Abstract
Purpose.
Glaucoma is a major cause of blindness in the world. Recent genome-wide association studies (GWAS) have identified common genetic variants for glaucoma, but still a significant heritability gap remains. We hypothesized that copy number variants (CNVs) might influence part of the susceptibility to glaucoma or its related quantitative endophenotypes.
Methods.
This study examined the association between CNVs and intraocular pressure (IOP), the major modifiable risk factor for primary open-angle glaucoma (POAG), in three population panels of European ancestry: the TwinsUK cohort (n = 1047), the Australian Twin Eye Study (n = 561), and the Wellcome Trust Case-Control Consortium 2 (WTCCC2)/Blue Mountains Eye Study (BMES) (n = 1660). We also used PCR-based assays to investigate a locus of interest that we found associated with IOP in a POAG case–control panel of European ancestry from London, United Kingdom.
Results.
We identified associations between IOP and two CNV regions in the TwinsUK cohort: 5q21.2 (P = 0.003) overlapping the gene RAB9BP1 and 12p13.3 (P = 0.03) overlapping the genes SLC2A14 and SLC2A3. The Australian Twin Eye Study and BMES both replicated the 5q21.2 CNV association and direction of effect (P = 0.001 and P = 0.02, respectively). A meta-analysis across all the cohorts showed that presence of a copy number change at this locus increased IOP by 1.56 mm Hg (P = 1.24 × 10−6). In the case–control study, the 5q21.2 CNV locus did not show association with high-pressure (≥21 mm Hg) POAG cases.
Conclusions.
The 5q21.2 CNV locus could represent a novel locus controlling IOP. Interestingly, this IOP locus is located in close vicinity to the previously widely replicated GLC1G linkage locus for glaucoma, for which subsequent studies have not reached consensus on the causal gene.
Keywords: copy number variation, intraocular pressure, 5q21.2
Copy number variation at chromosome 5q21.2 is associated with intraocular pressure.
Introduction
Glaucoma is a major cause of visual impairment and the most common cause of irreversible blindness in the world.1 Glaucoma is diagnosed clinically by its characteristic optic nerve head appearance (increased optic disc cupping, assessed clinically as an increased optic cup–disc ratio) resulting from retinal ganglion cell degeneration and loss of nerve axons passing through the optic nerve head. Typical features of glaucoma include peripheral visual field loss that slowly progresses to central visual field loss and eventually blindness. Primary open-angle glaucoma (POAG) is the most common form of glaucoma in Caucasian populations (accounting for approximately 50% of all the cases of glaucoma) and affects between 2% and 12% of the population over the age of 60 years.2
Glaucoma is caused by the interaction of environmental and genetic risk factors. Population-based familial aggregation studies show an approximately 10-fold increase in the risk of developing POAG among relatives of affected individuals.3 Intraocular pressure (IOP), the major modifiable risk factor for POAG,4 is also highly heritable, with heritability estimates ranging from 0.35 to 0.62 in different studies.5–7 Thus genetic factors seem to play a significant role in determining susceptibility to glaucoma and its underlying endophenotypes.
Linkage-based studies have identified and replicated two glaucoma-causing genes (MYOC and OPTN),8,9 while two more genes identified in linkage studies (WDR36 and NTF4) have had limited replication.10,11 Genome-wide association studies (GWAS) increased the number of potential glaucoma-causing genes having identified variants in CDKN2B-AS1, TMCO1, CAV1 and CAV2, SIX1/SIX6, SRBD1, and ELOVL5.12–15 In addition, GWAS on IOP and optic disc parameters such as optic disc area and vertical cup–disc ratio (VCDR) as endophenotypes for glaucoma have revealed other plausible candidate genes for glaucoma.16–18 Interestingly, many findings from the case–control design have been replicated in the endophenotype-based approach, such as the association of TMCO1 with IOP12,19 and the associations of CDKN2B and SIX1 with VCDR.12,16 While GWAS have been successful in identifying Single Nucleotide Polymorphisms (SNPs) in several genes that modify the risk of developing glaucoma, they explain only a fraction of the estimated heritability of glaucoma.
