The type III TGF-β receptor (TβRIII) undergoes ectodomain shedding, with surface TβRIII enhancing and soluble TβRIII inhibiting TGF-β signaling. TβRIII mutants with impaired or enhanced shedding are used to demonstrate that the ratio of soluble to membrane-bound TβRIII regulates TβRIII/TGF-β–mediated signaling and biology in vitro and in vivo.
Abstract
The type III transforming growth factor β (TGF-β) receptor (TβRIII), also known as betaglycan, is the most abundantly expressed TGF-β receptor. TβRIII suppresses breast cancer progression by inhibiting migration, invasion, metastasis, and angiogenesis. TβRIII binds TGF-β ligands, with membrane-bound TβRIII presenting ligand to enhance TGF-β signaling. However, TβRIII can also undergo ectodomain shedding, releasing soluble TβRIII, which binds and sequesters ligand to inhibit downstream signaling. To investigate the relative contributions of soluble and membrane-bound TβRIII on TGF-β signaling and breast cancer biology, we defined TβRIII mutants with impaired (ΔShed-TβRIII) or enhanced ectodomain shedding (SS-TβRIII). Inhibiting ectodomain shedding of TβRIII increased TGF-β responsiveness and abrogated TβRIII's ability to inhibit breast cancer cell migration and invasion. Conversely, expressing SS-TβRIII, which increased soluble TβRIII production, decreased TGF-β signaling and increased TβRIII-mediated inhibition of breast cancer cell migration and invasion. Of importance, SS-TβRIII–mediated increases in soluble TβRIII production also reduced breast cancer metastasis in vivo. Taken together, these studies suggest that the ratio of soluble TβRIII to membrane-bound TβRIII is an important determinant for regulation of TβRIII- and TGF-β–mediated signaling and biology.
INTRODUCTION
The transforming growth factor β (TGF-β) signaling pathway is a critical regulator of many cellular processes, including proliferation, differentiation, migration, invasion, and angiogenesis. In normal epithelia and premalignant lesions, the TGF-β signaling pathway functions to both maintain tissue homeostasis and suppress malignant initiation and progression. However, once transformation has occurred, cancer cells are able to subvert the actions of TGF-β to promote cancer progression (Siegel and Massague, 2003). During malignant progression, the production of TGF-β ligands in the tumor and stroma increases (Massague, 2008). However, most cancers develop resistance to the homeostatic effects of TGF-β, including TGF-β–induced growth inhibition (Elliott and Blobe, 2005), and respond instead with increased migration, invasion, and metastatic potential (Mooradian et al., 1992).
TGF-β signals through heteromeric cell-surface receptor complexes consisting of a type I and type II receptor that upon ligand binding recruit and phosphorylate the Smad family of transcriptional regulators. In addition, there are a number of TGF-β superfamily coreceptors, including the type III TGF-β receptor (TβRIII), which can modulate ligand presentation to the type II receptor (Wang et al., 1991; Bernabeu et al., 2009). TβRIII, also known as betaglycan, is the most abundantly expressed TGF-β receptor (Cheifetz et al., 1990). TβRIII has an essential role in regulating TGF-β signaling, mediated through its ability to bind TGF-β ligands with high affinity. TβRIII binds all three isoforms of TGF-β (Wang et al., 1991), as well as bone morphogenetic proteins (BMPs; Kirkbride et al., 2008) and inhibin (Lewis et al., 2000), through two distinct binding domains in its core protein, and basic fibroblast growth factor (bFGF), through its heparan sulfate glycosaminoglycan chains (Andres et al., 1992). Membrane-bound TβRIII presents ligand to TβRII to increase signaling (Lopez-Casillas et al., 1993). However, TβRIII can also undergo ectodomain shedding, releasing a soluble form that binds ligand in the extracellular space, thereby reducing ligand availability to the signaling receptors and inhibiting downstream signaling (Lopez-Casillas et al., 1994).
TβRIII has a role as a suppressor of cancer progression in multiple types of cancer, including breast cancer (Dong et al., 2007). Breast cancer is the most common malignancy and the second-most-common cause of cancer-related death in women in the United States (Siegel et al., 2012). In human breast cancers, TGF-β levels are frequently elevated and correlate with poor patient prognosis (Ghellal et al., 2000). TβRIII expression is decreased in breast cancer cell lines and human breast cancer patient specimens (Dong et al., 2007). Restoring TβRIII expression suppresses breast cancer progression by inhibiting migration, invasion, metastasis, and angiogenesis (Sun and Chen, 1997; Dong et al., 2007). Similar to the effects of restoring full-length TβRIII expression, treatment with ectopic soluble TβRIII inhibits tumor growth, angiogenesis, and metastasis in breast cancer models (Bandyopadhyay et al., 1999). These data suggest that the tumor-suppressive effects of TβRIII could be mediated, in part, by production of soluble TβRIII, which antagonizes the tumor-promoting effects of TGF-β signaling.
Although the role of soluble TβRIII can be investigated by the addition of recombinant soluble TβRIII, the mechanisms regulating ectodomain shedding and generation of soluble TβRIII remain undefined, making it more difficult to delineate the function of cell-surface TβRIII or the relative contribution of cell-surface TβRIII and soluble TβRIII to signaling and biology. Cleavage of many cell-surface proteins is carried out by a common machinery involving zinc-dependent metalloproteinases of the matrix metalloproteinase (MMP) and a disintegrin and metalloproteinase (ADAM) families. These proteases are regulated by several mechanisms, including protein kinase C activation, intracellular calcium levels, and other activated growth factor signaling pathways (Arribas and Borroto, 2002). However, TβRIII release is largely unaffected by phorbol myristate acetate, calcium ionophores, and other factors that induce cleavage of canonical transmembrane-shedding substrates (Arribas et al., 1997). TβRIII shedding can be reduced, but not blocked, with the panmetalloproteinase inhibitor TAPI-2, as well as with more specific inhibitors against MT1-MMP and MT3-MMP (Velasco-Loyden et al., 2004). However, studies with these inhibitors are complicated by their ability to alter the shedding of many other membrane proteins, making it difficult to use metalloprotease inhibition as a method to specifically determine the role of cell-surface TβRIII. Here we adopted a structure–function approach by creating TβRIII-shedding mutants to investigate the significance of TβRIII ectodomain shedding on TGF-β–mediated signaling and TβRIII-mediated biology during breast cancer progression in vitro and in vivo.
RESULTS
Mutations in the juxtamembrane region of TβRIII alter ectodomain shedding
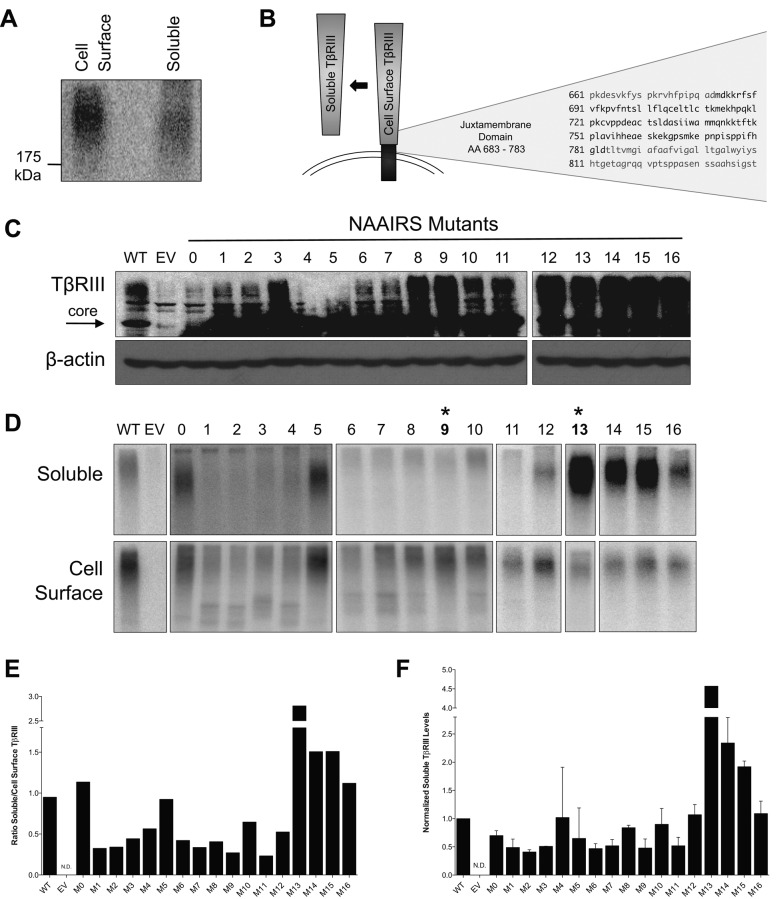
To investigate the significance of TβRIII ectodomain shedding on TGF-β–mediated signaling and biology, we set out to identify TβRIII mutants with altered ectodomain shedding. Human TβRIII is an 851–amino acid transmembrane proteoglycan with a large 766–amino acid extracellular domain, a single hydrophobic transmembrane region, and a short, 42–amino acid cytoplasmic domain (Lopez-Casillas et al., 1991; Wang et al., 1991). Endogenous soluble TβRIII has nearly the same electrophoretic mobility as full-length, membrane-bound TβRIII (Figure 1A), suggesting that cleavage occurs just proximal to the plasma membrane, consistent with what has been demonstrated with other shed receptors (Perez-Torres et al., 2008). A substitution mutagenesis approach replacing six amino acids of the endogenous TβRIII protein sequence with the asparagine-alanine-alanine-isoleucine-arginine-serine (NAAIRS) amino acid sequence was chosen, as this sequence can adopt multiple secondary structures, potentially minimizing structural changes (Hamad et al., 2002). To identify regions important for TβRIII shedding, we created a series of TβRIII NAAIRS mutants beginning 95 amino acids upstream of the transmembrane region and spanning the entire juxtamembrane region (Figure 1B and Supplemental Table S1).
