Structures of Drosophila Msps TOG4 and human ch-TOG TOG4 are presented. TOG4 departs from the other TOG structures and predicts novel α-tubulin engagement. Whereas TOG domains across the array have different tubulin-binding properties, cellular studies show that a fully functional TOG array is required for microtubule polymerase activity.
Abstract
XMAP215 family members are potent microtubule (MT) polymerases, with mutants displaying reduced MT growth rates and aberrant spindle morphologies. XMAP215 proteins contain arrayed tumor overexpressed gene (TOG) domains that bind tubulin. Whether these TOG domains are architecturally equivalent is unknown. Here we present crystal structures of TOG4 from Drosophila Msps and human ch-TOG. These TOG4 structures architecturally depart from the structures of TOG domains 1 and 2, revealing a conserved domain bend that predicts a novel engagement with α-tubulin. In vitro assays show differential tubulin-binding affinities across the TOG array, as well as differential effects on MT polymerization. We used Drosophila S2 cells depleted of endogenous Msps to assess the importance of individual TOG domains. Whereas a TOG1-4 array largely rescues MT polymerization rates, mutating tubulin-binding determinants in any single TOG domain dramatically reduces rescue activity. Our work highlights the structurally diverse yet positionally conserved TOG array that drives MT polymerization.
INTRODUCTION
Microtubules (MTs) are filamentous cytoskeletal polymers that facilitate directed cell migration, establish cell polarity, serve as intracellular transport tracks, and form the bipolar mitotic spindle. MTs are formed by the polymerization of αβ-tubulin heterodimers into structurally polarized, tube-like polymers. Tubulin heterodimers bind GTP and undergo polymerization-dependent GTP hydrolysis, which underlies the polymer's endogenous dynamic instability (Desai and Mitchison, 1997). Although the polymer has inherent dynamics, MT-associated proteins (MAPs) regulate the rates of polymerization and depolymerization and transitions between these states in space and time to facilitate cytoskeletal restructuring, as required during migration or mitotic spindle formation. Determining how MAPs mechanistically regulate MT dynamics is a key area of research in the cytoskeletal field. XMAP215 family proteins are master MT polymerases that promote MT growth and are critical for mitotic spindle structure and dynamics (Gard and Kirschner, 1987; Vasquez et al., 1994; Cullen et al., 1999; Tournebize et al., 2000; Kosco et al., 2001; Kawamura and Wasteneys, 2008; Cassimeris et al., 2009; Kronja et al., 2009; Zanic et al., 2013).
All XMAP215 family members have an N-terminal tumor overexpressed gene (TOG) domain array and a C-terminal region that links XMAP215 members to other MAPs (Spittle et al., 2000; Lee et al., 2001; Hsu and Toda, 2011; Li et al., 2011; van der Vaart et al., 2011). TOG domains range in size from 220 to 250 amino acids (Charrasse et al., 1998; Slep, 2009; Al-Bassam and Chang, 2011). Whereas the XMAP215 C-terminal domain differs between yeast and higher eukaryotes, the N-terminal TOG array feature is conserved and can promote MT polymerization in cell culture (Brittle and Ohkura, 2005; Currie et al., 2011). Yeast members Stu2 and Dis1 have two N-terminal TOG domains and a C-terminal coiled-coil domain and function as homodimers (Gard et al., 2004; Al-Bassam et al., 2006). In contrast, higher eukaryotic XMAP215 members are monomeric and contain a pentameric TOG array. TOG domains are bridged by linkers (60–100 amino acids in length) that have no predicted secondary structure but contain basic residues that promote MT association (Currie et al., 2011; Widlund et al., 2011).
TOG domain structures reveal a series of six Huntingtin, elongation factor 3, protein phosphatase 2A, target of rapamycin 1 (HEAT) repeats (HRs) that form a 60-Å oblong structure (Al-Bassam et al., 2007; Slep and Vale, 2007). The intra-HEAT loops on one face are highly conserved. A recent low-resolution structure of Stu2 TOG1 in complex with a polymerization-blocked mutant αβ-tubulin heterodimer confirmed that the intra-HEAT loop TOG face is the tubulin-interaction surface (Ayaz et al., 2012). In this structure, the first four N-terminal HRs engage β-tubulin, whereas the final two HRs engage α-tubulin, collectively binding a curved αβ-tubulin conformation believed to be its free, unpolymerized state (Rice et al., 2008; Pecqueur et al., 2012). Although TOG domain structures determined to date provide insight into the XMAP215 mechanism, these studies only report structures of TOG1, TOG2, and TOG3 from three diverse species: Saccharomyces cerevisiae Stu2 TOG1 and TOG2, Drosophila Mini spindles (Msps) TOG2, and Caenorhabditis elegans Zyg9 TOG3 (Al-Bassam et al., 2007; Slep and Vale, 2007; Ayaz et al., 2012). Zyg9 contains a trimeric TOG array that has diverged from other XMAP215 members. Although Zyg9’s TOG3 sequence is divergent, it is most closely related to TOG5 from XMAP215 family members that contain a pentameric TOG array, but whether Zyg9 TOG3 is functionally and structurally similar to TOG5 remains to be determined (Al-Bassam and Chang, 2011). TOG1 and TOG2 from S. cerevisiae, Schizosaccharomyces pombe, and Drosophila can bind tubulin heterodimers as assayed by gel filtration (Al-Bassam et al., 2006, 2012; Slep and Vale, 2007; Ayaz et al., 2012), but determining how the TOG array promotes MT polymerization requires knowledge of the TOG array's tubulin-binding stoichiometry. Determination of this stoichiometry is complicated by XMAP215–tubulin association and dissociation rates that are tuned to afford rapid, processive polymerization activity. Accordingly, studies have calculated quite different stoichiometries for XMAP215 members in complex with tubulin (Al-Bassam et al., 2006; Kerssemakers et al., 2006; Slep and Vale, 2007; Brouhard et al., 2008; Ayaz et al., 2012). Models derived from these studies range from a wrap-around model in which functional XMAP215 molecules, either a Stu2 dimer or a XMAP215 monomer, use their TOG array(s) to encapsulate a single tubulin heterodimer and deliver it to the MT plus end, to a templating model in which each TOG domain in the array uses similar tubulin-binding determinants to interact cooperatively with an array of tubulin heterodimers. Key to distinguishing these models is a determination of the structure, tubulin-binding, and MT polymerization activities of each TOG domain within the array. The structures of yeast TOG1, TOG2, Drosophila TOG2, and C. elegans TOG3 show a conserved TOG structure with little deviation in domain architecture within or across species. However, we lack a structural and functional characterization of the remaining TOG domains in the higher eukaryotic pentameric TOG array (TOGs 3–5), and we do not know whether these TOG domains are equivalent to TOG1 and 2 or structurally and functionally distinct.