We hypothesized that structural genetic variation such as copy number variants (CNVs) might influence part of the susceptibility to glaucoma or its underlying endophenotypes. While CNV analyses have so far largely been successful in detecting susceptibility genes for neurodevelopmental conditions,20–24 their role in other complex human diseases remains uncertain. Mutations affecting dosage of genes, including karyotypic abnormalities, are known to be associated with developmental forms of glaucoma.25,26 A genome-wide CNV scan on POAG by Davis et al.27 provided suggestive evidence that rare copy number variations are involved in POAG; however, further studies to confirm their role are warranted.
We measured IOP as an underlying endophenotype for POAG and investigated the association between CNVs and IOP in three population panels of European ancestry. We further investigated the role of a CNV locus of interest that we found associated with IOP in a case–control study comparing POAG patients (with elevated IOP) and control subjects with normal IOP.
Materials and Methods
We conducted our quantitative association studies in three populations. We first analyzed a panel of 1047 individuals (all of Caucasian ancestry) that was a subset of the TwinsUK Adult Twin Registry based at St. Thomas' Hospital in London28 for which both genotype and IOP information was available (Table 1). Twins largely volunteered unaware of the eye studies at the time of their enrollment and gave fully informed consent under a protocol reviewed by the St. Thomas' Hospital Local Research Ethics Committee. Research was conducted in accordance with the Declaration of Helsinki. Genotyping was carried out using HumanHap610-Quad array (Illumina Inc., San Diego, CA) at the Center for Inherited Disease Research (CIDR), Baltimore, MD.
Table 1. .
Study Characteristics of the Three Caucasian Population Panels Used for the Quantitative Analysis
|
Characteristics |
TwinsUK |
Australian Twins |
WTCCC2/BMES |
| Number of participants | 1047 | 561 | 1660 |
| Number of participants after QC | 992 | 467 | 1620 |
| Age, y, mean ± SD (range) | 56.19 ± 12.21 (16–82) | 24.83 ± 15.95 (5–79) | 64.27 ± 8.36 (49–91) |
| Male sex, % | 2.5 | 42.2 | 43 |
| IOP, mm Hg, mean ± SD (range) | 15.63 ± 3.06 (7–27) | 15.90 ± 2.96 (9–26) | 16.05 ± 2.65 (8–35) |
IOP was measured using the Ocular Response Analyzer (ORA; Reichert, Inc., Buffalo, NY), a noncontact air-puff tonometer that ejects an air impulse lasting 20 ms and monitors the time course changes of the cornea by an electro-optical collimation detector system. The mean IOP was calculated from four readings (two from each eye) for each participant.
The software package PennCNV (available in the public domain at http://openbioinformatics.org) was used to detect CNV regions for each sample using Log R Ratio (LRR) and B allele frequency (BAF) information.29 As a part of quality control (QC), samples having high variability in raw hybridization intensities (SD of LRR > 0.35) and those having a high number of CNV calls (>40) were excluded, leaving 992 samples for further analysis (Supplementary Figure S1). To make final CNV calls, we used the following criteria: (1) We merged neighboring CNVs when the distance between them was less than half the total distance from the start of the first CNV to the end of the last CNV; (2) we called only CNVs containing at least 10 SNPs; and (3) we ignored CNVs located in the centromeric and telomeric regions. Genes occurring within 100 Kb of the CNV calls were recognized using build 36 (hg18) of the human genome (available in the public domain, http://genome.ucsc.edu/). We filtered to consider only those genes that harbored CNVs in at least 1% of the subjects.
CNVs at a locus can be recurrent (and multiple), disrupting the gene sequence at different physical locations within a gene. We considered genes as functional units and tested the hypothesis that any copy number change (deletion or duplication) affecting the normal diploid state of the gene was associated with IOP levels. A linear regression model, adjusted for age and sex, was used to test for association between copy number changes affecting no, one, or both chromosomes and IOP level. A score test statistic as implemented in MERLIN30 was used to adjust for family structure.
