Abstract
The innate immune defence of multicellular organisms against microbial pathogens requires cellular collaboration. Information exchange allowing immune cells to collaborate is generally attributed to soluble protein factors secreted by pathogen-sensing cells. Cytokines, such as type I interferons (IFNs), serve to alert non-infected cells to the possibility of pathogen challenge1. Moreover, in conjunction with chemokines they can instruct specialized immune cells to contain and eradicate microbial infection. Several receptors and signalling pathways exist that couple pathogen sensing to the induction of cytokines, whereas cytosolic recognition of nucleic acids seems to be exquisitely important for the activation of type I IFNs, master regulators of antiviral immunity2. Cytosolic DNA is sensed by the receptor cyclic GMP-AMP (cGAMP) synthase (cGAS), which catalyses the synthesis of the second messenger cGAMP(2′-5′)3, 4, 5, 6, 7, 8. This molecule in turn activates the endoplasmic reticulum (ER)-resident receptor STING9, 10, 11, thereby inducing an antiviral state and the secretion of type I IFNs. Here we find in murine and human cells that cGAS-synthesized cGAMP(2′-5′) is transferred from producing cells to neighbouring cells through gap junctions, where it promotes STING activation and thus antiviral immunity independently of type I IFN signalling. In line with the limited cargo specificity of connexins, the proteins that assemble gap junction channels, most connexins tested were able to confer this bystander immunity, thus indicating a broad physiological relevance of this local immune collaboration. Collectively, these observations identify cGAS-triggered cGAMP(2′-5′) transfer as a novel host strategy that serves to rapidly convey antiviral immunity in a transcription-independent, horizontal manner.
On recognition of virus-derived nucleic acids, innate immune signalling initiates cell-autonomous antiviral effector mechanisms that aim to block viral propagation. Moreover, virus-infected cells alert non-infected neighbouring cells, a process largely attributed to the de novo expression and secretion of cytokines and chemokines. At the same time, a few reports have documented the phenomenon of cytokine-independent activation of bystander cells via gap junctions in the context of bacterial infection12, irradiation13 or DNA transfection14. However, the molecular mechanisms responsible for these effects remained elusive.
The finding that pattern sensing relies on a specific intermediate messenger molecule to activate a second receptor is unique in innate immunity, thus raising the question whether cGAMP(2′-5′)-mediated information transduction might provide organisms with an advantage over the use of a canonical, cell-autonomous signal transduction pathway15.
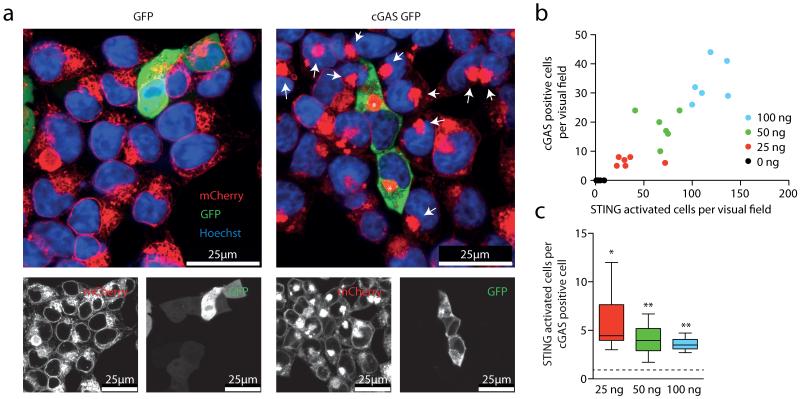
Activation of STING triggers its oligomerization into a supramolecular complex and its translocation from the ER to a perinuclear compartment16, a process that can be monitored at the single-cell level using fluorescence microscopy. To characterize the molecular mechanism of the cGAS–STING pathway better, we used HEK cells stably transduced with an amino-terminally mCherry-tagged STING construct (HEK STING)17. As expected, transient overexpression of cGAS–GFP in HEK STING cells led to phosphorylation of IRF3 and re-localization of STING to perinuclear complexes (Fig. 1a, asterisks and data not shown). Surprisingly, we also observed STING translocation in cells that lacked cGAS–GFP expression, but that were located adjacent to cGAS-expressing cells (Fig. 1a, arrows). In contrast, the cell-permeable STING activator CMA induced homogenous STING clustering (see below)17, indicating that stimulation of surrounding cells occurs via an event that is spatially and temporally linked to cGAS activity. Quantifying cGAS expression next to STING activation revealed an approximately fourfold higher number of STING-activated cells compared to cGAS-expressing cells (Fig. 1b, c).
Figure 1. cGAS overexpression activates STING in adjacent cells.
a, Confocal microscopy of HEK STING cells 20 h after transfection with GFP (left) or a cGAS–GFP (right). Asterisks and arrows highlight STING complexes in GFP-positive cells and bystander cells. b, c, HEK STING cells were transfected with varying amounts of cGAS–GFP as indicated. The number of GFP-positive cells is plotted against the number of activated HEK STING cells (y = 0.27x, R2 = 0.84) (b) and the respective ratio of STING-activated cells over cGAS-expressing cells is depicted. Data are depicted as box plots with whiskers indicating minimum and maximum (c). One representative experiment out of two independent experiments is shown. *P < 0.05, **P < 0.01.