FIGURE 1:
Mutations in the juxtamembrane domain of TβRIII alter ectodomain shedding. (A) Binding and cross-linking of transiently transfected, HA-tagged WT-TβRIII in COS7 cells. After [125I]TGF-β1 binding and cross-linking, cell lysate and conditioned medium were immunoprecipitated with an antibody against HA. (B) Schematic of TβRIII with the amino acid sequence of the juxtamembrane domain. (C) Western blots of WT-TβRIII and indicated NAAIRS mutants transiently transfected in COS7 cells. EV, empty vector. β-Actin was used as a loading control. (D) Binding and cross-linking of COS7 cells transiently transfected with the indicated constructs. Cells were grown in full growth medium for 20 h. After [125I]TGF-β1 binding and cross-linking, cell lysates and conditioned medium were immunoprecipitated with an antibody against HA. Asterisk denotes the mutants that were further used in these studies. Representative images from two independent experiments. (E) Quantification of D. Densitometric analysis was performed in ImageJ, and the ratio of soluble/cell-surface TβRIII was determined. (F) ELISA analysis of soluble TβRIII from COS7 cells transiently transfected with the indicated constructs. Media were conditioned for 24 h. Concentration of soluble TβRIII was determined from a standard curve. Soluble TβRIII levels were then normalized to TβRIII expression determined via Western blotting from control lysates. Data are from two independent experiments and shown as mean ± SEM normalized to WT-TβRIII.
We initially assessed and confirmed expression of all 17 mutants (M0–M16) in COS7 cells, which lack endogenous TβRIII (Figure 1C). To evaluate processing to the cell surface, ligand-binding ability, and ectodomain shedding, we performed iodinated TGF-β1 binding and cross-linking assays on both the conditioned media from COS7 cells and on COS7 cells expressing either controls (pDNR–empty vector [EV] or pDNR–wild type [WT]-TβRIII) or one of the membrane-proximal TβRIII NAAIRS mutants (M0–M16), followed by immunoprecipitation of TβRIII and soluble TβRIII. Whereas all TβRIII NAAIRS mutants trafficked to the cell surface and bound ligand, there were significant differences in the electrophoretic mobility of the mutants (Figure 1D), suggesting differential posttranslational processing of the TβRIII mutants. However, by directly comparing levels of ligand binding to soluble TβRIII in the conditioned media to levels of ligand binding to membrane-bound TβRIII on the cell surface, we could assess ectodomain shedding of each mutant independently of these factors. Four NAAIRS mutants (M1, M2, M9, and M11) consistently exhibited decreased ectodomain shedding compared with wild-type TβRIII controls (Figure 1, D and E). Of interest, three NAAIRS mutants (M13–M15) exhibited increased ectodomain shedding compared with wild-type TβRIII controls (Figure 1, D and E). These differences in shedding were independently confirmed by performing enzyme-linked immunosorbent assay (ELISA) analysis for soluble TβRIII (Figure 1F).
A single–amino acid substitution at M742 significantly inhibits TβRIII ectodomain shedding
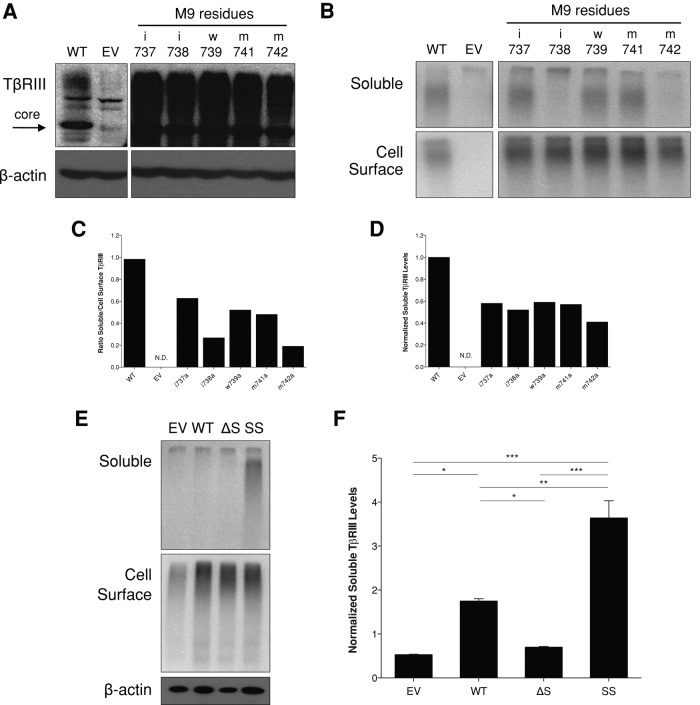
To further define the distinct amino acid residues regulating ectodomain shedding of TβRIII, we created individual alanine point mutations in TβRIII of each amino acid contained within the NAAIRS mutants M1, M2, M9, and M11 (Supplemental Table S2). Expression of each mutant was confirmed via Western blotting (Figure 2A and Supplemental Figure S1A). Ectodomain shedding of each mutant was evaluated via both [125I]TGF-β1 binding and cross-linking assays and ELISA. Perturbing any amino acid residue within the M9 mutant disrupted ectodomain shedding to some degree, and substituting methionine 742 to alanine (M742A) fully recapitulated the decreased ectodomain shedding phenotype of the M9 NAAIRS mutant (Figure 2, B–D). Mutation of isoleucine 738 to alanine (I738A) also significantly decreased ectodomain shedding (Figure 2, B–D). In contrast, no single point mutations within M1, M2, or M11 NAAIRS mutants significantly altered ectodomain shedding (Supplemental Figure S1, B and C).
FIGURE 2:
A single–amino acid substitution at M742 significantly inhibits ectodomain shedding. (A) Western blot showing expression of WT-TβRIII and M9 alanine point mutants transiently transfected in COS7 cells. β-Actin was used as a loading control. (B) Binding and cross-linking of COS7 cells transiently transfected with the indicated constructs. Cells were grown in full growth medium for 20 h. After [125I]TGF-β1 binding and cross-linking, cell lysates and conditioned medium were immunoprecipitated with an antibody against HA. Representative images from two independent experiments. (C) Quantification of B. Densitometric analysis was performed in ImageJ, and the ratio of soluble/cell-surface TβRIII was determined. (D) ELISA analysis of soluble TβRIII from COS7 cells transiently transfected with the indicated constructs. Media were conditioned for 24 h. Concentration of soluble TβRIII was determined from a standard curve. Soluble TβRIII levels were then normalized to TβRIII expression determined via Western blotting from control lysates. Representative data from two independent experiments. (E) Binding and cross-linking of monoclonal stable lentiviral MDA-MB-231 cell lines made with EV, WT-TβRIII (WT), ΔShed-TβRIII (ΔS) (M9 mutant), or Super-Shed TβRIII (SS; M13 mutant). After [125I]TGF-β1 binding and cross-linking, cell lysates and conditioned medium were immunoprecipitated with an antibody against the extracellular domain of TβRIII. β-Actin was used as a loading control. Representative data from three independent experiments. (F) ELISA analysis of soluble TβRIII from stable MDA-MB-231 cell lines. Media were conditioned for 24 h. Concentration of soluble TβRIII was determined from a standard curve of known amounts. Soluble TβRIII levels were then normalized to β-actin expression determined via Western blotting from control lysates. Data are from three independent experiments and shown as mean ± SEM. One-way ANOVA: p < 0.0001. Tukey's multiple-comparisons tests: *p < 0.05; **p < 0.001; ***p < 0.0001.
