Summary
A major challenge in obtaining a full molecular description of evolutionary adaptation is to characterize how transcription factor (TF) DNA binding specificity can change. To identify mechanisms of TF diversification, we performed detailed comparisons of yeast C2H2 ZF proteins with identical canonical recognition residues that are expected to bind the same DNA sequences. Unexpectedly, we found that ZF proteins can adapt to recognize new binding sites in a modular fashion whereby binding to common core sites remains unaffected. We identified two distinct mechanisms, conserved across multiple Ascomycota species, by which this molecular adaptation occurred. Our results suggest a route for TF evolution that alleviates negative pleiotropic effects by modularly gaining new binding sites. These findings expand our current understanding of ZF DNA binding and provide evidence for paralogous ZFs utilizing alternate modes of DNA binding to recognize unique sets of noncanonical binding sites.
Keywords: transcription factors, zinc fingers, DNA binding sites, modularity, binding modes
Introduction
Cross- and intra-species analyses of TF binding site motifs and chromatin immunoprecipitation (ChIP) profiles have identified considerable binding site turnover from yeast to humans, supporting the prominent role for cis-changes in the evolution of regulatory networks (Borneman et al., 2007; Bradley et al., 2010; Gasch et al., 2004; Wilson et al., 2008). However, recent work has highlighted flexibility to changes in TFs themselves (i.e., changes in trans) (Baker et al., 2011; Lynch and Wagner, 2008; Nakagawa et al., 2013)}. A major challenge in obtaining a full molecular description of evolutionary adaptation is to characterize how TFs can change.
In this study, we focused on C2H2 zinc finger (ZF) proteins – the largest structural class of TFs in eukaryotes – as a model protein family to investigate novel mechanisms of TF evolution. C2H2 ZF proteins (hereafter referred to as “ZF proteins”) bind DNA using arrays of ZF domains, each containing an alpha helix and two beta strands (Klug, 2010) (Figure 1D). Extensive experimental (Beerli et al., 1998; Choo and Klug, 1997; Enuameh et al., 2013; Persikov et al., 2014; Wolfe et al., 2000) and computational (Benos et al., 2002; Kaplan et al., 2005; Mandel-Gutfreund and Margalit, 1998; Persikov and Singh, 2011; Siggers and Honig, 2007) analyses have established a ZF DNA recognition code in which amino acids at 4 canonical ‘recognition’ positions (− 1, 2, 3, and 6, as in (Elrod-Erickson et al., 1996)) in each ZF domain mediate DNA base contacts (Figure 1, Figure S1A). This stereotyped binding mode has made ZFs an object of intense research for the design of artificial TFs and custom ZF nucleases for site-specific genome editing (Klug, 2010).
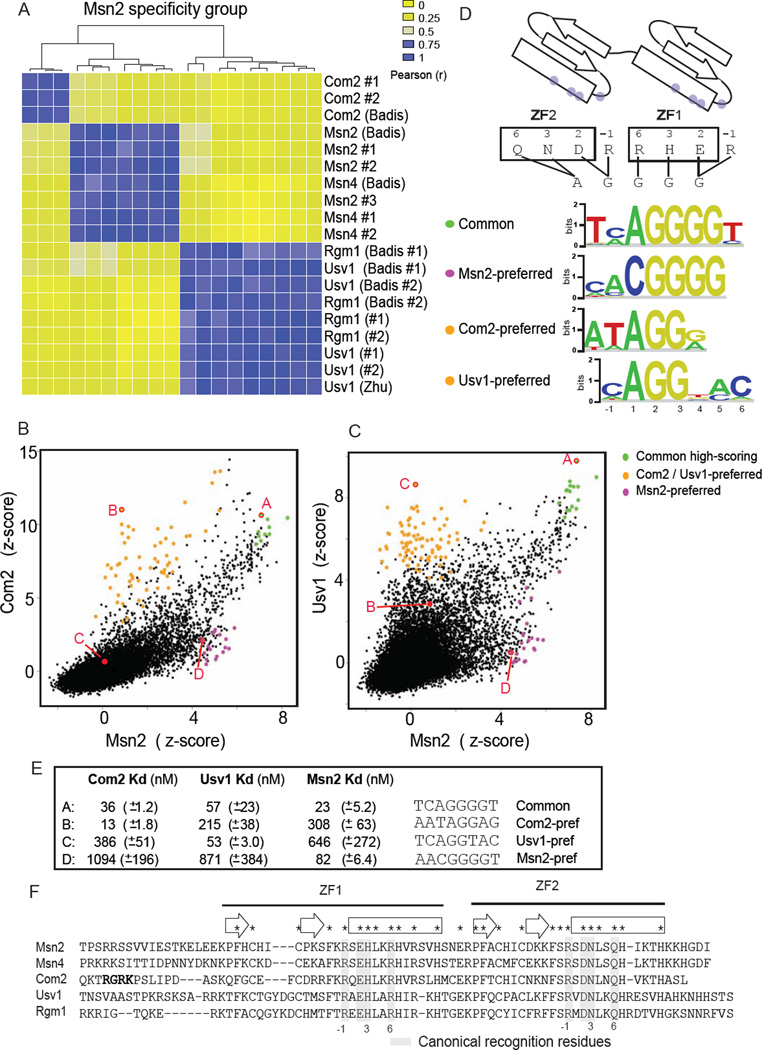
Figure 1. Common and TF-preferred DNA-binding specificities of Msn2-family members.
