Abstract
A pvsB-vctA-irgA triple deletion mutant of Vibrio parahaemolyticus can utilize enterobactin under iron-limiting conditions by inducing a previously undescribed receptor, PeuA (VPA0150), in response to extracellular alkaline pH and enterobactin. In silico analyses revealed the existence of a two-component regulatory system operon, peuRS, immediately upstream of peuA, which constitutes an operon with the TonB2 system genes. Both the peuRS and peuA-tonB2 operons were found to be upregulated under iron-limiting conditions in a ferric uptake regulator (Fur)-dependent manner. The involvement of peuA and peuRS in enterobactin utilization was analyzed by complementation experiments using deletion mutants. Primer extension analysis indicated that, under iron-limiting conditions, the transcription of peuA was initiated from the +1 site at pH 7.0 and from both the +1 and +39 sites at pH 8.0 in the presence of enterobactin. The +39 transcript was absent from the peuRS deletion mutant. Secondary structure prediction of their 5′-untranslated regions suggested that translation initiation is blocked in the +1 transcript, but not in the +39 transcript. Consistent with this, in vitro translation analysis demonstrated that production of PeuA was determined only by the +39 transcript. These studies establish a novel gene regulation mechanism in which the two-component regulatory system PeuRS enhances expression of the alternative +39 transcript that possesses non-inhibitory structure, allowing the peuA expression to be regulated at the translation stage.
Introduction
Iron is essential for the growth of nearly all forms of life, but its very limited solubility makes iron scarce. The predominant type (ferric iron) preferentially forms barely soluble hydroxide complexes under aerobic conditions and at neutral and alkaline pH [1]–[3]. To solubilize iron from these complexes and to acquire adequate levels, bacteria as well as other microorganisms frequently secrete siderophores, including catecholates, hydroxamates, and polycarboxylates [2], [4], all of which exhibit high affinity for ferric iron. In Gram-negative bacteria, ferric siderophore complexes thus formed in the extracellular milieu are conveyed into the bacterial cell by a high-affinity active transport system composed of an outer membrane receptor (OMR) coupled with both a TonB-ExbBD protein complex (known as a TonB system) and an ATP-binding cassette (ABC) transporter system [5], [6]. The TonB system transduces the proton-motive force of the cytoplasmic membrane to the OMRs [5], [6], which are therefore known as TonB-dependent receptors. It is well known that, in these processes, the siderophore specificity resides mainly in the OMRs [2], [7]. Furthermore, expression of the genes responsible for iron acquisition is regulated by the cellular pool of iron through a ferric uptake regulator (Fur), which is ubiquitous in Gram-negative bacteria and usually acts as a repressor with ferrous iron as a co-repressor [8], [9]. When the intracellular iron concentration increases, the Fur-Fe2+ complex binds to a consensus sequence, termed the Fur box, located in the promoter regions of the Fur target genes, thereby leading to repression of transcription initiation. In contrast, when iron becomes scarce in the cell, Fur is inactivated by release of the iron cofactor, and the target genes are transcribed to efficiently scavenge iron from the surroundings.
In addition to their own siderophores, some bacteria have evolved transport systems for ferric complex that use exogenous siderophores (xenosiderophores) produced by other bacterial or fungal species [10]. This strategy, called siderophore piracy [11], may be highly advantageous for survival and proliferation of these bacteria, because it allows them to escape any bacteriostatic or competitive effects caused by xenosiderophores likely to coexist under various environmental conditions [12]. In Pseudomonas aeruginosa, the ferric enterobactin (Ent) receptor PfeA is induced in the presence of Ent combined with iron-starvation via the PfeRS two-component regulatory system [13], [14]. This system typically comprises an inner membrane-integrated histidine sensor kinase and a cytoplasmic response regulator that together form a signal transduction pathway to regulate gene expression; environmental stimuli, including a wide range of physical and chemical signals, trigger autophosphorylation of a histidine sensor kinase, and its phosphoryl group is subsequently transferred to a response regulator, which activates or represses transcription of the target gene required for the appropriate physiological response [15]–[17].
Vibrio parahaemolyticus is a Gram-negative and halophilic human pathogen that naturally inhabits marine and estuarine environments. It is a significant cause of acute gastroenteritis worldwide, acquired through the consumption of raw or undercooked seafood [18]–[20]. Under iron-limiting conditions, this bacterium secretes its own siderophore, vibrioferrin [21], which is biosynthesized by four enzymes encoded by pvsABDE [22], and transports extracellular iron as ferric vibrioferrin back to the cell via two OMRs specific to ferric vibrioferrin and the ABC transporter complex, which are encoded by pvuA1-pvuA2 and pvuBCDE, respectively [22]–[24]. In addition to producing vibrioferrin, V. parahaemolyticus can utilize aerobactin [25], ferrichrome [26], and Ent [27] as xenosiderophores by expressing their cognate OMRs.
In this report, we show that the V. parahaemolyticus peuA gene encoding the ferric Ent receptor is responsible for Ent utilization under iron-limiting conditions at pH 8.0. We also present evidence that the expression of PeuA is determined by an alternative transcript (+39 transcript) of peuA that is induced under iron-limiting conditions via a two-component regulatory system encoded by peuRS in response to extracellular alkaline pH and Ent.
Materials and Methods
Bacterial Strains, Plasmids, Growth Conditions, and Primers
The bacterial strains and plasmids used in this study are listed in Table 1 and S1, respectively. Escherichia coli β2155 [29], which is a diaminopimelic acid auxotroph, was grown under routine conditions and maintained in Luria-Bertani (LB) medium containing 0.5% NaCl and 0.5 mM 2,6-diaminopimelic acid. V. parahaemolyticus RIMD2210633 [28] and its deletion mutants were incubated in LB medium containing 3% NaCl or in LB medium containing 3% NaCl and 100 mM Tris-HCl (LB-Tris medium) at pH 7.0 and 8.0. To impose iron limitation on V. parahaemolyticus strains, they were grown in LB-Tris medium containing 25 µM ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDA; Sigma-Aldrich) (LB-Tris/+EDDA medium). When required, the siderophore Ent (Sigma-Aldrich) was added to the LB-Tris/+EDDA medium at a final concentration of 5 µM (LB-Tris/+EDDA/+Ent medium). Antibiotics were added at the following concentrations: 10 µg/mL chloramphenicol and 10 µg/mL tetracycline. The oligonucleotide primers used in this study are listed in Table S2.
Table 1. Bacterial strains used in this study.
| Strain | Description | Reference or source |
| V. parahaemolyticus | ||
| RIMD2210633 | Clinical isolate of serotype O3:K6; wild-type strain | [28] |
| VPD5 | RIMD2210633 ΔpvsB (vibrioferrin-deficient mutant) | [24] |
| VPD54 | VPD5 ΔvctA ΔirgA | [27] |
| VPD55 | VPD5 ΔvctA ΔpeuA | This study |
| VPD56 | VPD5 ΔirgA ΔpeuA | This study |
| VPD57 | VPD5 ΔvctA ΔirgA ΔpeuA | This study |
| VPD72 | VPD5 ΔvctA ΔirgA ΔtonB1 | This study |
| VPD73 | VPD5 ΔvctA ΔirgA ΔtonB2 | This study |
| VPD74 | VPD5 ΔvctA ΔirgA ΔtonB3 | This study |
| VPD102 | VPD5 ΔvctA ΔirgA ΔpeuRS | This study |
| VPD107 | VPD5 ΔpvuA1 ΔpvuA2 ΔhutA ΔfhuA ΔiutA ΔvctA ΔirgA | This study |
| VPD108 | VPD5 ΔpvuA1 ΔpvuA2 ΔhutA ΔfhuA ΔiutA ΔvctA ΔirgA ΔpeuA | This study |
| VPD109 | VPD5 ΔpvuA1 ΔpvuA2 ΔhutA ΔfhuA ΔiutA ΔvctA ΔirgA ΔpeuRS | This study |
| VPD110 | VPD5 ΔvctA ΔirgA ΔVP0168 | This study |
| E. coli | ||
| β2155 | thrB1004 pro thi strA hsdS Δ(lacZ)ΔM15 (F′ Δ(lacZ)M15 lacI q traD36 proA + proB +) ΔdapA::erm(Emr), pir::RP4(::kan(Kmr) from SM10) | [29] |
Growth Assay
The growth assay was performed using a TVS062CA biophotorecorder (Advantec Toyo, Tokyo, Japan). Briefly, V. parahaemolyticus cells grown overnight in LB medium were diluted with LB-Tris/+EDDA or LB-Tris/+EDDA/+Ent medium to an optical density at 600 nm (OD600) of 0.005. The cultures were shaken at 70 rpm at 37°C, and the OD600 was measured every 3 h for 18 h.