Because the M9 NAAIRS, M11 NAAIRS, M742A, and I738A mutants all significantly reduced but did not abrogate ectodomain shedding, we created mutants that combined the M742A and I738A mutations within M9 and the L752A and V754A mutations within M11. However, neither of these mutants exhibited any further decrease in shedding compared with the M9 or M742A mutation alone (unpublished data). Therefore we used the M9 NAAIRS and the M742A mutants as our models for shedding-deficient TβRIII (ΔShed-TβRIII) and M13 as a “supershedder” (SS-TβRIII) to investigate the effects of altering the ratio of soluble to cell-surface TβRIII. To investigate whether the shedding properties of these mutants were retained in different cell contexts, we transiently expressed the mutants in human embryonic kidney epithelial cells (HEK-293) and mink lung epithelial cells (Mv1Lu). In all cases, the M9 NAAIRS and M742A mutants exhibited reduced ectodomain shedding, and the M13 NAAIRS mutant exhibited increased ectodomain shedding (Supplemental Figure S2A).
To investigate the effects of increased or decreased TβRIII ectodomain shedding in breast cancer, we used the well-characterized breast cancer cell line MDA-MB-231, in which we and others have already demonstrated the ability of TβRIII and soluble TβRIII to regulate its cancer biology, including migration and invasion in vitro and in vivo (Bandyopadhyay et al., 1999; Dong et al., 2007). MDA-MB-231 cells exhibit low endogenous TβRIII levels, consistent with loss of TβRIII expression during breast cancer progression. WT-TβRIII, ΔShed-TβRIII, SS-TβRIII, or an empty-vector DNA control was stably incorporated into MDA-MB-231 cells via lentiviral infection, and single clones were selected, expanded, and examined for TβRIII expression and ectodomain shedding via [125I]TGF-β1 binding and cross-linking assays. Monoclonal MDA-MB-231 cell lines with equivalent levels of cell-surface TβRIII were chosen for further study (Figure 2E), and the expected levels of soluble TβRIII production were confirmed via binding and cross-linking (Figure 2E) and ELISA (Figure 2F).
Effects of altered TβRIII ectodomain shedding on TGF-β signaling
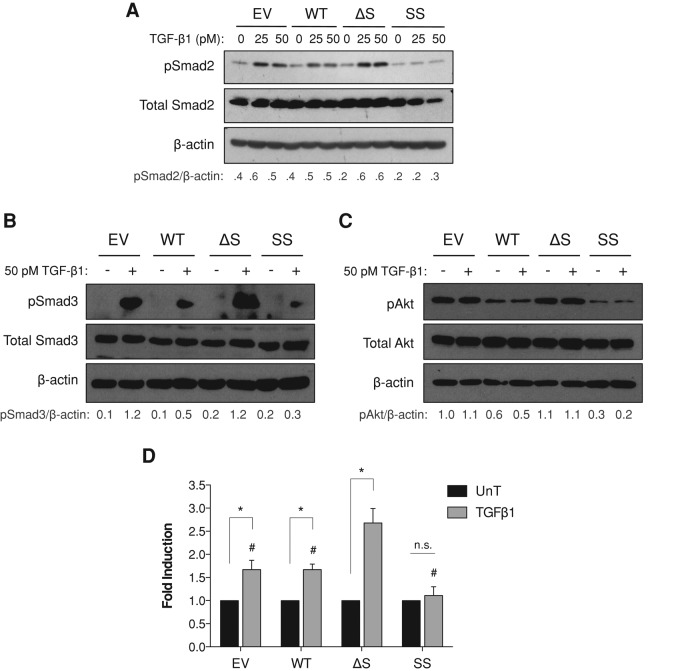
In many cell lines, including MDA-MB-231 cells, increasing TβRIII expression results in decreased TGF-β signaling, as measured by phosphorylation of Smad2 and Smad3 and transcription of TGF-β–responsive genes (Dong et al., 2007). Because increasing TβRIII expression can result in increased soluble TβRIII (Lopez-Casillas et al., 1991) and soluble TβRIII is sufficient to inhibit downstream TGF-β signaling (Arribas and Borroto, 2002), this TβRIII-mediated decrease in TGF-β responsiveness has been hypothesized to be the result of increased production of soluble TβRIII. To address this hypothesis directly and to examine the effects of high levels of cell-surface TβRIII in the absence of soluble TβRIII on TGF-β signaling, we used the stable MDA-MB-231 TβRIII ectodomain-shedding mutant cell lines. Consistent with prior studies, compared with empty vector control MDA-MB-231 cells, WT-TβRIII–expressing cells exhibited slightly decreased TGF-β1–mediated Smad2 and Smad3 phosphorylation (Figure 3, A and B). In contrast, in ΔShed-TβRIII cells, TGF-β1–mediated Smad phosphorylation was increased, suggesting that in the absence of significant ectodomain shedding, cell-surface TβRIII is able to present ligand and enhance signaling (Figure 3, A and B). Consistent with this hypothesis, relative to WT-TβRIII cells, TGF-β1–mediated Smad phosphorylation was further reduced in SS-TβRIII cells (Figure 3, A and B). Because TβRIII can also mediate noncanonical TGF-β signaling responses, we assessed activation of Akt signaling. Both basal and TGF-β1–stimulated Akt activation were similarly dependent on TβRIII ectodomain shedding (Figure 3C). A dose–response curve from 0 to 100 pM TGF-β1 demonstrated a consistent pattern of responsiveness at all concentrations tested (data not shown). To determine whether alterations in Smad phosphorylation were leading to changes in gene transcription, dual-luciferase reporter assays were performed using the TGF-β–responsive pE2.1 promoter. While there was no significant difference between WT-TβRIII and EV expressing cells, SS-TβRIII cells treated with TGF-β1 had decreased TGF-β–induced transcription (Figure 3D). Conversely, cells stably expressing ΔShed-TβRIII had significantly increased TGF-β1–mediated pE2.1 transcription (Figure 3D). To establish whether the observed changes in signaling were due to changes in soluble TβRIII levels, we treated cells with ectopic recombinant soluble TβRIII. Recombinant soluble TβRIII reduced TGF-β signaling in a dose-dependent manner in each cell line, with the exception of the SS-TβRIII–expressing cells (Supplemental Figure S3), perhaps because signaling is already maximally inhibited by the increased levels of soluble TβRIII produced by this cell line. Together these data demonstrate that the decrease in TGF-β responsiveness frequently observed when TβRIII is expressed requires TβRIII ectodomain shedding and generation of soluble TβRIII.
FIGURE 3:
Effects of altered TβRIII ectodomain shedding on TGF-β signaling. (A, B) Lentiviral stable MDA-MB-231 cell lines expressing EV, WT-TβRIII, ΔShed-TβRIII, or Super-Shed TβRIII were plated in full serum medium and allowed to condition for 20 h before treatment with the indicated concentrations of TGF-β1 for 30 min. Western blot analysis was performed with the indicated antibodies. Total Smad2, Smad3, and β-actin were used as loading controls. Quantification of densitometric analysis shown as ratio of phosphorylated Smad2/β-actin. Data are representative of at least three independent experiments. (C) Lentiviral stable MDA-MB-231 cell lines expressing EV, WT-TβRIII, ΔShed-TβRIII. or Super-Shed TβRIII were treated with reduced serum medium (1% FBS) that had been preconditioned from the corresponding cell line for 20 h. Cells were serum starved for 6 h before treatment with the indicated concentrations of TGF-β1 for 30 min. Western blot analysis was performed with the indicated antibodies. Total Akt and β-actin were used as loading controls. Quantification of densitometric analysis is shown as ratio of phosphorylated Akt/β-actin. Data are representative of at least three independent experiments. (D) Stable MDA-MB-231 cell lines were transfected with a pE2.1-responsive luciferase construct and a Renilla construct. The next day, cells were treated with serum-free medium that had been preconditioned from the corresponding cell line for 24 h and 50 pM TGF-β1. Cells were treated for 24 h. Results from four independent experiments are shown as pE2.1/Renilla activity and normalized to ligand untreated condition of each cell line. Two-way ANOVA, p < 0.001. *One-sample t test, p < 0.05; #two-tailed t-test, p < 0.05, relative to ΔS + TGF-β1.
We next examined the effects of the shedding mutants on mediating TGF-β2 signaling. Because TGF-β2 cannot bind to TβRII on its own (De Crescenzo et al., 2006), TGF-β2 requires TβRIII for presentation to TβRII and functional signaling. Consistent with this role, both WT-TβRIII and the ΔShed mutant increased TGF-β2 responsiveness in MDA-MB-231 cells (Supplemental Figure S4). However, the high ratio of soluble TβRIII relative to cell-surface TβRIII in the SS-TβRIII–expressing cells shifted the balance from TβRIII-mediated promotion of signaling to inhibition of signaling (Supplemental Figure S4).