Two additional cohorts of Caucasian ancestry, both from Australia, were used for the quantitative analysis (Australian Twin Eye Study and Wellcome Trust Case-Control Consortium 2 [WTCCC2]/Blue Mountains Eye Study [BMES]) (Table 1). The Australian Twin Eye Study comprises participants examined as part of the Twins Eye Study in Tasmania (TEST) or the Brisbane Adolescent Twins Study (BATS). Similarly to the TwinsUK samples, they were genotyped with HumanHap610-Quad array (Illumina) at CIDR. In most participants, the IOP was measured with the TONO-PEN XL (Reichert, Inc.) as outlined by Mackey et al.31 The BMES includes individuals recruited from a defined geographical region in the Blue Mountains (west of Sydney, Australia). The BMES protocol has been described in detail by Mitchell et al.32 Participants were genotyped with Human660W-Quad (Illumina) as part of the Wellcome Trust Case Control Consortium 2 (WTCCC2) at the Sanger Institute, Hinxton, United Kingdom. IOP was measured by applanation tonometry using a Goldmann tonometer (Haag-Streit, Bern, Switzerland). The CNV calling for both cohorts was carried out using PennCNV in a manner similar to that used with the TwinsUK cohort. Quality control measures and association analysis were implemented using procedures identical to those for the TwinsUK cohort. A fixed-effect meta-analysis of the three cohorts was performed using the package metan on Stata Statistical Software, 11 (StataCorp LP, College Station, TX).
We used PCR-based assays for experimental validation of copy number calls in a subset of the TwinsUK cohort. This was performed for a locus of interest that was associated with IOP in the PennCNV analysis. TaqMan (Applied Biosystems, Foster City, CA) PCR assay was designed to target a region common to the CNVs overlapping the open reading frame of the gene at the locus of interest (assay location, Chr5:104,519,101 [build 36]; amplicon length, 87). However, the probes did not target some relatively rare CNVs in the vicinity of the open reading frame of the gene, since the scarcity of DNA was an impediment to running multiple TaqMan PCR assays targeting these CNVs. Sample DNA concentration was normalized prior to running the assays. Copy number variation calling for the assays was implemented with the software Copy Caller v2.0 (Applied Biosystems) that used sample with known diploid state at this locus as a denominator for the comparison.
Finally, using the TaqMan PCR assays, we investigated whether CNVs at our locus of interest, identified in association with IOP in the previously described populations, also increased the risk of developing POAG in a case–control panel of European ancestry from London, United Kingdom. High-pressure (≥21 mm Hg) POAG cases (n = 302) with self-reported European ancestry were ascertained from eye clinics in South East London at the Princess Royal University Hospital, Orpington, and St. Thomas' Hospital. They had a clinical diagnosis of POAG based on an abnormal visual field in at least one eye on Humphrey 24–2 threshold testing with an associated cup-to-disc ratio of 0.6 or greater, and were either initiated into or were already receiving treatment. Control subjects (n = 538) were recruited from cataract clinics in the same hospitals. All were found to have healthy optic discs on clinical examination, with cup-to-disc ratios less than 0.6. Control participants were excluded if they were found to have an IOP ≥ 21 mm Hg. Formal visual field testing was not undertaken on these participants.
Results
In the TwinsUK cohort, 5.27 CNV calls were made per sample. A summary characteristic of the CNVs identified in this cohort is provided in Table 2.
Table 2. .
Summary Statistics for CNVs Detected in the TwinsUK Cohort
|
Characteristic |
Number |
| Total number of CNVs detected | 5223 |
| Average number of CNVs per sample | 5.27 |
| Number of deletions detected | 3172 |
| Number of duplications detected | 2051 |
| Average size of deletions in Kb (SD) | 76.96 (144.09) |
| Average size of duplications in Kb (SD) | 153.31 (372.53) |
We identified 45 genes that harbored CNVs in at least 1% of the subjects. Three genes were nominally associated with IOP: RAB9BP1 on 5q21.2 (P = 0.003) and SLCA2A14 and SLCA2A3 genes on 12p13.3 (P = 0.03 for both). Results of the top 10 associated genes identified in the analysis are reported in Table 3.
Table 3.