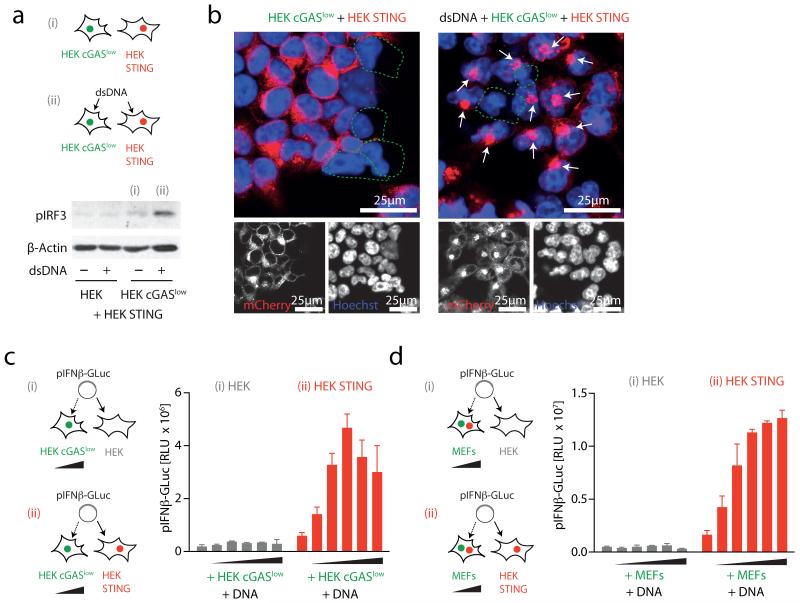
To assess the function of cGAS as a DNA receptor, we next generated monoclonal HEK cGAS cells with either high or low constitutive expression of cGAS. As expected, a cell clone with high cGAS expression (HEK cGAS*) induced spontaneous activation of STING and IRF3 phosphorylation in bystander cells (Supplementary Fig. 1 and data not shown). In contrast, a monoclonal cell line with low cGAS expression (HEK cGASlow) additionally required DNA stimulation to exert STING and subsequent IRF3 activation in bystander cells (Fig. 2a, b). Moreover, titrating the number of HEK cGASlow cells on top of STING competent cells in conjunction with DNA transfection revealed a dose-dependent increase in IFN-β promoter transactivation (Fig. 2c and Supplementary Fig. 2a). This bystander STING activation phenomenon was also observed when HEK STING cells were co-incubated with DNA-stimulated murine embryonic fibroblasts (MEFs) that are inherently competent for cGAS (Fig. 2d and Supplementary Fig. 2b). Of note, knockdown of cGAS in MEFs markedly decreased in trans activation of HEK STING cells following DNA stimulation (Supplementary Fig. 2c-f). Moreover, switching donor and recipient cells showed the same effect: HEK cGAS* but not unmodified HEK cells transactivated MEFs and the murine cell line LL171 in a STING-dependent fashion, indicating that cGAS-dependent STING activation in trans was conserved across species (see below). Notably, this phenomenon of bystander cell activation was not observed when expressing an RNA-polymerase-III-driven RIG-I stimulatory RNA molecule18: whereas cell-intrinsic RIG-I activation was observed under these conditions, no bystander activation could be detected (Supplementary Fig. 3).
Figure 2. Cytosolic DNA sensing via cGAS propagates STING activation in trans.
a, HEK or HEK cGASlow co-incubated with HEK STING cells were stimulated (4 h) as indicated and IRF3 phosphorylation was assessed. b, Confocal microscopy of HEK cGASlow co-incubated with HEK STING cells unstimulated or transfected with ISD (6 h). c, d, HEK cells and HEK STING cells were co-cultured with HEK cGASlow cells (c) or primary MEFs (d) (ratios ranging from 1:0.25 to 1:0.0156 HEK/HEK STING:cGASlow/MEFs) and transfected with pIFN-β-GLuc, whereas transactivation of the reporter was assessed after 20 h. Representative experiments of n = 2 (a and b) or mean and s.e.m. (biological duplicates) of one representative experiments out of six (c) or eight (d) are depicted. RLU, relative light unit.