DNA Manipulation and in silico Sequence Analysis
Chromosomal DNA was extracted with a Wizard genomic DNA purification kit (Promega), and plasmid DNA was routinely prepared with a High Pure Plasmid Isolation Kit (Roche), according to the manufacturer's instructions. Standard DNA manipulation was performed as described [30]. Homology searches were performed using the BLAST program of the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/) [31].
Preparation of Outer Membrane Protein (OMP)-Rich Fractions and Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Stationary-phase V. parahaemolyticus cells were inoculated at a final OD600 of 0.005–0.01 into LB-Tris, LB-Tris/+EDDA, and LB-Tris/+EDDA/+Ent media at pH 7.0 and 8.0, and the cultures were shaken at 37°C for 4 h. Sarkosyl-insoluble OMPs were prepared and analyzed by SDS-PAGE, as previously described [32]. Separated OMPs were electroblotted onto a wet polyvinylidene difluoride membrane, and the N-terminal amino acid sequence was determined using the Edman degradation method with a Procise 491 HT protein sequencer (Applied Biosystems).
Gene Deletion and Complementation
Gene deletions in the V. parahaemolyticus genome were constructed by allelic exchange using the suicide plasmid pXAC623, according to the procedure described by Kuroda et al. [33]. Briefly, DNA fragments with deletions in the peuA, peuRS, hutA, iutA, fhuA, and VP0168 genes were prepared by overlap extension PCR [34], as previously described [24]. The deleted gene fragments were ligated into appropriately digested pXAC623 to yield pXAC623ΔpeuA, pXAC623ΔpeuRS, pXAC623ΔhutA, pXAC623ΔiutA, pXAC623ΔfhuA, and pXAC623ΔVP0168 (Table S1), which were then transformed into E. coli β2155 to generate the respective donor strains. After filter mating between each donor strain and an appropriate V. parahaemolyticus strain, merodiploid recombinants were selected on LB plates containing chloramphenicol, but not diaminopimelic acid. Each merodiploid recombinant was spread on VDS-broth agar plates (1% polypeptone, 0.5% yeast extract, 30 mM NaCl, 55 mM KCl, 10% sucrose, and 2.5% agar) [33] and incubated at 25°C for 30 h, at which point sucrose-resistant and chloramphenicol-sensitive colonies were selected. The deletions were verified by PCR, using chromosomal DNA isolated from each deletion mutant (data not shown). To complement the peuA and peuRS deletion mutants, PCR amplicons containing the respective genes were ligated into appropriately digested pRK415 [35], and the resulting complementing plasmids were transformed into the respective V. parahaemolyticus mutant strains.
RNA Analysis
Stationary-phase V. parahaemolyticus cells were inoculated as described for the preparation of OMP-rich fractions, and the cultures were then shaken at 37°C until they reached OD600 0.3–0.6. Each cell pellet was treated with the RNAprotect Bacteria Reagent (Qiagen), according to the manufacturer's instructions, and total RNA was prepared from each cell sample using the RNeasy Mini kit (Qiagen) or TriPure Reagent (Roche), according to the manufacturer's instructions. Total RNA samples thus obtained were used for primer extension, reverse transcriptase (RT)-PCR, and RT-quantitative (q) PCR.
Primer extension analysis of peuA and peuR was performed with the oligonucleotide primers peuA-PE and peuR-PE (see Table S2), respectively, which had been 5′-labeled with Texas Red prior to use. Each labeled primer was annealed to 10 µg or 150 µg of total RNA and extended with avian myeloblastosis virus RT XL (TaKaRa Biochemicals, Shiga, Japan) at 50°C for 90 min. The primer extension products were separated on a sequencing gel using an SQ5500E DNA sequencer (Hitachi High-Tech, Tokyo, Japan) alongside the DNA sequence ladder of the control region synthesized using the same primers used for the primer extension analysis.
For RT-PCR analysis, total RNA samples prepared from RIMD2210633 cells grown in LB-Tris/+EDDA medium at pH 7.0 were treated with TURBO DNase (Ambion) to remove contaminating chromosomal DNA. ReverTra Ace RT (Toyobo, Osaka, Japan) and the gene-specific primer VPA0156-R or peuS-R (see Table S2) was used to synthesize cDNA. cDNA synthesis was performed by incubating 0.5 µg of DNase-treated RNA in a 20-µl reaction for 60 min at 42°C. One microliter of the cDNA reaction mixture was then used as a template for PCR with the specific PCR primer pairs (see Table S2). PCR conditions were as follows: after an initial denaturation for 2 min at 95°C, DNA was amplified for 30 cycles, with each cycle consisting of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. As a negative control, PCR omitting prior reverse transcription was performed directly for the same RNA template to confirm the absence of contaminating chromosomal DNA. PCR products were electrophoresed through 1.5% agarose gels, stained with ethidium bromide, and visualized with UV light.
For RT-qPCR analysis, total RNA samples were treated with TURBO DNase, and a 0.5-µg aliquot of RNA was reverse transcribed with the ReverTra Ace RT and random hexamer primers (TaKaRa Biochemicals) for 60 min at 37°C. qPCR was performed using the peuA-specific primer pair VppeuA-qF/VppeuA-qR (see Table S2) and the Thunderbird SYBR qPCR Mix (Toyobo) in a Chromo4 Real-Time PCR detection system (Bio-Rad) under the conditions specified in the manufacturer's protocol. Relative mRNA expression levels were determined by the comparative threshold cycle method, using the 16S rRNA expression level as an internal control. The RT-qPCR primers for 16S rRNA are listed in Table S2.
Preparation of DNA Templates and in vitro RNA Synthesis
Both a longer form of the truncated peuA fragment (+1 to +402) flanked by the T7 promoter and flag sequences (+1-peuA′-flag DNA) and a shorter form of the truncated peuA fragment (+39 to +402) flanked by T7 promoter and flag sequences (+39-peuA′-flag DNA) were amplified from V. parahaemolyticus chromosomal DNA by PCR. The primer pairs T7-VppeuA-F/T7-VppeuA-FLAG-R and T7-VppeuA-F/T7-VppeuA-FLAG-R (see Table S2) were used to amplify the longer form and the shorter form, respectively. A full-length fur fragment flanked by the T7 promoter and flag sequences (fur-flag DNA) was amplified by first-step PCR using V. parahaemolyticus chromosomal DNA and the primer pair T7-Vpfur-F/T7-Vpfur-FLAG-R (see Table S2), and subsequently by second-step PCR using the first-step amplicon and the primer pair UNIVERSAL/T7-Vpfur-FLAG-R (see Table S2). The amplified DNA fragments were purified by agarose gel electrophoresis and used as templates for in vitro RNA synthesis. The +1-peuA′-flag, +39-peuA′-flag, and fur-flag RNAs were synthesized from the +1-peuA′-flag, +39-peuA′-flag, and fur-flag DNAs, respectively, by in vitro transcription with T7 RNA polymerase (Roche). Following the in vitro transcription reaction, the reaction mixtures were treated with 1 U RQ1 DNase (Promega) and purified on a ProbeQuant G50 Micro column (GE Healthcare), followed by ethanol precipitation.
In vitro Translation Assay and Western Blotting
In vitro translation was performed using a PURESYSTEM classic II (BioComber, Tokyo, Japan). For the RNA template for in vitro translation, either a +1-peuA′-flag RNA (30 pmol)/fur-flag RNA (3 pmol) mixture or a +39-peuA′-flag RNA (30 pmol)/fur-flag RNA (3 pmol) mixture was used. The PURESYSTEM reaction mixture (20 µl) was incubated at 37°C for 2 h, and the reaction was then terminated by adding an equal volume of 2× SDS-PAGE sample buffer. The samples were separated on a 15% SDS-polyacrylamide gel, and the protein bands were transferred to a Clear Blot Membrane-P (Atto, Tokyo, Japan). The membrane was blocked with Tris-buffered saline with Tween 20 (TBST) containing 0.3% skim milk and incubated overnight at 4°C with mouse anti-FLAG M2 antibody (Sigma) diluted 1,000-fold with blocking solution. The membrane was then washed four times with TBST, incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-mouse secondary antibody (GE Healthcare), diluted 20,000-fold with blocking solution, and washed four times with TBST. Immunoreactive bands were detected with an ECL Select Western Blotting Detection Reagent (GE Healthcare) and visualized with a LAS-3000 gel imager (Fujifilm, Tokyo, Japan)
Northern Blotting
The reaction mixtures (2.5 µl) obtained after in vitro translation of +1-peuA′-flag and +39-peuA′-flag RNAs were separated by electrophoresis on a 5% polyacrylamide/8 M urea gel in Tris-borate-EDTA buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA, pH 8.3), blotted onto a Biodyne B positively charged nylon membrane (Pall Corporation), and fixed to the membrane by baking for 30 min at 80°C. The digoxigenin (DIG)-labeled peuA probe was prepared with a primer pair, VppeuA-F/VppeuA-R (see Table S2), internal to the peuA gene using a DIG PCR Probe Synthesis Kit (Roche). Hybridization was performed overnight at 65°C, and the hybridized DIG-labeled peuA probe was detected using a DIG Luminescent Detection Kit (Roche) with a LAS-3000 image analyzer.
