Abstract
DNA methylation is a crucial epigenetic mark in mammalian development, genomic imprinting, X-inactivation, chromosomal stability and suppressing parasitic DNA elements. DNA methylation in neurons has also been suggested to play important roles for mammalian neuronal functions, and learning and memory. In this review, we first summarize recent discoveries and fundamental principles of DNA modifications in the general epigenetics field. We then describe the profiles of different DNA modifications in the mammalian brain genome. Finally, we discuss roles of DNA modifications in mammalian brain development and function.
Keywords: DNA methylation, learning and memory, 5-methylcytosine, 5-hydroxymethylcytosine
1. Introduction
Epigenetics is a rapidly evolving branch of biology that studies how temporal and spatial cellular diversity is achieved from invariable genomic sequences. It addresses how a single genome with a defined number of genes can have radically different gene expression patterns and allows building a whole organism with diverse cell types. We now know that each cellular lineage has a distinct genomic distribution of epigenetic features, such as histone variants, histone modifications and DNA modifications, which engenders distinct gene expression profiles. Over the past decade, researchers have been successful in describing (i) the epigenetic landscape of embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs), as opposed to somatic cells, (ii) stages of epigenetic alterations during differentiation processes from ESCs or iPSCs to various cellular lineages, and (iii) the epigenetic landscape of cancer cells. Although there is an array of evidence supporting the importance of epigenetic regulation in neurons, there is far less information for the neuronal epigenome than for other well-studied cell types, such as ESCs or cancer cells.
A human brain consists of billions of neurons forming a complex network with precise spatio-temporal functions. Each neuron in the brain works as a functional unit that receives, integrates and transmits information. Neurons can alter their electrophysiological properties and responsiveness towards particular stimuli. Such neuronal plasticity requires sustained alteration of the local synaptic strength as well as sustained alteration of global gene expression in the nuclei, for timescales of hours, days or even years after the initial stimulus was present. Early studies have shown that long-term memory formation requires activation of transcription [1] and translation [2,3], but the molecular mechanisms by which neurons sustain the newly formed states for such a long time are unknown. The strength of individual neuronal connectivity is an important mechanism for neuronal plasticity, and constitutive macromolecular synthesis from nuclei is necessary to support the process [4]. Human memory often lasts for decades, whereas most mRNAs have half-lives of minutes to hours [5] and most proteins, including synaptic structural proteins, have half-lives of less than a few days [6]. Thus, it is unlikely that newly expressed protein factors or newly formed synaptic structures in response to neuronal activity are the sole mechanisms underlying the extreme stability of memory. A self-perpetuating nature or extremely long half-life is necessary for explaining the stability of memory [7]. DNA methylation is an attractive candidate for mediating long-term plasticity and memory. DNA methylation is chemically stable with a half-life of over a thousand years [8]. Mammalian DNA methyltransferase (Dnmt) 1 is active on hemimethylated cytosine–guanine dinucleotide (CpG) in double-strand DNA [9], which allows for the self-perpetuating nature of DNA methylation. The possibility of DNA modification being a key mechanism for neuronal plasticity has been considered since the late 1960s [7,10] and the disruption of contextual fear conditioning following injection of a methylation inhibitor was shown in the 1970s [11]. However, not until recent conceptual and technical advances in the epigenetics field have we been able to study epigenetic regulations in mammalian brains extensively and several important discoveries have been made. First, oxidation products of 5-methylcytosine (5-mC), such as 5-hydroxymethylcytosine [12,13] (5-hmC), 5-formylcytosine (5-fC) and 5-carboxylcytosine [14,15] (5-caC) were discovered in the mammalian genome along with the Ten-eleven translocation (Tet) protein family of modifying enzymes, which formed a conceptual basis for understanding regulatory mechanisms of DNA methylation (figure 1). Second, rapidly evolving sequencing technologies and novel epigenetic assays form a technical basis for achieving a more comprehensive picture of DNA methylation in the mammalian brain in relation to other epigenetic modifications. Notably, many of the recent novel findings regarding DNA modifications are characteristically prominent in the mammalian brain. In this review, we shall focus on DNA methylation and other novel DNA modifications. First, we revisit lessons from our classical understanding of DNA methylation in the mammalian system. Second, we discuss representative features of the neuronal epigenome. Third, we discuss the evidence for active regulation of DNA methylation and its potential function in the mammalian nervous system (figure 1).
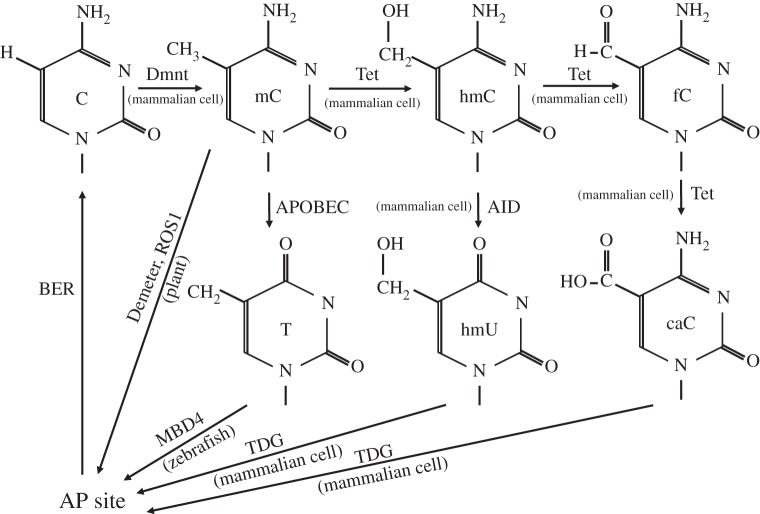
Figure 1.
DNA modification models in the mammalian genome. In mammalian cells, three DNA methyltransferase proteins generate 5-methylcytosine. 5-Methylcytosine is directly removed in plant genome, but the plant enzymes responsible for such processes are not conserved in the mammalian genome. 5-Methylcytosine can be deaminated by APOBEC proteins to generate T-G mismatch base pairs and the thymine is removed by MBD4, but the existence and relevance of this pathway has not been shown in mammalian cells. The 5-methyl group can be oxidized by Tet proteins to generate 5-hydroxymethycytosine, which is then deaminated by AID and generates 5-hydroxyuracil that is then removed by TDG. Tet proteins can further oxidize 5-hydroxymethylcytosine to generate 5-formylcytosine and 5-carboxylcytosine. 5-formylcytosine and 5-carboxylcytosine are readily recognized and removed by TDG in mammalian cells. BER, Base excision repair; AP site, apurinic site; Dnmt, DNA methyltransferase; Tet, ten-eleven translocation methylcytosine dioxygenase; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; MBD4, methyl-CpG-binding domain protein 4; TDG, thymine-DNA glycosylase; C, cytosine; mC, 5-methylcytosine; hmC, 5-hydroxymethylcytosine; fC, 5-formylcytosine; caC, 5-carboxylcytosine; T, thymine; hmU, 5-hydroxymethyluracil.
