012;16:649–656. doi: 10.1111/j.1582-4934.2011.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon DE, Bond LM, Sahlender DA, Peden AA. A targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic. 2010;11:1191–1204. doi: 10.1111/j.1600-0854.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- 27.Mashima H, Sato T, Horie Y, Nakagawa Y, Kojima I, Ohteki T, Ohnishi H. Interferon regulatory factor-2 regulates exocytosis mechanisms mediated by SNAREs in pancreatic aci- nar cells. Gastroenterology. 2011;141:1102–1113. doi: 10.1053/j.gastro.2011.05.051. [DOI] [PubMed] [Google Scholar]
- 28.Tsunemi M, Matsuura Y, Sakakibara S, Katsube Y. Crystal structure of an elastase- specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1 A res- olution. . Biochemistry. 1996;35:11570–11576. doi: 10.1021/bi960900l. [DOI] [PubMed] [Google Scholar]
- 29.O’Hanlon TP, Rider LG, Gan L, Fannin R, Paules RS, Umbach DM, Wein- berg CR, Shah RR, Mav D, Gourley MF, Miller FW. Gene expression profiles from discordant monozygotic twins suggest that molecular pathways are shared among multiple systemic autoimmune diseases. Arthritis Res. Ther. 2011;13:R69. doi: 10.1186/ar3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor MW, Tsukahara T, McClintick JN, Edenberg HJ, Kwo P. Cyclic changes in gene expression induced by Peg-interferon alfa-2b plus ribavirin in peripheral blood mono-cytes (PBMC) of hepatitis C patients during the first 10 weeks of treatment. J. Transl. Med. 2008;6:66. doi: 10.1186/1479-5876-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell LN, Cai L, Johnstone BH, Traktuev DO, March KL, Considine RV. A central role for hepatocyte growth factor in adipose tissue angiogenesis. Am. J. Physiol. Endocrinol. Metab. 2008;294:E336–344. doi: 10.1152/ajpendo.00272.2007. [DOI] [PubMed] [Google Scholar]
- 32.Min JK, Lee YM, Kim JH, Kim YM, Kim S W, Lee SY, Gho YS, Oh GT, Kwon YG. Hepatocyte growth factor suppresses vascular endothelial growth factor-induced expression of endothelial ICAM-1 and VCAM-1 by inhibiting the nuclear factor-kappaB pathway. Circ. Res. 2005;96:300–307. doi: 10.1161/01.RES.0000155330.07887.EE. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi M, Sugiura T, Abe M, Ishii K, Shirasuna K. Regulation of c-Met signaling by the tetraspanin KAI-1/CD82 affects cancer cell migration. Int. J. Cancer. 2007;121:1919–1929. doi: 10.1002/ijc.22887. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Guo XZ, Zhang WW, Lu ZZ, Zhang QW, Duan HF, Wang LS. KAI1 inhibits HGF-induced invasion of pancreatic cancer by sphingosine kinase activity. HBPD INT. 2011;10:201–208. doi: 10.1016/s1499-3872(11)60032-5. [DOI] [PubMed] [Google Scholar]
- 35.Latonen L, Jarvinen PM, Suomela S, Moore HM, Saarialho-Kere U, Laiho M. Ul- traviolet B radiation regulates cysteine-rich protein 1 in human keratinocytes. Photodermatol. Photoimmunol. Photomed. 2010;26:70–77. doi: 10.1111/j.1600-0781.2010.00488.x. [DOI] [PubMed] [Google Scholar]
- 36.Frey TG, Chan SH, Schatz G. Structure and orientation of cytochrome c oxidase in crystalline membranes Studies by electron microscopy and by labeling with subunit-specific antibodies. J. Biol. Chem. 1978;253:4389–4395. [PubMed] [Google Scholar]
- 37.Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 38.Zoller KE, MacNeil IA, Brugge JS. Protein tyrosine kinases Syk and ZAP-70 display distinct requirements for Src family kinases in immune response receptor signal transduc-tion. J. Immunol. 1997;158:1650–1659. [PubMed] [Google Scholar]
- 39.Lin JX, Li P, Liu D, Jin HT, He J, Rasheed MA, Rochman Y, Wang L, Cui K, Liu C, Kelsall BL, Ahmed R, Leonard WJ. Critical Role of STAT5 Transcription Factor Tetramerization for Cytokine Responses and Normal Immune Function. Immunity. 2012;36:586–599. doi: 10.1016/j.immuni.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan HF, Leng RX, Tao JH, Li XP, Ye DQ. Ets-1 a new player in the pathogenesis of systemic lupus erythematosus? Lupus. 2011;20:227–230. doi: 10.1177/0961203310389842. [DOI] [PubMed] [Google Scholar]
- 41.Heijmans J, Buller NV, Hoff E, Dihal AA, van der Poll T, van Zoelen MA, Bierhaus A, Biemond I, Hardwick JC, Hommes DW, Muncan V, van den Brink GR. Rage signalling promotes intestinal tumourigenesis. Oncogene. 2012;32(9):1202–6. doi: 10.1038/onc.2012.119. [DOI] [PubMed] [Google Scholar]
- 42.Selvaraj RK, Shanmugasundaram R, Klasing KC. Effects of dietary lutein and PUFA on PPAR and RXR isomer expression in chickens during an inflammatory response. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010;157:198–203. doi: 10.1016/j.cbpa.2010.06.172. [DOI] [PubMed] [Google Scholar]