Results
Identification of the OMR Gene Responsible for Alkaline pH-Dependent Utilization of Ent in V. parahaemolyticus
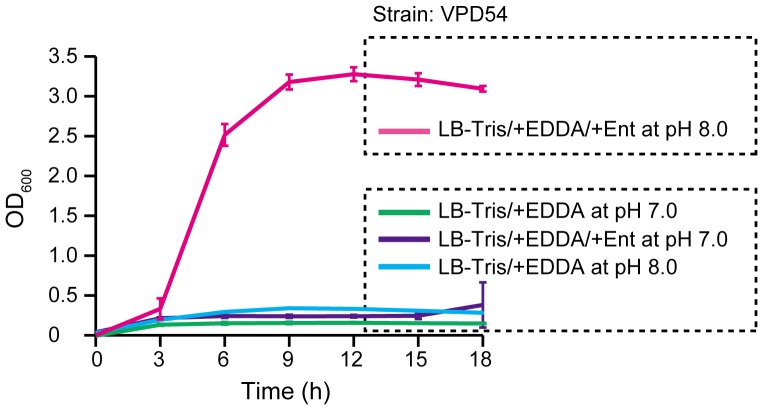
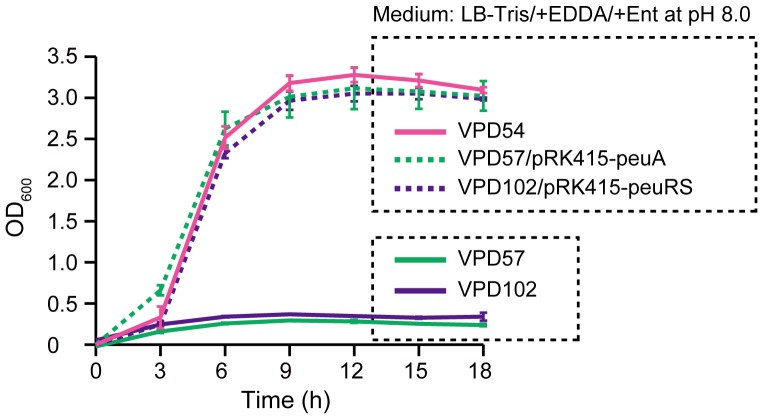
We previously reported that V. parahaemolyticus can utilize ferric Ent as an iron source via the VctA and IrgA receptors [27]. Although the VPD54 mutant with deletion of vctA and irgA that was generated from the VPD5 vibrioferrin-deficient mutant failed to grow in LB-Tris/+EDDA medium at pH 7.0 and 8.0, it showed normal growth when Ent was added to LB-Tris/+EDDA medium (LB-Tris/+EDDA/+Ent) at pH 8.0, but not at pH 7.0 (Figure 1). These data indicate that V. parahaemolyticus possesses a ferric Ent receptor gene that is specifically induced in response to extracellular alkaline pH and Ent. Several ferric siderophore receptors have already been identified and characterized in V. parahaemolyticus [23]–[27]; VPA0150 and VP0168 [28] have been documented as putative TonB-dependent receptors, but neither has been proven to be functional. To determine whether these receptors are involved in Ent utilization at pH 8.0, the two genes were deleted from the VPD54 mutant, and the resulting mutants, VPD57 and VPD110, were subjected to a growth assay. VPD110 grew in LB-Tris/+EDDA/+Ent medium at pH 8.0 (data not shown). In contrast, VPD57 showed no growth in LB-Tris/+EDDA/+Ent medium at pH 8.0, and the VPD57/pRK415-peuA complementing strain exhibited growth similar to the wild-type RIMD2210633 in the same medium (Figure 2). These data suggest that the VPA0150 gene encodes a receptor engaged in the uptake of ferric Ent at pH 8.0. In this paper, the VPA0150 gene is termed peuA (peu stands for V. parahaemolyticus Ent utilization).
Figure 1. Alkaline pH-dependent utilization of Ent in VPD54.
VPD54, which is a vctA and irgA deletion mutant generated from the VPD5 vibrioferrin-deficient mutant, was grown in LB-Tris/+EDDA medium (at indicated pH) at 37°C for 18 h with shaking at 70 rpm. When required, Ent was added at 5 µM. Cultures were monitored by measuring the OD600 every 3 h. Data are shown as means ± SD from 3 separate experiments.
Figure 2. Involvement of peuA and peuRS in Ent utilization.
The growth assay was performed as described in Figure 1. Data are shown as means ± SD from 3 separate experiments.
Identification of PeuA by SDS-PAGE and Determination of Its N-Terminal Amino Acid Sequence
To assess whether extracellular alkaline pH and Ent affect OMP profiles, OMP fractions prepared from V. parahaemolyticus mutants were analyzed by SDS-PAGE. The wild-type V. parahaemolyticus expresses several iron-repressible OMRs under iron-limiting conditions, as shown in Figure 3, lane 2. We used a VPD107 mutant lacking all of the known iron-repressible OMPs for this experiment, to eliminate potential interference by these OMPs. In LB-Tris/+EDDA medium at pH 7.0, VPD107 was unable to produce PeuA, regardless of Ent presence (Figure 3, lanes 3 and 4). In contrast, when VPD107 was grown in LB-Tris/+EDDA medium at pH 8.0, a faint protein band was detected (Figure 3, lane 5); the sequence of its first 10 N-terminal amino acids was determined to be NVQTDEHLVV. This sequence exactly matched the N-terminal sequence deduced from peuA (see Figure 4A), indicating that peuA indeed encodes the ferric Ent receptor. The production of PeuA in VPD107 was remarkably increased in LB-Tris/+EDDA+Ent medium at pH 8.0 (Figure 3, lane 6). These results indicate that PeuA is not produced in significant amounts, even under iron-limiting conditions at pH 8.0, unless Ent is present in the growth medium. In addition, VPD108, a peuA deletion mutant derived from VPD107, failed to produce PeuA in LB-Tris/+EDDA/+Ent medium at pH 8.0, whereas the complementing strain VPD108/pRK415-peuA restored the ability to produce PeuA (Figure 3, lanes 7 and 8).
Figure 3. SDS-PAGE analysis of Sarkosyl-insoluble OMPs of V. parahaemolyticus.
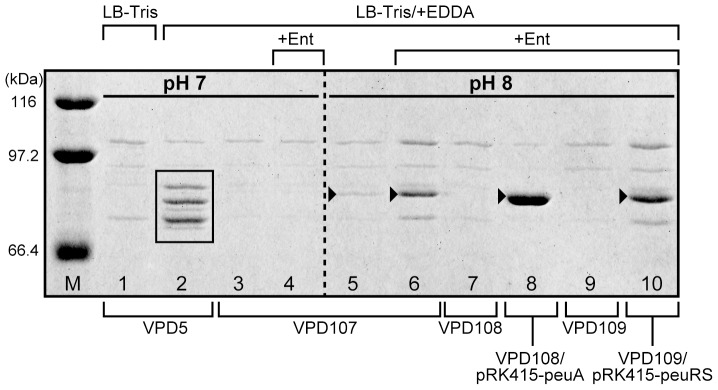
SDS-PAGE analysis was performed with VPD5, VPD107 (seven iron-repressible OMRs-deficient mutant derived from VPD5), VPD108 (peuA-deficient mutant derived from VPD107), VPD108/pRK415-peuA, VPD109 (peuRS-deficient mutant derived from VPD107), and VPD109/pRK415-peuRS. The OMP fractions were prepared from cells grown in LB-Tris medium at pH 7.0, LB-Tris/+EDDA media at pH 7.0 and 8.0, or LB-Tris/+EDDA/+Ent media at pH 7.0 and 8.0. Lanes 1–7 and 9–10 were loaded with 20 µg OMPs, and lane 8 was loaded with 3 µg OMPs. Electrophoresis was performed on 7.5% SDS-polyacrylamide gels (130 mm long) at a constant current of 15 mA at 4°C. The gel was stained with Coomassie Brilliant Blue. The figure shows only the relevant portions of the gel. The iron-repressible OMPs expressed by VPD5 at pH 7.0 under iron-limiting conditions are boxed in lane 2. Lane M, molecular weight marker proteins; closed arrowheads, PeuA.