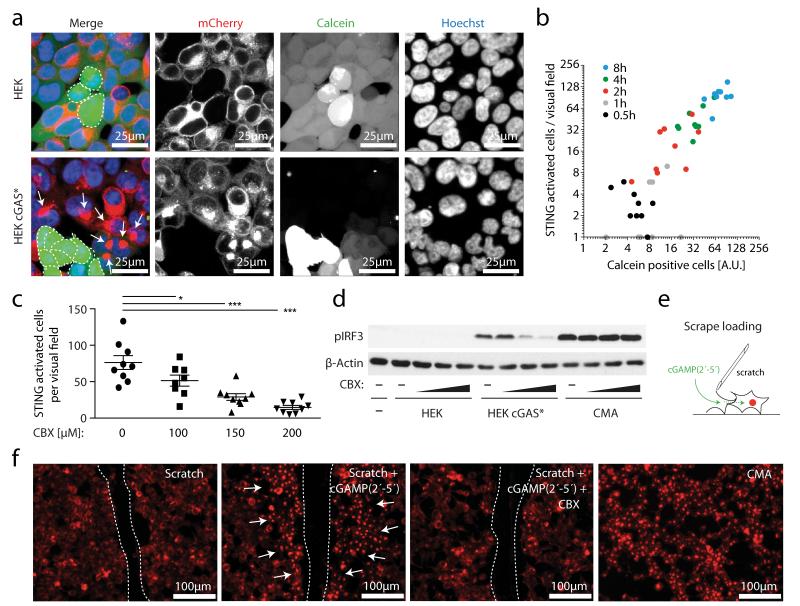
Separating donor and recipient cells via a trans-well system completely blunted bystander cell activation (Supplementary Fig. 4), indicating that a cell-to-cell contact-dependent transfer mechanism was responsible for conveying the IRF3 activating signal14. When we loaded HEK cGAS* cells with the low-molecular-weight dye calcein as a tracer, we observed transfer of calcein from HEK cGAS* cells into HEK STING cells that were in direct or indirect contact (Fig. 3a). Most notably, calcein transfer coincided with STING activation in the recipient cells, indicating a physical connection of signal transduction (Fig. 3a and Supplementary Videos 1 and 2). Indeed, quantitative analysis of calcein transfer and STING activation over time revealed a strong correlation of the two processes (Fig. 3b). These results indicated the involvement of gap junctions, which represent a well-established route of cell-contact-dependent intercellular communication. We therefore tested the impact of carbenoxolone (CBX), a well-characterized inhibitor of connexin function and thus gap junctions. CBX treatment potently inhibited calcein transfer from HEK cGAS* cells to HEK STING cells in a dose-dependent fashion and also blocked STING activation and IRF3 phosphorylation (Fig. 3c, d and Supplementary Fig. 5). Gap junctions allow the passage of small molecules below 1 kDa between cells, whereas larger biomolecules or proteins are precluded from this means of intercellular connection19. The fact that cGAS expression per se led to bystander STING activation indicated that cGAMP(2′-5′) was the second messenger molecule transported across gap junctions. To test this hypothesis we made use of scrape loading20, which is a well-established technique to study gap junction intercellular communication in vitro. In this assay a cellular monolayer is wounded by a cut allowing the extracellular space to gain access to the cytoplasm of lacerated cells, which are still coupled to neighbouring cells through gap junctions (Fig. 3e). Supplying cGAMP(2′-5′) to scratched HEK STING cells led to rapid and strong STING activation along the margins of the laceration and CBX treatment abrogated this effect (Fig. 3f and Supplementary Fig. 6).
Figure 3. cGAS-produced cGAMP(2′-5′) passes through gap junctions to trigger STING activation in bystander cells.
a, Confocal microscopy of HEK cells and HEK cGAS* cells loaded with calcein and added to HEK STING cells for 4 h. b, Co-culturing was performed as in a and after 0–8 h HEK STING cells were analysed by fluorescence microscopy for STING aggregation in nine independent visual fields. A dot-blot diagram correlating calcein-positive HEK STING cells with STING aggregate formation is presented (y = 1.27x, R2 = 0.85). AU, arbitrary units. c, HEK STING cells were co-cultured with HEK cGAS* in the presence of CBX as indicated for 4 h and studied for STING activation in nine independent visual fields. Data are presented as mean and s.e.m. d, IRF3 phosphorylation in HEK STING cells with HEK cells, with HEK cGAS* cells or with CMA in the presence or absence of CBX (100 μM, 150 μM and 200 μM) (3 h). e, The scrape loading technique. f, Fluorescence images of wounded HEK STING cells incubated with nothing, cGAMP(2′-5′) or cGAMP(2′-5′) with 150 μM CBX. HEK STING cells with CMA served as control (dashed line, scratch margins; arrows, STING complexes). One representative experiment out of two independent experiments is shown (a-d, f). *P < 0.05, ***P < 0.001.