- 43.Bingle CD, Vyakarnam A. Novel innate immune functions of the whey acidic protein family. Trends Immunol. 2008;29:444–453. doi: 10.1016/j.it.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Abd-Allah SH, el Shal AS, Shalaby SM, Pasha HF, Abou el Saoud AM, el Najjar AR, el Shahawy EE. PADI4 polymorphisms and related haplotype in rheumatoid arthritis patients. Joint Bone Spine. 2012;79:124–128. doi: 10.1016/j.jbspin.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Cronstein BN. Interleukin-6-a key mediator of systemic and local symptoms in rheumatoid arthritis. Bull NYU Hosp Jt Dis. 2007;65(Suppl 1):S11–15. [PubMed] [Google Scholar]
- 46.Dogliotti G, Galliera E, Licastro F, Porcellini E, Corsi MM. Serum neutrophil gelatinase-B associated lipocalin (NGAL) levels in Down’s syndrome patients. Immun Ageing. 2010;7 Suppl 1:S7. doi: 10.1186/1742-4933-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samadani U, Costa RH. The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression. Mol Cell Biol. 1996;16:6273–6284. doi: 10.1128/mcb.16.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin T, Perry A, Jiang J, Smith P, Curry J A, Unterholzner L, Jiang Z, Horvath G, Rathinam V A, Johnstone R W, Hornung V, Latz E, Bowie A G, Fitzgerald K A, Xiao T S. Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Ac- tivation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. RIP1 is an essential mediator of Toll like receptor 3-induced NF-kappa B activation. Nat. Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 50.Quek AM, et al. Autoimmune Epilepsy: Clinical Characteristics and Response to Immunotherapy. Arch Neurol. 2012;69:582–593. doi: 10.1001/archneurol.2011.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skeie GO, Bentsen PT, Freiburg A, Aarli JA, Gilhus NE. Cell-mediated immune response against titin in myasthenia gravis evidence for the involvement of Th1 and Th2 cells. Scand. J. Immunol. 1998;47:76–81. doi: 10.1046/j.1365-3083.1998.00260.x. [DOI] [PubMed] [Google Scholar]
- 52.Fong L, Ruegg CL, Brockstedt D, Engleman EG, Laus R. Induction of tissue-specific autoimmune prostatitis with prostatic acid phosphatase immunization implications for immunotherapy of prostate cancer. J. Immunol. 1997;159:3113–3117. [PubMed] [Google Scholar]
- 53.Tsunoda T, Takashima Y, Tanaka Y, Fujimoto T, Doi K, Hirose Y, Koyanagi M, Yoshida Y, Okamura T, Kuroki M, Sasazuki T, Shirasawa S. Immune-related zinc finger gene ZFAT is an essential transcriptional regulator for hematopoietic differentiation in blood islands. Proc. Natl Acad Sci USA. 2010;107:14199–14204. doi: 10.1073/pnas.1002494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gabr AA, Reed M, Newman DR, Pohl J, Khosla J, Sannes PL. Alterations in cytoskeletal and immune function-related proteome profiles in whole rat lung following intratracheal instillation of heparin. Respir Res. 2007;8:36. doi: 10.1186/1465-9921-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huijbers EJ, Ringvall M, Femel J, Kalamajski S, Lukinius A, Abrink M, Hellman L, Olsson AK. Vaccination against the extra domain-B of fibronectin as a novel tumor therapy. FASEB J. 2010;24:4535–4544. doi: 10.1096/fj.10-163022. [DOI] [PubMed] [Google Scholar]
- 56.Sarkar S, Song Y, Sarkar S, Kipen HM, Laumbach R J, Zhang J, Strickland PA, Gardner CR, Schwander S. Suppression of the NF-? B pathway by diesel exhaust particles impairs human antimycobacterial immunity. J. Immunol. 2012;188:2778–2793. doi: 10.4049/jimmunol.1101380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O-Sullivan I, Chopra A, Carr J, Kim T S, Cohen E P. Immunity to growth factor receptor-bound protein 10 a signal transduction molecule inhibits the growth of breast cancer in mice. Cancer Res. 2008;68:2463–2470. doi: 10.1158/0008-5472.CAN-07-5685. [DOI] [PubMed] [Google Scholar]
- 58.Liu Z, Li N, Diaz LA, Shipley M, Senior RM, Werb Z. Synergy between a plasminogen cascade and MMP-9 in autoimmune disease. J Clin Invest. 2005;115:879–887. doi: 10.1172/JCI23977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tripp R A, Barskey A, Goss L, Anderson LJ. Substance P receptor expression on lymphocytes is associated with the immune response to respiratory syncytial virus infection. J. Neuroimmunol. 2002;129:141–153. doi: 10.1016/s0165-5728(02)00169-8. [DOI] [PubMed] [Google Scholar]
- 60.Silva JA, Ferrucci DL, Peroni LA, Abrahao PG, Salamene AF, Rossa-Junior C, Carvalho HF, Stach-Machado DR. Sequential IL-23 and IL-17 and increased Mmp8 and Mmp14 expression characterize the progression of an experimental model of periodontal disease in type 1 diabetes. J. Cell Physiol. 2012;227:2441–2450. doi: 10.1002/jcp.22979. [DOI] [PubMed] [Google Scholar]
- 61.Mashiach E, Nussinov R, Wolfson H J. FiberDock a web server for flexible induced-fit backbone refinement in molecular docking. Nucleic Acids Res. 2010;38:W457–461. doi: 10.1093/nar/gkq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machin- ery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodolog- ical). 1995;57 pp:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publishers Web site along with the published article.