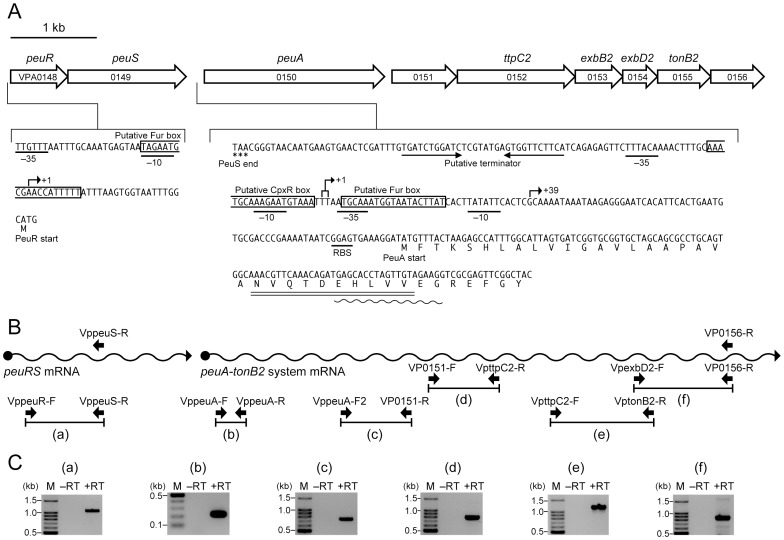
Figure 4. Genetic map and operon structure of VPA0148–VPA0156 locus.
(A) Genetic map of the peuA gene and the flanking genes. Thick arrows indicate genes and their orientations. The –35 and –10 promoter elements and putative Fur box sequences in the promoter regions of peuR (VPA0148) and peuA (VPA0150) are indicated. The transcription start sites for peuR (+1) and peuA (+1 and +39) are indicated by right-angled arrows. The putative terminator signal between the peuS and peuA genes, the predicted RBS for the peuA gene, the start codons for peuR and peuA genes, and the stop codon for peuS are also indicated. The amino acid sequence consistent with the N-terminal sequence determined for the iron-repressible OMR induced in LB-Tris/+EDDA and LB-Tris/+EDDA/+Ent media at pH 8.0 (see Figure 3) is indicated by a double underline. (B) Schematic representation of mRNAs transcribed from the VPA0148-VPA0156 genes and the primer pairs used for RT-PCR. For preparation of cDNAs by RT, VPpeuS-R and VP0156-R were used. (C) RT-PCR analysis of RT-PCR products. +RT and –RT, RT-PCR was performed with and without reverse transcriptase, respectively. M, 100-bp DNA ladder.
In silico Sequence Analyses of peuA and Its Adjacent Genes
A map of peuA and the neighboring genes is shown in Figure 4A, accompanied by partial nucleotide and deduced amino acid sequences [28]. PeuA shared amino acid similarity with many ferric hydroxamate-type siderophore receptors, such as FhuE (25% identity over 751 amino acids) of E. coli [36]; however, it displayed lower sequence similarity with FepA (20% identity over 729 amino acids) and PfeA (23% identity over 226 amino acids), the ferric Ent receptors of E. coli [37] and P. aeruginosa [13], respectively. A putative Fur box sequence resembling the consensus binding site for the Fur protein in E. coli [38] was detected in the promoter region of peuA (Figure 4A), and indeed this region cloned on pUC19 was positive in the Fur titration (FURTA) in vivo assay [39] (data not shown), indicating that the cloned region harbor the binding site of the E coli Fur protein. Moreover, a tandem repeat of 5′-A(N)3 GCAAA(N)4 GTAAA-3′ (the conserved nucleotides are underlined), termed the CpxR-box [40], [41], which is typical of the CpxR-binding site, was identified in the promoter region of peuA (Figure 4A).
Homology searches revealed the existence of putative two-component regulatory system genes (VPA0148-0149), collectively named peuRS, immediately upstream of peuA (Figure 4A). PeuR and PeuS showed amino acid sequence similarity to components of the CpxAR signaling system [42], such as the P. aeruginosa PfeR response regulator (36% identity over 225 amino acids) [14] and the E. coli CpxA histidine sensor kinase (24% identity over 447 amino acid residues) [43]. The consensus amino acid sequences in the conserved domains of the response and sensor components [13], [15] were also determined for PeuR and PeuS. In addition, they displayed striking structural features, including the presence of invariant amino acid residues, aspartic acid-9 in PeuR and histidine-244 in PeuS, both of which probably serve as phosphorylation sites. The hydropathy profile revealed that PeuS contains two transmembrane-spanning regions and an intervening 115-amino acid extracytoplasmic loop domain that is exposed to the cytoplasmic space. These features of PeuRS are illustrated in Figure S1. Moreover, the cloned promoter region of peuR also showed a FURTA-positive phenotype (data not shown), indicative of the presence of the Fur binding site.
Identification of peuA/VPA0151-0156 and peuRS as Iron-Repressible Operons by RT-PCR
The VPA0151-0156 genes, including the ttpC2-tonB2 system genes, are located downstream of the peuA gene, and the open reading frames of the VPA0151-0156 genes have overlapping stop and start codons, an arrangement typical of transcription unit boundaries in prokaryotic genomes [44]. Although there is a 53-bp gap, including an inverted repeat, immediately downstream of peuA, this region contains no potential promoter sequences for the downstream tonB2 operon (data not shown). As expected, RT-PCR using the primer pairs designed to cover the respective intergenic regions of the peuA and VPA0151-0156 genes produced extension bands of the expected size for total RNA prepared from the wild-type strain grown in LB-Tris/+EDDA at pH 7.0 (Figure 4B and C), indicating that these genes are co-transcribed in an iron-regulated operon.
The stop codon of peuR overlaps with the start codon of peuS, and no definitive promoter sequence was detected upstream of peuS. An RT-PCR product of the expected size was also detected for total RNA prepared from the wild-type strain grown in LB-Tris/+EDDA at pH 7.0 when the primer pair VppeuR-F/VppeuS-R was used to amplify the intergenic region between the peuRS genes (Figure 4B and C), indicating that these genes also comprise an iron-regulated operon.
Involvement of the peuRS Genes in Ent Utilization at pH 8.0
To investigate the requirement of peuRS for Ent utilization at pH 8.0 under iron-limiting conditions, we generated the peuRS mutant VPD102 from VPD54, which possesses the peuA gene but not the vctA and irgA genes, for use in a growth assay. While VPD102 abolished the Ent-mediated growth observed for VPD54 in LB-Tris/+EDDA medium at pH 8.0, the complementing strain VPD102/pRK415-peuRS grew well in the same medium (Figure 2). Furthermore, to examine the effect of peuRS on PeuA production by SDS-PAGE, the VPD109 mutant with the peuRS operon deleted was constructed from VPD107. VPD109 showed a complete lack of PeuA, even in LB-Tris/+EDDA/+Ent medium at pH 8.0 (Figure 3, lane 9); however, the complementing strain VPD109/pRK415-peuRS restored the ability to produce PeuA in the same medium (Figure 3, lane 10). Taken together, these data highlight a possible role of the PeuRS two-component regulatory system in the production of PeuA induced by alkaline pH and Ent.
Iron-Repressible Transcription of peuR
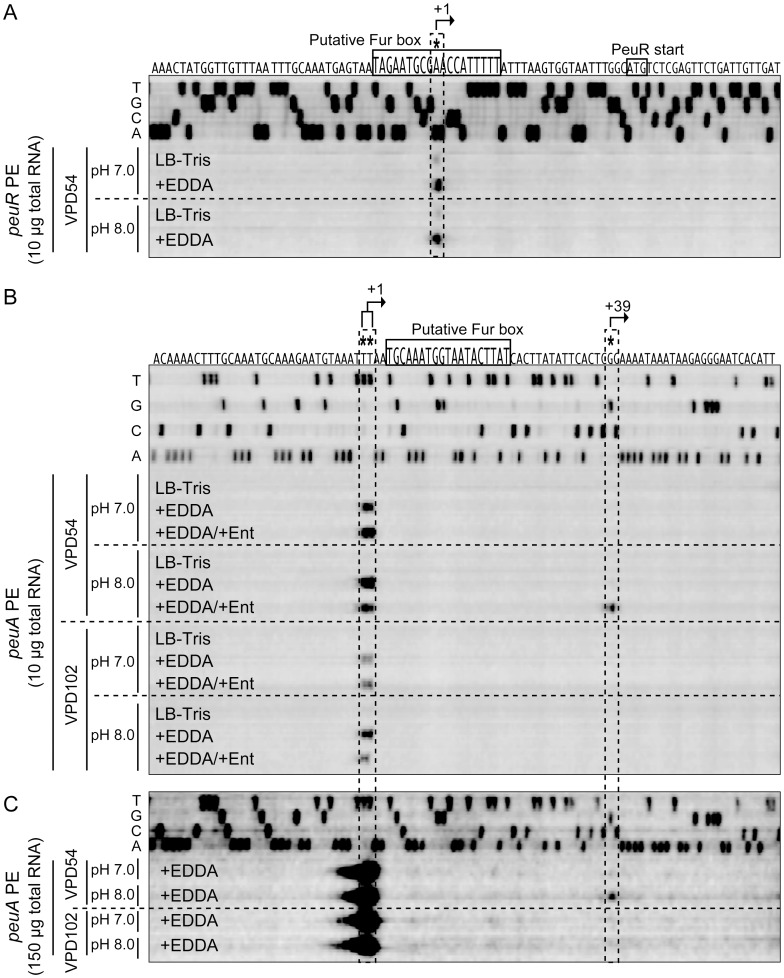
To determine the transcriptional start site for peuR, and to test whether its expression is iron-regulated, primer extension analysis was also performed for total RNA samples of VPD54 cells grown in LB-Tris and LB-Tris/+EDDA media. The transcription of peuR was unambiguously derepressed in LB-Tris/+EDDA medium, independent of pH (Figure 5A), and the transcription start site (+1) of peuR was determined to be 29 nucleotides upstream of its start codon (Figure 4A). It is evident from these data that the peuRS operon is constitutively expressed under iron-limiting conditions.