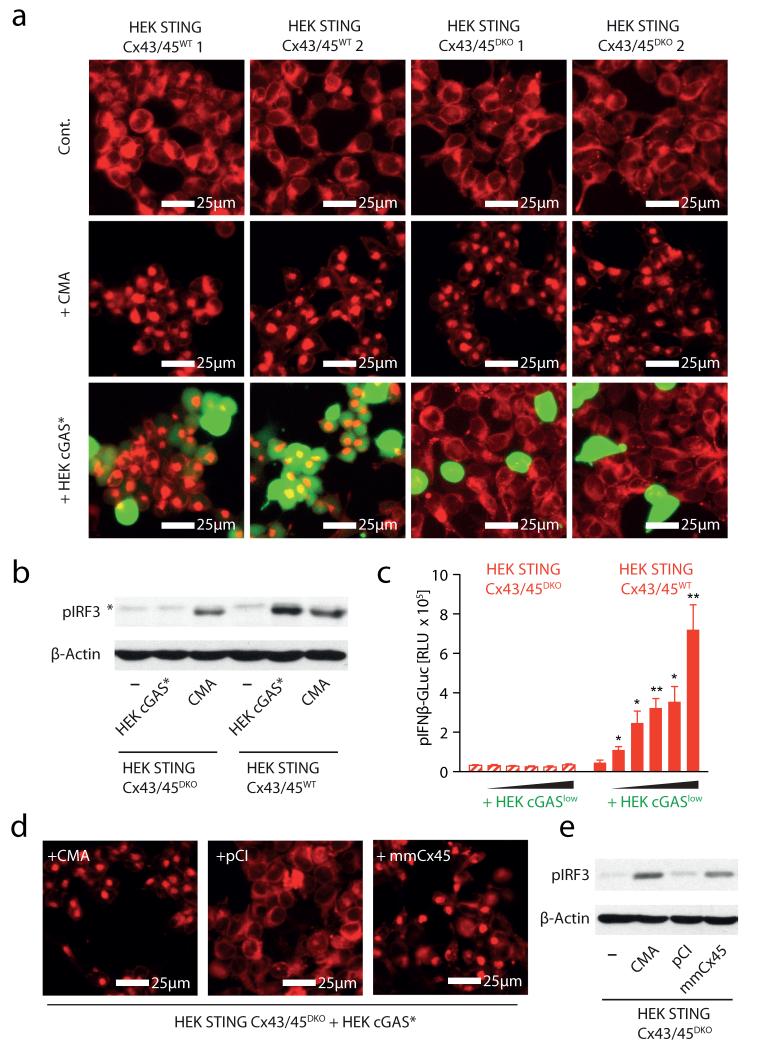
Gap junctions are formed by connexin proteins that assemble into clusters of hundreds of intercellular channels, thus physically connecting the cytoplasm of neighbouring cells. The connexin family of proteins consists of 20 members in the mouse and 21 members in the human system, with overlapping yet distinctive cellular distribution patterns19. HEK cells have been reported to express connexins 43 (CX43) and 45 (CX45)21, 22. Consequently, to investigate the mechanism of gap-junction-mediated transfer of cGAMP(2′-5′), we simultaneously targeted CX43 and CX45 in HEK STING cells using the CRISPR/Cas9 system23, 24. We thus generated two CX43/CX45-competent (wild type (WT)) and two CX43/CX45-deficient (double knockout (DKO)) HEK STING cell lines (HEK STING CX43/45WT cells and HEK STING CX43/45DKO cells) (Supplementary Fig. 7). HEK STING CX43/45DKO cells showed normal responsiveness towards CMA, indicating that STING responses were intact in these cells (Fig. 4a, b). However, co-culture of HEK STING CX43/45DKO cells with calcein-loaded HEK cGAS* cells did not result in dye transfer and, at the same time, STING activation, IRF3 phosphorylation and transactivation of an IFN-β reporter were completely abrogated (Fig. 4a-c). In line with this, DNA-stimulated MEFs only transactivated gap-junction-competent HEK STING CX43/45WT cells but not HEK STING CX43/45DKO cells (data not shown). Moreover, scrape loading of HEK STING CX43/45DKO cells with cGAMP (2′-5′) did not give rise to extended patches of STING-activated cells (Supplementary Fig. 8a). Reconstitution of CX45 and CX43 in HEK STING CX43/45DKO cells restored bystander STING activation and the resulting phosphorylation of IRF3 (Fig. 4d, e and Supplementary Fig. 8b; data not shown) and, at the same time, overexpression of five out of six additional distinct human connexin family members restored bystander cell activation in HEK STING CX43/45DKO cells as well (Supplementary Fig. 8b). This mutual potential for complementation is in line with the notion that connexins have only limited cargo specificity and thus can functionally compensate for other family members19.
Figure 4. Connexin 43 and 45 mediate cGAMP(2′-5′) transfer in HEK STING cells.
a, Fluorescence microscopy of HEK STING CX43/45WT and HEK STING CX43/45DKO cells left untreated, stimulated with CMA or co-cultured with calcein-loaded HEK cGAS* cells (ratio HEK/HEK STING:HEK cGAS* = 1:0.25) after 8 h. b, Phosphorylation of IRF3 in HEK STING CX43/45WT and HEK STING CX43/45DKO cells left untreated, co-incubated with HEK cGAS* cells or stimulated with CMA for 4 h (asterisk indicates nonspecific band). c, HEK STING CX43/45WT and HEK STING CX43/45DKO cells were co-cultured with HEK cGASlow cells (ratios from 1:0.5 to 1:0.0312 HEK STING CX43/45WT or HEK STING CX43/45DKO cells:cGASlow) transfected with pIFN-β-GLuc and luciferase activity was assessed after 20 h. Mean and s.e.m. (biological duplicates) of one representative experiment out of three independent experiments is shown. d, Fluorescence microscopy of HEK STING CX43/45DKO cells co-cultured with HEK cGAS* cells and stimulated with CMA, transfected with empty vector (pCI) or an expression vector for murine CX45 (mmCX45, 20 h). e, Co-cultures from d were analysed for phosphorylation of IRF3. One representative experiment out of two independent experiments is shown (a, b, d, e). *P < 0.05, **P < 0.01.