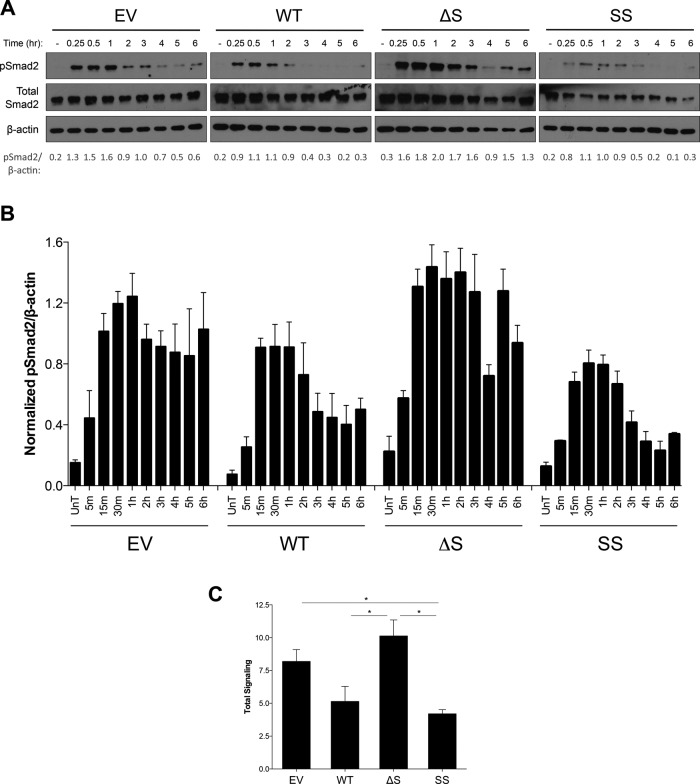
To investigate the effects of TβRIII ectodomain shedding on the kinetics and duration of TGF-β signaling, we performed time-course experiments. In the absence of significant soluble TβRIII (EV and ΔShed-TβRIII), Smad2 phosphorylation peaked at ∼1 h, and signaling persisted out to 6 h (Figure 4A). In contrast, in cells with higher levels of soluble TβRIII (WT-TβRIII and SS-TβRIII), Smad2 phosphorylation peaked earlier (∼ 30 min), and signaling persisted only out to 2–3 h (Figure 4, A and B). Similar results were obtained by transient transfection of these TβRIII shedding mutants in the normal human epithelial cell line HEK293 (Supplemental Figure S5). Of interest, we also consistently observed evidence of a biphasic signaling pattern in the ΔShed-TβRIII cell line, with signal diminishing between 3 and 4 h but then returning to nearly peak levels at 5-h posttreatment (Figure 4, A and B). Integrating the signaling that occurred over the 6-h period demonstrated a 50% reduction in total signaling in WT-TβRIII–expressing cells compared with ΔShed-TβRIII (Figure 4C). These data establish an important role for the ratio of soluble and cell-surface TβRIII in regulating the kinetics and magnitude of TGF-β signaling.
FIGURE 4:
TβRIII ectodomain shedding regulates the kinetics and magnitude of TGF-β signaling in MDA-MB-231 cells. (A) Lentiviral stable MDA-MB-231 cell lines expressing EV, WT-TβRIII, ΔShed-TβRIII, or Super-Shed TβRIII were plated in full serum medium and allowed to condition for 20 h before treatment with 50 pM of TGF-β1 for the indicated times. Western blot analysis was performed with indicated antibodies. Total Smad2 and β-actin were used as loading controls. Quantification of densitometric analysis is shown below as levels of phosphorylated Smad2/β-actin. Representative data from four independent experiments. (B) Summary of time-course experiment data. Densitometric analysis of phosphorylated Smad2/β-actin. Data for at least independent experiments for each time point shown as mean ± SEM. One-way ANOVA p < 0.05 for 15-m, 30-m, 1-h, 2-h, 3-h, 4-h, 5-h, and 6-h time points. (C) Integrated signaling over 6-h time course. Densitometric analysis of phosphorylated Smad2/β-actin of each experiment plotted as line graphs, with area under the curve calculated for each cell line. Data from four independent experiments shown as mean ± SEM. One-way ANOVA, p < 0.05. *Two-tailed t-test, p < 0.05.
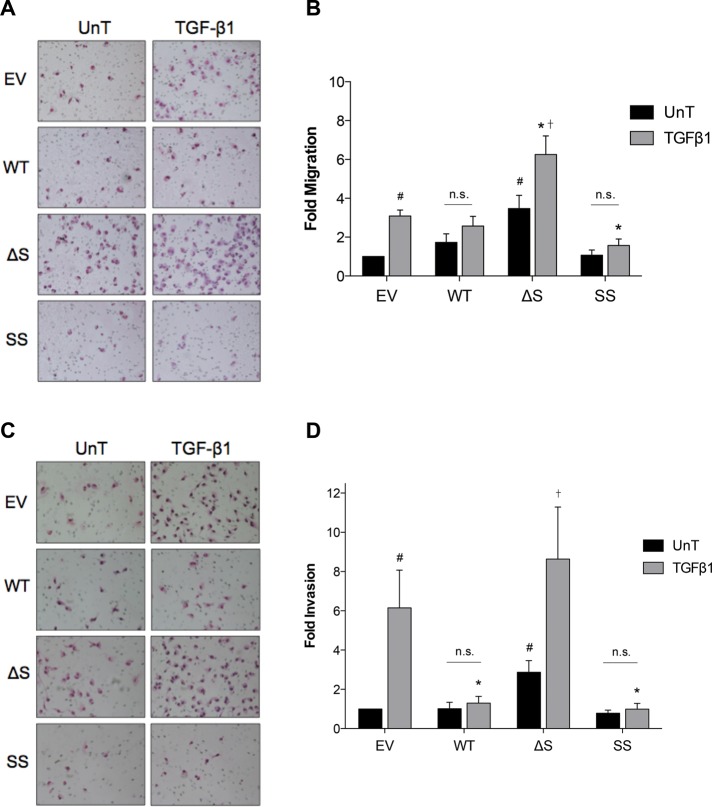
Effects of altered TβRIII ectodomain shedding on TGF-β–mediated migration and invasion
TβRIII inhibits epithelial and cancer cell motility and invasion in multiple human cancers, including breast cancer (Turley et al., 2007; Finger et al., 2008; Mythreye and Blobe, 2009; Lambert et al., 2011). Increasing wild-type TβRIII expression in MDA-MB-231 cells inhibited TGF-β–induced Transwell migration and invasion (Dong et al., 2007). Similarly, plating MDA-MB-231 cells in conditioned media from COS7 cells overexpressing TβRIII also reduced TGF-β–induced invasion (Dong et al., 2007), suggesting that the production of soluble TβRIII is one mechanism for TβRIII-mediated inhibition of cell motility and invasion. To determine directly the contribution of TβRIII ectodomain shedding and soluble TβRIII production on TβRIII-mediated inhibition of migration and invasion, we used the monoclonal stable MDA-MB-231 cell lines expressing the TβRIII shedding mutants. Because these experiments are usually performed in serum-free media, we first examined the effect of serum on TβRIII ectodomain shedding. In the absence of serum, ectodomain shedding was potently inhibited (Supplemental Figure S6). Therefore, for these experiments, cells were plated in media that had been preconditioned for 24 h, to reflect more accurately the differential levels of ectodomain shedding. We next investigated whether expression of the TβRIII shedding mutants altered the proliferation rates of the stable cell lines, using [3H]thymidine incorporation assays. Over 24 h—the time frame for both the invasion and migration assays—there was a decrease in the basal proliferation rates of WT-TβRIII, ΔShed-TβRIII, and SS-TβRIII compared with EV control cells of ∼40, 30, and 50%, respectively (Supplemental Figure S7). TGF-β treatment also slightly reduced proliferation rates in all four cell lines (Supplemental Figure S7). To reflect these changes, migration and invasion results were normalized to the proliferative index of each corresponding condition.
Whereas TGF-β1 stimulated migration of the MDA-MB-231 cells expressing EV, cells expressing WT-TβRIII were unresponsive to ligand (Figure 5, A and B). Conversely, cells expressing ΔShed-TβRIII demonstrated a threefold increase in basal migration and an increase in ligand-mediated migration relative to EV control cells (Figure 5, A and B). Expressing SS-TβRIII had an even more potent effect on motility than WT-TβRIII, producing a significant decrease in ligand-induced Transwell migration (Figure 5, A and B).
FIGURE 5:
Effects of altered TRIII ectodomain shedding on TGF-β–mediated migration and invasion. Stable MDA-MB-231 cell lines were plated in medium that had been preconditioned from the corresponding cell line for 24 h in (A, B) fibronectin-coated Transwell chambers or (C, D) Matrigel-coated Transwell chambers in either the absence (UnT) or presence of 50 pM TGF-β1. Cells were allowed to migrate or invade for 24 h. (A) Representative images of migrated cells. (B) Summary of four experiments. Data normalized to EV UnT and shown as mean ± SEM. Two-way ANOVA for cell line and treatment, p < 0.05. Tukey's multiple-comparisons tests: #p < 0.05 relative to EV UnT; *p < 0.05 relative to EV + TGF-β1; †p < 0.05 relative to ΔS UnT. (C) Representative images of invaded cells. (D) Summary of five experiments. Data normalized to EV UnT and shown as mean ± SEM. Two-way ANOVA for interaction, cell line, and treatment, p < 0.05. Tukey's multiple-comparisons tests: #p < 0.05 relative to EV UnT; *p < 0.05 relative to EV + TGF-β1; †p < 0.05 relative to ΔS UnT.