Figure 5. Primer extension analyses of total RNA from VPD54 or VPD102 to determine the transcription start sites of peuR (A) and peuA (B and C).
Total RNAs were isolated from VPD54 (vctA- and irgA-deficient mutant derived from VPD5) and VPD102 (peuRS-deficient mutant derived from VPD54) grown at pH 7.0 and 8.0 in LB-Tris, LB-Tris/+EDDA, and LB-Tris/+EDDA/+Ent media. The amounts of total RNA and primers used for reverse transcription were as follows: (A) 10 µg VppeuR-PE, (B) 10 µg VppeuA-PE, and (C) 150 µg VppeuA-PE. The same primers used for primer extension analysis were used to generate the sequence ladders (A, C, G, T). The transcription start sites and putative Fur boxes are indicated at the top of panels A and B (also see Figure 3A).
Induction of an Alternative Transcript of peuA under Iron-Limiting Conditions in Response to Extracellular Alkaline pH and Ent
Primer extension analyses were performed with total RNA samples of both VPD54 and its peuRS deletion mutant VPD102. Two primer extension products were detected, when total RNA samples prepared from VPD54 grown in LB-Tris/+EDDA media at pH 7.0 and 8.0 or LB-Tris/+EDDA/+Ent medium at pH 7.0 were used as the templates. However, the growth of VPD54 was not promoted under the same conditions. Two probable transcription start sites of peuA were mapped to 105 bp and 104 bp upstream of the peuA ATG start codon (Figure 5B); hereafter, the transcripts from the +1 and +2 sites are collectively referred to as the +1 transcript. Another transcription start site for peuA (+39 transcript) was detected 67 bp upstream of its start codon, when VPD54 was grown in LB-Tris/+EDDA/+Ent medium at pH 8.0 (Figure 5B). In addition, the growth of VPD54 was promoted under the same conditions. These results suggested that the +1 transcript is not responsible for the growth of VPD5 as shown in Figure 1. Interestingly, the primer extension product of the +39 transcript was not detected when VPD102 was grown in the same medium at pH 7.0 (Figure 5B). However, when primer extension analysis was performed using a 15-fold excess of RNA, a small amount of the primer extension product of the +39 transcript was also detected in VPD54 cells grown in LB-Tris/+EDDA medium at pH 8.0, even when Ent was absent from the medium (Figure 5C). This is consistent with the expression of PeuA in VPD107 grown under the same conditions (Figure 3, lane 5), and this low level of PeuA expression in VPD107 under iron-limiting conditions may contribute to the initial uptake of ferric Ent for stimulation of PeuS prior to induction of peuA expression at pH 8.0. However, no extension band was detected for total RNA prepared from VPD54 grown in LB-Tris medium, suggesting that the putative Fur box detected in the promoter region of peuA (Figure 4A) was functional for the iron-repressive regulation of peuA. Collectively, these findings indicate that peuA mRNA is transcribed as the +1 transcript under iron-limiting conditions irrespective of Ent and pH, while the +39 transcript responsible for the expression of PeuA is expressed in trace amounts in LB-Tris/+EDDA medium at pH 8.0 and is significantly increased by the presence of Ent under the same conditions. These data also suggested that the increase in the +39 transcript was absolutely dependent on the PeuRS two-component regulatory system. In addition, two sets of the −35 and −10 promoter sequences are properly positioned for transcription from the +1 and +39 sites (Figure 4A).
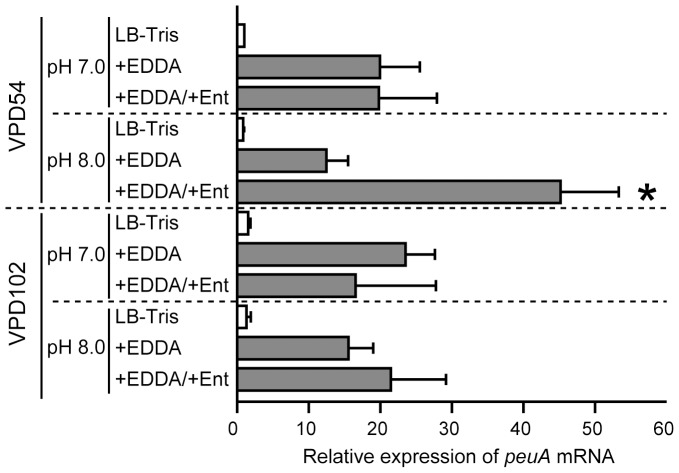
To confirm the iron-regulated expression of peuA, RT-qPCR analysis was performed. At pH 7.0 and 8.0, the transcription of peuA was strongly induced in VPD54 and VPD102 in LB-Tris/+EDDA medium (10- to 20-fold increases compared to the level in LB-Tris medium) (Figure 6). Moreover, in LB-Tris/+EDDA medium at pH 7.0, VPD54 produced peuA mRNA at similar levels in the presence and absence of Ent; however, the addition of Ent to LB-Tris/+EDDA medium at pH 8.0 conspicuously increased the level of peuA mRNA (Figure 6). Considering the results of primer extension analysis (Figure 5B), these data suggested that the increase in the peuA mRNA was due to transcription from the +39 site. No such effect of Ent on peuA transcription at pH 8.0 was observed for the peuRS-deletion mutant VPD102 (Figure 6), implying that the peuRS operon is responsible for the transcription of peuA from the +39 site.
Figure 6. Relative levels of peuA mRNA, assessed by RT-qPCR.
Total RNA samples were prepared from VPD54 (vctA- and irgA-deficient mutant derived from VPD5) and VPD102 (peuRS-deficient mutant derived from VPD54) grown at pH 7.0 and 8.0 in LB-Tris, LB-Tris/+EDDA, and LB-Tris/+EDDA/+Ent media. Data are shown as means ± SD from 3 separate experiments. An asterisk indicates P<0.05 compared to other samples.
mRNA Secondary Structure Prediction
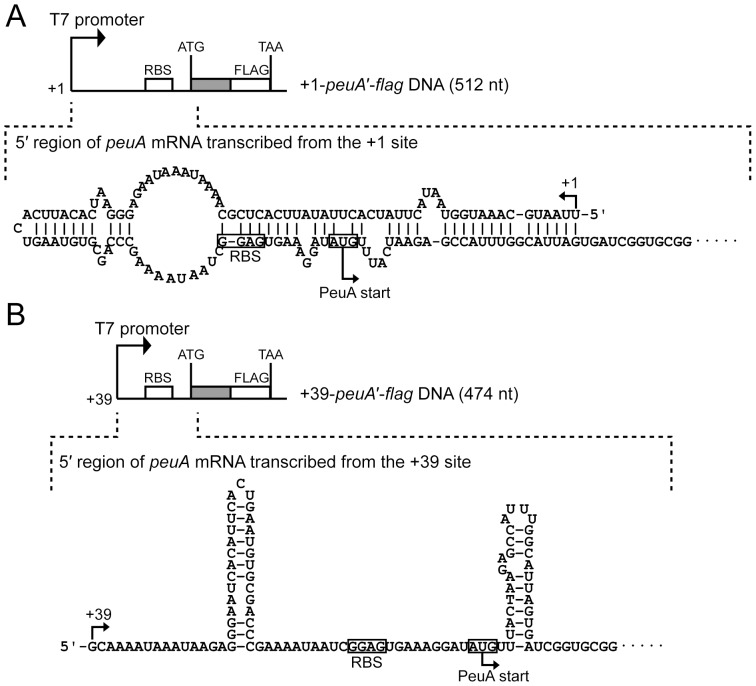
The results shown in Figure 3 indicate that remarkable amounts of PeuA are produced under iron-limiting conditions in response to an extracellular pH of 8.0, but not 7.0, and Ent. This corresponds with appearance of the +39 transcript, in addition to +1 transcription, under the same growth conditions. These findings led us to hypothesize that these transcripts might contain regulatory signals in their 5′-untranslated regions (UTRs) that couple transcription to translation. Thus, to better define the nature of these transcripts, the secondary structures of their 5′-UTRs were predicted using the CENTROIDFOLD program (http://www.ncrna.org/centroidfold/) [45]. Figure 7A shows that the first 40 nucleotides of the 5′-UTR of the +1 transcript are folded into the secondary structure with the ribosomal binding site (RBS) and the start codon of peuA to block initiation of translation. As opposed to the +1 transcript, the +39 transcript does not form an inhibitory structure in its 5′-UTR, thus allowing initiation of translation (Figure 7B). Therefore, we hypothesized that PeuA production might be dependent on the translation of the +39 peuA transcript.