Studies have highlighted the differential contribution of TOG domains across the array to XMAP215 family MT polymerase function. Initial structure–function studies involving Xenopus XMAP215 demonstrated that a construct spanning TOGs 1–5 colocalized with MTs when expressed in XL177 cells, whereas constructs embodying either TOG1-2 or TOG3-4 did not (Popov et al., 2001). Both the TOG1-2 construct and the TOG3-4 construct (each containing a C-terminal basic linker region) bound Taxol-stabilized MTs in an in vitro cosedimentation experiment. The TOG1-2 construct bound to asters and largely rescued spindle formation in extracts depleted of endogenous XMAP215, whereas the TOG3-4 construct did not. A subsequent in vitro study using full-length XMAP215 probed the role of the TOG domains in tubulin-binding activity and MT polymerase activity by mutating two TOG tubulin-binding determinants to alanine systematically across the TOG array (Widlund et al., 2011). This study demonstrated that XMAP215’s first four TOG domains contribute to the full-length protein's tubulin-binding activity, with a greater contribution from TOG1 and TOG2 than from TOG3 and TOG4, whereas TOG5 did not contribute significantly. Each TOG domain's contribution to the full-length protein's tubulin-binding activity correlated with its contribution to MT polymerase activity, with TOG1 and TOG2 providing the greatest contribution. Indeed, a “Bonsai” construct containing TOG1-2 fused to three tandemly repeated basic MT-binding K-loops from the kinesin KIF1A largely rescued MT growth rates in vitro, suggesting that although TOG1-2 is mechanistically critical, C-terminal MT/tubulin-binding determinants play a key role in the molecule's activity. A subsequent investigation probing the role of XMAP215’s TOG domains on mitotic spindle length found a direct correlation between each TOG domain's effect on MT polymerization activity and its effect on mitotic spindle length (Reber et al., 2013). Cellular studies involving Drosophila Msps demonstrated that the first four TOG domains in the pentameric array could partially rescue MT dynamics when endogenous Msps was depleted (Brittle and Ohkura, 2005; Currie et al., 2011). However, rescue activity was compromised when a conserved residue in TOG1 was mutated (E190K; Brittle and Ohkura, 2005). Msps TOG pairs, either TOG1-2 or TOG3-4, showed limited ability to rescue MT polymerization rates, and an artificially arrayed construct, TOG1-2-1-2, failed to enhance MT polymerization rates beyond that observed with the TOG1-2 construct (Currie et al., 2011). Studies involving the Arabidopsis XMAP215 family member MOR1 demonstrated that a temperature-sensitive mutation in TOG1 (E195K; mor1-1) resulted in a disorganized MT array and reduced MT growth rates (Whittington et al., 2001). Expressing MOR1 TOG1-2 ectopically partially rescued the mor1-1 phenotype. MOR1 TOG1-2 displayed MT binding activity in vitro but had a low affinity for free tubulin heterodimers (Lechner et al., 2012). In yeast, XMAP215 members contain an N-terminal TOG pair and a C-terminal coiled coil that mediates dimerization. Studies involving S. cerevisiae Stu2 demonstrated that Stu2 requires homodimerization for in vivo activity and that TOG1 is key for Stu2-dependent MT stabilization and anaphase spindle elongation (Al-Bassam et al., 2006). Both Stu2 TOG1 and TOG2 have tubulin heterodimer–binding activity and use similar tubulin-binding determinants across their intra-HEAT loops (Al-Bassam et al., 2007; Slep and Vale, 2007; Ayaz et al., 2012). Studies involving S. pombe Alp14 showed that dimerization is essential for rescue activity and that mutations in TOG2 were more detrimental toward rescue activity than mutations in TOG1 (Al-Bassam et al., 2012). Overall, studies collectively point to a key tubulin-binding/MT polymerase role for TOG1 and TOG2. However, for XMAP215 family members with a pentameric array, TOGs 3–5 function with TOGs 1 and 2 to yield wild-type tubulin-binding and MT polymerase activities.
Here we investigate how the arrayed TOG domains in the XMAP215 family collectively promote MT polymerization, comparing and contrasting the functional and structural properties of TOG domains in the array. We carried out structural, biochemical, and in vivo studies on the Drosophila XMAP215 family member Msps and supported our structural findings with a parallel investigation of the human member, ch-TOG. We report the first structures of TOG4 from both Msps and ch-TOG, revealing a TOG architecture quite distinct from that of TOG1 and TOG2. We find that the full TOG array is required for Msps polymerization activity and that it uses architecturally distinct yet positionally conserved TOG domains, all with different tubulin-binding affinities and MT polymerization activities.
RESULTS
TOG domains in the XMAP215 array display position-dependent conservation
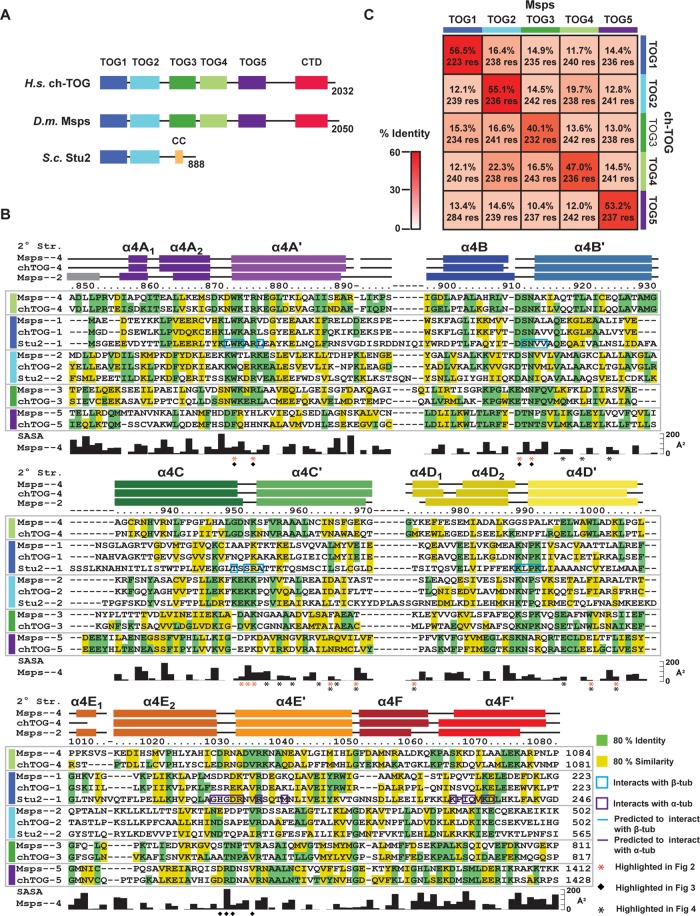
TOG domains in the XMAP215 family are arranged in a pentameric array in higher eukaryotes, including human and Drosophila, and a dimeric array in yeast (Figure 1A). To investigate the similarity of TOG domains in the XMAP215 family array, we performed an alignment across 13 diverse species. We mapped residues that were 80% identical (green) and 80% similar (yellow) for each specific TOG domain in the array (i.e., residues conserved across TOG1, residues conserved across TOG2, etc.). Figure 1B displays the conservation, mapped onto three species: human ch-TOG, Drosophila Msps, and S. cerevisiae Stu2, with all TOG domains from these species aligned. Although each specific TOG domain in the array shows a high degree of cross-species conservation, most of this cross-species conservation is not maintained across the different TOG domains within a species. TOG domains comprise six HRs (A–F) that form a solenoid-like structure (Al-Bassam et al., 2007; Slep and Vale, 2007). We note greater conservation across the array in HR A and B than in HRs C–F, suggesting that TOG domains may have divergent structures and/or functions in their C-terminal region.
FIGURE 1:
TOG domains with unique and universally conserved determinants comprise the XMAP215-family TOG array. (A) Domain architecture of XMAP215 family members ch-TOG, Msps, and Stu2 (CC, coiled coil; CTD, C-terminal domain). (B) Sequence alignment of TOG domains from Msps, ch-TOG, and Stu2. Class-specific TOG conservation has been mapped based on a multispecies alignment: residues with 80% identity (highlighted green) and 80% similarity (highlighted yellow) are indicated for each individual TOG class (TOG1, TOG2,…, TOG5). Residue numbers are for Msps TOG4. Msps TOG4 solvent-accessible surface area (SASA) is plotted. Stu2 TOG1 residues that interact with α- and β-tubulin are boxed in blue and purple, respectively. (C) Identity matrix, comparing sequence identity between Msps and ch-TOG TOG domains.