(A) Hierarchical clustering of pairwise binding profile comparisons for the Msn2-specificity-group proteins. Comparisons were performed for published datasets ((Badis et al., 2008) (Zhu et al., 2009)) and duplicate PBMs from this study. (B) Comparison of Msn2 and Com2 binding to all possible (32,896) 8-bp sequences. Z-scores are transformed 8-mer median signal intensities (see Experimental Procedures). TF-preferred sites bound preferentially by Msn2 or Com2 are in magenta or orange, respectively. Common sites bound significantly (PBM E-score > 0.48) by both proteins are highlighted. 8-bp sequences (labeled A, B, C and D) assayed for binding in EMSAs are in red. (C) Comparison for Msn2 and Usv1 (details as in B, except that Usv1-preferred sites are in orange). (D) Binding schematic for Msn2-family proteins and binding site motifs for TF-preferred and common sites. Canonical recognition residues are indicated (grey dots). Proposed interaction map for canonical residues and DNA bases of STRE is shown (see also Figure S1). (E) Dissociation binding constants (mean and standard deviation) for Com2, Usv1 and Msn2 to selected DNA sequences are listed. (F) Protein sequence alignment for Msn2 specificity group proteins. Highlighted are conserved residues (*), canonical ZF DNA-contacting residues (grey bars, canonical numbering scheme), Com2 N-terminal RGRK motif (bold), and ZF secondary structure elements: beta-strands (empty arrows) and alpha-helices (empty boxes).
Despite the appeal of a simple recognition code based on a few canonical residues, additional features of ZF proteins can affect DNA binding specificity, including inter-domain interactions (Isalan et al., 1997; Liu and Stormo, 2008; Wolfe et al., 1999), the inter-domain linker sequence (Handel et al., 2009), ZF docking geometry (Siggers and Honig, 2007), and residues outside the canonical recognition residues (Persikov and Singh, 2011). Residues in the loop between the beta strands (i.e., beta-turn) can also affect DNA binding affinity and footprinting pattern (Shiraishi et al., 2005). Deviations from a simple recognition code provide an explanation for studies demonstrating that while binding specificity of ZF arrays can target particular sequences, off-target binding will occur (Lam et al., 2011; Ramirez et al., 2008). Binding plasticity is an impediment to DNA binding predictions and design; however, we speculated that it might provide an opportunity to identify mechanisms that perturb or broaden TF-DNA binding specificity. We reasoned that analysis of binding variability between related ZFs might identify mechanisms for TF diversification and provide insights into TF evolution.
Analyzing the DNA binding of yeast C2H2 ZF proteins, we observed widespread differences among ZFs with identical canonical recognition residues. In addition to high affinity binding to sites that conform to the canonical ZF recognition rules, we identified binding to noncanonical sites that has been conserved throughout fungal evolution. Unexpectedly, we found that ZF proteins can gain new DNA binding specificities in a modular fashion whereby binding to common sites is unaffected. Furthermore, we demonstrate that this molecular adaptation occurs via at least two distinct mechanisms conserved across multiple Ascomycota species. Our results support a model of TF evolution in which the binding to only a subset of DNA binding sites is altered – this allows for evolution of novel regulatory functions among paralogous TFs while alleviating negative pleiotropic effects.
Results
Yeast C2H2 proteins exhibit DNA-binding diversity beyond a simple recognition code
To evaluate the DNA binding diversity in a group related ZFs, we focused on the simplest system available – the ZF proteins from Saccharomyces cerevisiae that bind DNA via only two adjacent ZF domains (ZFs) (Figure 1D). We compared the DNA binding of ZF proteins with identical canonical recognition residues; according to the canonical recognition code, these ZF proteins should bind the same DNA sites. We subdivided 28 proteins with two adjacent ZFs into 10 ‘specificity groups’ such that proteins in each group have identical recognition residues (Table S1).
High-resolution universal protein binding microarray (uPBM) data were available for 24 proteins in 8 specificity groups (Badis et al., 2008; Gordan et al., 2011; Zhu et al., 2009). The uPBM data provide unbiased and comprehensive binding profiles of each ZF protein to all 32,896 possible 8-bp sequences. We quantified the DNA-binding similarity between proteins by correlating the binding profiles over the 500 top-scoring 8-bp sequences from each experiment (see Experimental Procedures). Clustering the pairwise comparisons showed clear divisions between proteins within the same specificity groups (Figures 1A and S1B–S1E). These observations demonstrate that for this model system of two-ZF proteins mechanisms exist that perturb the DNA binding specificity from that predicted by a simple model based on canonical recognition residues.
In the S. cerevisiae lineage a whole-genome duplication (WGD) event occurred leaving many yeast genes with close paralogs (Wapinski et al., 2007). We found that the DNA-binding specificities for the majority of paralogs (6/8) are highly correlated (e.g., Msn2 and Msn4, Figures 1 and S1G). In contrast, with the exception of Mig proteins and Ygr067c / Yml081w, homologs that arose prior to the WGD exhibit DNA binding differences. These results suggest that DNA binding differences that deviate from a simple recognition code are the norm, rather than the exception, even for these short C2H2 ZF proteins.