Figure 7. Schematic representation of the +1-peuA′-flag (A) and +39-peuA′-flag (B) DNA fragments.
Each of these DNA fragments includes a nucleotide sequence corresponding to the peuA 5′-UTR from the +1 or +39 sites and the nucleotide sequence for the N-terminal 99 amino acid residues (in gray), in addition to a T7 promoter and a FLAG tag preceding the stop codon (TAA). The secondary structures of the 5′-UTRs of the +1 transcript (A) and the +39 transcript (B) of peuA are shown, both of which were predicted by the CentroidFold software (http://www.ncrna.org/centroidfold/). The RBS and start codon of peuA mRNA are boxed in the secondary structures.
The +39 Transcript is Responsible for PeuA Production
To test the above hypothesis, an in vitro translation assay was performed. The RNA templates for in vitro translation, i.e., +1-peuA′-flag (Figure 7A), +39-peuA′-flag (Figure 7B), and fur-flag RNAs, were constructed by in vitro transcription using DNA templates containing the T7 promoter. The in vitro translation products were analyzed by SDS-PAGE followed by western blotting using anti-FLAG IgG. The PeuA′-FLAG product was not detected when a mixture of +1-peuA′-flag RNA and fur-flag RNA was used for in vitro translation, even though Fur-FLAG, a positive control for in vitro translation, was detected; however, when a mixture of +39-peuA′-flag RNA and fur-flag RNA was used as the template, a significant amount of PeuA′-FLAG was detected along with Fur-FLAG (Figure 8A). Simultaneously, the +1-peuA′-flag RNA and +39-peuA′-flag RNA in each reaction mixture for in vitro translation were validated by northern blotting using a DIG-labeled peuA probe (Figure 8B). These data are concordant with the secondary structures predicted for the +1 and +39 transcripts of peuA.
Figure 8. In vitro translation of peuA mRNA.
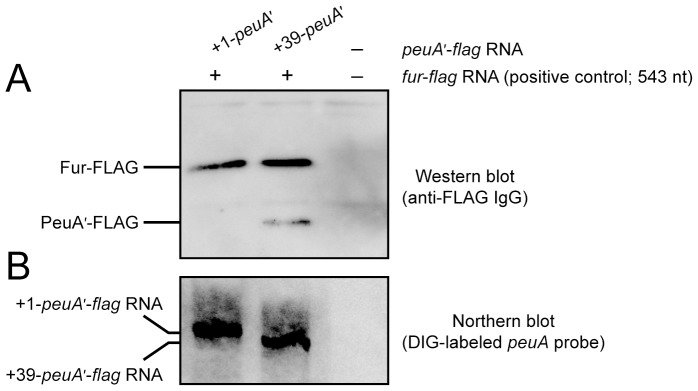
(A) In vitro translation analysis of the +1 and +39 peuA transcripts labeled with the FLAG tag. The +1-peuA′-flag RNA and +39-peuA′-flag RNA were first synthesized by in vitro transcription, as described in the MATERIALS AND METHODS, and a mixture containing either the +1-peuA′-flag RNA (30 pmol)/fur-flag RNA (3 pmol) or the +39-peuA′-flag RNA (30 pmol)/fur-flag RNA (3 pmol) as the template was subjected to in vitro translation. The FLAG-fused proteins translated were separated on 15% SDS-polyacrylamide gels, and were detected by western blotting using anti-FLAG IgG. (B) Confirmation of the presence of +1-peuA′-flag RNA and +39-peuA′-flag RNA in the reaction mixture for in vitro translation. These RNA fragments were detected in the reaction mixture by northern blotting using a DIG-labeled peuA probe.
TonB Specificity of PeuA
V. parahaemolyticus contains up to three TonB clusters encoding TonB1 and TonB2 systems on the small chromosome and a TonB3 system on the large chromosome, and the TonB2 system is located downstream of peuA. To determine which TonB systems are involved in the transport of ferric Ent via PeuA, a set of isogenic tonB deletion mutants were constructed from VPD54. A growth assay showed that only the VPD73 mutant deficient in tonB2 lost the ability to grow in LB-Tris/+EDDA/+Ent medium at pH 8.0; however, the VPD72 and VPD74 mutants deficient in tonB1 and tonB3, respectively, grew as well as their parental strain, VPD54, in the same medium (Figure S2). These observations show that the TonB2 system functions as an energy modulator for PeuA.
Distribution of Orthologs of the V. parahaemolyticus PeuRSA-VPA0151-VPA0156 Genes Among Other Vibrio Species
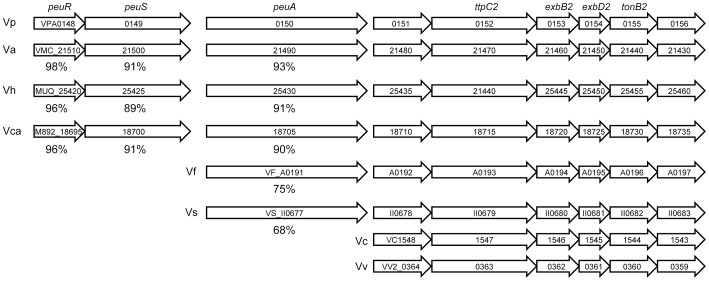
Using BLAST analyses, we examined whether orthologs to the V. parahaemolyticus peuRSA-VPA0151–VPA0156 locus genes are distributed among the whole-genome sequences of other Vibrio species. Although the VPA0151–VPA0156 (the ttpC2-tonB2 system genes) cluster was identified in all Vibrio species examined, peuRS and peuA orthologs were identified only in V. alginolyticus, V. harveryi, and V. campbellii, which belong to the same phylogenetic group (the V. harveyi group) as V. parahaemolyticus [20] (Figure 9). However, all peu orthologs are absent from V. chorelae and V. vulnificus, and V. fischeri and V. splendidus possess the peuA ortholog, but not the peuRS orthologs.
Figure 9. Distribution of the Vibrio parahaemolyticus VPA0148–VPA0156 orthologs in other Vibrio species for which whole-genomic sequences have been reported.
Arrows represent genes and their orientations. The numbers below the genes indicate percent amino acid sequence similarities to V. parahaemolyticus PeuRS and PeuA. Vp, V. parahaemolyticus; Va, V. alginolyticus; Vh, V. harveyi; Vca, V. campbellii; Vf, V. fischeri; Vs, V. splendidus; Vc, V. cholerae; Vv, V. vulnificus.
Discussion
Based on the results obtained in this study, we propose a mechanism for enhanced expression of the V. parahaemolyticus peuA through the action of PeuRS in response to extracellular alkaline pH and Ent (Figure S3). Our data showed that peuA is transcribed in a polycistronic mRNA with the VPA0151–VPA0156 genes under iron-limiting conditions (Figure 4B). However, at neutral pH, translation of the +1 peuA transcript appeared to be inhibited by the formation of a secondary structure in its 5′-UTR that blocks the RBS and start codon of peuA (Figure 7A); in contrast, the VPA0151-VPA0156 mRNA responsible for the TonB2 system was expected to be translated normally under the same conditions, because the RBSs and start codons of their genes are available for ribosomal access. This expectation is supported by the fact that the V. parahaemolyticus PvuA1 is exclusively dependent on the energy transduced by the TonB2 system [24] and remains fully functional as the ferric vibrioferrin receptor under iron-limiting conditions at neutral pH. When grown under iron-limiting conditions at pH 8.0, V. parahaemolyticus expressed the peuA-VPA0151–VPA0156 polycistronic mRNA from the +39 start site that was induced via the two-component regulatory system, PeuRS (Figure 5B). No inhibitory secondary structure was identified in the 5′-UTR of the +39 transcript; in other words, PeuA is synthesized owing to the translation of the +39 transcript (Figure 7B). Moreover, as shown in Figures 3, 5, and 6, the levels of PeuA and the +39 transcript expressed under iron-limiting conditions were more markedly elevated in the presence than in the absence of Ent. These observations indicate that Ent functions as a potent inducer for transcription from the +39 site, with the aid of PeuRS.