To quantitate the differences between TOG domains within and across species, we analyzed and compared human ch-TOG, Drosophila Msps, and S. cerevisiae Stu2. We first calculated the identity between TOG domains within a species based on the alignment presented in Figure 1B (Supplemental Figure S1, A–C). Consistent with what we observed in our sequence alignment, identity ranged from a low of 11.7% (Msps TOG3 vs. TOG5) to a high of 21.8% (TOG2 vs. TOG4 for both Msps and ch-TOG). We next compared identity across species (Figure 1C and Supplemental Figure S1, D and E). When ch-TOG and Msps TOG domains were compared, nonequivalent TOG domains had identities that ranged from 10.4 to 22.3% (Figure 1C). Surprisingly, TOG domains at equivalent positions retained much higher sequence identity, spanning 40.1% (TOG3) to 56.5% (TOG1). This reveals that TOG domains have retained positional conservation across species more than pan-array conservation within a species. Similar trends are evident when Msps and ch-TOG are compared with Stu2. Stu2 TOG1 and TOG2 are most identical to Msps and ch-TOG TOG1 and TOG2, respectively (20.7–26.4% identity), whereas Msps and ch-TOG TOG3-5 show significantly lower identity to Stu2 TOG1 or TOG2 (9.2–15.8%). These analyses indicate that TOG domains within an array have position-specific conservation. We asked whether this correlated with position-specific TOG structure and polarized TOG array activity.
Msps TOG4 comprises six HEAT repeats and forms an oblong structure with conserved intra-HEAT loops
XMAP215-family TOG domain structures determined include Stu2 TOG1, Stu2 TOG2, Msps TOG2, and Zyg-9 TOG3 (Al-Bassam et al., 2007; Slep and Vale, 2007; Ayaz et al., 2012). These XMAP215-family TOG structures show a similar TOG domain architecture in the arrangement of HRs and the conformation of the intra-HEAT loops that form the tubulin-binding surface. Whether all TOG domains in the array have a similar domain architecture remains open. We cloned and purified all five individual TOG domains from Msps and analyzed their secondary structure and domain stability using circular dichroism (CD). All five individual Msps TOG domains produced CD spectra with minima at 208 and 222 nm, in accord with an α-helical domain (Supplemental Figure S2A). We then monitored CD signal at 208 and 222 nm as the protein samples were heated from 20 to 94°C in order to determine each TOG domain's thermal stability. Msps TOG1, TOG2, TOG3, and TOG4 displayed cooperative unfolding curves with melting temperatures defined by the curve inflection point of 46.5, 64.5, 50.5, and 46.5°C, respectively (Supplemental Figure S2, B–E). The TOG5 CD melt profile was nearly linear, with no apparent inflection point (Supplemental Figure S2F), suggesting that TOG5, as defined by the region exclusively spanning the six predicted HEAT repeats, is not stable and may require flanking regions to promote domain stability as observed for CLASP's second TOG domain (Leano et al., 2013).
To further investigate structural differences across the Msps TOG domains, we screened Msps TOG3, TOG4, and TOG5 for crystallization. We obtained crystals of Msps TOG4 and focused our efforts on structure determination of this domain. Native and single-wavelength anomalous diffraction (SAD) data sets were collected on native and selenomethionine (SeMet)-substituted isomorphous TOG4 crystals, respectively. The Msps TOG4 structure was refined to 1.65-Å resolution with R and Rfree values of 15.7 and 18.7, respectively. Crystallographic and refinement information is presented in Table 1.
TABLE 1:
Crystallographic data, phasing, and refinement statistics.
| Structure | Msps TOG 4 | ch-TOG TOG 4 | ||
|---|---|---|---|---|
| Crystal | Native | SeMet | Native | SeMet |
| PDB code | 4QMH | 4QMI | 4QMJ | |
| Space group | P1 | P1 | P212121 | P43212 |
| Unit cell | ||||
| a, b, c (Å) | 31.8, 32.6, 59.8 | 31.7, 32.4, 60.1 | 64.4, 74.3, 93.7 | 79.1, 79.1, 68.5 |
| α, β, γ (deg) | 100.1, 95.2, 110.0 | 100.1, 95.1, 109.7 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 1.12714 | 0.97957 | 1.00000 | 0.97926 |
| dmin (Å) | 1.65 (1.71–1.65) | 1.90 (1.97–1.90) | 1.90 (1.97–1.90) | 2.5 (2.59–2.50) |
| Number of observations: measured/unique | 45402 (3809)/25054 (2355) | 64094 (5383)/33239 (3229) | 255235 (22145)/35954 (3515) | 106775 (9490)/14453 (1438) |
| Redundancy | 1.9 (1.6) | 2.0 (1.7) | 7.1 (6.3) | 7.4 (6.6) |
| Completeness (%) | 95.3 (88.9) | 96.6 (93.8) | 100.0 (100.0) | 99.9 (99.7) |
| I/σ | 33.0 (5.7) | 29.5 (14.7) | 22.9 (4.7) | 19.9 (3.3) |
| Rsym (%)a | 2.1 (11.7) | 2.2 (4.8) | 8.2 (42.8) | 9.5 (49.7) |
| Resolution (Å) at which anomalous completeness >85% for I/σI > 5, > 3, > 2 | — | 2.0, 1.9, 1.9 | — | 3.4, 3.1, 2.9 |
| SAD phasing: overall log-likelihood gain/figure of merit | — | 160751/0.741 (0.726) | — | 79522/0.36 (0.20) |
| Figure of meritb centrics/acentrics | — | 0 (0)/0.75 (0.75) | — | 0.12 (0.11)/0.42 (0.22) |
| Figure of meritb after density modification centrics/acentrics | — | 0 (0)/0.83 (0.74) | — | 0.61 (0.24)/0.66 (0.35) |
| Figure of meritb | 0.89 (0.85) | — | 0.87 (0.84) | 0.81 (0.83) |
| Refinement (Å) | 29.9–1.65 (1.69–1.65) | — | 48.7–1.90 (1.95– 1.90) | 28.0–2.50 (2.65–2.50) |
| R valuec | 15.7 (21.0) | — | 17.5 (21.0) | 20.8 (20.8) |
| Rfreed | 18.7 (30.2) | — | 21.5 (28.5) | 27.6 (29.4) |
| rmsd bond lengths (Å) | 0.006 | — | 0.007 | 0.008 |
| rmsd bond angles (deg) | 1.00 | — | 1.07 | 1.20 |
| Mean B (min/max) (Å2) | 18.2 (4.6/74.6) | — | 21.4 (5.7/78.6) | 37.9 (17.9/131.9) |
| Number of protein atoms/solvent atoms | 1805/401 (2 SO4) | — | 3541/360 | 1790/48 |
Values in parentheses are for the highest-resolution shells unless otherwise denoted.
aRsym = ΣhΣi ∣Ii(h) − < I(h) >∣/ΣhΣi Ii(h), where Ii(h) is the integrated intensity of the ith measurement and < I(h) > is the mean of all measurements of I(h) for Miller indices h.
bFigure of merit is the weighted mean of the cosine of the deviation from αbest.
cR value = Σ(∣Fobs∣ − k∣Fcalc∣)/Σ∣Fobs∣.
dRfree is calculated using a 10% subset of the data that is removed randomly from the original data and excluded from refinement.