Msn2-family proteins bind both common and TF-preferred DNA sequences
To examine in more detail the nature of the binding differences between related ZFs, we focused on the Msn2 specificity group (Msn2/Msn4, Com2, and Rgm1/Usv1). Msn2 and Msn4 proteins are major stress-response mediators and bind to the stress response element (STRE) AGGGG in stress-response gene promoters (Martinez-Pastor et al., 1996). We compared the binding profiles of paralog representatives and identified: (1) ‘common’ sites – high affinity sites common to both TFs (green points, Figure 1B and 1C); and (2) ‘TF-preferred’ sites – sites bound preferentially by one TF (orange and magenta points, Figure 1B and 1C). Sequence motifs generated from these distinct sets of sites illustrate the nature of the binding differences (Figure 1D). Common sites recognized by all Msn2 specificity group members match the AGGGG-type STRE reported as an Msn2 and Msn4 target site. Binding to AGGGG is explained by a simple recognition model based on canonical residues and known residue-base preferences (Figure 1D and S1). In contrast, TF-preferred sites differ significantly from the AGGGG common site, with distinct differences at unique base positions throughout the motifs (Figure1D). These results highlight that TF-preferred sites are recognized in addition to the common sites recognized by all members.
To evaluate the magnitude of the specificity differences we determined equilibrium binding constants (Kd) for select DNA sites by electrophoretic mobility shift assay (EMSA) (Figures 1E and S2). Binding experiments for Com2, Usv1 and Msn2 demonstrated high affinity (i.e., lower nanomolar) binding to the common and their ‘preferred’ sites. In contrast, binding to the preferred sites of the other proteins was significantly lower (e.g., Com2 bound 10.7-fold more weakly to the Usv1-preferred site than its own). These results demonstrate that the PBM data correspond well with traditional equilibrium binding affinities, as has been shown in previous studies (Siggers et al., 2011). Furthermore, the data show that binding affinities to the common and TF-preferred sites are of comparable magnitude.
To determine whether the TF-preferred sites are functionally relevant, we tested for enrichment of the TF-preferred sites in genomic regions bound in vivo. TF-preferred sites specific to the Usv1/Rgm1 paralogs, and not bound by other Msn2-group proteins, are significantly enriched (P < 1×10−5, Fisher’s exact test) in regions bound by Rgm1 during growth in complete medium (Wang et al., 2011), supporting that the TF-preferred sites are utilized in vivo (see Experimental Procedures).
Msn2-family DNA-binding differences are conserved throughout fungal evolution
We next examined whether the DNA binding preferences were conserved in orthologs from other species, or whether they occurred only in S. cerevisiae. We performed uPBM experiments for 18 Msn2-family orthologs from five other Ascomycota fungi, including three species that diverged before the WGD (Candida albicans, Kluyveromyces waltii, Kluyveromyces lactis) and two that diverged afterwards (Saccharomyces castellii, Candida glabrata) (Table S2). Clustering the binding profiles revealed that binding by the orthologs from the five other fungal species fell into the same 3 specificity groups that we previously identified (Figure S1F). Thus, the TF-preferred binding identified for the Msn2 family orthologs has been conserved since the last common ancestor of C. albicans and S. cerevisiae, approximately 300 million years ago, and therefore likely represents a selected and functionally relevant deviation from the canonical binding mode.
Com2-preferred binding is mediated in a modular fashion and requires an N-terminal basic motif
We sought to determine the mechanism by which the Com2 protein recognizes the Com2-preferred sites. We identified a conserved RGRK motif N-terminal to ZF1 in the Com2 orthologs that is not present in the other Msn2-family members (Figure 1F and S3). Strikingly, mutating the RGRK motif to RGEE (Com2 RK→EE, Figure S3, Table S2) selectively abrogated the Com2-preferred binding behavior (compare Figure 2A and 2B) but did not affect binding to the common sites. Therefore, an intact RGRK motif is required for the Com2-preferred binding. An N-terminal truncated version of Com2 (Com2 ΔN-term, Figure S3) missing the entire N-terminus flanking ZF1 produced nearly identical results to the Com2 RK→EE mutant (Figure S2B).
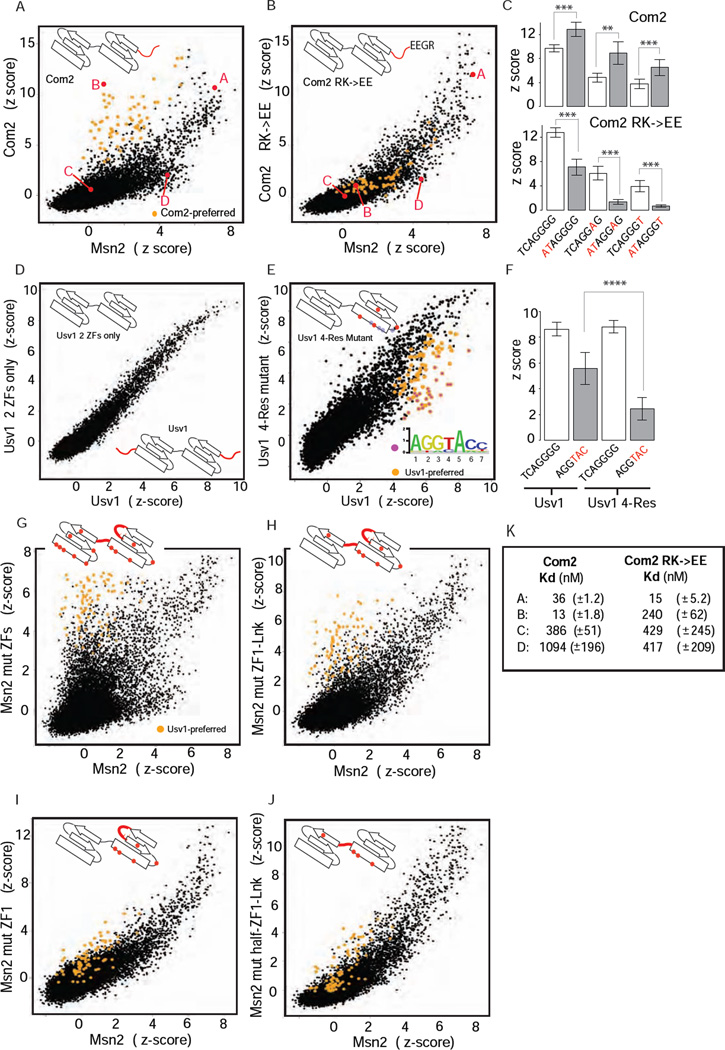
Figure 2. Binding specificities of ZF mutants.