2. DNA modifications in the mammalian genome
5-mC is one of the earliest discovered epigenetic modifications in the mammalian genome [16]. 5-mC mostly appears in the CpG context in the mammalian genome. Under physiological conditions, cytosine is spontaneously deaminated to become uracil [17], which is rapidly repaired back to cytosine. When 5-mC is deaminated, 5-mC becomes thymine, which is one of the four bases of DNA. Mammalian cells have repair mechanisms that preferentially remove a thymine base, rather than the guanine base on the opposite strand from a T-G mismatch pair [18]. Owing to the imperfect surveillance system and pervasive DNA methylation, the mammalian genome has lost CpG dinucleotides and gained TpG/CpA dinucleotides throughout evolution [19,20]. Certain regions of the mammalian genome, however, have preserved their CpG dinucleotides in spite of such evolutionary pressure [21]. These genomic regions lack DNA methylation in germ cells, so spontaneous deamination can easily be repaired without error [21]. Such genomic regions are called CpG islands and have been suggested to be proximal to important regulatory regions [22].
While histone modifications and their modifying enzymes are well conserved from yeasts to humans, DNA methylation exhibits surprising diversity in regulatory mechanisms and genome-wide profiles over various organisms. In fact, the distributions, regulatory mechanisms and potential functions of DNA methylation are extremely diverse in eukaryotic genomes. Many model organisms, such as Saccharomyces cerevisiae and Caenorhabditis elegans, lack DNA methylation [23]. Drosophila melanogaster has only trace amounts of non-CpG methylation during embryonic stages, and lose most methylation by adulthood [24]. On the other hand, honeybees and silk moths have around 10% CpG methylation over gene bodies, but not within intergenic regions [25]. Plants, such as Arabidopsis thaliana, have a significant amount of non-CpG methylation because they have non-CpG-targeted methyltransferases [26]. Moreover, plants have genes encoding glycosylases that directly remove 5-mC. Neither orthologues of such plant non-CpG-targeted methyltransferases, nor mC-specific glycosylases, exist in the mammalian genome [27]. Distinct regulatory mechanisms and profiles of DNA methylation in non-mammalian model organisms made mammalian DNA methylation studies more challenging. The recent discovery of methylcytosine dioxigenases, or Tet protein family together with 5-mC oxidation derivatives such as 5-hmC, was a conceptual breakthrough for the DNA methylation field, and a number of models for DNA demethylation pathways in mammalian genome have been proposed [28]. The amount of 5-hmC is the highest in neurons and ESCs [29]. Although the independent function of 5-hmC is still under debate, at least during mammalian embryogenesis it becomes clear that 5-hmC serves as an intermediate of DNA demethylation through DNA replication-dependent dilution or sequential oxidation followed by thymidine-DNA glycosylase (TDG)-mediated DNA repair [30,31]. Potential functions of 5-hmC are described in §5b.
3. Approaches to study DNA modifications
There are three standard approaches most often used in epigenetic studies. The first approach is to generate genetic models of writers, readers or erasers of epigenetic modifications to reveal the roles of specific DNA modifications in biological contexts of interest. For example, neurodevelopmental or behavioural phenotypes have been observed in genetic models of DNA methyltransferases, which suggest the importance of DNA methylation dynamics in brain functions.
The second approach is to obtain genome-wide profiles of epigenetic modifications to investigate the relationship with other epigenetic marks or nearby transcriptional activities. For example, 5-mC is enriched at promoters of repressed genes [32], and the H3K4me3/H3K27me3 bivalent histone mark appears at promoters of developmentally poised genes [33]. 5-mC can be profiled using various assays, including whole-genome bisulfite sequencing (WGBS), reduced representation bisulfite sequencing (RRBS), methylated DNA immunoprecipitation or array-based methylation assays. 5-hmC, 5-fC and 5-caC profiles can be achieved by antibody pull-down sequencing, chemical capture sequencing or chemically/enzymatically modified versions of WGBS [34] (table 1). Thanks to the genetic models and profiling studies for DNA modification in mammalian brain, it is now clear that precise regulation of DNA methylation is especially important where frequent de novo methylation and demethylation occur and that disturbance of DNA methylation regulation causes functional defects.
Table 1.
Assays for DNA modification profiling.
| methods with single base-pair resolution | covered region | context | DNA modification with signal | DNA modification without signal | current cost per sample |
|---|---|---|---|---|---|
| WGBS | whole genome | CpG and CpH | 5-mC + 5-hmC | C + 5-fC + 5-caC | ∼$10K |
| RRBS | between two CCGG sites that are 150–400 bp apart (mostly CpG islands) | CpG and CpH | 5-mC + 5-hmC | C + 5-fC + 5-caC | <$500 |
| methylation microarray (Illumina 450 k) | targeted regions (only human) | CpG only | 5-mC + 5-hmC | C + 5-fC + 5-caC | <$300 |
| bisulfite sequencing with padlock probes | targeted regions | CpG and CpH | 5-mC + 5-hmC | C + 5-fC + 5-caC | <$500 |
| capture bisulfite sequencing (Agilent Sureselect) | targeted regions | CpG and CpH | 5-mC + 5-hmC | C + 5-fC + 5-caC | <$500 |
| TAB-seq | whole genome | CpG and CpH | 5-hmC | C + 5-mC + 5-fC + 5-caC | ∼$15K |
| oxBS-seq | whole genome | CpG and CpH | 5-mC | C + 5-hmC + 5-fC + 5-caC | ∼$15K |
| fCAB-seq | whole genome | CpG and CpH | 5-mC + 5-hmC + 5-fC | C + 5-caC | ∼$15K |
| enrichment-based methods | modification before pull down | pull-down methods | pull-down base | ||
|---|---|---|---|---|---|
| MeDIP-seq | no modification | methyl-CpG-binding domain protein or anti-5-mC antibody | 5-mC | ||
| GLIB-seq | β-glucosultransferase-mediated glucose addition | biotin–streptavidin interaction | 5-hmC | ||
| hME-Seal-seq | β-glucosultransferase-mediated glucose addition | biotin–streptavidin interaction | 5-hmC | ||
| fC-Seal-seq | β-glucosultransferase-mediated glucose addition, NaBH4-mediated reduction | biotin–streptavidin interaction | 5-fC | ||
| 5-hmC antibody pull-down-seq | no modification | anti-5-hmC antibody | 5-hmC | ||
| 5-fC antibody pull-down-seq | no modification | anti-5-fC antibody | 5-fC | ||
| 5-caC antibody pull-down-seq | no modification | anti-5-caC antibody | 5-caC | ||
The third approach is to interrogate the functions of epigenetic modification directly by inducing or removing epigenetic modifications into targeted regions of the genome. Although a number of profiling studies showed the inverse correlation of DNA methylation and nearby transcriptional activity, whether DNA methylation alone is necessary and sufficient for suppressing gene expression is not quite clear yet. Only a few studies directly demonstrated the causality of DNA methylation on transcriptional repression and there is still no report of manipulated DNA methylation to study its causal role in the mammalian nervous system. Interestingly, the lessons from other systems suggest that DNA methylation has a more complex role than being a simple suppressor for the transcription activity, which will be reviewed in §4.