We also evaluated the effects of altered TβRIII ectodomain shedding on MDA-MB-231 cell invasion via Matrigel-coated Transwell invasion assays. TGF-β–mediated invasion was significantly inhibited in both the WT-TβRIII– and SS-TβRIII–expressing cells (Figure 5, C and D). In contrast, compared with EV, there was an increase in both basal and TGF-β–mediated invasion in ΔShed-TβRIII–expressing cells (Figure 5, C and D). Together these data demonstrate that ectodomain shedding is required for TβRIII-mediated inhibition of TGF-β–induced breast cancer cell migration and invasion.
Effects of altered TβRIII ectodomain shedding on metastatic breast cancer growth
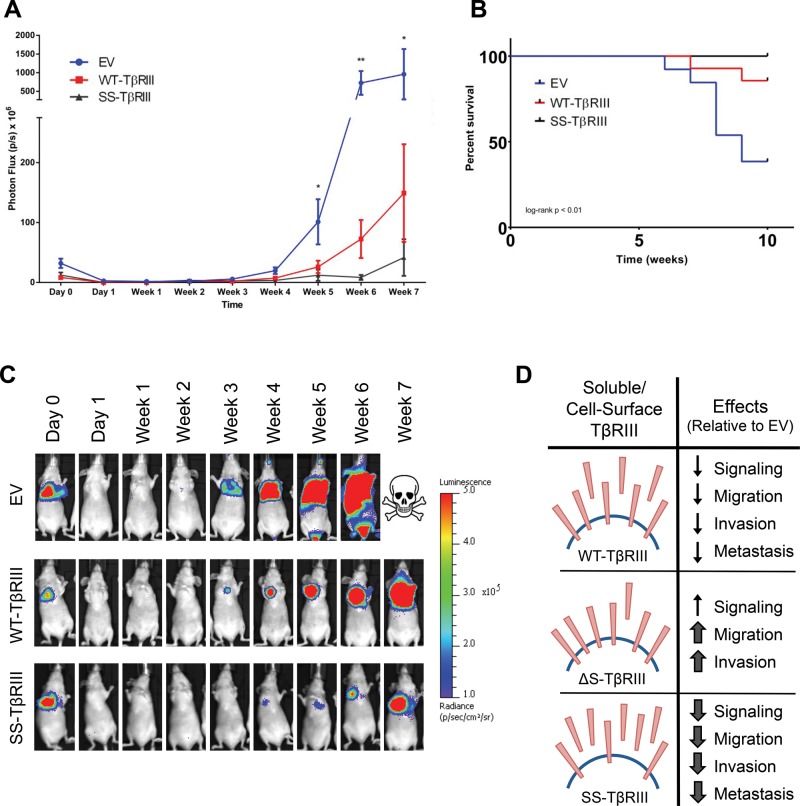
Our lab and others previously showed that TβRIII can inhibit breast cancer migration and invasion, both important precursors to metastasis, and here we provide direct evidence that this effect is mediated by production of soluble TβRIII. In addition, TβRIII and ectopic soluble TβRIII have been demonstrated to decrease metastatic potential in vivo, supporting a potential metastasis suppressor role for soluble TβRIII (Bandyopadhyay et al., 2002; Dong et al., 2007). To examine the effect of varying levels of endogenously produced soluble TβRIII to cell-surface TβRIII on tumor metastasis, we used the MDA-MB-231-4175 cell line, a variant of the highly metastatic MDA-MB-231 cell line that has been selected for a high tropism to the lung (Minn et al., 2005), in a tail-vein injection assay. MDA-MB-231-4175 cells stably expressing luciferase were lentivirally infected with WT-TβRIII, SS-TβRIII, or an empty-vector DNA control. Stable, pooled colonies were examined for TβRIII expression and ectodomain shedding via [125I]TGF-β1 binding and cross-linking assays and ELISA (Supplemental Figure S8). Mice injected with MDA-MB-231-4175 cells expressing EV control exhibited metastatic lesions that were larger than those of the mice injected with WT-TβRIII or SS-TβRIII cells, with a statistically significant difference in photon flux starting at week 5 and becoming more prominent at week 6 (Figure 6A). The photon flux plateaued at week 7 due to a number of mice dying or reaching humane endpoints. Of importance, SS-TβRIII mice had significantly delayed metastatic growth and smaller lesions compared with WT-TβRIII mice (Figure 6, A and C). In addition, by week 8, five of 13 mice injected with the EV expression cells had died or reached humane endpoints, whereas only one of the 14 mice injected with WT-TβRIII–expressing cells died during this time period, and all 10 of the SS-TβRIII mice survived. At 10 wk, there was a significant difference in survival among the EV, WT-TβRIII, and SS-TβRIII cell expression groups (Figure 6B). Hematoxylin and eosin stains of the lung tissue from the mice injected with EV-expressing cells showed partial to complete replacement of lung tissue with tumors. In contrast, lung samples from the mice injected with the WT-TβRIII cells showed tumor nodules interspersed by large amounts of normal lung tissue (unpublished data). Lung samples from mice in the SS-TβRIII were not examined, as none of the mice reached humane endpoints.
FIGURE 6:
Effects of TβRIII ectodomain shedding on MDA-MB-231-4175 metastatic growth. MDA-MB-231-4175 cells expressing EV (N = 13), WT-TβRIII (N = 14), or SS-TβRIII (N = 10) were injected into 6-wk-old athymic mice via tail-vein injection. (A) Initial and weekly bioluminescence of lung metastatic lesions expressed as photon flux of mice after initial injection. Kruskal–Wallis analysis: *p < 0.05, **p < 0.01. (B) Kaplan–Meier survival curve of mice injected with cells expressing EV, WT-TβRIII, or SS-TβRIII. Log-rank Mantel–Cox test: p < 0.01. (C) Representative images of bioluminescence scans of each cohort of mice at each time point. (D) Schematic summary of the effects of altered ratios of soluble and cell-surface TβRIII on TGF-β–mediated signaling and biology in the context of breast cancer.
DISCUSSION
Here we demonstrated that mutating the juxtamembrane region of TβRIII can alter its ectodomain shedding and that inhibiting production of soluble TβRIII results in increase in TGF-β responsiveness, increase in duration of TGF-β signaling, and decrease in TβRIII's ability to inhibit TGF-β–mediated migration and invasion (Figure 6D). Of importance, we also demonstrate that the amount of endogenous ectodomain shedding TβRIII is inversely correlated with metastatic potential in vivo (Figure 6D).
Ectodomain shedding of transmembrane proteins is a common phenomenon that often contributes to regulation of signal transduction. Ligands and growth factors, including TGF-α, can be activated for autocrine signaling by release from the membrane (Teixido et al., 1990), and cell-surface signaling receptor levels can be altered to either increase or decrease cellular responsiveness. Recent studies demonstrated that the type I TGF-β receptor is released from the membrane by TNF α–converting enzyme, resulting in decreased TGF-β signaling (Liu et al., 2009). After this shedding event, a secondary γ-secretase cleavage releases the intracellular domain (ICD) of TβRI, which can then accumulate in the nucleus and interact with transcriptional regulators to alter TGF-β–induced gene transcription and subsequent tumor cell invasion (Mu et al., 2011). In addition, it was reported that TβRIII is also a substrate for γ-secretase cleavage and that its ICD is stable after this event (Blair et al., 2011); however, it is unknown whether the ICD is involved in downstream signal transduction. If the TβRIII-ICD functions similarly to the TβRI-ICD, it will be interesting to determine whether any of the alterations in signaling and biology reported here are regulated by increased or decreased activity of the ICD resulting from the ΔShed- or Super-Shed-TβRIII mutation. Further, TβRIII interacts with several proteins in its cytoplasmic domain that contribute to its regulation of signaling, including β-arrestin2 and GIPC (Blobe et al., 2001; Chen et al., 2003), and these functions may be altered in some way by the shedding mutants as a result of potentially increased or decreased stability on the cell surface.
Although we have established discrete mutations in the TβRIII juxtamembrane region that regulate TβRIII shedding, we have not elucidated how these mutations result in increased or decreased ectodomain shedding. The M9 NAAIRS mutation and M742A substitution could decrease shedding either by disrupting a potential metalloprotease consensus sequence or altering the structure of TβRIII so that it is unable to physically interact with or be recognized as a substrate by the appropriate proteases. Knowledge regarding the precise site of endogenous TβRIII cleavage and structural information regarding the extracellular domain of TβRIII would both be useful in determining how these alterations regulate TβRIII shedding. We used the protease specificity prediction server (PROSPER) to perform in silico analysis of the TβRIII juxtamembrane domain sequence (Song et al., 2012). The region containing the M9 NAAIRS mutant revealed several potential cleavage sites at A740, M741, and M742 by MMP3, MMP9, and cathepsin, suggesting that the M9 and M742A mutants could be disrupting these recognition sequences. The region containing the NAAIRS M13–M15 mutants, which increased TβRIII ectodomain shedding, is highly proline rich. Owing to the unique biochemical and structural properties of proline, mutating this region may alter the structure of the extracellular domain of TβRIII to make it more accessible to an endogenous, constitutive sheddase. Indeed, although there were no protease consensus sites within the M13–M15 region, when the M13 supershedder sequence containing the NAAIRS substitution was queried in the PROSPER software, two novel consensus sites for MMP9 and elastase were introduced at that site. Whether these proteases are responsible for shedding TβRIII is being explored.