(A), (B) Binding profile comparisons (as in Figure 1) for Com2 and Com2 RK→EE mutant relative to Msn2. Com2-preferred sites (as in Figure 1B) are highlighted (orange). 8-bp sequences (labeled A, B, C and D) assayed in EMSAs are in red. (C) Comparison of Com2 and Com2 RK→EE mutant binding to select binding sites. DNA base differences from the common site TCAGGGG (Figure 1D) are in red. Scores are mean z-scores for the eight different 8-mers containing the 7-mer sites shown (error bars = 1 standard deviation (SD)). (**) P < 10−3, (***) P < 10−4, unpaired Student’s t-test (D), (E) Binding profile comparisons for Usv1 two-ZF and 4-Res mutants shown relative to Usv1. Usv1-preferred sites (as in Figure 1D) are highlighted (orange); the subset of sites preferentially bound by Usv1 relative to Usv1 4-Res mutant are highlighted (magenta). Approximate positions of canonical recognition residues (grey dots) and mutated residues (red dots) are illustrated in ZF cartoons. (F) Comparison of Usv1 and Usv1 4-Res mutant binding to two binding sequences: (i) TCAGGGG common site (Figure 1D), and (ii) AGGTAC – a Usv1-preferred site (Figure 1D) that was bound poorly by Usv1 4-Res mutant. Scores are mean z-scores for the eight different 8-mers containing TCAGGGG (columns 1 and 3) and for the 48 different 8-mers containing AGGTAC (columns 2 and 4) (error bars = 1 SD). (****) P < 10−15, unpaired t-test. (G)–(J) Binding specificities of Msn2 wild-type and mutant proteins. Binding profile comparison for Msn2 mutants relative to Msn2. Usv1-preferred sites (as in Figure1D) are highlighted (orange). (K) Dissociation binding constants (mean and standard deviation) for Com2 (as in Figure 1E, for comparison) and Com2 RK→EE mutant to select DNA sequences are listed. Probe sequences are as in Figure 1E.
To determine if the Com2 RK→EE adversely affected the overall binding affinity, in addition to altering the binding specificity, we determined equilibrium binding constants (Kds) for the mutant protein to the select DNA sequences analyzed previously (Figure 2B and 2K). We found that the Com2 RK→EE mutant maintains its high affinity to the common site sequence (15 nM) but has an 18.5-fold reduction in affinity for the Com2-preferred site (13 nM to 240 nM, Figure 2K). These results demonstrate that binding to the Com2-preferred sites is a completely modular activity that can be specifically removed without affecting binding to the common site. Furthermore, the modular nature of binding suggests that Com2 uses two distinct binding modes to recognize different sequences.
The Com2 N-terminal basic region enhances binding to AT-rich sites
A general feature of RXR and RXXR peptide motifs is to select for AT-rich DNA sequences via DNA minor groove contacts (Rohs et al., 2009). Com2-preferred sites show a strong preference for an AT dinucleotide at positions −2 and −1 (Figure 1D). To test if Com2-preferred binding, mediated by the RGRK motif, operates by selective stabilization to sites with AT sequences 5’ to the AGGGG core, we compared the binding of Com2 and the Com2 RK→EE mutant to three different ‘core’ sequences (AGGGG, AGGAG, AGGGT) with either a 5’ TC dinucleotide (seen in the common sites) or a 5’ AT dinucleotide (see in Com2-preferred sites). As predicted, the Com2 N-terminal region enhanced the binding affinity to all sequences with the AT dinucleotide at positions −2 and −1, but did not affect the relative preference for the AGGGG, AGGAG or AGGGT core sequences (Figure 2C). These results, in conjunction with the Kd values (Figure 2K), demonstrate that the Com2 N-terminal region enhances binding to sites with an AT dinucleotide 5’ to the common AGGGG core.
Noncanonical residues in Usv1 ZF1 are involved in binding to Usv1-preferred sites
We next examined the mechanism of Usv1/Rgm1-preferred binding. In contrast to Com2, removal of the N- and C-terminal sequence outside the ZF domains (Table S2) had no effect on Usv1 binding (Figure 2D); therefore, Usv1-preferred binding operates by a different mechanism. We focused on residues within ZF1, which is predicted to interact with DNA bases at positions 4, 5 and 6 that varied in the Usv1-preferred sites (Figure 1D). In ZF1, we identified four residues (−5, −2, 5, 8 canonical numbering) conserved in Usv1, Rgm1 and their orthologs but not in the other Msn2-group members (Table S2, Usv1 4-Res). Mutating these four residues to their Msn2 counterparts significantly and selectively weakened binding to Usv1-preferred sites, while not affecting binding to common sites (Figure 2E, 2F). These results demonstrate that residues in ZF1, distinct from the canonical recognition residues, are involved in the selective binding to Usv1-preferred sites.