4. Potential roles of DNA methylation in mammals
The prevailing view for role of DNA methylation in the mammalian genome is its repressive function for nearby transcriptional activity. There are, however, still uncertainties and complexities concerning the function of DNA methylation in mammals. First, most genes regardless of their expression level have unmethylated promoters, suggesting that promoter methylation is not a primary mechanism for transcriptional regulation. Second, the function of DNA methylation outside promoters, such as in gene bodies and enhancer regions, is not well understood. While one early study reported that gene body methylation reduces transcription elongation efficiency [35], a more recent report argues that unmethylated gene body CpG islands are cryptic promoters that might impede the transcriptional efficiency of the original promoter [36]. The repressive effect of DNA methylation in the promoter region can either be direct, by blocking binding of transcription factors [37], or indirect, by recruiting methyl-binding domain proteins which contain transcription repressive domains [38]. Schubeler et al. [39] used recombination-mediated cassette exchange (RMCE) to ask whether DNA methylation is sufficient for gene repression. When a fully methylated DNA construct containing an H2 enhancer element (from human β-globin), human β-globin promoter and eGFP, was inserted into the designated genomic locus using Cre-recombination, expression of eGFP was significantly repressed through histone deacetylation, as opposed to its unmethylated counterpart [39]. Raynal et al. [40], on the contrary, showed that DNA methylation was not sufficient to suppress gene expression in the presence of histone deacetylase (HDAC) inhibitor. Interestingly, when the HDAC inhibitor was removed, cells treated only with HDAC inhibitor rapidly re-suppressed GFP expression, whereas cells treated with HDAC inhibitor plus the DNA demethylating drug decitabine showed sustained GFP expression for months. Thus, although DNA methylation is not sufficient to mediate transcriptional suppression, DNA methylation is important for maintaining stable suppression. Likewise, Bouhassira et al. [41] showed that DNA methylation is not necessary for gene silencing, but is important for maintaining epigenetic memory of suppression. Specific DNA sequence recognition technologies have been significantly improved with the discovery of the transcription activator-like effector nuclease (TALEN) and Cas9 systems, and have been successfully applied to local epigenome editing [42]. Using local epigenome editing, we can now inarguably show the effects of local epigenome change on other epigenetic modifications as well as the transcriptional activity of nearby genes. These valuable new tools will lead to a much better understanding of the functions of DNA methylation.
5. Landscape of DNA modifications in mammalian brain
A number of profiling studies accumulated genome-wide distributions of DNA methylation and hydroxymethylation, histone modifications and transcription factor binding sites in various cell types. Although there is considerable evidence for the importance of epigenetic regulation in neurons, there is far less information about the neuronal epigenetic landscape than in other well-studied cell types. There are at least three reasons for the relatively few studies of neuronal epigenetics. First, with the currently available tools, most epigenetic analyses require a large number of cells. Unlike cancer cells or ESCs, neurons are not mitotic, so they cannot be expanded in vitro. Second, the nervous system exhibits significant heterogeneity of cell types in vivo. For example, neurons reside with supporting glial cells, which might have completely different epigenetic features than neurons [43]. On average, half of the cells in the mammalian brain are glial cells; thus half of the brain epigenome information is typically coming from glial cells [43]. Moreover, there are hundreds of different neuronal cell types with diverse morphologies, functions and gene expression profiles [44]. Third, neurons exhibit significant heterogeneity with respect to their physiological state. For example, only a small subset of neurons in dentate gyrus is activated in a specific context and expresses activity-dependent genes at any given time [45]. Thus, in order to study neuronal epigenetics, we need to synchronize neuronal signalling. Although it is possible to activate or suppress neuronal activity of cells simultaneously in vitro, endogenous synchronization of neuronal activity in large populations in vivo is rare because neurons have highly specific synaptic connections that are differentially engaged by the same stimulus. It is precisely this temporal and spatial specificity of neuronal responses that underlie some of the most fundamental properties of neural circuitry and function. It is not currently possible to circumvent this heterogeneity entirely, but efforts to minimize cell-type and activation-state variability in the intact brain have been made and have resulted in important insights from genome-wide DNA modification profiling studies. In this section, we review findings from profiling studies on different DNA modifications in the mammalian nervous system (figure 2).
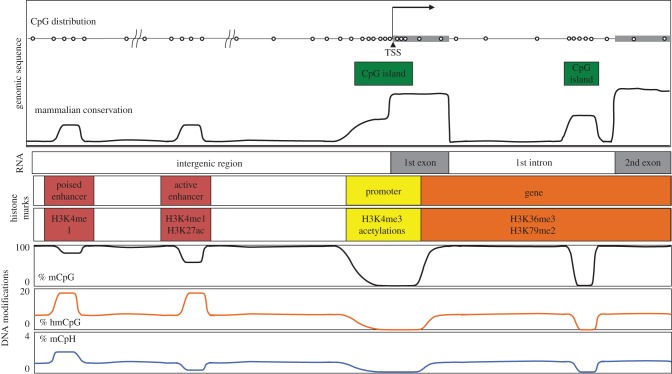
Figure 2.