A number of Gram-negative bacteria, including P. aeruginosa [46], Bordetella spp. [47], V. cholerae [48], V. anguillarum [49], V. parahaemolyticus [27], and Neisseria gonorrhoease [50], are known to utilize Ent as a xenosiderophore, which induces the cognate ferric Ent receptors under iron-limiting conditions. Such a wide distribution of this system in bacteria may be explained by the fact that Ent has an exceptionally high affinity for ferric iron, and its production by bacterial species is wider than previously thought [51]; Ent has been reported to be synthesized and excreted by most enterics [52], as well as two Gram-positive Streptomyces species [53]. Moreover, ferric Ent is more stable at alkaline pH than ferric hydroxamate-type siderophores [54]. In the pathogens described above, except for P. aeruginosa, the AraC-like or LysR-like transcriptional regulators operate to induce these ferric Ent receptors. P. aeruginosa has been reported to utilize Ent through the two-component regulatory system PfeRS [13], [14]; however, expression of the ferric Ent receptor (PfeA) in this species is enhanced in response to Ent under iron-limiting conditions [46], and only a single set of promoter sequences (−10 and −35) are present in the region upstream of pfeA [14]. Another unique two-component regulatory system, operating through heme-dependent regulation, has been described in the Gram-positive bacterium Corynebacterium diphtheriae for the expression of a heme oxygenase gene responsible for the utilization of heme as an iron source [55]. In these systems, the signal molecules likely interact with the sensors, leading to activation of the response regulators.
The E. coli CpxAR two-component regulatory system is well known to be involved in counteracting extracellular stresses, including alkaline pH exposure [56], [57]. Extracellular signals cause a conformational change in CpxA, stimulating the autophosphorylation of a conserved histidine residue. Once this residue is phosphorylated, CpxA acts as a kinase and phosphorylates a conserved aspartate residue on CpxR. Phosphorylated CpxR acts on its target gene as a transcriptional activator [42]. Considering that PeuRS is homologous to members of the Cpx signaling system, it seems likely that the conformation of PeuS is altered to initiate the signal transduction cascade in response to an extracellular alkaline pH and Ent, although it is not known whether these stimuli interact with PeuS separately or cooperatively. The activated PeuS phosphorylates PeuR, and the resulting phosphorylated PeuR is expected to bind to the peuA promoter region to induce transcription from the +39 site. However, it remains unclear whether additional factor(s) are required to transduce the signals of extracellular alkaline pH and Ent.
V. parahaemolyticus has also been reported to utilize Ent through two other ferric Ent receptors, VctA and IrgA [27]. In this study, these receptors were ascertained to operate under iron-limiting conditions at pH 8.0 (see Figure S4). Therefore, it is likely that the PeuA-mediated Ent utilization system is substituted and/or supplemented by Ent utilization via VctA and IrgA, and vice versa, signifying that, in bacteria, the expression of multiple siderophore receptors may be a common strategy or a backup system to capture the iron essential for survival and proliferation. Moreover, from evolutionary and ecological points of view, it is of interest that the peuRSA cluster is restrictively distributed in the phylogenetic group that includes V. alginolyticus, V. harveyi, and V. campbellii in addition to V. parahaemolyticus, all of which live in marine or estuarine environments at a pH of approximately 8.1 [58], often in association with plankton or animals, including fish and shellfish [19], [20]. However, it is uncertain whether V. parahaemolyticus and the other species naturally encounter Ent. Alternatively, the authentic ligand for PeuA and PeuS could be another siderophore structurally similar to Ent that is produced by microorganisms inhabiting the same niches as V. parahaemolyticus.
In conclusion, our study establishes that under iron-limiting conditions, the V. parahaemolyticus two-component regulatory system PeuRS functions in concert with extracellular alkaline pH and Ent for the induction of peuA transcription at the +39 site, leading to production of PeuA. Further studies are needed to clarify the molecular mechanisms by which the PeuRS two-component system is activated in response to extracellular alkaline pH and Ent to induce transcription beginning at the +39 site.
Supporting Information
Amino acid sequences of PeuR (A) and PeuS (B). Consensus amino acid residues in the conserved regions are boxed, and invariant amino acid residues (proposed to be important for the function of PeuRS) are indicated by asterisks. In panel B, transmembrane (TM) helices proposed by the HMMTOP transmembrane topology prediction server (http://www.enzim.hu/hmmtop/index.php) are underlined. (C) A hydropathy plot of PeuS. The hydropathic index was calculated by the method of Kyte and Doolittle using a window of 21 amino acid residues. Solid bars correspond to the TM helices shown in panel B.
(PDF)
TonB specificity of PeuA in Ent utilization. The growth assay was performed as described in Figure 1. Data are shown as means ± SD from 3 separate experiments.
(PDF)
Proposed expression mechanism for V. parahaemolyticus PeuA ferric Ent receptor under iron-limiting conditions in response to extracellular alkaline pH and Ent. Thick arrows and wavy arrows represent the open reading frames and the direction of transcription and mRNAs, respectively. (A) Under iron-limiting conditions at pH 7.0, peuA is co-transcribed with VPA0151-VPA0156 from the +1 transcription start site. However, the transcript from the +1 site forms a secondary structure within its 5′-UTR, leading to inhibition of translation of the peuA mRNA, although the remaining VPA0151–VPA0156 mRNA is translated. (B) Under iron-limiting conditions at pH 8.0 in the absence of Ent, transcription of the peuA/VPA0151–VPA0156 operon from the +39 site also occurs to a slight extent, combined with normal transcription beginning at the +1 site. The presence of Ent under iron-limiting conditions at pH 8.0 is proposed to result in induction of transcription from the +39 site, and thereby leads to enhanced expression of the ferric Ent receptor PeuA, because the RBS and start codon of peuA in the +39 transcript are available for translation initiation. The peuA gene, therefore, is optimally expressed under iron-limiting conditions in response to extracellular alkaline pH and Ent. In addition, the two-component regulatory system, PeuRS, is proposed to be necessary to activate peuA transcription in response to these signals.
(PDF)
Growth assays of the VPD54, VPD55, VPD56, and VPD57 mutants in LB-Tris/+EDDA/+Ent medium at pH 8.0. The growth assay was performed as described in Figure 1. Data are shown as means ± SD from 3 separate experiments.
(PDF)
Plasmids used in this study.
(PDF)
PCR primers used in this study.