Usv1 binding preferences can be engineered onto Msn2 in a modular fashion
To better understand the modularity and possible evolutionary path to the observed TF-preferred binding, we set out to engineer the Com2- and Usv1-preferred binding activity onto Msn2. Adding the Com2 N-terminal region onto Msn2 (Table S2, Msn2 ZFs/Com2 N-term) did not result in Com2-preferred binding for Msn2 (data not shown), suggesting that additional amino acid positions within the ZF domains are required to stabilize or permit the Com2-preferred binding mode. In contrast, an extensive set of mutations across the Msn2 ZFs – changing the Msn2 residues to their Usv1 counterparts (Table S2, Msn2 mut ZFs) – led to strong Usv1-preferred binding for Msn2 (compare Figure 1C and 2G). When mutations were restricted to ZF1 and the inter-domain linker region, less pronounced but clearly present Usv1-preferred binding remained (Figure 2H). Therefore, while full Usv1-preferred binding requires residues from ZF2, partial Usv1-preferred binding can be obtained with residues from only ZF1 and the linker region. Further restrictions to the mutated residues in Msn2 largely abrogated the Usv1-preferred binding (Figure 2I, 2J). These results demonstrate that: (1) Usv1-preferred binding can be added in a modular fashion without affecting binding to the common sites; (2) full Usv1-preferred binding requires residues distributed across both ZF domains and the inter-ZF linker. These results demonstrate a second novel mechanism to expand C2H2 TF binding in a modular fashion.
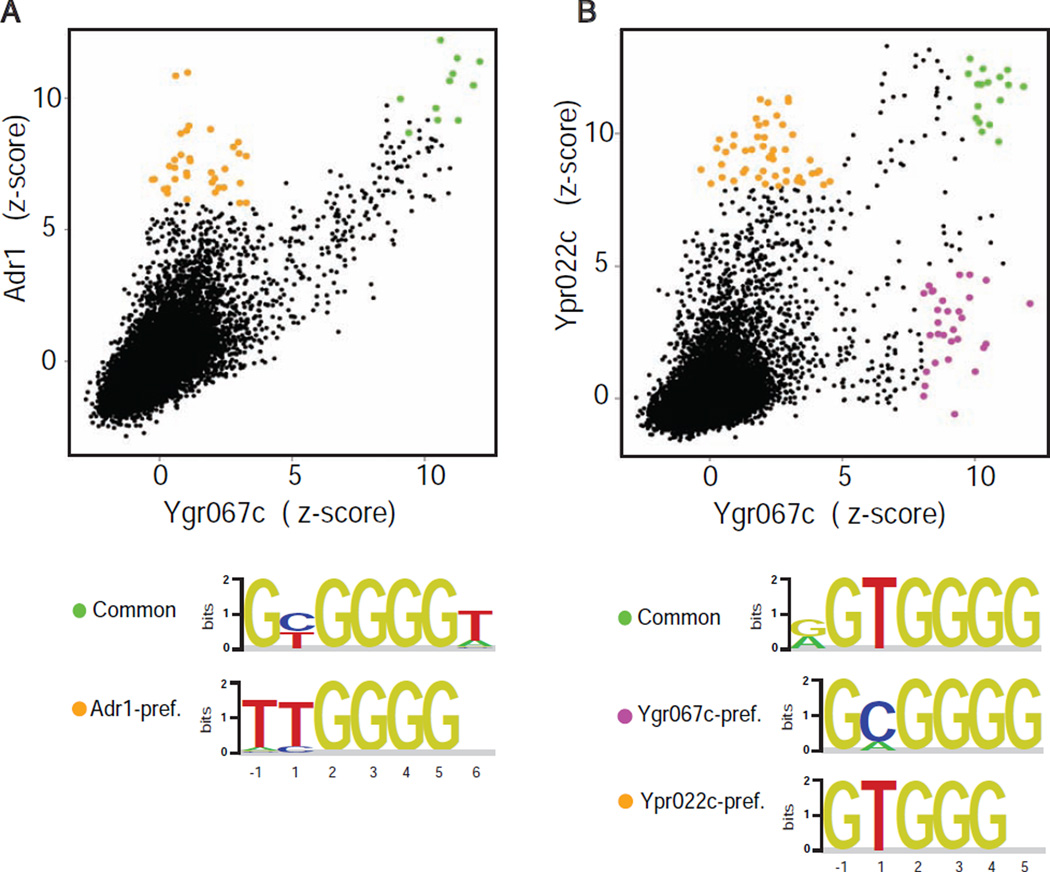
Adr1 specificity group proteins exhibit binding similarities with Msn2 group proteins
Our initial survey of ZF DNA binding profiles identified binding variability within multiple ZF specificity groups (Figure S1B–S1E, Table S1). We examined the nature of the binding variability in the other large specificity group, hereafter referred to as the Adr1 group (Table S1, top row) (Ypr022c, Rsf2, Yml081w/Ygr067c, Adr1). As seen for the Msn2 group, comparisons of Adr1 versus Ygr067c (Figure 3A) and Ypr022c versus Ygr067c (Figure 3B) revealed both common and TF-preferred sites. Furthermore, additional features of the TF-preferred binding in the Adr1 group resemble those seen for the Msn2 group, suggesting similar mechanisms might be operating. For example, both the Com2- and Adr1-preferred sites are distinguished by the preference for a Thy at position −1 (Figure 1 and 3). Provocatively, studies have shown that an N-terminal proximal accessory region (PAR) in Adr1 is necessary for high-affinity binding to the 5’-TTGGAG UAS1 element that resembles the Adr1-preferred sites (Schaufler and Klevit, 2003; Thukral et al., 1991). This suggests that the N-terminal PAR domain in Adr1 may mediate Adr1-preferred binding in a manner analogous to the Com2 N-terminal region described here. Future studies should clarify whether the TF-preferred binding in the Adr1-group is similarly modular, and whether the mechanisms used are identical to those of Com2 and Usv1, or if they represent additional ways to diversify ZF binding.