Summary of the Top 10 Genes Identified in the Analysis Testing the Association of Copy Number Change With IOP in the TwinsUK Cohort
|
Gene |
Locus |
Frequency, % |
Beta |
SE |
P
Value |
| RAB9BP1 | 5q21.2 | 1.4 | 2.04 | 0.699 | 0.003 |
| SLC2A14 | 12p13.31 | 2.2 | −1.266 | 0.588 | 0.031 |
| SLC2A3 | 12p13.32 | 2.4 | −1.266 | 0.588 | 0.031 |
| GOLGA8A | 15q14 | 8 | 0.47 | 0.307 | 0.126 |
| KIAA1267 | 17q21.31 | 2.6 | −0.611 | 0.475 | 0.198 |
| LOC644246 | 17q21.32 | 2.6 | −0.611 | 0.475 | 0.198 |
| FAM86DP | 3p12.3 | 3.8 | 0.564 | 0.461 | 0.222 |
| OR4K1 | 14q11.2 | 2.2 | 0.672 | 0.565 | 0.234 |
| OR4K2 | 14q11.3 | 2.2 | 0.672 | 0.565 | 0.234 |
| OR4K5 | 14q11.4 | 2.2 | 0.672 | 0.565 | 0.234 |
The frequency of the RAB9BP1 locus CNV was 1.4% in the TwinsUK cohort, and copy number change at this locus increased IOP by 2.04 mm Hg (95% CI: 0.67–3.41 mm Hg). The median length of the CNVs at this locus was 55.7 Kb. In almost all the individuals with CNV at this locus, only the RAB9BP1 gene was involved, except in one individual who had a large 4.5-Mb deletion involving other genes at the locus as well (C5orf30, GIN1, NUDT12, PAM, PPIP5K2, SLCO4C1, SLCO6A1).
Copy number change at the 12p13.3 locus had a frequency of 2.2% and had a more moderate effect on IOP, decreasing it by 1.26 mm Hg (95% CI: 0.1–2.42 mm Hg). The CNVs at this locus overlapped the glucose-transporter genes SLC2A14 and SLC2A3.
The association for the RAB9BP1 locus was replicated in the Australian twin cohort (P = 0.001) and the WTCCC2/BMES cohort (P = 0.02). The frequency of the CNV in the two cohorts was 2.6% and 1.1%, respectively, with the presence of copy number change at this locus increasing the IOP by 2.03 and 1.09 mm Hg, respectively. Table 4 provides a summary of the RAB9BP1 locus findings in the three cohorts. A combined fixed-effect meta-analysis of the three yielded a highly significant association (P = 1.24 × 10−6). However, the association of the 12p13.3 locus was not replicated in either the Australian twin (P = 0.70) or the WTCCC2/BMES cohort (P = 0.10).
Table 4. .
Summary of RAB9BP1 CNV Findings in the Three Cohorts Used in the Quantitative Analysis
|
Study Cohort |
Frequency, % |
Beta |
SE |
P
Value |
| TwinsUK | 1.4 | 2.04 | 0.7 | 0.003 |
| Australian Twins | 2.6 | 2.03 | 0.63 | 0.001 |
| WTCCC2/BMES | 1.1 | 1.09 | 0.47 | 0.02 |
| Meta-analysis | 1.45 | 1.56 | 0.33 | 1.24 × 10−6 |
TaqMan assays validated 15 of 20 (75%) of the RAB9BP1 CNV events identified initially in the TwinsUK discovery study (Supplementary Table S1). Fifteen of the 16 deletions (93.8%) at this locus validated, whereas the four duplications at this locus failed to validate. Following adjustment for the results of the validation study, the association strength and significance for the RAB9BP1 locus in the TwinsUK cohort differed little from the previous estimates (adjusted beta = 1.94 and P = 0.01). For the Australian twin cohort and the BMES, all the RAB9BP1 CNV events that were identified in the PennCNV analysis were deletions.