Schematic for genome and epigenome of mammalian neurons. CpG (shown as open circles) dinucleotide sequences are unevenly distributed throughout the genome; CpG dinucleotides are largely depleted in most genomic regions. Half of the promoters and some enhancers have an unusually high density of CpG dinucleotides, which are called CpG islands. Enhancers and promoters are defined by the composition of histone marks and show characteristic 5-mC or 5-hmC features. Poised enhancers, marked by H3K4me1 without H3K27ac have an intermediate level of 5-mCpG and a high level of 5-hmC and 5-mCpH. Active enhancers, which are marked by H3K4me1 and H3K27ac have lower or similar levels of 5-mCpG as poised enhancers, and low levels of 5-mCpH. Promoters, which are marked by H3K4me3 with histone acetylation, such as H3K27ac, are largely devoid of all DNA modifications. (Online version in colour.)
(a). 5-Methylcytosine in the CpG context
With few exceptions, most mammalian cells have high CpG methylation levels; 80–90% of cytosine within the CpG context is methylated [46]. By contrast, most CpG islands, where CpG dinucleotides are present at relatively higher frequency, are devoid of DNA methylation regardless of cell type [22]. Different genomic annotations have characteristic methylation levels and potentially distinct functions. First, half of the promoters in the mammalian genome overlap with CpG islands and are largely unmethylated [47]. Methylation levels in the promoter region are inversely correlated with gene expression levels both in ESCs and neurons [32]. Second, gene bodies in the mammalian genome are highly methylated [25]. Interestingly, gene body methylation is inversely correlated with gene expression in neurons but positively correlated with gene expression in ESCs [32]. Third, distal regulatory elements, such as enhancers or insulators, have intermediate levels of CpG methylation and are differentially methylated according to cell type [48]. For example, neuron-specific active enhancers are more hypomethylated in neurons than in other cell types [46]. Genome-wide CpG methylation can be altered upon neuronal activity and most of the methylation changes occur within genomic regions, apart from promoters, many of which are thought to be located at distal regulatory elements [32]. Fourth, intergenic regions are almost always completely methylated [48]. The large domains of hypomethylation observed in some types of cancer cells have not been reported in neurons, but small hypomethylated regions appear more frequently in neurons than in other cell types [46,49].
(b). 5-Hydroxymethylcytosine
The presence of 5-hmC in the mammalian genome was first reported in the 1970s using chromatographical separation followed by spectrophotometric measurements [50]. This observation has since been questioned because the amount of 5-hmC in that study was more than two orders of magnitude higher than what we now know to be the actual amount, and 5-mC was not detected in the assays. More than three decades later, 5-hmC was identified in ESCs and Purkinje neurons from two independent laboratories using more precise detection by mass spectroscopy [12,13]. Although 5-hmC was first confirmed in ESCs and cerebellar Purkinje neurons, the amount of 5-hmC in the brain is much higher in the hypothalamus, cerebral cortex and hippocampus [51]. The amount of 5-hmC in these brain regions is up to 0.7% of all cytosines, whereas 5-mC is 4% of all cytosines. Since 5-hmC appears to be methylcytosine in bisulfite sequencing, it is important to remember that one-sixth of ‘methylated cytosines’ in the WGBS in previous studies of the mammalian brain is in fact 5-hmC rather than 5-mC. Promoters with CpG islands are devoid of 5-hmC as well as 5-mC. Enhancers are the genomic annotation that have the highest amount of 5-hmC, regardless of their activity [52].
Recent studies showed that 5-hmC is the first step of the Tet-TDG-mediated DNA demethylation pathway, which is followed by further oxidation to 5-fC and 5-caC and DNA repair back to C. It is, however, still under debate why 5-hmC levels are orders of magnitude higher than its oxidation products 5-fC and 5-caC, if 5-hmC is an intermediate to 5-fC and 5-caC. Moreover, 5-hmC distributions are tissue specific, and gene body 5-hmC shows a positive correlation with transcriptional activity in brain, which suggests a biological role for 5-hmC [53]. A few potential functions have been suggested but there is no consensus currently. Khare et al. [54] reported that exonal 5-hmC density determines whether the exon is included or excluded when the pre-mRNA is spliced in mouse and human frontal cortex. Although population-level data support the 5-hmC effect for alternative splicing, it is not clear whether it holds true for individual cells, because the amount of 5-hmC is relatively low (approx. 10%), even in neurons. Second, Mellén et al. [55] showed that methyl-CpG-binding protein 2 (MeCP2) binds to 5-hmC as well as 5-mC in brain. They suggested that when MeCP2 is bound to 5-hmC, it facilitates nearby gene expression, which can explain the long standing paradox of global gene expression increases following knockdown of MeCP2 expression. Although MeCp2 binding to 5-hmC was independently validated, there is still no plausible explanation for how MeCP2 can either activate or repress depending on its binding to 5-mC or 5-hmC.
(c). 5-Methylcytosine in the non-CpG context
Unlike the plant genome where enzymes for generating and erasing non-CpG methylation have been well characterized, non-CpG methylation in the mammalian genome has not been studied extensively. Lister et al. [56] showed that non-CpG methylation is abundant in human ESCs and human iPSCs as opposed to fibroblasts, which exhibit hardly any non-CpG methylation. Lister et al. also reported that non-CpG methylation is depleted at tissue-specific enhancers, suggesting its importance either as a repressor of enhancer activity, or prohibition by enhancer activity. Xie et al. [57] later reported a similar level of non-CpG methylation in the mouse frontal cortex. The discrepancy between abundant non-CpG methylation in ESCs and neurons, but minimal methylation in intermediately differentiated cells, was resolved by a recent intensive whole methylation study by Lister et al. [58]. The authors did whole-genome bisulfite sequencing for sorted neurons from human and mice. They found that both in the human and mouse brain, non-CpG methylation accumulates throughout development and is maintained during the lifespan of the organism. It was, however, not clearly shown how non-CpG methylation is regulated. There are anecdotal observations of non-CpG methylation in the mammalian genome. Ramsahoye et al. [59] found that although the enzymatic activity of Dnmt3a is mostly biased towards CpG methylation, Dnmt3a also has weak methylation activity towards the non-CpG context. Thus, it is likely that Dnmt3a generates non-CpG methylation, but it is still completely unknown why neurons and ESCs have such a high amount of non-CpG methylation compared with other tissues. The function of non-CpG methylation is suggested to be repressive. It has also been suggested that inactive enhancers have higher amounts of non-CpG methylation than other genomic regions. Indeed, Lomvardas et al. [60] also observed that inactive enhancers are enriched with non-CpG methylation, suggesting that non-CpG methylation in enhancers might also be important in mammalian neurons. Guo et al. [61] showed that non-CpG methylation can help nearby CpG methylation recruit methyl-binding proteins and suppress transcriptional activities in vivo. They showed that non-CpG methylation is blocked in the absence of Dnmt3a during early development, and thus certain genes become de-suppressed.