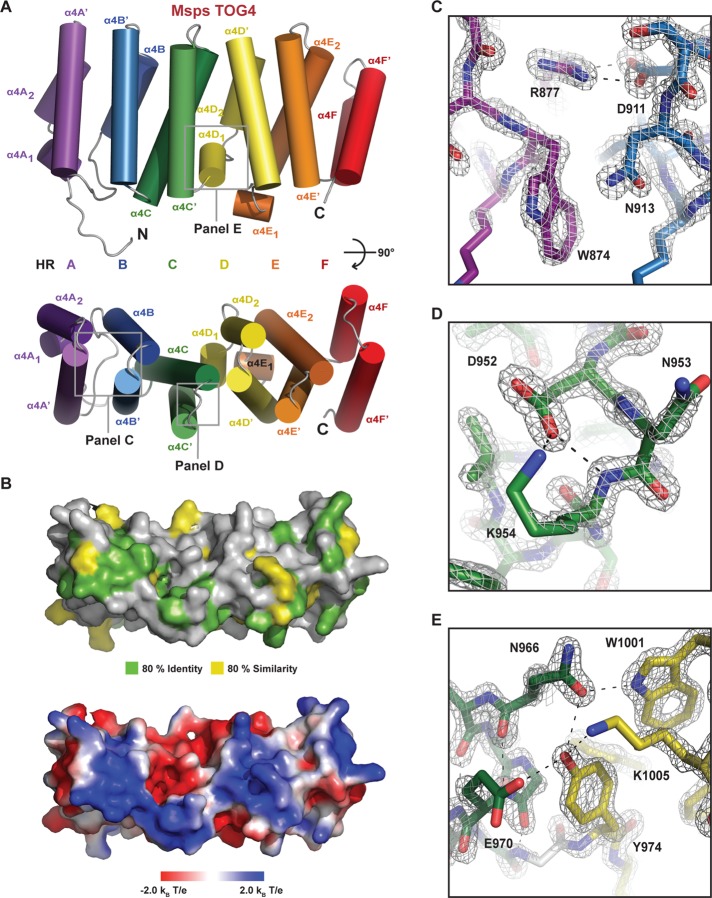
Like other TOG domains, the Msps TOG4 structure is helical, composed of six HRs that form an oblong structure of dimensions 15 × 30 × 55 Å (Figure 2A). HRs are defined by an antiparallel pair of helices in which the first helix undergoes a bend near its N-terminus. HRs are labeled A–F, with paired helices denoted X and X′ (Slep and Vale, 2007). If a helix is split, we delineate it with subscripts: X1 and X2. The number that precedes the HR indicates the specific TOG domain–for example, TOG4. As observed in other TOG domains, the TOG4 HRs are structurally divided into two triads: HR A–C and HR D–F (Leano et al., 2013). Within a triad, the HRs pack linearly against one another with a slight twist, common in HR-containing proteins (Andrade et al., 2001). Between TOG4 HR C and D, there is an offset afforded by α4D1 that effectively positions α4D′ alongside α4C (Figure 2A, bottom). Whereas the HR A–C triad has a right-handed twist, the HR D–F triad has a right-handed twist between repeats D and E but a dramatic change to a left-handed twist between HR E and F.
FIGURE 2:
Structure of Msps TOG4. (A) Cartoon representation of the Msps TOG4 domain. TOG4 consists of six HRs, A–F. (B) Msps TOG4 in surface representation (oriented as in A, bottom) mapping TOG4-specific conservation (top) as in Figure 1B and electrostatics (bottom). (C–E) The 2Fo − Fc electron density map (2σ) illustrating the HR A-loop W874 residue and the R877-D911 salt bridge (C), the HR C-loop D952–K954 interaction (D), and the buried HR C-D interaction network (E).
Msps TOG4 has conserved, intra-HEAT determinants in the first HR triad that parallel those used by Stu2 TOG1 to bind β-tubulin. Mapping TOG4-specific conservation onto the Msps TOG4 structure using the contours in Figure 1B maps the highest degree of invariance and similarity to the intra-HEAT loops (Figure 2B, top). This surface has a net positive charge (Figure 2B, bottom), which would complement the negatively charged tubulin surface on the MT exterior. Previous work mutating residues on this face of TOG1 and TOG2 abrogated the ability of the TOG domains to bind tubulin (Al-Bassam et al., 2007; Slep and Vale, 2007; Ayaz et al., 2012). In the recent Stu2 TOG1–αβ-tubulin complex structure, the TOG1 intra-HEAT loop face engages the αβ-tubulin heterodimer: HR A–D binds β-tubulin, and HR E–F binds α-tubulin (Ayaz et al., 2012). Similar tubulin-binding determinants are found on the TOG4 intra-HEAT surface, including a conserved, surface-exposed tryptophan (W874) in the intra-HEAT A loop and a lysine residue (K954) in the intra-HEAT C loop (Figure 2, C and D). The intra-HEAT A loop tryptophan (W874) is conserved across all Msps TOG domains except TOG5, where a phenylalanine is positioned. W874 is stabilized by a conserved salt bridge formed between aspartate D911 and arginine R877 (Figure 2C). In the intra-HEAT C loop, K954 is stabilized by a hydrogen bond between its backbone amide and side-chain amine with the aspartate D952 side-chain carboxyl (Figure 2D). The highest degree of surface-exposed intra-HEAT conservation maps to the HR A–C triad, consistent with the idea that these TOG4 HR A–C triad determinants engage β-tubulin as observed in the Stu2-TOG1–αβ-tubulin structure. At the interface of the two HR triads, Msps TOG4 uses a set of TOG4-specific conserved determinants to form a buried hydrogen-bond network involving HR C N966 and E970 and HR D Y974, W1001, and K1005 (Figure 2E). HR C and HR D are bridged by the α4D1 helix, which directs Msps Y974 (ch-TOG M974) into the domain's core. Surprisingly, the presence of α4D1 and its bulky side chains wedge the two HR triads apart into an architecture not observed in XMAP215-family TOG domains determined to date, as we will describe in detail.
Human ch-TOG TOG4 is structurally identical to Drosophila Msps TOG4
Before comparing and contrasting structural differences between TOG domains within a species’ array, we asked whether the structure of TOG4 was conserved across species and chose human ch-TOG TOG4 as a comparative target. We determined the structure of human ch-TOG TOG4 in two space groups (Figure 3, A–C). The structure of SeMet-substituted TOG4 was determined to 2.5-Å resolution (space group P43212; one TOG4 molecule in the asymmetric unit) and refined to R and Rfree values of 20.8 and 27.6, respectively. A native structure was also determined to 1.9-Å resolution (space group P212121; two TOG4 molecules in the asymmetric unit) and refined to R and Rfree values of 17.5 and 21.5, respectively. Crystallographic details are presented in Table 1.
FIGURE 3:
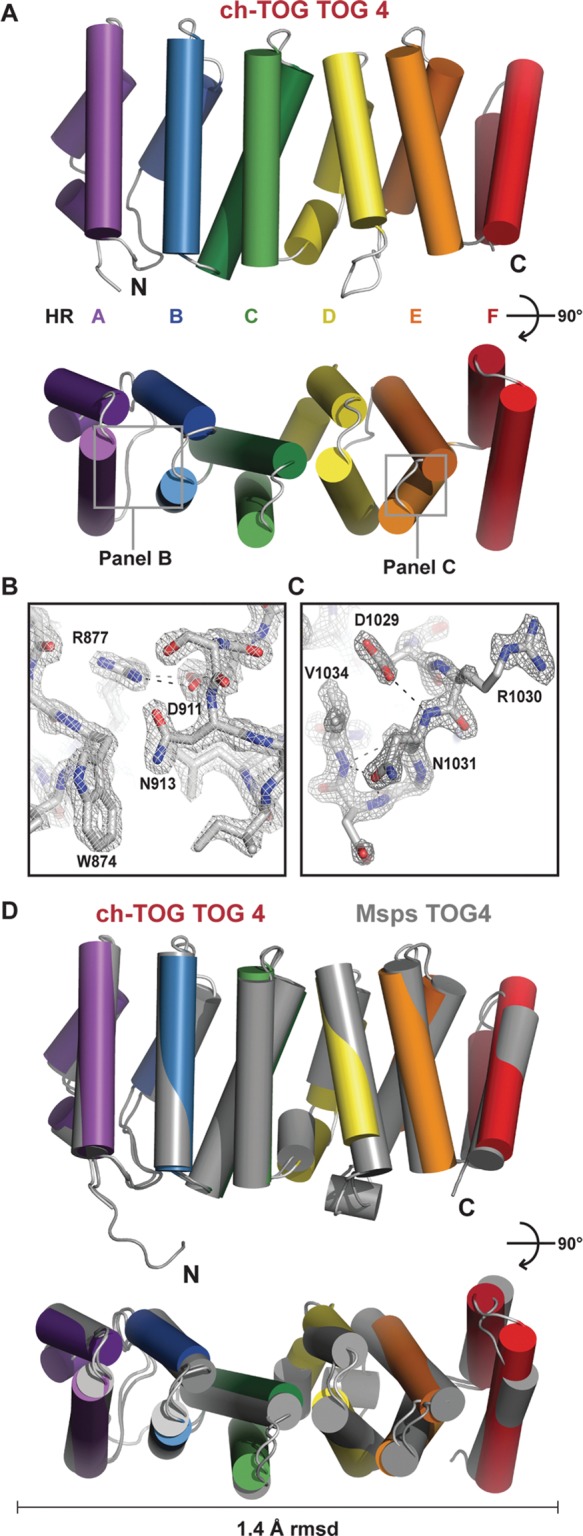
Structure of ch-TOG TOG4. (A) Cartoon representation of ch-TOG TOG4. (B, C) The 2Fo − Fc electron density map (2σ) of the ch-TOG TOG4 structure showing conserved determinants in the HR A (B) and HR E (C) loops. (D) Pairwise alignment of ch-TOG TOG4 (color) and Msps TOG4 (gray) showing structural conservation with a 1.4-Å rmsd over 226 Cα atoms.