Figure 3. Binding specificity for Adr1 specificity group proteins.
(A)–(B) Pairwise binding profile comparisons for Adr1 and Ypr022C relative to Ygr067c. Z-scores are as in Figure 1B,C. Common and TF-preferred sites are highlighted. Binding motifs are shown for highlighted TF-preferred and common sites (base numbering as in Figure 1D, with base congruence defined by ZF binding schematic (see Figure S1)).
Discussion
In this work, we sought to uncover new mechanisms by which related TFs can diversify their DNA binding. Analyzing a model system of two-ZF proteins in yeast, we identified two distinct mechanisms by which ZF proteins can gain binding specificity in a modular fashion. We propose that the modular binding of these proteins comes from using multiple modes of DNA binding (Figure 4, discussed more below). Analysis of orthologs from other Ascomycota species demonstrated that this modular diversification strategy has been conserved since the divergence of C. albicans and S. cerevisiae and is likely a functionally important adaptation. Modular evolution of TF binding specificity, in which protein changes mediate binding to new sites while not affecting the binding to a core set of common sites, provides an elegant solution to the problem of widespread negative pleiotropic effects – modularity allows a TF to gain novel regulatory functions while avoiding potential negative consequences from loss of regulation from the core sites.
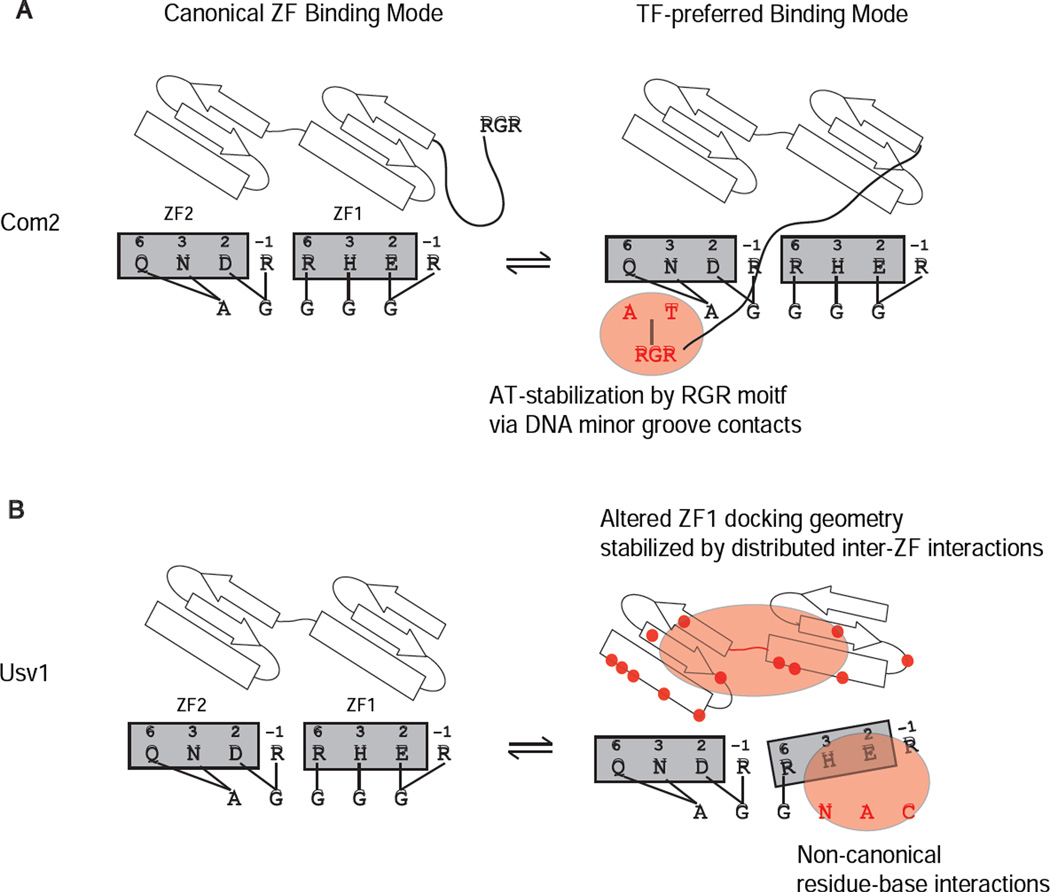
Figure 4. Models for TF-specific binding modes.