6. Potential functional roles of DNA methylation in the mammalian brain
(a). DNA methylation in brain development
In the mammalian genome, there are three genes encoding Dnmt, which transfers a methyl group from S-adenosylmethionine (SAM) to the cytosine nucleotide in the CpG context of double-stranded DNA. Dnmt1, the earliest member of the Dnmt family to be identified, has preferential activity towards unmethylated CpG base pairs with methylated CpG (hemimethylated CpG dinucleotides), whereas Dnmt3a and Dnmt3b have de novo methyltransferase activity for cytosine in the CpG context [9]. Homozygous knock out of Dnmt1 in ESCs does not cause any notable abnormalities, but embryos fail to develop past mid-gestation [62]. Dnmt3a homozygous knockout mice die at around four weeks of age, whereas Dnmt3b homozygous knockout mice are embryonic lethal with multiple tissue defects that include cardiac anomaly and hepatic hypotrophy [63]. In humans, homozygous hypomorphic mutations or compound heterozygous hypomorphic mutations of Dnmt3b cause ICF syndrome, which results in immunodeficiency, centromere instability, facial anomalies and mental retardation [63]. These genetic mutation studies clearly stress the importance of DNA methylation in normal mammalian development.
DNA methylation also has been postulated to play important roles in brain development and neuronal function. In order to study brain-specific functions of DNA methylation, a late-stage or inducible knockout of a gene in specific cell types, without affecting general developmental processes, is necessary since disruption of DNA methylation in prenatal stages results in neurodevelopmental defects. A series of conditional knock out studies by Fan and co-workers [64–68] showed various neurodevelopmental defects caused by Dnmt deletions and global hypomethylation in the mammalian nervous system. Mice with a Dnmt1 deletion in excitatory post-mitotic neurons using the calmodulin-kinase IIα promoter-driven Cre (CamK-Cre) system exhibited no overt phenotype. Mice with Dnmt1 deletion in neural stem cells using Nestin-Cre system, however, showed global hypomethylation in the nervous system and died immediately after birth due to respiratory failure [65]. Dnmt1-deleted neural stem cells became globally hypomethylated and underwent precocious astroglial differentiation due to activation of the JAK-STAT pathway [66]. Mice with Dnmt1 deletion in telencephalic excitatory neurons and astroglial lineages using the Emx promoter-driven Cre system had a normal lifespan, but developed severe cortical and hippocampal degeneration with morphological and electrophysiological defects in affected neurons. Mice lacking Dnmt3a in the nervous system were born healthy, but degenerated in adulthood and died prematurely showing a loss of motor neurons and morphological defects in the neuromuscular junctions in the diaphragm [69]. Wu et al. [70] explained the mechanism by which Dnmt3a deficiency causes neurodevelopmental defects using in vitro neuronal differentiation of neural stem cells. Interestingly, the induction of Dnmt3a-mediated de novo methylation occurs in genes that are active during neuronal differentiation. De novo methylation blocks binding of the polycomb repressive complex and facilitates the activation of gene expression. In addition to the Dnmt defects, mutations of the methylation reader MeCP2 also cause severe neurodevelopmental defects and result in Rett syndrome. MeCP2 is the primary member of a methyl-CpG-binding domain protein family and is highly expressed in mature neurons [71]. According to a report from the International Rett Syndrome Foundation, Rett syndrome can be caused by over 1000 different mutations in the X-linked MeCP2 gene. Interestingly, Rett syndrome patients develop normally until 6–18 months of age, and then gradually regress as autistic symptoms, seizure, ataxia and stereotypical hand wringing behaviour emerge [72]. Male mice with a hemizygous (X'Y) MeCP2 gene mutation appear to be normal at birth, but start to show severe neurological symptoms at approximately five to six weeks of age, typically resulting in premature death between six and 12 weeks. Female mice with heterozygous (X'X) MeCP2 gene mutation start to show behavioural symptoms after several months [73,74].
(b). DNA methylation in mature neuronal function
Establishment of cell-type-specific methylation during mammalian development suggests that proper methylation and recognition of methylation during neuronal development are crucial. But it is surprising that post-mitotic neurons still retain the capacity for de novo, activity-dependent and/or sustained methylation [75,76]. It is now clear that (i) mammalian neurons express enzymes with methylating or demethylating activities, (ii) the mammalian neuronal methylome can change within a few hours, and (iii) disrupting such changes by blocking enzyme activity results in various behavioural phenotypes.
Both DNA methyltransferases and demethylases are abundantly expressed and actively regulated in mature neurons. Using in situ hybridization, Goto et al. [26] showed that Dnmt1 is abundant not only in rapidly dividing embryonic tissues, but also in most mature neurons in the adult mouse brain. Feng et al. [76] used lacZ knock-in lines to achieve the spatio-temporal expression maps of Dnmt3a and Dnmt3b. Unlike Dnmt3b whose temporal expression pattern is restricted to a narrow window of early neurogenesis, Dnmt3a has bimodal expression patterns: at E13.5, Dnmt3a expression is highly localized to ventricular and subventricular zones, whereas at E17.5 and six months old, Dnmt3a is strongly expressed in NeuN-expressing mature neurons in most central nervous system regions. Dnmt3a expression levels in the CA1 area of the mouse brain were significantly upregulated by fear conditioning [77] and in the dentate gyrus by electroconvulsive stimulation (ECS) [32], suggesting potential roles of de novo methylation in mature neurons. Tet enzymes, key for the DNA demethylation pathway, are also highly expressed in mature neurons [78].
Evidence that DNA methylation can be dynamically regulated comes from a number of observations of population-wide demethylation in non-dividing neurons, where DNA methylation cannot be diluted. Levenson et al. [79] used pharmacological inhibitors to investigate the function of Dnmt enzymes in long-term potentiation (LTP) and found that blockade of Dnmt induced significant reduction of methylation levels of Reelin and Bdnf promoters in mice hippocampal slices after only 20 min of drug treatment [79]. This study shows that neurons have highly active DNA methyltransferases and demethylases in equilibrium in certain regions of the genome and that local neuronal activity can perturb this equilibrium, resulting in changes in local methylation.