The ch-TOG TOG4 structure aligns well to the Msps TOG4 structure, with a Cα root-mean-square deviation (rmsd) of 1.4 Å across 226 residues, indicative of high structural conservation (Figure 3D). The ch-TOG TOG4 structures we determined in different space groups showed nearly identical conformations, with low pairwise Cα rmsd values ranging from 0.7 to 0.8 Å (Supplemental Figure S3). This suggests that TOG4 is structurally static both within and across species. ch-TOG TOG4 and Msps TOG4 HR A–F structurally align with minor differences in the terminal region of HR D, the absence of α4E1 in ch-TOG, and an extension of ch-TOG's α4F′ helix. Many of the TOG4 intra-HEAT loop residues are conserved between Drosophila and humans and use similar flanking residues to buttress their rotamer arrangement. In the ch-TOG TOG4 structure, the conserved HR A loop tryptophan, W874, is solvent exposed and stabilized by an R877-D911 salt bridge, as observed in Msps TOG4 (Figure 3B). Similarly, the conserved HR E loop aspartate, D1029, is stabilized by a hydrogen bond with the asparagine N1031 backbone amide (Figure 3C).
TOG4 is structurally distinct
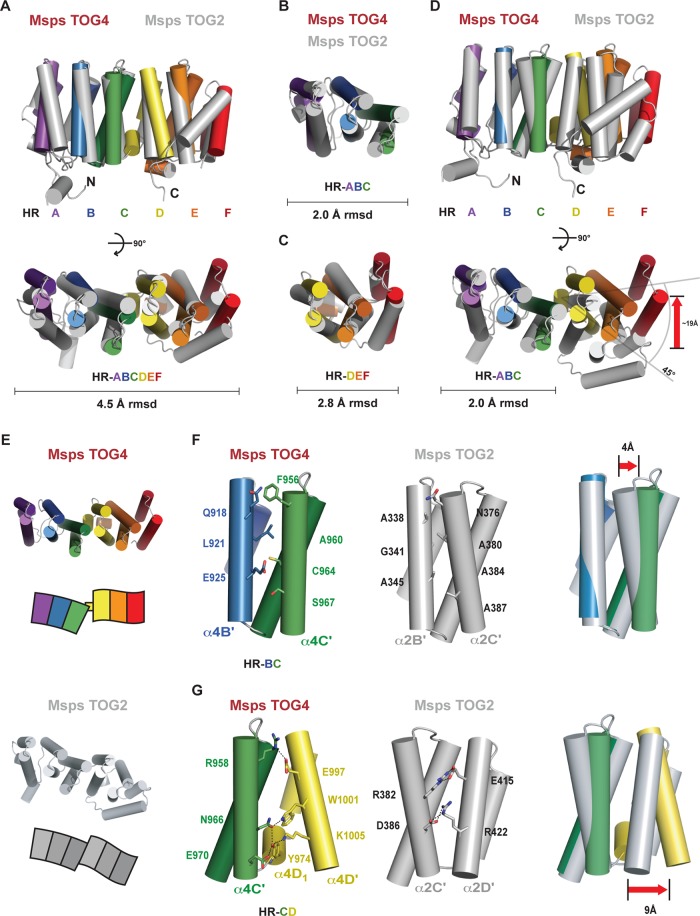
XMAP215-family TOG structures determined to date (S. cerevisiae TOG1 and TOG2, Drosophila TOG2, and C. elegans TOG3) are structurally similar (Supplemental Figure S4). We next asked how TOG4 compares to these structures by focusing on the intraspecies comparison of Drosophila Msps TOG2 and TOG4 (Figure 4A). Surprisingly, Msps TOG2 and Msps TOG4 align quite poorly, with an rmsd of 4.5 Å across 226 Cα atoms (Supplemental Figure S4A). The individual HR triads align better; the HR A–C triads align with an rmsd of 2.0 Å, and the HR D–F triads align with an rmsd of 2.8 Å (Figure 4, B and C, and Table 2). Because the HR A–C triads align reasonably well and contain the highly conserved β-tubulin–binding determinants (including the HR A loop tryptophan), we aligned Msps TOG2 and TOG4 over this conserved, functional reference region to see how the C-terminal HR triads were comparatively positioned. In this alignment, TOG4, like TOG2, retains a relatively flat surface across its tubulin-binding face. However, the TOG4 HR D–F triad, while maintained in this plane, is dramatically reoriented ∼45° relative to TOG2’s HR D–F triad, effectively positioning HR F 19 Å away from its TOG2 counterpart (Figure 4, D and E; see red arrow in D). The differential positioning of the HR D–F triad is mirrored by a lower degree of conservation between the TOG2 and TOG4 HR D–F intra-HEAT loop sequences (Figure 1B). This contrasts with the sequence conservation in the HR A–C triad intra-HEAT loops that exists across all TOG domains in the array. Together these differences suggest that if the TOG4 HR D–F triad engages tubulin, we anticipate that its binding mode will be distinct from the tubulin-binding mode of TOG1 and TOG2.
FIGURE 4:
Msps TOG4 is structurally distinct from Msps TOG2. (A) Pairwise alignment of Msps TOG4 (color) and Msps TOG2 (gray) across the full domain yields a 4.5-Å rmsd. (B, C) Pairwise alignment of Msps TOG4 and Msps TOG2 across the first HR triad (B; 2.0 Å rmsd) and the second HR triad (C; 2.8 Å rmsd). (D) Superpositioning of Msps TOG4 and Msps TOG2 based on the first HR triad alignment shown in B, highlighting the 45° difference in the orientation of each domain's second HR triad and the 19-Å differential placement of HR F. (E) Comparative views of Msps TOG4 and Msps TOG2 structures oriented as in D; models shown below. (F) The Msps TOG4 α4B′-α4C′ interface contains bulky, conserved residues, whereas the Msps TOG2 α2B′-α2C′ interface has conserved residues with no or small side chains. (G) Msps TOG4 contains the α4D1 helical insert that forms an extensive interaction network at the base of HR C and HR D, absent in Msps TOG2.