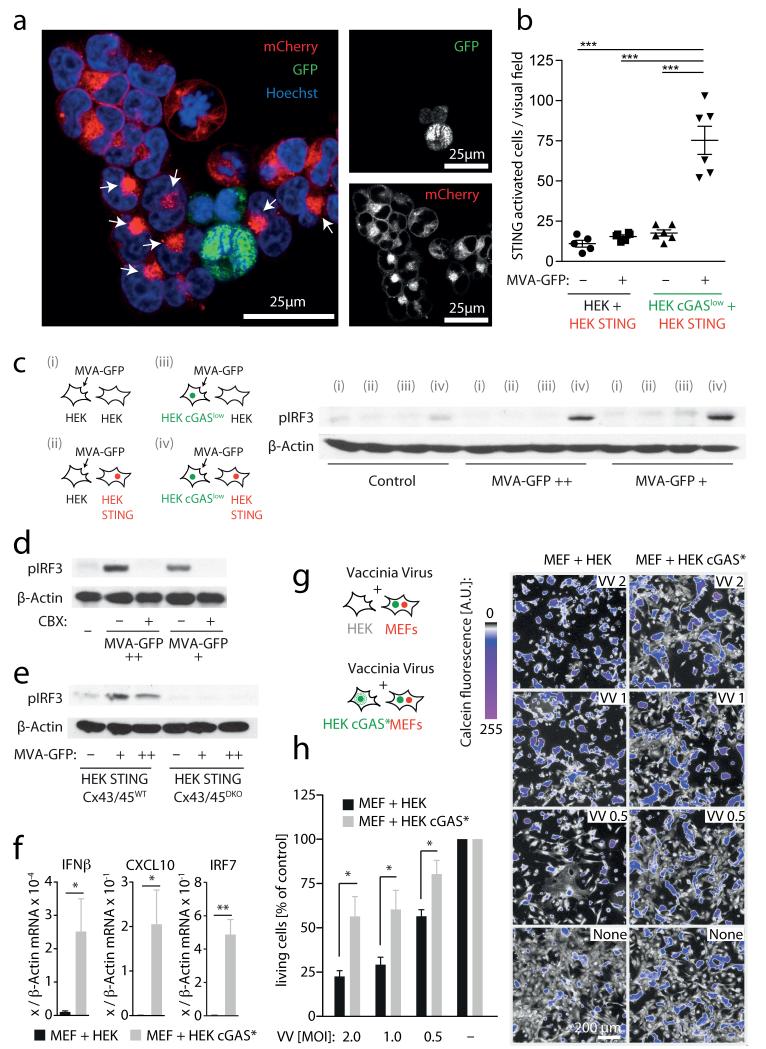
We next sought to address the physiological relevance of this phenomenon in the context of infection with a DNA virus known to activate STING. With this aim, we infected HEK cGASlow cells with a replication-incompetent vaccinia virus strain encoding nucleus-targeted GFP (MVA–GFP, modified vaccinia Ankara). Three hours after infection, HEK cGASlow cells were washed and co-cultured with HEK STING cells. HEK STING cells adjacent to MVA-infected cells showed prominent STING clustering (Fig. 5a, b) as well as robust IRF3 phosphorylation (Fig. 5c). Of note, this MVA-initiated antiviral signalling was dependent on both cGAS expression in donor cells as well as STING expression in recipient cells. Consistent with our earlier results, MVA-initiated bystander antiviral response was blocked by CBX (Fig. 5d) and was absent in HEK STING cells deficient for CX43/45 (Fig. 5e). Similarly, co-culturing MVA-infected MEFs induced upregulation of IFN-β in HEK STING cells, but not in unmodified HEK cells, and required functional gap junctions (Supplementary Fig. 9). Finally, we tested whether this bystander STING activation conferred antiviral protection. To this end, we made use of a replication-competent strain of vaccinia virus (Western Reserve) that induces rapid cell death in primary MEFs. Co-incubating MEFs with HEK cGAS* cells induced strong expression of antiviral genes in MEFs (Fig. 5f) and led to a marked increase in viral resistance of MEFs infected with vaccinia virus, as observed by an increase in cell viability (Fig. 5g, h).
Figure 5. Vaccina virus triggers STING-dependent antiviral immunity in bystander cells.
a, b, HEK cGASlow cells or HEK cells were infected with MVA–GFP, washed and then loaded onto HEK STING cells for 8 h. a, Confocal microscopy of HEK STING cells co-incubated with MVA-infected HEK cGAS cells (arrows, STING activation in bystander cells). b, Quantification of STING activation in response to MVA-infected HEK cGASlow cells or MVA-infected HEK cells. c, HEK cells or HEK cGASlow cells were MVA-infected or left untreated, added onto HEK cells or HEK STING cells and studied for IRF3 phosphorylation (viral particles per ml: ++ = 3.2 × 107; + = 1.6 × 107). d, e, Experiments as in a in the presence or absence of CBX 150 μM (d) or using HEK STING CX43/45WT and HEK STING CX43/45DKO cells as responder cells (e). f, HEK cGAS* cells were co-incubated with MEFs and after 14 h mouse Ifnb mRNA, mouse Cxcl10 mRNA and mouse Irf7 mRNA were assessed by qPCR. g, h, HEK cGAS* cells were co-incubated with MEFs and after 12 h vaccinia virus was added (multiplicity of infection (m.o.i.): 2-0.5). Twenty-four hours later cell survival was analysed. Visual fields of one representative experiment are depicted (g, right panel) and mean and s.e.m. of three independent experiments are summarized (h). VV, vaccinia virus. One representative experiment out of two (a, b, e) or three (c, d) independent experiments are shown or mean and s.e.m. of n = 5 independent experiments is presented (f). *P < 0.05, **P < 0.01, ***P < 0.001.