Given that we and others have demonstrated an important role for TβRIII and soluble TβRIII in regulating breast cancer progression, through both effects on breast cancer cells (Sun and Chen, 1997; Dong et al., 2007) and the tumor microenvironment, including the local immune response (Hanks et al., 2013) and angiogenesis (Bandyopadhyay et al., 1999; Dong et al., 2007), understanding the regulation and mechanism of TβRIII shedding and how this process might be altered during cancer progression could yield insight into targeting TGF-β signaling in cancer patients. Although TβRIII does not appear to be a substrate of the canonical ectodomain shedding machinery (Arribas et al., 1997), determining what factors do regulate TβRIII shedding could be quite informative. Ectodomain shedding of other cell surface receptors can be activated by numerous triggers, including ultraviolet irradiation, inflammation, and growth factor stimulation (Seo et al., 2007; Killock and Ivetic, 2010). In addition to decreased TβRIII expression in human cancers, the balance of cell-surface and soluble TβRIII could be altered by aberrant processes or signaling within the tumor that potentially could be targeted therapeutically. Our studies suggest that increasing TβRIII ectodomain shedding and soluble TβRIII levels could be beneficial in reducing the protumorigenic effects of TGF-β in established cancers. Indeed, receptor trap molecules based partially on soluble TβRIII have been described (Verona et al., 2008) and could be developed for this indication.
In addition to TGF-β ligands, TβRIII binds other TGF-β superfamily members, including multiple BMPs, and can enhance binding of these ligands to signaling receptors (Kirkbride et al., 2008). Studies using the shedding mutants defined here demonstrate that soluble TβRIII is able to sequester BMP and reduce downstream signaling and BMP-mediated biology, whereas ΔShed-TβRIII enhances BMP-mediated signaling and biology (Gatza et al., 2014). These studies suggest that TβRIII ectodomain shedding plays a critical role in regulating BMP signaling as well as TGF-β. Future studies will determine whether soluble and cell-surface TβRIII have differential effects on other TβRIII-binding proteins, including inhibin and bFGF.
Whereas the role of TβRIII in breast cancer cell migration and invasion is well established, its effects on cell growth and proliferation are less defined. One study performed in MDA-MB-231 cells found that TβRIII knockdown decreased cell growth (Criswell et al., 2008), whereas another reported that treatment with recombinant soluble TβRIII induced apoptosis and inhibited cell growth (Lei et al., 2002). We found that all versions of TβRIII expressed (WT, Δ-Shed, or SS) reduced proliferation relative to EV-expressing cells after 24 h (Supplemental Figure S7); however, further studies are underway to determine whether altering TβRIII ectodomain shedding in our model system affects longer-term cell growth and survival. These data will be important for interpreting in vivo results.
TGF-β levels are frequently elevated in human breast cancers (Ghellal et al., 2000), and most human breast cancers become resistant to the antiproliferative effects of TGF-β, despite an intact core signaling pathway (Riggins et al., 1997). These data support an important role for the TGF-β signaling pathway in mammary carcinogenesis. Accordingly, several strategies are being explored for targeting TGF-β signaling in breast cancer patients (Connolly et al., 2012). However, the highly contextual nature and dichotomous functions of TGF-β signaling during breast cancer progression suggest that further definition of this pathway is required to safely and effectively target the TGF-β signaling pathway. Because we demonstrated that there is increased TGF-β responsiveness when ectodomain shedding is inhibited, our work suggests that anti–TGF-β therapies may be more effective on tumors with a low ratio of soluble/cell-surface TβRIII. Understanding the distinct roles of soluble and membrane-bound TβRIII and how they interact to regulate TGF-β–mediated signaling and biology at different ratios will be useful when considering using TGF-β–targeted therapies for cancer and other diseases. The work presented here contributes to this understanding by defining differential effects of a low versus high ratio of soluble and cell-surface TβRIII on TGF-β signaling, TGF-β-mediated migration and invasion, and metastatic potential of breast cancer cells.
MATERIALS AND METHODS
Cell culture and reagents, transfections, lentivirus production, and infections
COS7, HEK293, MDA-MB-231-4175, and 293FT cells were maintained in DMEM (Life Technologies, Carlsbad, CA) with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA). HMEC cells were maintained in DMEM supplemented with 10% FBS and 0.01 mg/ml recombinant human insulin. MDA-MB-231 cells were grown in MEM (Life Technologies) supplemented with 10% FBS, sodium pyruvate (Life Technologies), and nonessential amino acids (Life Technologies). All cells were incubated at 37°C with 5% CO2. For transfections, cells were plated to be ∼70% confluent the next day (between 2 × 105 and 3 × 105 cells, depending on cell line) on six-well dishes. The next day, cells were transfected with either FuGENE 6 (Roche, Basel, Switzerland) or X-treme Gene 9 (Roche) transfection reagent and 3 μg of plasmid DNA at a ratio of 2.5:1. Experiments were performed on transfected cells 48–72 h posttransfection, as indicated. For lentivirus production, 293FT cells were plated in 10-cm dishes. Cells were transfected with Lipofectamine 2000 (Invitrogen) at a ratio of 3:1 to DNA, 6 μg of TβRIII wild-type, mutant, or empty vector (pSMPUW-Neo expression vector; (Cell Biolabs, San Diego, CA) and 3 μg each of three third-generation lentiviral packaging plasmids (AddGene, Cambridge, MA) in Opti-MEM (Life Technologies). Medium was changed 6 h after transfection. Forty-eight hours later, medium was collected, spun down to clear, and filtered through a 0.45-μm-pore membrane. Viral medium was aliquoted and stored at −80°C. For infections, viral medium was added to cells in normal growth medium at a ratio of either 1:10 or 1:100 in the presence of Polybrene at 6 μg/ml. To create stable lentiviral-expressing cell lines, 48 h postinfection, medium was changed, and complete growth medium containing 2 mg/ml G418 (KSE Scientific, Durham, NC) was added as a selection agent. After selection, stable lentiviral cell lines were maintained in complete growth medium containing 0.5 mg/ml G418.
Site-directed mutagenesis
Primers for NAAIRS and alanine mutants were designed as in Supplemental Tables S1 and S2, respectively. Mutagenesis PCRs, digests, and transformations were performed using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). NAAIRS and alanine mutants were made in a pDNR-CMV Donor Vector (Clontech, Mountain View, CA) containing an N-terminally hemagglutinin (HA)-tagged wild-type TβRIII. Sequence analysis of all mutants was performed twice for verification.
Binding and cross-linking
Twenty-five thousand cells were plated on six-well dishes. Cells were transfected 18–20 h later. Twenty-four hours later, medium was removed and replaced with 1 ml of complete growth medium. Unless otherwise stated, media were conditioned for 18–20 h before being removed, and both cells and conditioned media were incubated with [125I]TGF-β1 (PerkinElmer, Waltham, MA) at 100 and 25 pM, respectively, in the presence of bovine serum albumin and protease inhibitors for 3 h at 4°C. After incubation, ligand was chemically cross-linked using 0.5 mg/ml disuccinimidyl suberate and quenched with 1 M glycine. Cells were lysed with RIPA buffer supplemented with protease inhibitors, and ligand–receptor complexes were pulled down by immunoprecipitation overnight at 4°C with either an antibody against HA (Roche) or a polyclonal antibody against the extracellular domain of TβRIII (R&D Systems, Minneapolis, MN). The resulting complexes were separated via SDS–PAGE, and dried gels were exposed to an autoradiograph. Images were acquired with phosphorimaging and analyzed using ImageJ (National Institutes of Health, Bethesda, MD).
Enzyme-linked immunosorbent assay
Twenty-five thousand cells were plated in six-well dishes and transfected the next day. After 24 h, medium was removed and replaced with 1 ml of full growth medium. Culture medium was allowed to condition for 24 h (unless otherwise indicated) and then was removed and immediately spun down at 4°C to pellet dead cells and debris. Cleared medium was aliquoted and immediately placed at –80°C. For the TβRIII ELISA, capture antibody (R&D Systems) was immobilized onto an E1A/R1A plate (Corning, Corning, NY) overnight. After washing, samples were loaded onto plate and incubated at room temperature for 2 h. Then detection antibody (R&D systems) was applied and incubated for 2 h, followed by streptavidin–horseradish peroxidase (HRP) R&D systems) incubation for 30 min. Finally, Fast OPD substrate (Sigma-Aldrich, St. Louis, MO) was added, 3 M HCl was applied to stop reaction 30 min later, and optical absorbance at 490 nm was recorded immediately.