TABLE 2:
Pairwise alignment of TOG domains.
| Protein 1 | Protein 2 | rmsd (Å) | Number of residues aligned | Percentage identity |
|---|---|---|---|---|
| Msps TOG4 | chTOG TOG4 Native Protomer A | 1.4 | 226 | 45 |
| Msps TOG4 | chTOG TOG4 | 1.3 | 223 | 46 |
| Native Protomer B | ||||
| chTOG TOG4 | chTOG TOG4 | 0.5 | 224 | 100 |
| Native Protomer A | Native Protomer B | |||
| chTOG TOG4 Native Protomer A | chTOG TOG4 SeMet | 0.8 | 227 | 97 |
| chTOG TOG4 Native Protomer B | chTOG TOG4 SeMet | 0.7 | 224 | 97 |
| Msps TOG2 | Msps TOG4 | |||
| HR-A | HR-A | 1.5 | 33 | 18 |
| HR-B | HR-B | 1.2 | 33 | 27 |
| HR-C | HR-C | 1.4 | 38 | 18 |
| HR-D | HR-D | 1.7 | 32 | 25 |
| HR-E | HR-E | 2.0 | 41 | 15 |
| HR-F | HR-F | 2.6 | 28 | 18 |
| HR-AB | HR-AB | 1.8 | 74 | 20 |
| HR-BC | HR-BC | 1.7 | 73 | 22 |
| HR-CD | HR-CD | 1.9 | 72 | 21 |
| HR-DE | HR-DE | 2.2 | 76 | 18 |
| HR-EF | HR-EF | 2.5 | 67 | 15 |
| HR-ABC | HR-ABC | 2.0 | 113 | 19 |
| HR-BCD | HR-BCD | 2.3 | 107 | 22 |
| HR-CDE | HR-CDE | 2.4 | 116 | 19 |
| HR-DEF | HR-DEF | 2.8 | 104 | 18 |
| HR-ABCD | HR-ABCD | 2.7 | 148 | 20 |
| HR-ABCDE | HR-ABCDE | 3.4 | 197 | 21 |
| HR-ABCDEF | HR-ABCDEF | 4.5 | 226 | 20 |
| Stu2 TOG1 | Msps TOG4 | |||
| HR-ABC | HR-ABC | 2.5 | 102 | 19 |
| HR-DEF | HR-DEF | 2.8 | 95 | 12 |
| HR-ABCDEF | HR-ABCDEF | 4.2 | 202 | 15 |
Two regions likely underlie the conformational differences between TOG4 and TOG2. The first site occurs between helices αB′ and αC′ (Figure 4F). The TOG2 α2B′ and α2C′ helices pack in close proximity via TOG2-specific conserved alanine and glycine residues, including A338, G341, and A345 in α2B′ and A380, A384, and A387 in α2C′. In contrast, the TOG4 α4B′-α4C′ interface contains large side chains, including Q918 and L921 in α4B′ and F956 in α4C′. In TOG4, this results in a 4-Å displacement of the α4C′ N-terminal region relative to TOG2 α2C′ (Figure 4F, right, red arrow). The second site occurs between HR C and HR D at the HR triad junction (Figure 4G). A major difference between TOG2 and TOG4 at this junction is the presence of the TOG4-specific α4D1 helix that bridges HR C and HR D and comparatively expands the interface between the triads. As discussed, Msps α4D1 Y974 expands the core, forming a buried hydrogen bond network with N966 and E970 from α4C′ as well as K1005 from α4D′. Msps α4D′ also places the TOG4-specific conserved residue W1001 between HR C and HR D, where it forms a hydrogen bond with the N966 side chain (also observed in ch-TOG TOG4). In contrast, TOG2 lacks a helical insert between HR C and HR D. As a result, the C-terminal regions of TOG2 α2C′ and α2D′ come in close proximity, stabilized by a D386-R422 salt bridge. Effectively, bulky side chains and the α4D1 helical insert displace the TOG4 HR D C-terminal region 9 Å relative to TOG2’s HR D (Figure 4G, right, red arrow). These two sites underlie a major structural perturbation of the TOG architecture that positions TOG4 HR F 19 Å from the homologous position of TOG2 HR F. Thus the position-specific conservation of TOG domains in the array (Supplemental Figure S1) mirrors position-specific structure: we find that the spatial arrangement of HRs in Drosophila Msps TOG2 and yeast Stu2 TOG2 is conserved, just as the spatial arrangement of HRs in Drosophila Msps TOG4 and human ch-TOG TOG4 is conserved. However, the spatial arrangement of HRs in TOG4 is distinct from that in TOG2. This indicates that TOG domains in the XMAP215 family have conserved, position-specific architectures within the array.
The TOG4 structure suggests differential tubulin-binding modes along the TOG array
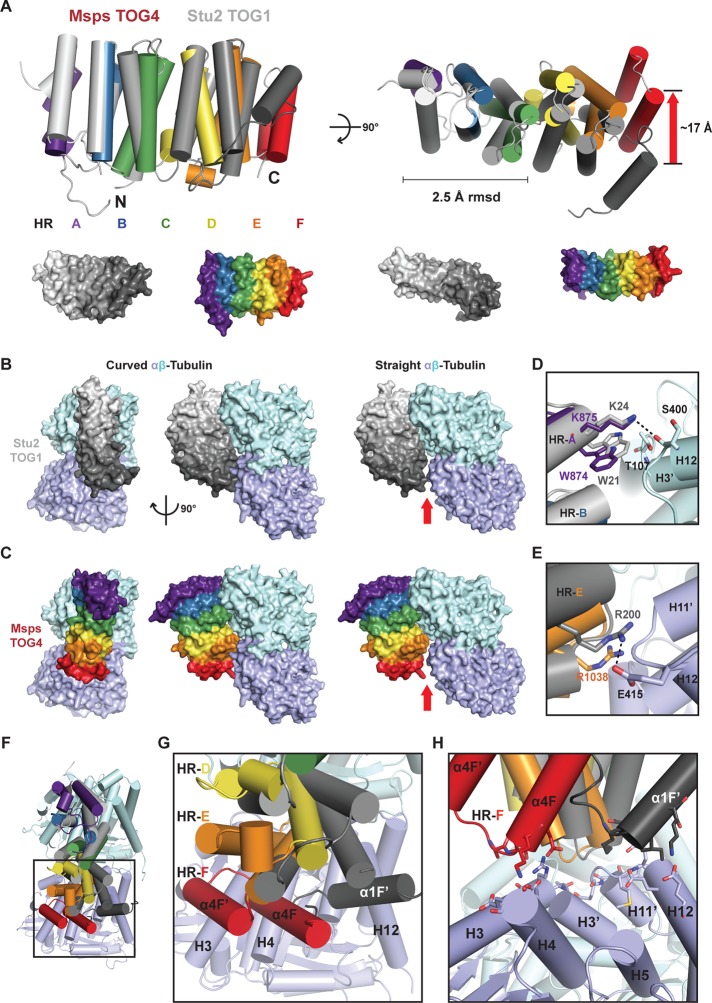
To investigate how TOG4 might interact with tubulin, we superimposed Msps TOG4 onto the structure of the Stu2 TOG1–αβ-tubulin complex (Ayaz et al., 2012). Whereas Msps TOG4 and Stu2 TOG1 align poorly, with an rmsd of 4.2 Å across 202 Cα residues (Supplemental Figure S4A and Table 2), the HR A–C triad intra-HEAT loops used to engage β-tubulin retain a high degree of identity between Msps TOG4 and Stu2 TOG1 (33%). We therefore docked Msps TOG4 onto the Stu2 TOG1–tubulin complex by structurally aligning the TOG4 and TOG1 HR A–C triads, which align reasonably well, with an rmsd of 2.5 Å over this region (Figure 5A). This HR A–C triad alignment enabled us to generate a model of Msps TOG4 bound to tubulin (Figure 5C). When Stu2 TOG1 and Msps TOG4 are aligned over the HR A–C triads, the TOG4 HR D–F triad shows a dramatically different architecture than Stu2 TOG1 HR D–F, culminating in a 17-Å differential positioning of HR F, similar to the architectural change evident in the TOG2–TOG4 comparison presented earlier (Figures 4D and 5A, red arrows).