Binding schematics depict key features of the canonical ZF binding mode and the TF-specific binding modes that facilitate the binding to the Msn2-family common and TF-preferred sequences. Models are presented for (A) Com2 and (B) Usv1 (same model applies for Rgm1). Key features proposed for the TF-specific binding modes are highlighted. Residues mutated in Msn2 and Usv1 constructs (Figure 2) that affected DNA binding are highlighted (red dots).
The ability of individual TFs to bind DNA via multiple binding modes has been reported for a number TFs (reviewed in (Siggers and Gordan, 2013)), and likely represents a general mechanism for TF binding diversification. A recent study has described multiple binding modes for forkhead TFs and provided evidence that the distinct binding modes have arisen repeatedly and independently in the course of forkhead evolution (Nakagawa et al., 2013). Interestingly, the DNA sequences bound by distinct forkhead modes are completely different (i.e., 5’-GTAAACAA vs 5’-GACGC), whereas in this study we find that different binding modes exhibit different preferences over only a portion of the binding site (Figures 1 and 3), suggesting different mechanisms are operating. Future studies using similar approaches will be highly informative to delineate both the mechanisms and the evolutionary history of TF diversification by the gain and loss of alternate binding modes. In addition to altering a TF’s target sites, alternate binding modes provide a mechanism for DNA allostery whereby DNA sequence differences can affect protein structure and result in differences in cofactor recruitment and transcriptional activity (reviewed in (Siggers and Gordan, 2013)). Our data supports a model in which the ZF proteins adopt different conformations based on the DNA sequence. Future studies are needed to investigate whether the alternate binding modes and target sequences identified for the Msn2-family might relate to differential cofactor recruitment and transcriptional activity.
C2H2 ZFs have been studied as a paradigm of modularity in DNA binding and as a powerful scaffold for designing synthetic ZFs (Klug, 2010). For protein engineers, the appeal of this family has been the ability to model and manipulate DNA binding specificity by altering a small set of canonical residues. While it is widely appreciated that residues outside of the canonical recognition positions affect DNA binding, the mechanisms by which these residues alter binding remain unclear (Lam et al., 2011; Persikov et al., 2014; Persikov and Singh, 2011; Ramirez et al., 2008). Here we present two novel mechanisms for altering ZF DNA binding specificity that operate via stabilization of alternate binding modes. Importantly, these alternate modes bind DNA with comparable affinity, operate in a modular fashion (i.e., can be selectively abrogated), and do not involve changes in ZF number or canonical residue identity. One mechanism involves a basic RGRK motif found N-terminal to Com2 that stabilizes the binding to AT-rich DNA sites (Figure 1 and 2). We propose that the RGRK motif works similarly to other RXR motifs in which Arg residues project into the DNA minor groove and stabilize AT-rich sequences (Rohs et al., 2009) (Figure 4A). The D. melanogaster Trl (GAGA) ZF protein also uses a basic region N-terminal to the ZF to mediate minor groove interactions; however, its role in binding specificity remains unknown (Omichinski et al., 1997). Analysis of existing ZF structures suggest that the RGRK motif in Com2 could reach and interact with the AT base pairs (as in Figure 4) via the minor groove. The second mechanism we identified operates via a distributed set of amino acids throughout the two ZFs and inter-ZF linker of Usv1 (Figure 1, 2). We propose a model in which an altered binding mode, or docking geometry, of ZF1 is stabilized by inter-domain residue-residue contacts (Figure 4B). In this altered binding mode alternate DNA-base contacts are made, either by the residues at the canonical recognition positions or by alternate, noncanonical recognition residues.
Our results that ZF proteins can switch between alternate binding modes to recognize different DNA sites (Figure 4) has implications for predicting and designing ZF DNA binding. First, the ability to selectively abrogate binding to a subset of the DNA sequences presents a conceptual complication to representing the DNA binding specificity by single binding models such as a position weight matrix (PWM) (Enuameh et al., 2013; Gupta et al., 2014; Jolma et al., 2013; Zhao and Stormo, 2011). Second, the ability to switch between different binding modes and bind different DNA sites could lead to off-target binding in a ZF design experiment - one might optimize residues to bind select target sites only to find that an alternate mode permitted binding to undesirable sites. Multiple binding modes further motivates the utility of selection assays and the inability to simply ‘stich-together’ ZF domains of characterized specificity (Beerli et al., 1998; Choo and Klug, 1997; Enuameh et al., 2013; Klug, 2010; Persikov et al., 2014; Wolfe et al., 2000) and suggests that studies aimed at finding ways to inhibit alternate binding modes may minimize off-target binding of synthetic ZF proteins.
Experimental Procedures
GST-tagged proteins were expressed and purified from bacteria, or made by in vitro transcription translation (IVT), all ZF constructs listed in Table S3. PBM experiments and analysis were carried out as previously described (Berger and Bulyk, 2006, 2009), all PBM data provided in Tables S3 and S4. Genome analyses were performed using custom Perl scripts (available on request), clustering performed using R statistical package. (see Extended Experimental Procedures for full details).
Supplementary Material
Acknowledgements
We thank Ilan Wapinski, Anton Aboukhalil, Steve Gisselbrecht and Bo Jiang for helpful discussions. We thank Aviv Regev and Ilan Wapinski for kindly providing yeast cDNA. This work was supported by NIH/NHGRI grant # R01 HG003985 (M.L.B.) and lab startup funds from Boston University (T.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
TS and JR cloned TF ZF domains, BB performed protein expression, purification and EMSA binding assays, TS, JR and BB performed PBM experiments, TS performed data analysis, TS and MLB designed the study and the experiments, TS and MLB wrote the manuscript.