For the case–control study, the Copy Caller software identified a copy number state for the RAB9BP1 locus with >95% confidence in 819 of the 840 samples analyzed. A copy number change (all deletions) at the RAB9BP1 locus was detected in 16 of the 819 samples (frequency = 1.95%). Nine of the deletions were detected in controls (frequency = 1.67%), while the remaining seven deletions (frequency = 2.31%) were detected in cases. The difference in the frequency of the CNV in the cases and controls was not statistically significant (P = 0.60).
Discussion
The 5q21.2 CNV locus identified and replicated in three population-based cohorts could represent a novel locus controlling IOP. The protein coded for by RAB9BP1 is not known, but there is evidence that rare variants at or near this locus may be relevant to IOP and glaucoma. Several studies have reported linkage loci (GLC1G and GLC1M) for glaucoma and IOP encompassing the region 5q21-32.33–37 Davis et al.27 demonstrated that two POAG patients carried large CNVs within the GLC1G/GLC1M linkage locus (involving the genes DMXL1 and DTWD2) while none of the controls in their study had CNVs at that locus, thus suggesting that rare CNVs contained in this region might cause POAG.
These studies, however, failed to unequivocally identify the causal gene at this locus; candidate gene sequencing of the GLC1G linkage locus by Monemi et al.33 suggested that a mutation in the WDR36 gene segregates with the phenotype, but subsequent independent studies failed to replicate their finding or identify any other causal variants in this region.34,36,38 While linkage and CNV findings in this region so far have implicated large regions encompassing many genes, our study implicates a much smaller region overlapping the gene RAB9BP1. Moreover, given that the RAB9BP1 CNV locus (5q21.2) is very near the centromeric end of the GLC1G locus (5q21.3), it is also worth investigating whether RAB9BP1 copy number change explains any part of the GLC1G linkage signal. The relative locations of the RAB9BP1 CNV locus, the GLC1G linkage locus, the GLC1M linkage locus, and the CNV locus in GLC1G/GLC1M are provided in the Figure.
Figure.
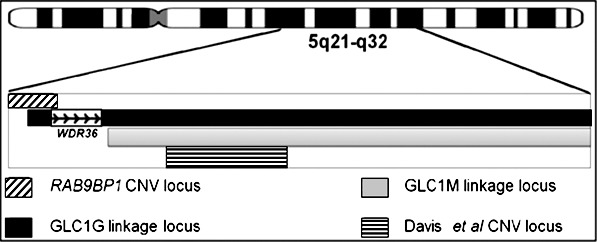
Relative positions of the glaucoma susceptibility loci reported in the 5q21-32 region by various studies and the RAB9BP1 CNV locus reported in our study. From top to bottom: the RAB9BP1 CNV locus found associated with IOP in our study; the GLC1G linkage locus with the candidate gene WDR36 reported at this locus33; the GLC1m linkage locus36; and a CNV locus associated with POAG within these linkage loci.27 In the figure, the extent of the RAB9BP1 CNV locus represents the largest CNV at this locus reported in our study (4.5 Mb). As can be seen, it is located near the centromeric end of the GLC1G linkage locus.
Studies have raised the possibility that the 5q region might harbor common genetic factors regulating both IOP and glaucoma risk.27,33–38 Our case–control study, however, failed to show an association for the RAB9BP1 locus with high-pressure POAG cases, which could be a reflection of the fact that while IOP is an important quantitative trait for POAG, the two phenotypes are not equivalent. Our finding is more consistent with the hypothesis that RAB9BP1 CNV locus controls IOP but not necessarily POAG, the pathogenesis of which may also involve other mechanisms such as those affecting susceptibility to optic nerve head changes.
CNVs at this locus are present in just 1% to 3% of the general population, which poses a problem in terms of analytical power for association and replication analyses. Larger sample sizes might be required to detect or replicate associations for that locus. Moreover, the case–control samples used in our study have been collected in a clinical setting, with the cases obtained from a glaucoma clinic and the controls obtained from a cataract clinic. It is possible that biases in reporting IOP measurements in clinical as opposed to research settings might have confounded our ability to detect an association; it is often a practice in glaucoma clinics to report higher IOP values (owing to confirmation bias of high IOPs measured in primary care at the time of referral) than cataract clinics (considering that high IOP might be a contraindication for cataract surgery). Finally, the TaqMan assay targeted most but not all CNVs that overlapped with the RAB9BP1 gene; failure to detect some of the CNVs lying in the vicinity of the RAB9BP1 gene, putatively disrupting gene regulatory elements, but not overlapping the open reading frame of the gene, could also have affected our ability to detect significant associations in the case–control study.