FIGURE 5:
TOG4 is predicted to form TOG4-specific contacts with tubulin. (A) Msps TOG4 (color) and Stu2 TOG1 (gray) aligned pairwise over the first HR triad (HR A–C) with a 2.5-Å rmsd over HR A–C, highlighting the differential arrangement of each domain's second HR triad and the 17-Å shift in HR F (red arrow). Lower images show identical orientations in surface representation. (B) Stu2 TOG1 bound to curved αβ-tubulin (left; Ayaz et al., 2012) vs. straight tubulin (modeled at right). (C) Msps TOG4 was docked onto the Stu2 TOG1–αβ-tubulin structure and model presented in B by aligning the first HR triads as in A. (D) Msps TOG4 conserved HR A residues W874 and K875 can interact with β-tubulin residues T107 and S400, respectively, as observed in the Stu2 TOG1–αβ-tubulin structure. (E) Msps TOG4 conserved HR E residue R1038 is within 5 Å of α-tubulin E415 and is likely to reposition and form a salt bridge as observed with Stu2 TOG1 R200. (F) Superposition of the Stu2 TOG1–αβ-tubulin structure and the Msps TOG4–αβ-tubulin model reveals major differences in HR F's interaction with α-tubulin. (G, H) Zoom view of the region boxed in F (G) and shown after a 90° rotation about the x-axis (H).
Our model is consistent with the idea that Msps TOG4 preferentially binds curved tubulin. In the Stu2 TOG1–αβ-tubulin structure, TOG1 binds curved tubulin, a quaternary conformation observed in nonpolymerized tubulin (Rice et al., 2008; Pecqueur et al., 2012). This contrasts with the straight tubulin conformation, determined from zinc-induced tubulin sheets believed to represent the structure of tubulin in MT protofilaments (Nogales et al., 1998; Lowe et al., 2001). When the straight tubulin structure is docked onto the Stu2 TOG1–αβ-tubulin complex by structurally aligning β-tubulin, a gap is evident between TOG1 HR E–F and α-tubulin (Figure 5B, red arrow). Similarly, in our TOG4–αβ-tubulin model, TOG4’s HRs fully engage αβ-tubulin in the curved but not in the straight conformation (Figure 5C, red arrow), suggesting that tubulin maintains a curved conformation when bound to TOG1 or TOG4.
Stu2 TOG1 binds αβ-tubulin using intra-HEAT loop residues; HR A–D bind β-tubulin and HR E–F bind α-tubulin (Figure 1B; see Stu2 TOG1 α- and β-tubulin–binding residues boxed in blue and purple, respectively). Because we aligned Stu2 TOG1 and Msps TOG4 over the first HR triad, we observe, as expected, that HR A-C intra-HEAT loop residues conserved between Stu2 TOG1 and Msps TOG4 are equivalently positioned to interact with β-tubulin. Key interactions with β-tubulin involve Stu2 TOG1 HR A residues W21 and K24, which engage β-tubulin residues T107 and S400, respectively. Our Msps TOG4 model suggests a similar mode of interaction; HR A residues W874 and K875 are positioned to interact with β-tubulin residues T107 and S400 (Figure 5D). We also see conserved interactions that extend into the second HR triad. Stu2 TOG1 HR E residue R200 forms a salt bridge with α-tubulin residue E415. Although Msps HR E is positioned differently from Stu2 TOG1 HR E, the Msps TOG4 HR E R1038 side chain is positioned within range to form a homologous salt bridge with α-tubulin E415 (Figure 5E).
Whereas the TOG4 N-terminal HRs position intra-HEAT loop determinants in conformations that would facilitate TOG–tubulin interactions homologous to those observed in the Stu2 TOG1–αβ-tubulin structure, major differences are evident in the position of the HR F intra-HEAT loop (Figure 5, C and F–H). Stu2 TOG1 HR F is positioned to interact with the α-tubulin H12 helix. In contrast, Msps TOG4 HR F is positioned to interact with the α-tubulin H3 and H4 helices, ∼17 Å away from the H12 helix. Collectively this model suggests that Msps TOG4 binds the αβ-tubulin heterodimer in a unique manner, with some TOG–tubulin interaction determinants common to the Stu2 TOG1 tubulin interaction and others distinct to TOG4. These findings suggest that different TOG domains in the array make specific, nonequivalent interactions with tubulin.
Msps TOG1-2 and TOG3-4 constructs show dramatically different tubulin-binding and MT polymerization activities
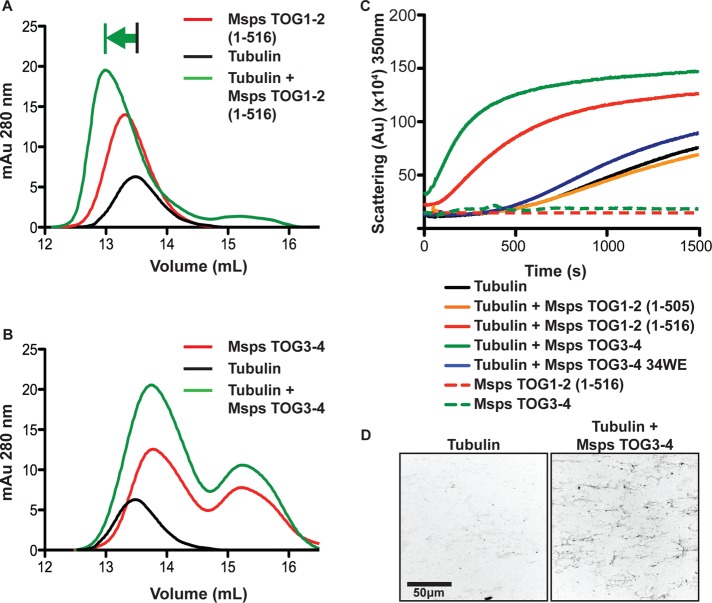
To investigate the tubulin-binding activity of different TOG domains in the array, we analyzed the ability of paired TOG domains to bind tubulin, using a gel filtration shift assay (Campbell and Slep, 2011). Previous work analyzing the ability of Msps TOG1 and TOG2 to bind and shift tubulin over gel filtration found that a detectable shift could be obtained with a TOG1-2 construct but not with either TOG domain individually or in trans (Slep and Vale, 2007). We first analyzed the ability of a Msps TOG1-2 construct (40 μM) to bind tubulin (20 μM) and reproduced the previously reported binding result, observing earlier peak elution, indicative of relatively stable complex formation (Figure 6A, green arrow). We then analyzed the ability of Msps TOG3-4 to bind tubulin. TOG3-4 itself produced a bimodal elution profile due to a degradation product that eluted later and contained individual TOG3 and TOG4 domains. We increased the total amount of protein load so that the amount of TOG3-4 was ∼40 μM. Of interest, no detectable peak shift was evident when TOG3-4 was incubated and run with tubulin (Figure 6B). This indicates that the TOG domains in the Msps TOG1-2 and TOG3-4 constructs have significantly different tubulin-binding properties and that if TOG3 and TOG4 each binds tubulin, their tubulin-binding affinities do not facilitate the stable interaction observed between Msps TOG1-2 and tubulin over gel filtration. Both Msps TOG1-2 and Msps TOG3-4 failed to cosediment with Taxol-stabilized MTs (Supplemental Figure S5, A–C), suggesting that any interaction between the TOG3-4 construct and the MT polymer is transient.