We provide a clear delineation of a unique in trans innate immune signalling mechanism that comprises cGAMP(2′-5′) being produced by cGAS in the sensing cell, which is delivered through gap junctions to bystander cells, leading to remote STING activation and subsequent antiviral immunity (Supplementary Fig. 10). Compared to the transcriptionally regulated paracrine activation of bystander cells, for example, via type I IFNs, the gap-junction-dependent transfer of cGAMP(2′-5′) might provide several key advantages to the host. Foremost, type I IFN-dependent induction of antiviral immunity in bystander cells takes considerably longer, given the requirement of a de novo transcription and translation event within the sensing cell. In addition, many viruses block type I IFN induction within the infected cell at multiple interdependent levels, whereas cGAS-dependent cGAMP(2′-5′) synthesis only requires host ATP and GTP, thus exhibiting only a minimal target for virus-encoded inhibitory mechanisms. As such, a virus-infected, potentially compromised cell can still propagate and even amplify antiviral immunity relying on bystander cells connected through gap junctions.
Although bystander activation and signal amplification might prove beneficial for the restriction of viral infection, it might at the same time aggravate disease manifestations in STING-dependent autoimmune syndromes (for example, Aicardi–Goutières syndrome)25 and as such potentially provide a novel target for therapeutic strategies.
Methods
Reagents and plasmids
DNA oligonucleotides corresponding to the 45-base-pair (bp) long dsDNA interferon stimulatory DNA (ISD) (sense sequence 5′-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3′)26 were obtained from Metabion and annealed in PBS. 10-carboxymethyl-9-acridanone, dynasore, poly-l-lysine and carbenoxolone were from Sigma-Aldrich. Calcein-AM and Hoechst were from Invitrogen. Recombinant murine IFN-α was from PBL interferon source. Cyclic di-GMP was obtained from Biolog. cGAMP(2′-5′) was generated via in vitro enzymatic assay and purified as described previously7. Expression plasmids encoding for human GFP-tagged cGAS27 and for an IFN-inducing stimulatory shRNA molecule18 were previously described. Expression plasmid encoding for GFP is based on pEFBOS. Plasmids encoding human connexins were obtained from Thermo Scientific (Precision LentiORFs). Murine connexin expression constructs were provided by K. Willecke.
Cell culture
HEK cells (HEK 293T cells throughout the study), primary mouse embryonic fibroblasts (MEFs) and LL171 cells28 (L929 cells containing a stable IFN-stimulated response element-luciferase reporter plasmid (ISRE-Luc)) were cultured in DMEM supplemented with 10% (v/v) FCS, sodium pyruvate (all Life Technologies) and ciprofloxacin (Bayer Schering Pharma). The HEK cell line stably expressing murine STING, which contains an N-terminal mCherry-tag, was previously described17, whereas HEK cells expressing murine cGAS were generated by retroviral transduction using the pRP system. No mycoplasma contamination was detected in regular screenings of our cell lines.
Cell stimulation and transient transfection
Direct stimulation of HEK cells, HEK STING cells or HEK cGAS (3.5 × 105 cells ml−1 in 96-well plates) was performed by transfecting cyclic di-GMP (2 μg ml−1) or dsDNA (1.33 μg ml−1) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. CMA was added to the cells at a final concentration of 500 μg ml−1. LL171 cells (0.15 × 106 per ml) were stimulated with recombinant IFN-α at a final concentration of 250 U ml−1. DNA-stimulated co-cultures of HEK cells and HEK STING cells with MEFs and HEK cGAS*/cGASlow cells were performed in a 96-well format and transfected with Lipofectamine and 200 ng of reporter plasmid, which served as stimulus and reporter at the same time. Unless otherwise indicated, transient overexpression of cDNA constructs was performed using GeneJuice (Novagen) according to the manufacturer’s instructions. In some titration experiments pCI empty vector was used as stuffer plasmid. Unless otherwise indicated, cells were analysed 3–4 h after direct stimulation or 14–20 h after transfection of expression plasmids.
Immunoblotting
Cells were lysed in 1× Laemmli buffer and denatured at 95 °C for 5 min. Cell lysates were separated by 10% or 8% (for CX43) SDS–PAGE and transferred onto nitrocellulose membranes. Blots were incubated with anti-phospho-IRF3 (Cell Signaling Technology), anti-cGAS (Sigma), anti-CX45 (G-7) (Santa Cruz Biotechnology) or anti-CX43 (gift from K. Willecke) as primary and anti-rabbit-IgG-HRP and anti-mouse-IgG-HRP as secondary antibody or β-actin-IgG-HRP (all Santa Cruz Biotechnology).
Luciferase reporter assays
LL171 cells were lysed in 5× passive lysis buffer (Promega) for 10 min at room temperature. The total cell lysate was incubated with firefly luciferase substrate at a 1:1 ratio. Transactivation of a transiently expressed IFN-β Gaussia luciferase construct (pIFN-β-GLuc) was assessed in the supernatants 14–20 h after transfection with coelenterazine (2.2 μM) as substrate. Luminescence was measured on an EnVision 2104 Multilabel Reader (Perkin Elmer).