Our study suggests that there may be a role for CNVs in regulating IOP; however, as the associated CNVs found in the study were rare, their population-wide risk contribution is only modest. The effect size we report for the RAB9BP1 CNV (1.56 mm Hg) is consistent with other loci identified through GWAS for many traits and diseases. These effect sizes are suggestive of plurigenic architecture of IOP, and we believe that the RAB9BP1 is only one among a number of genetic factors causing similar changes in IOP. Our finding also suggests that the regulation of IOP is genetically heterogeneous. We believe that our locus 5q21.2 CNV finding represents a novel locus influencing regulation of IOP; however, with various studies reporting different loci within this region (5q21-32), there appears to be a complex mechanism by which genes in this region influence susceptibility to glaucoma. Further investigation through sequencing studies might be able to provide more significant clues regarding the causal gene at this locus.
Supplementary Material
Acknowledgments
We thank all study participants, their relatives, and staff at the recruitment centers for their invaluable contributions. We thank Toby Andrew, Margarida Lopes, Samantha Fahy, and Diana Kozareva for their contributions. Australian Twins thanks Grant Montgomery, Nicholas Martin, Scott Gordon, Dale Nyholt, Sarah Medland, Brian McEvoy, Margaret Wright, Anjali Henders, and Megan Campbell for ascertaining and processing genotyping data; Jane MacKinnon, Shayne Brown, Lisa Kearns, Jonathan Ruddle, Paul Sanfilippo, Sandra Staffieri, Olivia Bigault, Colleen Wilkinson, Jamie Craig, Yaling Ma, and Julie Barbour for assisting with clinical examinations; and Camilla Day and staff at the Center for Inherited Disease Research. BMES thanks Elena Rochtchina from the Centre for Vision Research, Department of Ophthalmology and Westmead Millennium Institute, University of Sydney (New South Wales, Australia); John Attia, Rodney Scott, and Elizabeth G. Holliday from the University of Newcastle (Newcastle, New South Wales, Australia); Jing Xie and Andrea J. Richardson from the Centre for Eye Research Australia, University of Melbourne; Michael T. Inouye, Medical Systems Biology, Department of Pathology and Department of Microbiology and Immunology, University of Melbourne (Victoria, Australia); Ananth Viswanathan, Moorfields Eye Hospital (London, UK); Paul J. Foster, National Institute for Health Research Biomedical Research Centre for Ophthalmology, UCL Institute of Ophthalmology, and Moorfields Eye Hospital (London); Peter McGuffin, MRC Social Genetic and Developmental Psychiatry Research Centre, Institute of Psychiatry, King's College (London, UK); Fotis Topouzis, Department of Ophthalmology, School of Medicine, Aristotle University of Thessaloniki, AHEPA Hospital (Thessaloniki, Greece); and Xueling Sim, National University of Singapore.