(PDF)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by a Grant-in-Aid for Young Scientists (B) (25870987) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Andrews SC, Robinson AK, Rodriguez-Quinones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237. [DOI] [PubMed] [Google Scholar]
- 2. Braun V, Hantke K, Koster W (1998) Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst 35: 67–145. [PubMed] [Google Scholar]
- 3. Ratledge C, Dover LG (2000) Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54: 881–941. [DOI] [PubMed] [Google Scholar]
- 4. Hider RC, Kong X (2010) Chemistry and biology of siderophores. Nat Prod Rep 27: 637–657. [DOI] [PubMed] [Google Scholar]
- 5. Noinaj N, Guillier M, Barnard TJ, Buchanan SK (2010) TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64: 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Postle K, Larsen RA (2007) TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20: 453–465. [DOI] [PubMed] [Google Scholar]
- 7. Koster W (2001) ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res Microbiol 152: 291–301. [DOI] [PubMed] [Google Scholar]
- 8. Escolar L, Perez-Martin J, de Lorenzo V (1999) Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181: 6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hantke K (2001) Iron and metal regulation in bacteria. Curr Opin Microbiol 4: 172–177. [DOI] [PubMed] [Google Scholar]
- 10. Miethke M, Marahiel MA (2007) Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71: 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schubert S, Fischer D, Heesemann J (1999) Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J Bacteriol 181: 6387–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brickman TJ, Armstrong SK (2009) Temporal signaling and differential expression of Bordetella iron transport systems: the role of ferrimones and positive regulators. Biometals 22: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dean CR, Poole K (1993) Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol Microbiol 8: 1095–1103. [DOI] [PubMed] [Google Scholar]
- 14. Dean CR, Neshat S, Poole K (1996) PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa . J Bacteriol 178: 5361–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Albright LM, Huala E, Ausubel FM (1989) Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet 23: 311–336. [DOI] [PubMed] [Google Scholar]
- 16. Hoch JA, Varughese KI (2001) Keeping signals straight in phosphorelay signal transduction. J Bacteriol 183: 4941–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rowley G, Spector M, Kormanec J, Roberts M (2006) Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol 4: 383–394. [DOI] [PubMed] [Google Scholar]
- 18. Broberg CA, Calder TJ, Orth K (2011) Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect 13: 992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Igbinosa EO, Okoh AI (2008) Emerging Vibrio species: an unending threat to public health in developing countries. Res Microbiol 159: 495–506. [DOI] [PubMed] [Google Scholar]
- 20. Thompson FL, Iida T, Swings J (2004) Biodiversity of vibrios. Microbiol Mol Biol Rev 68: 403–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamamoto S, Okujo N, Yoshida T, Matsuura S, Shinoda S (1994) Structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus . J Biochem 115: 868–874. [DOI] [PubMed] [Google Scholar]
- 22. Tanabe T, Funahashi T, Nakao H, Miyoshi S, Shinoda S, et al. (2003) Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus . J Bacteriol 185: 6938–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Funahashi T, Moriya K, Uemura S, Miyoshi S, Shinoda S, et al. (2002) Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus . J Bacteriol 184: 936–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanabe T, Funahashi T, Okajima N, Nakao H, Takeuchi Y, et al. (2011) The Vibrio parahaemolyticus pvuA1 gene (formerly termed psuA) encodes a second ferric vibrioferrin receptor that requires tonB2 . FEMS Microbiol Lett 324: 73–79. [DOI] [PubMed] [Google Scholar]
- 25. Funahashi T, Tanabe T, Aso H, Nakao H, Fujii Y, et al. (2003) An iron-regulated gene required for utilization of aerobactin as an exogenous siderophore in Vibrio parahaemolyticus . Microbiology 149: 1217–1225. [DOI] [PubMed] [Google Scholar]
- 26. Funahashi T, Tanabe T, Shiuchi K, Nakao H, Yamamoto S (2009) Identification and characterization of genes required for utilization of desferri-ferrichrome and aerobactin in Vibrio parahaemolyticus . Biol Pharm Bull 32: 359–365. [DOI] [PubMed] [Google Scholar]
- 27. Tanabe T, Funahashi T, Shiuchi K, Okajima N, Nakao H, et al. (2012) Characterization of Vibrio parahaemolyticus genes encoding the systems for utilization of enterobactin as a xenosiderophore. Microbiology 158: 2039–2049. [DOI] [PubMed] [Google Scholar]
- 28. Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, et al. (2003) Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae . Lancet 361: 743–749. [DOI] [PubMed] [Google Scholar]
- 29. Demarre G, Guérout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marlière P, et al. (2005) A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156: 245–255. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. vol. 1 to 3, 3rd ed.Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 31. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamamoto S, Akiyama T, Okujo N, Matsu-ura S, Shinoda S (1995) Demonstration of a ferric vibrioferrin-binding protein in the outer membrane of Vibrio parahaemolyticus . Microbiol Immunol 39: 759–766. [DOI] [PubMed] [Google Scholar]
- 33. Kuroda T, Mizushima T, Tsuchiya T (2005) Physiological roles of three Na+/H+ antiporters in the halophilic bacterium Vibrio parahaemolyticus . Microbiol Immunol 49: 711–719. [DOI] [PubMed] [Google Scholar]
- 34. Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2: 924–932. [DOI] [PubMed] [Google Scholar]
- 35. Keen NT, Tamaki S, Kobayashi D, Trollinger D (1988) Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70: 191–197. [DOI] [PubMed] [Google Scholar]
- 36. Hantke K (1983) Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol Gen Genet 191: 301–306. [DOI] [PubMed] [Google Scholar]
- 37. Lundrigan MD, Kadner RJ (1986) Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J Biol Chem 261: 10797–10801. [PubMed] [Google Scholar]
- 38.Braun V, Hantke K (1991) Genetics of bacterial iron transport. In: Winkelman G, editor. Handbook of microbial iron chelates. Boca Raton, Fla: CRC Press. pp. 107–138. [Google Scholar]
- 39. Stojiljkovic I, Baumler AJ, Hantke K (1994) Fur regulon in gram-negative bacteria: Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol 236: 531–545. [DOI] [PubMed] [Google Scholar]
- 40. Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J (1997) Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev 11: 1169–1182. [DOI] [PubMed] [Google Scholar]
- 41. Yamamoto K, Ishihama A (2006) Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli . Biosci Biotechnol Biochem 70: 1688–1695. [DOI] [PubMed] [Google Scholar]
- 42. Vogt SL, Raivio TL (2012) Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett 326: 2–11. [DOI] [PubMed] [Google Scholar]
- 43. Weber RF, Silverman PM (1988) The Cpx proteins of Escherichia coli K12: Structure of the CpxA polypeptide as an inner membrane component. J Mol Biol 203: 467–478. [DOI] [PubMed] [Google Scholar]
- 44. Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J (2000) Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci USA 97: 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sato K, Hamada M, Asai K, Mituyama T (2009) CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res 37: W277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dean CR, Poole K (1993) Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa . J Bacteriol 175: 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Anderson MT, Armstrong SK (2004) The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J Bacteriol 186: 7302–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mey AR, Wyckoff EE, Oglesby AG, Rab E, Taylor RK, et al. (2002) Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect Immun 70: 3419–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Naka H, Crosa JH (2012) Identification and characterization of a novel outer membrane protein receptor FetA for ferric enterobactin transport in Vibrio anguillarum 775 (pJM1). Biometals 25: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hollander A, Mercante AD, Shafer WM, Cornelissen CN (2011) The iron-repressed, AraC-like regulator MpeR activates expression of fetA in Neisseria gonorrhoeae . Infect Immun 79: 4764–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raymond KN, Dertz EA, Kim SS (2003) Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci USA 100: 3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Earhart CF (2004) Iron uptake via the enterobactin. In: Crosa JH, Mey AR, Payne SM, editors. Iron transport in bacteria. Washington, DC: American Society for Microbiology. pp. 133–146. [Google Scholar]
- 53. Fiedler HP, Krastel P, Muller J, Gebhardt K, Zeeck A (2001) Enterobactin: the characteristic catecholate siderophore of Enterobacteriaceae is produced by Streptomyces species. FEMS Microbiol Lett 196: 147–151. [DOI] [PubMed] [Google Scholar]
- 54. Valdebenito M, Crumbliss AL, Winkelmann G, Hantke K (2006) Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int J Med Microbiol 296: 513–520. [DOI] [PubMed] [Google Scholar]
- 55. Schmitt MP (1999) Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J Bacteriol 181: 5330–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Danese PN, Silhavy TJ (1998) CpxP, a stress-combative member of the Cpx regulon. J Bacteriol 180: 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakayama S, Watanabe H (1995) Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J Bacteriol 177: 5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dore JE, Lukas R, Sadler DW, Church MJ, Karl DM (2009) Physical and biogeochemical modulation of ocean acidification in the central North Pacific. Proc Natl Acad Sci USA 106: 12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequences of PeuR (A) and PeuS (B). Consensus amino acid residues in the conserved regions are boxed, and invariant amino acid residues (proposed to be important for the function of PeuRS) are indicated by asterisks. In panel B, transmembrane (TM) helices proposed by the HMMTOP transmembrane topology prediction server (http://www.enzim.hu/hmmtop/index.php) are underlined. (C) A hydropathy plot of PeuS. The hydropathic index was calculated by the method of Kyte and Doolittle using a window of 21 amino acid residues. Solid bars correspond to the TM helices shown in panel B.
(PDF)
TonB specificity of PeuA in Ent utilization. The growth assay was performed as described in Figure 1. Data are shown as means ± SD from 3 separate experiments.
(PDF)
Proposed expression mechanism for V. parahaemolyticus PeuA ferric Ent receptor under iron-limiting conditions in response to extracellular alkaline pH and Ent. Thick arrows and wavy arrows represent the open reading frames and the direction of transcription and mRNAs, respectively. (A) Under iron-limiting conditions at pH 7.0, peuA is co-transcribed with VPA0151-VPA0156 from the +1 transcription start site. However, the transcript from the +1 site forms a secondary structure within its 5′-UTR, leading to inhibition of translation of the peuA mRNA, although the remaining VPA0151–VPA0156 mRNA is translated. (B) Under iron-limiting conditions at pH 8.0 in the absence of Ent, transcription of the peuA/VPA0151–VPA0156 operon from the +39 site also occurs to a slight extent, combined with normal transcription beginning at the +1 site. The presence of Ent under iron-limiting conditions at pH 8.0 is proposed to result in induction of transcription from the +39 site, and thereby leads to enhanced expression of the ferric Ent receptor PeuA, because the RBS and start codon of peuA in the +39 transcript are available for translation initiation. The peuA gene, therefore, is optimally expressed under iron-limiting conditions in response to extracellular alkaline pH and Ent. In addition, the two-component regulatory system, PeuRS, is proposed to be necessary to activate peuA transcription in response to these signals.
(PDF)
Growth assays of the VPD54, VPD55, VPD56, and VPD57 mutants in LB-Tris/+EDDA/+Ent medium at pH 8.0. The growth assay was performed as described in Figure 1. Data are shown as means ± SD from 3 separate experiments.
(PDF)
Plasmids used in this study.
(PDF)
PCR primers used in this study.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.