qPCR
RNA from cells was reverse transcribed using the RevertAid First Strand cDNA Synthesis kit (Fermentas) and quantitative PCR analysis was performed on an ABI 7900HT. All gene expression data are presented as relative expression to murine β-actin (murine cells) or human GAPDH (human cells). For human transcripts, the sequences were as follows: GAPDH forward 5′-GAGTCAACGGATTTGGTCGT-3′, GAPDH reverse 5′-GACAAGCTTCCCGTTCTCAG-3′; IFNB forward 5′-CAGCATCTGCTGGTTGAAGA-3′, IFNB reverse 5′-CATTACCTGAAGGCCAAGGA-3′; CXCL10 forward 5′-TCTGAATCCAGAATCGAAGG-3′, CXCL10 reverse 5′-CTCTGTGTGGTCCATCCTTG-3′. Primer sequences for murine IFN-β were as previously described18. For additional murine transcripts, the sequences were as follows: cGAS (Mb21d1) forward 5′-ACCGGACAAGCTAAAGAAGGTGCT-3′, cGAS reverse 5′-GCAGCAGGCGTTCCACAACTTTAT-3′; Sting (Tmem173) forward 5′-CACCTCTCTGAGCCTCAACC-3′, Sting reverse 5′-CCATCCACACAGGTCAACAG-3′; Irf7 forward 5′-GAAGACCCTGATCCTGGTGA-3′, Irf7 reverse 5′-CCAGGTCCATGAGGAAGTGT-3′; Cxcl10 forward 5′-AAGTGCTGCCGTCATTTTCT-3′, Cxcl10 reverse 5′-GTGGCAATGATCTCAACACG-3′; β-actin (Actb) forward 5′-AGCCATGTACGTAGCCATCC-3′, β-actin reverse 5′-CTCTCAGCTGTGGTGGTGAA-3′.
Epifluorescence and confocal fluorescence microscopy
For epifluorescence microscopy HEK STING cells were seeded at a density of 2.5 × 104 cells per well in poly-l-lysine-coated 96-well plates. HEK cGAS* cells (8,000 per well) pre-incubated with 2 μg ml−1 calcein-AM for 20–60 min at 37 °C were added on top of HEK STING cells. Calcein was chosen as the tracer of choice because of its comparable physiochemical properties to cGAMP(2′-5′). Images were collected using a Zeiss Observer.Z1 inverted microscope with ×1 (assessment of cell viability after vaccinia virus infection) or ×20 (STING activity assay in HEK cells) long-distance objective 8 h after stimulation or co-culture if not otherwise indicated. Image evaluation was performed as follows: STING aggregates were counted using the ImageJ plugin ‘Cell Counter’, whereas calcein dye transfer was quantified by measuring the area of the images with a green fluorescence value in between two arbitrary thresholds. The thresholds were chosen such as to exclude non-receiving acceptor cells as well as the very bright donor cells. Therefore, the resulting area measurements correspond only to acceptor cells having received calcein from neighbouring cells. Confocal microscopy was performed with living cells on a Leica SP5 SMD confocal microscope with a ×63 water-immersion objective at 37 °C. For nucleus staining, cells were incubated with Hoechst 34580 dye (10 μg ml−1) 30 min before imaging.
Scrape loading
HEK STING cells were seeded at a density of 2.5 × 105 cells ml−1 in 96-well plates. After 16 h cGAMP(2′-5′) was added to the medium to a final concentration of 50 μg ml−1. Monolayers of cells were manually wounded by six scratches per well using an 18G needle. Images were acquired after 4–8 h.
MVA and vaccinia virus infection assay
HEK cells, HEK cGASlow cells (both 3.2 × 105 cells per 12 wells) or MEFs (1.6 × 105 cells per 12 wells) were infected—if not otherwise indicated—with 3.2 × 107 or 1.6 × 107 viral particles per ml (for HEK cells) and 3.2 × 106 or 1.6 × 106 virus particles per ml (for MEFs) of MVA NP-S-GFP (MVA-GFP), which targets GFP to the nucleus of infected cells29. By applying these concentrations of viral particles, a homogenous expression of GFP was observed in more than 80% of recipient cells at the highest concentration. Three hours after infection, cells were washed three times and added onto HEK cells, HEK STING cells, HEK STING CX43/CX45WT and HEK STING CX43/CX45DKO cell lines, respectively (3.5 × 104 cells per 96-well or 4 × 105 cells per 12-well). In some experiments HEK STING cells were pre-treated with 150 μM CBX before co-culturing. After 6–8 h HEK cell co-cultures were either visualized by epifluorescence and confocal fluorescence microscopy or lysed for assessment of IRF3 phosphorylation by immunoblot. Co-cultures of infected MEFs and HEK cells or HEK STING cells were incubated overnight and induction of human IFN-β was measured via qPCR. Before infection with vaccinia virus (m.o.i. 2, 1 and 0.5) MEFs (1.5 × 104 cells per 96-well) were co-cultured with HEK cells or HEK cGAS* cells (5,000 per well) for 12 h. Cell survival in infected co-cultures was determined 24 h later via an epifluorescence-microscopy-based viability assay using calcein as a marker for viable cells. For these studies, calcein was added 1 h before microscopic analysis. Evaluation of images was performed using ImageJ by determining the surface area of monolayers of viable cells. In parallel induction of Ifnb, Cxcl10 and Irf7 in MEF–HEK cell or MEF–HEK STING cell co-cultures was quantified via qPCR.
siRNA experiments
siRNAs (Mission siRNA) against murine STING, described before4, murine cGAS and control siRNA were purchased from Sigma and transfected into MEFs and LL171 using Lipofectamine 2000 (Invitrogen) at a final concentration of 50 nM: mmSTING 5′-CGAAAUAACUGCCGCCUCAdTdT-3′; mmcGAS#1 5′-GAUUGAGCUACAAGAAUAUdTdT-3′, mmcGAS#2 5′-GAGGAAAUCCGCUGAGUCAdTdT-3′; MissionsiRNA Universal Negative Control 1. Forty-eight hours after transfection cells were used for further experiments and knockdown of the indicated genes was verified by qPCR.