The importance of promoter methylation for transcriptional activity is best studied at the Bdnf locus. Bdnf has multiple transcription starting sites and the DNA methylation level at these transcription start sites is indeed important for local transcription activity in the context of fear conditioning [80]. However, when Martinowich et al. [81] activated cultured embryonic hippocampal neurons in vitro with high KCl in the medium they found that activation of Bdnf IV exon expression occurred with demethylation of the Bdnf IV promoter in conjunction with histone acetylation at the same locus. Guo et al. [32] asked whether there are genome-wide methylation changes in dentate granule neurons upon synchronized neuronal activation by ECS in vivo. This study used methyl-sensitive cut counting technology to profile methylation levels of CpG in the CCGG context, which is around 1% of all CpGs in the mouse genome. Surprisingly, 1.4% of all CCGG sites showed altered DNA methylation 4 h after ECS [32].
The abundance of 5-hmCs and non-CpG methylation is additional evidence of dynamic DNA modifications in mammalian neurons. 5-hmC was first discovered as an oxidation product of 5-mC in Purkinje neurons [13] and ESCs [12], which led to discoveries of further oxidation products, 5-fC and 5-caC, and the identification of the Tet-TDG demethylation pathway [14,15]. Non-CpG methylation is a by-product of CpG-specific DNA methyltransferase Dnmt3a and is also abundant in ESCs and neurons [59]. Further studies have shown that 5-hmC and non-CpG methylation have critical functions in ESCs and neurons based on their high abundance in these cell types. The presence of high levels of 5-hmC and non-CpG methylation supports the notion of a highly dynamic nature of DNA methylation and demethylation in mammalian neurons.
Emerging evidence suggests that blocking dynamic regulation of DNA methylation in mammalian brain results in various neuropsychiatric malfunctions. Miller et al. [77] asked whether DNA methyltransferase activity is important for contextual fear conditioning by infusing a pharmacological blocker of Dnmt proteins immediately after contextual fear conditioning. This experiment was reminiscent of a classic experiment which showed the necessity of transcription and translation in mammalian long-term memory consolidation [3]. Mice infused with a Dnmt blocker immediately after fear-conditioning training showed significantly impaired expression of a learned response in later probe tests, compared to control mice. In control animals, associative learning involving contextual stimuli induced a dramatic increase in DNA methylation in the promoter region of the memory suppressor gene PP1 within 2 h along with slightly reduced Pp1 expression. Conversely, infusion of a Dnmt blocker resulted in significant demethylation in the same region associated with 100% increase of Pp1 1 h after infusion. Feng et al. [64] used the CamK-Cre system to knock out Dnmt3a or Dnmt1 in post-mitotic neurons after birth. Dnmt3a or Dnmt1 single knockout mice had normal total 5-mC amounts and showed normal phenotypes. Dnmt3a/Dnmt1 conditional double knockout mice had a normal lifespan, but had smaller hippocampi and showed defects in learning and contextual memory consolidation. Dnmt3a/Dnmt1 conditional double knockout neurons had 15–20% lower 5-mC and 5-hmC compared to the wild-type animals and showed attenuated LTP and enhanced long-term depression (LTD), which could account for the learning and memory consolidation deficit.
Proper recognition of 5-mC is important for mature neuronal function. Although MeCP2 defects are the primary cause of the devastating neurodevelopmental disorder, Rett syndrome, MeCP2 is also important for mature neuronal functions. MeCP2 elimination in mature neurons results in reduced spontaneous excitatory synaptic transmission (mEPSC), which is mimicked by HDAC inhibitor treatment [82]. Induction of MeCP2 defects in the adult mouse brain causes immediate onset of Rett syndrome-like symptoms as early as one week after the induction [83]. The neurological defects are accompanied by shrinkage of brain size, retraction of dendritic arbours and reduction of dendritic spinal density, potentially due to post-translational dysregulation of a number of synaptic proteins. The striking reversal of Rett symptoms by rescuing MeCP2 only in the microglial population, however, calls for further studies for the importance of MeCP2 function in mature neurons [84].
Tet1 knockout mice show grossly normal development and anatomy [85], so a targeted knockout is not necessary when interrogating brain-specific functions related to Tet1. Zhang et al. [86] showed that Tet1 knockout mice show hippocampal adult neurogenesis defects along with spatial learning and short-term memory impairments, but exhibit normal long-term memory. By contrast, Rudenko et al. [87] showed that Tet1 knockout mice exhibited normal memory acquisition, evaluated by Pavlovian fear conditioning and the Morris water maze test. Interestingly, the authors found significant impairments in contextual fear and spatial reference memory extinction in the Tet1 knockout animals. The Schaffer collateral-CA1 pathway in Tet1 knockout animals exhibited stronger and longer lasting LTD than in wild-type animals, as opposed to slightly decreased LTP, which can potentially explain the abnormalities in memory extinction. In contrast, Kaas et al. [78] showed that Tet1 is downregulated by neuronal activity and that overexpression of the Tet1 catalytic domain also induces hippocampus-mediated long-term associative memory formation.
7. Conclusion
Epigenetics has become one of the most heavily investigated domains in biological science over the past decade. The field of epigenetics is still young and technological advancement in this field has great potential to explain several mechanisms underlying brain function. A number of new technologies have emerged in recent years and many of them have yet to be applied to the mammalian nervous system. Although the epigenetics field has expanded due to standardization of sequencing technology, bioinformatics and statistical modalities, the high cost of sequencing experiments and labour-intensive process of bioinformatics prohibits the inclusion of many investigators in this field. Finally, we must remember that cell type may have a distinct epigenetic profile based on its state of activation. And each state is composed of a number of epigenetic features. For example, there are scores of histone modifications, five cytosine modifications in DNA and hundreds of crucial transcription factors in each cell type, all of which can be dynamically modulated. Unlike invariable genomic sequences, the epigenome reflects the state of the biological system at a particular moment in time. Therefore, to minimize heterogeneity and confounds arising from the unknown physiological state of individual neurons that contribute to population analyses, single-cell epigenomics are critical to control for many of these variables.
Neuroepigenetics is a recently developed subset of the epigenetics field in general and is poised to generate many surprising findings. First, many exciting discoveries in epigenetics turn out to be largely brain-specific. For example, the amount of non-CpG methylation as well as 5hmC is highest in neurons. Second, cell division is potentially the largest contributor that can reset epigenetic modifications. Since neurons do not divide, we can test hypotheses regarding causality directly. Third, the brain is a heterogeneous organ consisting of a number of different cell types. Moreover, even the same type of neurons can alter their methylome or histone modification radically upon electrical activity or during memory consolidation [32,88]. Epigenetics is a field that studies the mechanism by which the same genotype can give rise to different cellular phenotypes. Neuroepigenetics is focused on how cells with same genome and similar lineage can have distinct epigenome, gene expression profiles, and thus electrophysiological phenotypes.