Supported by the Wellcome Trust, the European Union MyEuropia Marie Curie Research Training Network, Guide Dogs for the Blind Association, the European Community's FP7 (HEALTHF22008201865GEFOS), ENGAGE (HEALTHF42007201413), the FP-5 GenomEUtwin Project (QLG2CT200201254), US National Institutes of Health/National Eye Institute (1RO1EY018246), National Institutes of Health Center for Inherited Disease Research, and the National Institute for Health Research Comprehensive Biomedical Research Centre award to Guy's and St. Thomas' National Health Service Foundation Trust partnering with King's College London (TwinsUK); and Fight for Sight and The Worshipful Company of Spectacle Makers (AN). PGH is the recipient of a Fight for Sight ECI award; CJH is a National Institute for Health Research Senior Research fellow. TEST and BATS (Australian Twins) were supported by an Australian National Health and Medical Research Council (NHMRC) Enabling Grant (2004–2009, 350415, 2005–2007) Project Grant (350415); Clifford Craig Medical Research Trust; Ophthalmic Research Institute of Australia; American Health Assistance Foundation; Peggy and Leslie Cranbourne Foundation; Foundation for Children; Jack Brockhoff Foundation; National Institutes of Health/National Eye Institute (RO1EY01824601 [2007-2010]); Pfizer Australia Senior Research Fellowship (DAM); and Australian NHMRC Career Development Award (SM). Genotyping was funded by an NHMRC Medical Genomics grant and US National Institutes of Health/National Eye Institute (1RO1EY018246). Australian sample imputation analyses were carried out on the Genetic Cluster Computer, which is financially supported by the Netherlands Scientific Organization (NWO48005003). BMES was supported by the Australian National Health and Medical Research Council (NH&MRC), Canberra, Australia (974159, 211069, 457349, 512423, 475604, 529912); the Centre for Clinical Research Excellence in Translational Clinical Research in Eye Diseases; NH&MRC research fellowships (358702, 632909 [Jie Jin Wang]; 1028444 [Paul Nigel Baird]); and the Wellcome Trust, United Kingdom, as part of Wellcome Trust Case Control Consortium 2 (Ananth Viswanathan, Peter McGuffin, Paul Mitchell, Fotis Topouzis, Paul Foster) for genotyping costs of the entire BMES population (085475B08Z, 08547508Z, 076113). The Centre for Eye Research Australia receives Operational Infrastructure Support funding from the Victorian government.
Disclosure: A. Nag, None; C. Venturini, None; P.G. Hysi, None; M. Arno, None; E. Aldecoa-Otalora Astarloa, None; S. MacGregor, None; A.W. Hewitt, None; T.L. Young, None; P. Mitchell, None; A.C. Viswanathan, None; D.A. Mackey, None; C.J. Hammond, None
References
- 1. Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006; 90: 262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Friedman DS, Jampel HD, Munoz B, West SK. The prevalence of open-angle glaucoma among blacks and whites 73 years and older: the Salisbury Eye Evaluation Glaucoma Study. Arch Ophthalmol. 2006; 124: 1625–1630 [DOI] [PubMed] [Google Scholar]
- 3. Wolfs RC, Klaver CC, Ramrattan RS, van Duijn CM, Hofman A, de Jong PT. Genetic risk of primary open-angle glaucoma. Population-based familial aggregation study. Arch Ophthalmol. 1998; 116: 1640–1645 [DOI] [PubMed] [Google Scholar]
- 4. Sommer A. Glaucoma risk factors observed in the Baltimore Eye Survey. Curr Opin Ophthalmol. 1996; 7: 93–98 [DOI] [PubMed] [Google Scholar]
- 5. Charlesworth J, Kramer PL, Dyer T, et al. The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest Ophthalmol Vis Sci. 2010; 51: 3509–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Koolwijk LM, Despriet DD, Pardo, van Duijn CM, et al. Genetic contributions to glaucoma: heritability of intraocular pressure, retinal nerve fiber layer thickness, and optic disc morphology. Invest Ophthalmol Vis Sci. 2007; 48: 3669–3676 [DOI] [PubMed] [Google Scholar]
- 7. Carbonaro F, Andrew T, Mackey DA, Spector TD, Hammond CJ. Heritability of intraocular pressure: a classical twin study. Br J Ophthalmol. 2008; 92: 1125–1128 [DOI] [PubMed] [Google Scholar]
- 8. Stone EM, Fingert JH, Alward WL, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997; 275: 668–670 [DOI] [PubMed] [Google Scholar]
- 9. Rezaie T, Child A, Hitchings R, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002; 295: 1077–1079 [DOI] [PubMed] [Google Scholar]