The authors declare that they have no competing financial interests.
References
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CR, Tuch BB, Johnson AD. Extensive DNA-binding specificity divergence of a conserved transcription regulator. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7493–7498. doi: 10.1073/pnas.1019177108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos PV, Lapedes AS, Stormo GD. Probabilistic code for DNA recognition by proteins of the EGR family. Journal of molecular biology. 2002;323:701–727. doi: 10.1016/s0022-2836(02)00917-8. [DOI] [PubMed] [Google Scholar]
- Berger MF, Bulyk ML. Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins. Methods Mol Biol. 2006;338:245–260. doi: 10.1385/1-59745-097-9:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Bradley RK, Li XY, Trapnell C, Davidson S, Pachter L, Chu HC, Tonkin LA, Biggin MD, Eisen MB. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS biology. 2010;8:e1000343. doi: 10.1371/journal.pbio.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Current opinion in structural biology. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- Enuameh MS, Asriyan Y, Richards A, Christensen RG, Hall VL, Kazemian M, Zhu C, Pham H, Cheng Q, Blatti C, et al. Global analysis of Drosophila Cys2-His2 zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome research. 2013;23:928–940. doi: 10.1101/gr.151472.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan R, Murphy KF, McCord RP, Zhu C, Vedenko A, Bulyk ML. Curated collection of yeast transcription factor DNA binding specificity data reveals novel structural and gene regulatory insights. Genome biology. 2011;12:R125. doi: 10.1186/gb-2011-12-12-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Christensen RG, Bell HA, Goodwin M, Patel RY, Pandey M, Enuameh MS, Rayla AL, Zhu C, Thibodeau-Beganny S, et al. An improved predictive recognition model for Cys2-His2 zinc finger proteins. Nucleic acids research. 2014 doi: 10.1093/nar/gku132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Molecular therapy : the journal of the American Society of Gene Therapy. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Kaplan T, Friedman N, Margalit H. Ab initio prediction of transcription factor targets using structural knowledge. PLoS computational biology. 2005;1:e1. doi: 10.1371/journal.pcbi.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annual review of biochemistry. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Lam KN, van Bakel H, Cote AG, van der Ven A, Hughes TR. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic acids research. 2011;39:4680–4690. doi: 10.1093/nar/gkq1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stormo GD. Context-dependent DNA recognition code for C2H2 zinc-finger transcription factors. Bioinformatics. 2008;24:1850–1857. doi: 10.1093/bioinformatics/btn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution; international journal of organic evolution. 2008;62:2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- Mandel-Gutfreund Y, Margalit H. Quantitative parameters for amino acid-base interaction: implications for prediction of protein-DNA binding sites. Nucleic acids research. 1998;26:2306–2312. doi: 10.1093/nar/26.10.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) The EMBO journal. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1310430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omichinski JG, Pedone PV, Felsenfeld G, Gronenborn AM, Clore GM. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nature structural biology. 1997;4:122–132. doi: 10.1038/nsb0297-122. [DOI] [PubMed] [Google Scholar]
- Persikov AV, Rowland EF, Oakes BL, Singh M, Noyes MB. Deep sequencing of large library selections allows computational discovery of diverse sets of zinc fingers that bind common targets. Nucleic acids research. 2014;42:1497–1508. doi: 10.1093/nar/gkt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov AV, Singh M. An expanded binding model for Cys2His2 zinc finger protein-DNA interfaces. Physical biology. 2011;8:035010. doi: 10.1088/1478-3975/8/3/035010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nature methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufler LE, Klevit RE. Mechanism of DNA binding by the ADR1 zinc finger transcription factor as determined by SPR. Journal of molecular biology. 2003;329:931–939. doi: 10.1016/s0022-2836(03)00550-3. [DOI] [PubMed] [Google Scholar]
- Shiraishi Y, Imanishi M, Morisaki T, Sugiura Y. Swapping of the beta-hairpin region between Sp1 and GLI zinc fingers: significant role of the beta-hairpin region in DNA binding properties of C2H2-type zinc finger peptides. Biochemistry. 2005;44:2523–2528. doi: 10.1021/bi047797h. [DOI] [PubMed] [Google Scholar]
- Siggers T, Duyzend MH, Reddy J, Khan S, Bulyk ML. Non-DNA-binding cofactors enhance DNA-binding specificity of a transcriptional regulatory complex. Mol Syst Biol. 2011;7:555. doi: 10.1038/msb.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers T, Gordan R. Protein-DNA binding: complexities and multi-protein codes. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers TW, Honig B. Structure-based prediction of C2H2 zinc-finger binding specificity: sensitivity to docking geometry. Nucleic acids research. 2007;35:1085–1097. doi: 10.1093/nar/gkl1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral SK, Morrison ML, Young ET. Alanine scanning site-directed mutagenesis of the zinc fingers of transcription factor ADR1: residues that contact DNA and that transactivate. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:9188–9192. doi: 10.1073/pnas.88.20.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Mayhew D, Chen X, Johnston M, Mitra RD. Calling Cards enable multiplexed identification of the genomic targets of DNA-binding proteins. Genome research. 2011;21:748–755. doi: 10.1101/gr.114850.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavare S, Odom DT. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. Journal of molecular biology. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annual review of biophysics and biomolecular structure. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Stormo GD. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nature biotechnology. 2011;29:480–483. doi: 10.1038/nbt.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.