CRISPR/Cas9-mediated knockout cell-line generation
HEK STING cells were transfected in duplicates with 150 ng of a Cas9 expression plasmid together with 25 ng of two U6-gRNA expression plasmids specific for early coding exons of the human GJA1 and GJC1 genes. After 2 days, genome editing at both loci was verified by a T7EI endonuclease assay as described30. Limiting dilution cloning was performed by plating on average 0.8 cells in each well of six 96-well plates. After 10 days, growing clones were selected by bright-field microscopy and split to obtain two clonal duplicates. In one of the duplicate plates, 8,000 HEK cGAS* per well were co-seeded and cells were analysed with an epifluorescence microscope after 8 h to assess STING activation. Two clones not responsive as well as two responsive control clones were selected and the corresponding replicate cells were expanded for further experiments.
Deep-sequencing-based genotyping of connexin-deficient cells
Genomic DNA was isolated using a direct lysis buffer (0.2 mg ml−1 proteinase K, 1 mM CaCl2, 3 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 10 mM Tris pH 7.5). The loci of interest were PCR-amplified and subsequently, using a secondary PCR, Illumina-compatible linkers and barcode sequences were added to the amplicons. The products of individual clones were pooled, gel- and silica-column purified, precipitated, quantified using a NanoDrop photospectrometer and used for an Illumina MiSeq 250 bases single read run using the v2 chemistry. FastQ data were analysed to call the allelic genotypes of the clones analysed.
Statistical analysis
On the basis of previous experience and expectations of biological effects, experiments that were assessed for statistical significance were typically performed 3-6 times (luciferase assays, qPCR studies) or 5–15 times (visual fields for microscopy studies). If not stated otherwise, data are presented as arithmetic means + s.e.m. and statistical analyses of the generally normally distributed data (Shapiro–Wilk normality test) were based on paired or unpaired t-tests, as appropriate. If unequal variances were observed for unpaired sample sets (F test for unequal variance), an unpaired t-test with Welch’s correction was performed. Statistical analyses of normalized data were performed using a one-sample t-test. All data calculations were performed using GraphPad Prism.
Supplementary Material
Acknowledgements
We thank M. Pellegrin for providing us with LL171 cells; W. Kastenmüller for MVA NP-S-GFP; J. Bennink for vaccinia virus; K.-P. Hopfner for recombinant cGAS; and K. Willecke for connexin expression constructs and anti-CX43 antibody. J.L.S.-B. is supported by the Studienstiftung des Deutschen Volkes. I.H. is supported by a BONFOR-funded thesis project. A.A., E.L. and V.H. are supported by the excellence cluster ImmunoSensation. V.H. is supported by grants from the German Research Foundation (SFB670 and SFB704) and the European Research Council (ERC 243046).
References
- 1.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasper CA, et al. Cell-cell propagation of NF-κB transcription factor and MAP kinase activation amplifies innate immunity against bacterial infection. Immunity. 2010;33:804–816. doi: 10.1016/j.immuni.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Hamada N, Matsumoto H, Hara T, Kobayashi Y. Intercellular and intracellular signaling pathways mediating ionizing radiation-induced bystander effects. J. Radiat. Res. 2007;48:87–95. doi: 10.1269/jrr.06084. [DOI] [PubMed] [Google Scholar]
- 14.Patel SJ, King KR, Casali M, Yarmush ML. DNA-triggered innate immune responses are propagated by gap junction communication. Proc. Natl Acad. Sci. USA. 2009;106:12867–12872. doi: 10.1073/pnas.0809292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ablasser A, Hornung V. DNA sensing unchained. Cell Res. 2013;23:585–587. doi: 10.1038/cr.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laird DW. Life cycle of connexins in health and disease. Biochem. J. 2006;394:527–543. doi: 10.1042/BJ20051922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juul MH, Rivedal E, Stokke T, Sanner T. Quantitative determination of gap junction intercellular communication using flow cytometric measurement of fluorescent dye transfer. Cell Adhes. Commun. 2000;7:501–512. doi: 10.3109/15419060009040307. [DOI] [PubMed] [Google Scholar]
- 21.Butterweck A, Gergs U, Elfgang C, Willecke K, Traub O. Immunochemical characterization of the gap junction protein connexin45 in mouse kidney and transfected human HeLa cells. J. Membr. Biol. 1994;141:247–256. doi: 10.1007/BF00235134. [DOI] [PubMed] [Google Scholar]
- 22.Langlois S, Cowan KN, Shao Q, Cowan BJ, Laird DW. Caveolin-1 and -2 interact with connexin43 and regulate gap junctional intercellular communication in keratinocytes. Mol. Biol. Cell. 2008;19:912–928. doi: 10.1091/mbc.E07-06-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gall A, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Civril F, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uzé G, et al. Domains of interaction between alpha interferon and its receptor components. J. Mol. Biol. 1994;243:245–257. doi: 10.1006/jmbi.1994.1651. [DOI] [PubMed] [Google Scholar]
- 29.Kastenmüller W, et al. Peripheral prepositioning and local CXCL9 chemokine-mediated guidance orchestrate rapid memory CD8+ T cell responses in the lymph node. Immunity. 2013;38:502–513. doi: 10.1016/j.immuni.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat. Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.