- 10. Fingert JH. Primary open-angle glaucoma genes. Eye (Lond). 2011; 25: 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miu Y, Allingham RR. Molecular genetics in glaucoma. Exp Eye Res. 2011; 93: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011; 43: 574–578 [DOI] [PubMed] [Google Scholar]
- 13. Fan BJ, Wang DY, Pasquale LR, Haines JL, Wiggs JL. Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest Ophthalmol Vis Sci. 2011; 52: 1788–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010; 42: 906–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meguro A, Inoko H, Ota M, Mizuki N, Bahram S. Genome-wide association study of normal tension glaucoma: common variants in SRBD1 and ELOVL5 contribute to disease susceptibility. Ophthalmology. 2010; 117: 1331–1338, e5 [DOI] [PubMed] [Google Scholar]
- 16. Ramdas WD, van Koolwijk LM, Ikram MK, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010; 6: e1000978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Macgregor S, Hewitt AW, Hysi PG, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010; 19: 2716–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khor CC, Ramdas WD, Vithana EN, et al. Genome-wide association studies in Asians confirm the involvement of ATOH7 and TGFBR3, and further identify CARD10 as a novel locus influencing optic disc area. Hum Mol Genet. 2011; 20: 1864–1872 [DOI] [PubMed] [Google Scholar]
- 19. van Koolwijk LM, Ramdas WD, Ikram MK, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012; 8: e1002611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007; 316: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008; 455: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008; 40: 880–885 [DOI] [PubMed] [Google Scholar]
- 23. Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009; 459: 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011; 43: 838–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nishimura DY, Searby CC, Alward WL, et al. A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am J Hum Genet. 2001; 68: 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakata R, Usui T, Mimaki M, Araie M. Developmental glaucoma with chromosomal abnormalities of 9p deletion and 13q duplication. Arch Ophthalmol. 2008; 126: 431–432 [DOI] [PubMed] [Google Scholar]
- 27. Davis LK, Meyer KJ, Schindler EI, et al. Copy number variations and primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2011; 52: 7122–7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moayyeri A, Hammond CJ, Hart DJ, Spector TD. The UK Adult Twin Registry (TwinsUK Resource). Twin Res Hum Genet. 2013; 16: 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007; 17: 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002; 30: 97–101 [DOI] [PubMed] [Google Scholar]
- 31. Mackey DA, Mackinnon JR, Brown SA, et al. Twins eye study in Tasmania (TEST): rationale and methodology to recruit and examine twins. Twin Res Hum Genet. 2009; 12: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitchell P, Smith W, Attebo K, Healey PR. Prevalence of open-angle glaucoma in Australia. The Blue Mountains Eye Study. Ophthalmology. 1996; 103: 1661–1669 [DOI] [PubMed] [Google Scholar]
- 33. Monemi S, Spaeth G, DaSilva A, et al. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005; 14: 725–733 [DOI] [PubMed] [Google Scholar]
- 34. Kramer PL, Samples JR, Monemi S, Sykes R, Sarfarazi M, Wirtz MK. The role of the WDR36 gene on chromosome 5q22.1 in a large family with primary open-angle glaucoma mapped to this region. Arch Ophthalmol. 2006; 124: 1328–1331 [DOI] [PubMed] [Google Scholar]
- 35. Rotimi CN, Chen G, Adeyemo AA, et al. Genomewide scan and fine mapping of quantitative trait loci for intraocular pressure on 5q and 14q in West Africans. Invest Ophthalmol Vis Sci. 2006; 47: 3262–3267 [DOI] [PubMed] [Google Scholar]
- 36. Pang CP, Fan BJ, Canlas O, et al. A genome-wide scan maps a novel juvenile-onset primary open angle glaucoma locus to chromosome 5q. Mol Vis. 2006; 12: 85–92 [PubMed] [Google Scholar]
- 37. Lee MK, Woo SJ, Kim JI, et al. Replication of a glaucoma candidate gene on 5q22.1 for intraocular pressure in mongolian populations: the GENDISCAN Project. Invest Ophthalmol Vis Sci. 2010; 51: 1335–1340 [DOI] [PubMed] [Google Scholar]
- 38. Fan BJ, Ko WC, Wang DY, et al. Fine mapping of new glaucoma locus GLC1M and exclusion of neuregulin 2 as the causative gene. Mol Vis. 2007; 13: 779–784 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