Acknowledgement
We thank K. Christian for comments.
Funding statement
The work in the authors' laboratories was supported by National Institutes of Health, Maryland Stem Cell Research Fund, Simons Foundation. J.S. was supported by a fellowship from Samsung.
References
- 1.Agranoff BW, Davis RE, Casola L, Lim R. 1967. Actinomycin D blocks formation of memory of shock-avoidance in goldfish. Science 158, 1600–1601. ( 10.1126/science.158.3808.1600) [DOI] [PubMed] [Google Scholar]
- 2.Barondes SH, Cohen HD. 1966. Puromycin effect on successive phases of memory storage. Science 151, 594–595. ( 10.1126/science.151.3710.594) [DOI] [PubMed] [Google Scholar]
- 3.Flexner LB, Flexner JB, Roberts RB. 1967. Memory in mice analyzed with antibiotics. Antibiotics are useful to study stages of memory and to indicate molecular events which sustain memory. Science 155, 1377–1383. ( 10.1126/science.155.3768.1377) [DOI] [PubMed] [Google Scholar]
- 4.Bailey CH, Kandel ER, Si K. 2004. The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron 44, 49–57. ( 10.1016/j.neuron.2004.09.017) [DOI] [PubMed] [Google Scholar]
- 5.Shyu AB, Greenberg ME, Belasco JG. 1989. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 3, 60–72. ( 10.1101/gad.3.1.60) [DOI] [PubMed] [Google Scholar]
- 6.Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. 2012. Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942 ( 10.1126/science.1217421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crick F. 1984. Memory and molecular turnover. Nature 312, 101 ( 10.1038/312101a0) [DOI] [PubMed] [Google Scholar]
- 8.Shen JC, Rideout WM, 3rd, Jones PA. 1994. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22, 972–976. ( 10.1093/nar/22.6.972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okano M, Xie S, Li E. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219–220. ( 10.1038/890) [DOI] [PubMed] [Google Scholar]
- 10.Griffith JS, Mahler HR. 1969. DNA ticketing theory of memory. Nature 223, 580–582. ( 10.1038/223580a0) [DOI] [PubMed] [Google Scholar]
- 11.Lloyd RL, Lewis DJ. 1979. Possible effects of the synthetic amino acid, L-ethionine, on memory in rats. Physiol. Behav. 22, 1015–1019. ( 10.1016/0031-9384(79)90348-2) [DOI] [PubMed] [Google Scholar]
- 12.Tahiliani M, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. ( 10.1126/science.1170116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kriaucionis S, Heintz N. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. ( 10.1126/science.1169786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He YF, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. ( 10.1126/science.1210944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. ( 10.1126/science.1210597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotchkiss RD. 1948. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 175, 315–332. [PubMed] [Google Scholar]
- 17.Duncan BK, Miller JH. 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287, 560–561. ( 10.1038/287560a0) [DOI] [PubMed] [Google Scholar]
- 18.Waters TR, Swann PF. 1998. Kinetics of the action of thymine DNA glycosylase. J. Biol. Chem. 273, 20 007–20 014. ( 10.1074/jbc.273.32.20007) [DOI] [PubMed] [Google Scholar]
- 19.Swartz MN, Trautner TA, Kornberg A. 1962. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J. Biol. Chem. 237, 1961–1967. [PubMed] [Google Scholar]
- 20.Bird AP. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8, 1499–1504. ( 10.1093/nar/8.7.1499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. 1985. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell 40, 91–99. ( 10.1016/0092-8674(85)90312-5) [DOI] [PubMed] [Google Scholar]
- 22.Deaton AM, Bird A. 2011. CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022. ( 10.1101/gad.2037511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bestor TH. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402. ( 10.1093/hmg/9.16.2395) [DOI] [PubMed] [Google Scholar]
- 24.Lyko F, Ramsahoye BH, Jaenisch R. 2000. DNA methylation in Drosophila melanogaster. Nature 408, 538–540. ( 10.1038/35046205) [DOI] [PubMed] [Google Scholar]
- 25.Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919. ( 10.1126/science.1186366) [DOI] [PubMed] [Google Scholar]
- 26.Finnegan EJ, Kovac KA. 2000. Plant DNA methyltransferases. Plant Mol. Biol. 43, 189–201. ( 10.1023/A:1006427226972) [DOI] [PubMed] [Google Scholar]
- 27.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536. ( 10.1016/j.cell.2008.03.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu SC, Zhang Y. 2010. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620. ( 10.1038/nrm2950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. 2010. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5, e15367 ( 10.1371/journal.pone.0015367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue A, Zhang Y. 2011. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194 ( 10.1126/science.1212483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu TP, et al. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610. ( 10.1038/nature10443) [DOI] [PubMed] [Google Scholar]
- 32.Guo JU, et al. 2011. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345–1351. ( 10.1038/nn.2900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein BE, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. ( 10.1016/j.cell.2006.02.041) [DOI] [PubMed] [Google Scholar]
- 34.Wu H, Zhang Y. 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68. ( 10.1016/j.cell.2013.12.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. 2004. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 11, 1068–1075. ( 10.1038/nsmb840) [DOI] [PubMed] [Google Scholar]
- 36.Maunakea AK, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257. ( 10.1038/nature09165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iguchi-Ariga SM, Schaffner W. 1989. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 3, 612–619. ( 10.1101/gad.3.5.612) [DOI] [PubMed] [Google Scholar]
- 38.Nan X, Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BM, Eisenman RN. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389. ( 10.1038/30764) [DOI] [PubMed] [Google Scholar]
- 39.Schubeler D, Lorincz MC, Cimbora DM, Telling A, Feng Y-Q, Bouhassira EE, Groudine M. 2000. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell Biol. 20, 9103–9112. ( 10.1128/MCB.20.24.9103-9112.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raynal NJ, Si J, Taby RF, Gharibyan V, Ahmed S, Jelinek J, Estecio MRH, Issa J-PJ. 2012. DNA methylation does not stably lock gene expression but instead serves as a molecular mark for gene silencing memory. Cancer Res. 72, 1170–1181. ( 10.1158/0008-5472.CAN-11-3248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng YQ, Desprat R, Fu H, Olivier E, Lin CM, Lobell A, Gowda SN, Aladjem MI, Bouhassira EE. 2006. DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet. 2, e65 ( 10.1371/journal.pgen.0020065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeder ML, et al. 2013. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 31, 1137–1142. ( 10.1038/nbt.2726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cahoy JD, et al. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278. ( 10.1523/JNEUROSCI.4178-07.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters A, Jones EG. 1984. Cerebral cortex, vol. 1. Cellular Components of the Cerebral Cortex. New York, NY: Plenum Press. [Google Scholar]