Western blotting
Cells were plated and transfected as described. For HEK293 cells, 24 h after transfection, medium was removed and replaced with serum-reduced (5% FBS) medium. After 18–20 h, cells were treated with TGF-β1 or TGF-β2 (R&D Systems) at the concentrations and time periods indicated. For MDA-MB-231 cells, medium was removed and replaced with full serum medium overnight before ligand treatment. After ligand treatment, cells were lysed directly in 2× sample buffer, and 20% of the lysate was loaded onto 10% SDS–PAGE gels. After electrophoresis, protein was transferred onto a polyvinylidene fluoride membrane, which was blocked in 10% low-fat milk in phosphate-buffered saline plus 0.5% Tween-20 (PBS-T) for 1 h. Blots were probed overnight at 4°C with antibodies in 5% milk/PBS-T against phosphorylated Smad2 (Cell Signaling, Danvers, MA), Total Smad2 (Cell Signaling), phosphorylated Smad3 (Cell Signaling), Total Smad3 (Cell Signaling), p21 (Cell Signaling), phosphorylated Akt (Cell Signaling), Total Akt (Cell Signaling), β-actin (Sigma-Aldrich), HA (Roche), or TβRIII-ECD (R&D Systems) and then probed for 1 h with HRP–conjugated secondary antibodies (Cell Signaling; Amersham, Piscataway, NJ). Bands were visualized using ECL (Western Lightning ECL Pro; PerkinElmer) and exposure to film, and densitometry was quantified using ImageJ software.
Thymidine incorporation assay
Fifteen hundred cells were plated in 96-well plates, in either the absence or presence of 50 pM TGF-β1 (each condition in triplicate), in preconditioned media (conditioned for 24 h from cells expressing EV or corresponding TβRIII mutants). At 20 h after plating, 1 μCi of [3H]thymidine (PerkinElmer) was added to each well and allowed to incubate for 4 h. After incorporation, cells were washed 2× with cold PBS and 1× with cold 10% trichloroacetic acid (TCA) and then rocked at 4°C for 1 h in 10% TCA. Then cells were washed 1× with 10% TCA, and 0.2 N NaOH was added to cells to lyse overnight. The next day, lysates were added to 2 ml of scintillation fluid (Ultima-Gold; Perkin Elmer), and the amount of incorporation was determined by scintillation counting.
Transwell migration assay
Twenty-five thousand cells in serum-free conditioned medium (24 h from corresponding cell lines) were plated in the upper chamber of a 50 μg/ml fibronectin–coated Transwell with 8-μm pores (Corning). Cells were either left untreated or treated with 50 pM TGF-β1 (each condition in duplicate). Medium containing serum was used as a chemoattractant in the bottom well, and cells were allowed to migrate for 24 h. After migration, the cells remaining on the top of the filter were removed by gently washing with a cotton swab, and migrated cells on the bottom of the filter were fixed in methanol and stained with hematoxylin and eosin (H&E). Filters were cut out, mounted onto slides, and examined microscopically. Three random fields of cells were chosen and counted.
Matrigel invasion assay
Seventy-five thousand cells in serum-free conditioned medium (24 h from corresponding cell lines) were plated in the upper chamber of a Matrigel-coated Transwell with 8-μm pores (BD Biosciences, San Jose, CA). Cells were either left untreated or treated with 50 pM TGF-β1 (each condition in duplicate). Medium containing serum was used as a chemoattractant in the bottom well, and cells were allowed to invade for 24 h. After invasion, the cells remaining on the top of the filter were removed by gently washing with a cotton swab, and invaded cells on the bottom of the filter were fixed in methanol and stained with H&E. Filters were cut out, mounted onto slides, and examined microscopically. Three random fields of cells were chosen and counted.
Dual-luciferase reporter assay
One hundred thousand cells were plated in six-well plates. At 24 h later, cells were transfected with 2.3 μg of pE2.1 luciferase reporter plasmid and 0.2 μg of pRL-SV40 Renilla plasmid. The next day, cells were treated with 24 h preconditioned serum-free media and 50 pM of TGF-β1 or TGF-β2. At 20 h later, cells were lysed, and the dual-luciferase reporter assay (Promega, Madison, WI) was performed per kit instructions.
In vivo metastasis assay
MDA-MB-231-4175 cells stably expressing EV control, WT-TβRIII, or SS-TβRIII lentiviral constructs were cultured in DMEM plus 10% FBS for 24 h. Of each cell line, 1 × 106 cells were diluted in 100 μl of PBS and injected via tail vein into 6-wk-old athymic nu/nu outbred mice (Duke University, Durham, NC). Mice were intraperitoneally injected with d-luciferin potassium salt (Gold Biotechnology, St. Louis, MO) at a concentration of 150 mg/kg 10 min before imaging at indicated time points. Mice were anesthetized using isoflurane, and the Xenogen IVIS Kinetic system and Living Image acquisition software were used to capture and analyze bioluminescence data. Total photon flux was calculated by measuring the visible size of flux at the preset minimum and maximum radiance.
Statistical analysis
Data are presented as mean ± SEM. One-way or two-way analyses of variance (ANOVAs) were performed, followed by either a one-sample Student's t test for values compared with a normalized control or either Tukey's test or a two-tailed Student's t test for comparing two experimental values. Mantel–Cox log-rank test was used to assess Kaplan–Meier curve significance. p < 0.05 is considered significant.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants R01-CA136786 (G.C.B.) and R01-CA136786S1 (J.L.E.), Komen for the Cure Grant SAC100002 (G.C.B.), Department of Defense Breast Cancer Research Program Predoctoral Fellowship BC-093966 (J.L.E.), and the Duke Medical Scientist Training Program, T32-GM007171 (J.J.H.). We thank T. How for technical assistance.
Abbreviations used:
- EV
empty vector
- ICD
intracellular domain
- MMP
matrix metalloproteinase
- ΔShed-TβRIII
nonshedding TβRIII mutant
- SS-TβRIII
super-shedding TβRIII mutant
- TβRIII
type III TGF-β receptor
- TGF-β
transforming growth factor β
- WT-TβRIII
wild-type TβRIII
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-09-0524) on June 25, 2014.
*These authors contributed equally to this work.
J.L.E., J.J.H., C.E.G., J.C., and M.S. performed the experiments. J.L.E., J.J.H., and G.C.B. wrote the manuscript. All authors edited and approved the final manuscript.
REFERENCES
- Andres JL, DeFalcis D, Noda M, Massague J. Binding of two growth factor families to separate domains of the proteoglycan betaglycan. J Biol Chem. 1992;267:5927–5930. [PubMed] [Google Scholar]
- Arribas J, Borroto A. Protein ectodomain shedding. Chem Rev. 2002;102:4627–4638. doi: 10.1021/cr010202t. [DOI] [PubMed] [Google Scholar]
- Arribas J, Lopez-Casillas F, Massague J. Role of the juxtamembrane domains of the transforming growth factor-alpha precursor and the beta-amyloid precursor protein in regulated ectodomain shedding. J Biol Chem. 1997;272:17160–17165. doi: 10.1074/jbc.272.27.17160. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Lopez-Casillas F, Malik SN. Antitumor activity of a recombinant soluble betaglycan in human breast cancer xenograft. Cancer Res. 2002;63:4690–4695. [PubMed] [Google Scholar]
- Bandyopadhyay A, Zhu Y, Cibull LB. A soluble transforming growth factor b type III receptor suppresses tumorigenicity and metastasis of human breast MDA-MB-231 cells. Cancer Res. 1999;59:5041–5046. [PubMed] [Google Scholar]
- Bernabeu C, Lopez-Novoa JM, Quintanilla M. The emerging role of TGF-beta superfamily coreceptors in cancer. Biochim Biophys Acta. 2009;1792:954–973. doi: 10.1016/j.bbadis.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Blair CR, Stone JB, Wells RG. The type III TGF-beta receptor betaglycan transmembrane-cytoplasmic domain fragment is stable after ectodomain cleavage and is a substrate of the intramembrane protease gamma-secretase. Biochim Biophys Acta. 2011;1813:332–339. doi: 10.1016/j.bbamcr.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobe GC, Liu X, Fang SJ, How T, Lodish HF. A novel mechanism for regulating transforming growth factor beta (TGF-beta) signaling. Functional modulation of type III TGF-beta receptor expression through interaction with the PDZ domain protein, GIPC. J Biol Chem. 2001;276:39608–39617. doi: 10.1074/jbc.M106831200. [DOI] [PubMed] [Google Scholar]
- Cheifetz S, Hernandez H, Laiho M, ten Dijke P, Iwata KK, Massague J. Distinct transforming growth factor-beta (TGF-beta) receptor subsets as determinants of cellular responsiveness to three TGF-beta isoforms. J Biol Chem. 1990;265:20533–20538. [PubMed] [Google Scholar]
- Chen W, Kirkbride KC, How T, Nelson CD, Mo J, Frederick JP, Wang XF, Lefkowitz RJ, Blobe GC. Beta-arrestin 2 mediates endocytosis of type III TGF-beta receptor and down-regulation of its signaling. Science. 2003;301:1394–1397. doi: 10.1126/science.1083195. [DOI] [PubMed] [Google Scholar]
- Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-beta targeted cancer therapy. Int J Biol Sci. 2012;8:964–978. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell TL, Dumont N, Barnett JV, Arteaga CL. Knockdown of the transforming growth factor-beta type III receptor impairs motility and invasion of metastatic cancer cells. Cancer Res. 2008;68:7304–7312. doi: 10.1158/0008-5472.CAN-07-6777. [DOI] [PubMed] [Google Scholar]
- De Crescenzo G, et al. Three key residues underlie the differential affinity of the TGFbeta isoforms for the TGFbeta type II receptor. J Mol Biol. 2006;355:47–62. doi: 10.1016/j.jmb.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Dong M, How T, Kirkbride KC, Gordon KJ, Lee JD, Hempel N, Kelly P, Moeller BJ, Marks JR, Blobe GC. The type III TGF-beta receptor suppresses breast cancer progression. J Clin Invest. 2007;117:206–217. doi: 10.1172/JCI29293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RL, Blobe GC. Role of transforming growth factor beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Finger EC, Turley RS, Dong M, How T, Fields TA, Blobe GC. TbetaRIII suppresses non-small cell lung cancer invasiveness and tumorigenicity. Carcinogenesis. 2008;29:528–535. doi: 10.1093/carcin/bgm289. [DOI] [PubMed] [Google Scholar]
- Gatza CE, Elderbroom JE, Oh SY, Starr M, Nixon A, Blobe GC. The balance of cell surface and soluble type III TGFbeta receptor regulates BMP signaling in normal and cancerous mammary epithelial cells. Neoplasia. 2014;16:489–500. doi: 10.1016/j.neo.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghellal A, Li C, Hayes M, Byrne G, Bundred N, Kumar S. Prognostic significance of TGF beta 1 and TGF beta 3 in human breast carcinoma. Anticancer Res. 2000;20:4413–4418. [PubMed] [Google Scholar]
- Hamad NM, Banik SS, Counter CM. Mutational analysis defines a minimum level of telomerase activity required for tumourigenic growth of human cells. Oncogene. 2002;21:7121–7125. doi: 10.1038/sj.onc.1205860. [DOI] [PubMed] [Google Scholar]
- Hanks BA, et al. Type III TGF-beta receptor downregulation generates an immunotolerant tumor microenvironment. J Clin Invest. 2013;123:3925–3940. doi: 10.1172/JCI65745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killock DJ, Ivetic A. The cytoplasmic domains of TNFalpha-converting enzyme (TACE/ADAM17) and L-selectin are regulated differently by p38 MAPK and PKC to promote ectodomain shedding. Biochem J. 2010;428:293–304. doi: 10.1042/BJ20091611. [DOI] [PubMed] [Google Scholar]
- Kirkbride KC, Townsend TA, Bruinsma MW, Barnett JV, Blobe GC. Bone morphogenetic proteins signal through the transforming growth factor-beta type III receptor. J Biol Chem. 2008;283:7628–7637. doi: 10.1074/jbc.M704883200. [DOI] [PubMed] [Google Scholar]
- Lambert KE, Huang H, Mythreye K, Blobe GC. The type III transforming growth factor-beta receptor inhibits proliferation, migration, adhesion in human myeloma cells. Mol Biol Cell. 2011;22:1463–1472. doi: 10.1091/mbc.E10-11-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X, Bandyopadhyay A, Le T, Sun L. Autocrine TGFbeta supports growth and survival of human breast cancer MDA-MB-231 cells. Oncogene. 2002;21:7514–7523. doi: 10.1038/sj.onc.1205966. [DOI] [PubMed] [Google Scholar]
- Lewis KA, Gray PC, Blount AL, Mac Conell LA, Wiater E, Bilezikjian LM, Vale W. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature. 2000;404:411–414. doi: 10.1038/35006129. [DOI] [PubMed] [Google Scholar]
- Liu C, Xu P, Lamouille S, Xu J, Derynck R. TACE-mediated ectodomain shedding of the type I TGF-beta receptor downregulates TGF-beta signaling. Mol Cell. 2009;35:26–36. doi: 10.1016/j.molcel.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Casillas F, Cheifetz S, Doody J, Andres JL, Lane WS, Massague J. Structure and expression of the membrane proteoglycan betaglycan, a component of the TGF-beta receptor system. Cell. 1991;67:785–795. doi: 10.1016/0092-8674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- Lopez-Casillas F, Payne HM, Andres JL, Massague J. Betaglycan can act as a dual modulator of TGF-beta access to signaling receptors: mapping of ligand binding and GAG attachment sites. J Cell Biol. 1994;124:557–568. doi: 10.1083/jcb.124.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Casillas F, Wrana JL, Massague J. Betaglycan presents ligand to the TGF beta signaling receptor. Cell. 1993;73:1435–1444. doi: 10.1016/0092-8674(93)90368-z. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooradian DL, McCarthy JB, Komanduri KV, Furcht LT. Effects of transforming growth factor-beta 1 on human pulmonary adenocarcinoma cell adhesion, motility, and invasion in vitro. J Natl Cancer Inst. 1992;84:523–527. doi: 10.1093/jnci/84.7.523. [DOI] [PubMed] [Google Scholar]
- Mu Y, et al. TRAF6 ubiquitinates TGFbeta type I receptor to promote its cleavage and nuclear translocation in cancer. Nat Commun. 2011;2:330. doi: 10.1038/ncomms1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mythreye K, Blobe GC. The type III TGF-beta receptor regulates epithelial and cancer cell migration through beta-arrestin2-mediated activation of Cdc42. Proc Natl Acad Sci USA. 2009;106:8221–8226. doi: 10.1073/pnas.0812879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Torres M, Valle BL, Maihle NJ, Negron-Vega L, Nieves-Alicea R, Cora EM. Shedding of epidermal growth factor receptor is a regulated process that occurs with overexpression in malignant cells. Exp Cell Res. 2008;314:2907–2918. doi: 10.1016/j.yexcr.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Riggins GJ, Kinzler KW, Vogelstein B, Thiagalingam S. Frequency of Smad gene mutations in human cancers. Cancer Res. 1997;57:2578–2580. [PubMed] [Google Scholar]
- Seo M, Lee MJ, Heo JH, Lee YI, Kim Y, Kim SY, Lee ES, Juhnn YS. G Protein betagamma subunits augment UVB-induced apoptosis by stimulating the release of soluble heparin-binding epidermal growth factor from human keratinocytes. J Biol Chem. 2007;282:24720–24730. doi: 10.1074/jbc.M702343200. [DOI] [PubMed] [Google Scholar]
- Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Song J, Tan H, Perry AJ, Akutsu T, Webb GI, Whisstock JC, Pike RN. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS One. 2012;7:e50300. doi: 10.1371/journal.pone.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Chen C. Expression of transforming growth factor beta type III receptor suppresses tumorigenicity of human breast cancer MDA-MB-231 cells. J Biol Chem. 1997;272:25367–25372. doi: 10.1074/jbc.272.40.25367. [DOI] [PubMed] [Google Scholar]
- Teixido J, Wong ST, Lee DC, Massague J. Generation of transforming growth factor-alpha from the cell surface by an O-glycosylation-independent multistep process. J Biol Chem. 1990;265:6410–6415. [PubMed] [Google Scholar]
- Turley RS, Finger EC, Hempel N, How T, Fields TA, Blobe GC. The type III transforming growth factor-beta receptor as a novel tumor suppressor gene in prostate cancer. Cancer Res. 2007;67:1090–1098. doi: 10.1158/0008-5472.CAN-06-3117. [DOI] [PubMed] [Google Scholar]
- Velasco-Loyden G, Arribas J, Lopez-Casillas F. The shedding of betaglycan is regulated by pervanadate and mediated by membrane type matrix metalloprotease-1. J Biol Chem. 2004;279:7721–7733. doi: 10.1074/jbc.M306499200. [DOI] [PubMed] [Google Scholar]
- Verona EV, Tang Y, Millstead TK, Hinck AP, Agyin JK, Sun L-Z. Expression, purification and characterization of BGERII: a novel pan-TGFβ inhibitor. Protein Eng Des Sel. 2008;21:463–473. doi: 10.1093/protein/gzn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Lin HY, Ng-Eaton E, Downward J, Lodish HF, Weinberg RA. Expression cloning and characterization of the TGF-beta type III receptor. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.