- 45.Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. 2012. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385. ( 10.1038/nature11028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. 2013. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet 45, 1198–1206. ( 10.1038/ng.2746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vavouri T, Lehner B. 2012. Human genes with CpG island promoters have a distinct transcription-associated chromatin organization. Genome Biol. 13, R110 ( 10.1186/gb-2012-13-11-r110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. ( 10.1038/nature11247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berman BP, et al. 2012. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 44, 40–46. ( 10.1038/ng.969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. 1972. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 126, 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, Müller M, Biel M, Carell T. 2010. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. 49, 5375–5377. ( 10.1002/anie.201002033) [DOI] [PubMed] [Google Scholar]
- 52.Yu M, et al. 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380. ( 10.1016/j.cell.2012.04.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo JU, et al. 2014. Genome-wide antagonism between 5-hydroxymethylcytosine and DNA methylation in the adult mouse brain. Front. Biol. 9, 66–74. ( 10.1007/s11515-014-1295-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khare T, et al. 2012. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 19, 1037–1043. ( 10.1038/nsmb.2372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430. ( 10.1016/j.cell.2012.11.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lister R, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322. ( 10.1038/nature08514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. 2012. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831. ( 10.1016/j.cell.2011.12.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lister R, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 ( 10.1126/science.1237905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramsahoye BH, et al. 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA 97, 5237–5242. ( 10.1073/pnas.97.10.5237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413. ( 10.1016/j.cell.2006.06.035) [DOI] [PubMed] [Google Scholar]
- 61.Guo JU, et al. 2014. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17, 215–222. ( 10.1038/nn.3607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. ( 10.1016/0092-8674(92)90611-F) [DOI] [PubMed] [Google Scholar]
- 63.Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. ( 10.1016/S0092-8674(00)81656-6) [DOI] [PubMed] [Google Scholar]
- 64.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. 2010. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430. ( 10.1038/nn.2514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan G, et al. 2001. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci. 21, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan G, et al. 2005. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 132, 3345–3356. ( 10.1242/dev.01912) [DOI] [PubMed] [Google Scholar]
- 67.Golshani P, Hutnick L, Schweizer F, Fan G. 2005. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Related Syst. 3, 227–233. ( 10.1017/S1472928807000222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, Silva AJ, Schweizer FE, Fan G. 2009. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum. Mol. Genet. 18, 2875–2888. ( 10.1093/hmg/ddp222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. 2007. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev. Dyn. 236, 1663–1676. ( 10.1002/dvdy.21176) [DOI] [PubMed] [Google Scholar]
- 70.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329, 444–448. ( 10.1126/science.1190485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skene PJ, Illingworth RS, Webb S, Kerr ARW, James KD, Turner DJ, Andrews R, Bird AP. 2010. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468. ( 10.1016/j.molcel.2010.01.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amir RE, Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188. ( 10.1038/13810) [DOI] [PubMed] [Google Scholar]
- 73.Chen RZ, Akbarian S, Tudor M, Jaenisch R. 2001. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 27, 327–331. ( 10.1038/85906) [DOI] [PubMed] [Google Scholar]
- 74.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. 2001. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 27, 322–326. ( 10.1038/85899) [DOI] [PubMed] [Google Scholar]
- 75.Goto K, Numata M, Komura J-I, Ono T, Bestor TH, Kondo H. 1994. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differ. Res. Biol. Divers. 56, 39–44. ( 10.1007/s002580050019) [DOI] [PubMed] [Google Scholar]
- 76.Feng J, Chang H, Li E, Fan G. 2005. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 79, 734–746. ( 10.1002/jnr.20404) [DOI] [PubMed] [Google Scholar]
- 77.Miller CA, Sweatt JD. 2007. Covalent modification of DNA regulates memory formation. Neuron 53, 857–869. ( 10.1016/j.neuron.2007.02.022) [DOI] [PubMed] [Google Scholar]
- 78.Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming G, King JR, Song H, Sweatt JD. 2013. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79, 1086–1093. ( 10.1016/j.neuron.2013.08.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levenson JM, Roth TL, Lubin FD, Miller CA, Huang I-C, Desai P, Malone LM, Sweatt JD. 2006. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 281, 15 763–15 773. ( 10.1074/jbc.M511767200) [DOI] [PubMed] [Google Scholar]
- 80.Lubin FD, Roth TL, Sweatt JD. 2008. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 28, 10 576–10 586. ( 10.1523/JNEUROSCI.1786-08.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302, 890–893. ( 10.1126/science.1090842) [DOI] [PubMed] [Google Scholar]
- 82.Nelson ED, Kavalali ET, Monteggia LM. 2006. MeCP2-dependent transcriptional repression regulates excitatory neurotransmission. Curr. Biol. 16, 710–716. ( 10.1016/j.cub.2006.02.062) [DOI] [PubMed] [Google Scholar]
- 83.Nguyen MV, Du F, Felice CA, Shan X, Nigam A, Mandel G, Robinson JK, Ballas N. 2012. MeCP2 is critical for maintaining mature neuronal networks and global brain anatomy during late stages of postnatal brain development and in the mature adult brain. J. Neurosci. 32, 10 021–10 034. ( 10.1523/JNEUROSCI.1316-12.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SBG, Guyenet PG, Kipnis J. 2012. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature 484, 105–109. ( 10.1038/nature10907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dawlaty MM, et al. 2011. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175. ( 10.1016/j.stem.2011.07.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang RR, et al. 2013. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245. ( 10.1016/j.stem.2013.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull K, Jaenisch R, Tsai L-H. 2013. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79, 1109–1122. ( 10.1016/j.neuron.2013.08.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Graff J, Woldemichael BT, Berchtold D, Dewarrat G, Mansuy IM. 2012. Dynamic histone marks in the hippocampus and cortex facilitate memory consolidation. Nat. Commun. 3, 991 ( 10.1038/ncomms1997) [DOI] [PubMed] [Google Scholar]