FIGURE 6:
Paired Msps TOG domains show differential tubulin-binding and MT polymerization activities in vitro. (A) Msps TOG1-2 (40 μM) binds and shifts tubulin (20 μM) to an earlier elution peak over gel filtration. (B) Msps TOG3-4 (40 μM) fails to shift tubulin (20 μM) over gel filtration. (C) Light scattering curves of tubulin (15 μM) polymerized at 37°C alone or in the presence of Msps TOG1-2 or TOG3-4 constructs (1 μM). TOG constructs alone showed no scattering activity. (D) Images of in vitro MT polymerization in the absence (left) and presence (right) of 1 nM Msps TOG34. Tubulin (20 μM) was doped with 10% rhodamine-labeled tubulin. Polymerization reactions were terminated after 180 s by diluting the samples into buffer containing 20 μM Taxol at 25°C. Scale bar, 50 μm. Gel filtration elution profiles, light scattering curves, and tubulin microscopy images shown are from individual experiments and are representative of experiments done in triplicate.
We next analyzed how Msps TOG1-2 and TOG3-4 individually affected MT polymerization in vitro, using a 90° light scattering assay. Tubulin alone (15 μM) showed standard polymerization behavior, with polymerization-dependent scattering apparent after ∼400-s lag time (Figure 6C). Tubulin in the presence of Msps TOG1-2 (residues 1–505, 1 μM) did not alter tubulin polymerization behavior, as previously observed (Slep and Vale, 2007). In contrast, a Msps TOG1-2 construct that contained an 11–amino acid C-terminal extension (residues 1–516, 1 μM) promoted MT polymerization after only ∼100-s lag time and showed faster bulk polymerization kinetics than observed with tubulin alone. The linker region bridging TOG2 and TOG3 has been shown to promote MT lattice association in cell culture, and we hypothesize that a series of basic residues in this C-terminal extension work in concert with the TOG domains to drive MT polymerization. We next investigated how a Msps TOG3-4 construct (with no N- or C-terminal extension, 1 μM) affected MT polymerization behavior. Surprisingly, Msps TOG3-4 drove polymerization even faster than the Msps TOG1-2 (1–516) construct, with only ∼50-s lag time, followed by a growth rate slightly faster than the TOG1-2 (1–516)–induced polymerization rate. TOG pairs alone showed no light scattering activity over time. We next analyzed the bulk tubulin polymerization activity of a Msps TOG3-4 construct in which the conserved HR A loop tryptophan in both Msps TOG3 and TOG4 was mutated to glutamate (TOG3-4 34WE), a mutation shown to ablate tubulin binding in other TOG domains but not affect TOG structure (Slep and Vale, 2007; Leano et al., 2013). We found that the Msps TOG3-4 double mutant did not dramatically alter MT polymerization kinetics from what we observed with the tubulin-alone control. Together these data suggest that Msps TOG3 and TOG4 use conserved tubulin-binding intra-HEAT loop determinants to drive MT polymerization. Although Msps TOG1-2 (1–505) is not sufficient to drive MT polymerization but requires a C-terminal extension (1–516), Msps TOG3-4 has potent MT polymerization activity mediated by its TOG domains’ tubulin-binding determinants. To confirm that Msps TOG3-4 promoted MT polymerization, we analyzed the ability of rhodamine-labeled tubulin to form MTs after 120 s at 37°C in the presence and absence of Msps TOG3-4, using a fluorescence microscopy-based assay. At this time point, more MTs were observed when tubulin was incubated with Msps TOG3-4 than in its absence (Figure 6D).
TOG3 and TOG4 promote MT colocalization in Drosophila S2 cells
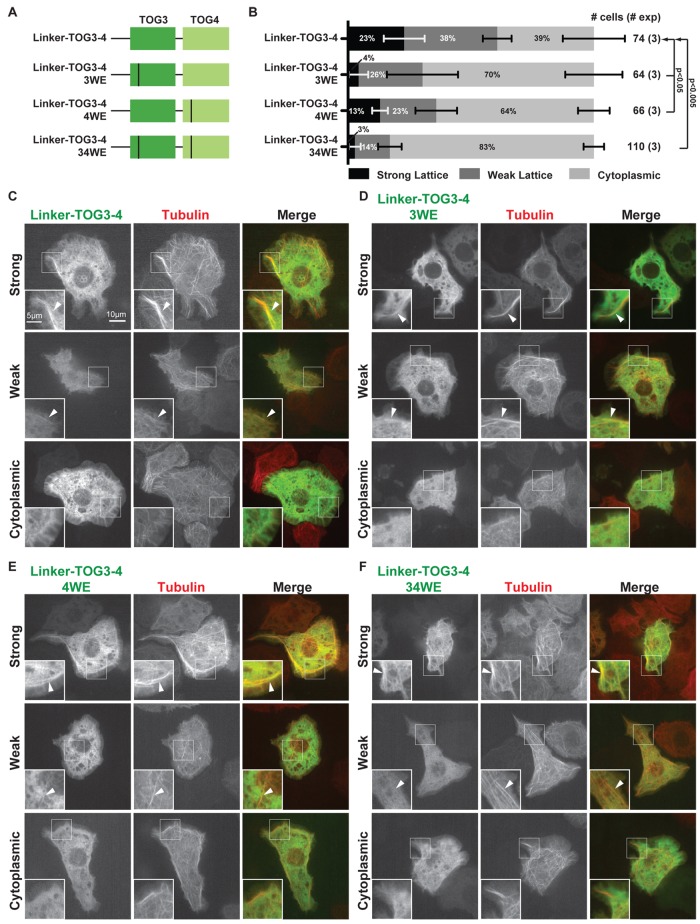
Whereas Msps TOG3-4 did not interact with tubulin heterodimers over gel filtration or cosediment with Taxol-stabilized MTs, the ability of Msps TOG3-4 to promote microtubule polymerization in vitro and the dependence of this activity on the HR A-loop tryptophans in both TOG3 and TOG4 suggest that Msps TOG3-4 can bind tubulin, at least transiently. Previous work demonstrated that a Msps TOG3-4 construct containing the TOG2-3 linker colocalized with MTs in Drosophila S2 cells (Currie et al., 2011). We therefore examined whether TOG3 and TOG4 contributed to this construct's MT colocalization activity by selectively mutating each TOG domain's HR A-loop tryptophans to glutamate and scoring MT colocalization in S2 cells. The linker–TOG3-4 construct (residues 498–1079) colocalized with MTs in 61% of cells examined, confirming previous findings (Figure 7, A–C; Currie et al., 2011). When either the TOG3 or TOG4 HR A-loop tryptophan was mutated (3WE and 4WE), MT colocalization decreased and was observed in only 30 and 36% of cells examined, respectively (Figure 7, B, D, and E). When the HR A-loop tryptophans in both TOG3 and TOG4 were mutated (34WE), MT colocalized decreased further, observed in only 17% of cells examined (Figure 7, B and F). This suggests that TOG3 and TOG4 work cooperatively with other Msps MT-association determinants (such as the TOG2-3 linker) to stably bind MTs, but without these additional determinants, the TOG3-4-tubulin interaction is transient. Attempts to obtain a soluble linker–TOG3-4 construct for in vitro tubulin/MT-binding experiments were not successful.
FIGURE 7:
TOG3 and TOG4 contribute to Msps MT lattice localization in cell culture. (A) Msps tRFP constructs analyzed; vertical line represents an HR A-loop W → E mutation. (B) Distribution of Msps 498–1079 tRFP lattice localization in Drosophila S2 cells cotransfected with α-tubulin GFP. Msps localization was binned into three categories: strong MT lattice localization (black), weak MT lattice localization (dark gray), and cytoplasmic localization (light gray). Error bars represent SD of the mean. Numbers at right list the total number of cells examined and the number of independent experiments performed. Representative images from each category are displayed (C–F). Mutating individual (D–E) or paired (F) conserved HR A-loop tryptophans resulted in an increase in cytoplasmic localization. Scale bars, 10 μm (images), 5 μm (insets). Statistical significance was determined using an unpaired t test to calculate two-tailed p values for the cytoplasmic bin.
Msps-dependent MT polymerization activity requires a fully functional TOG array
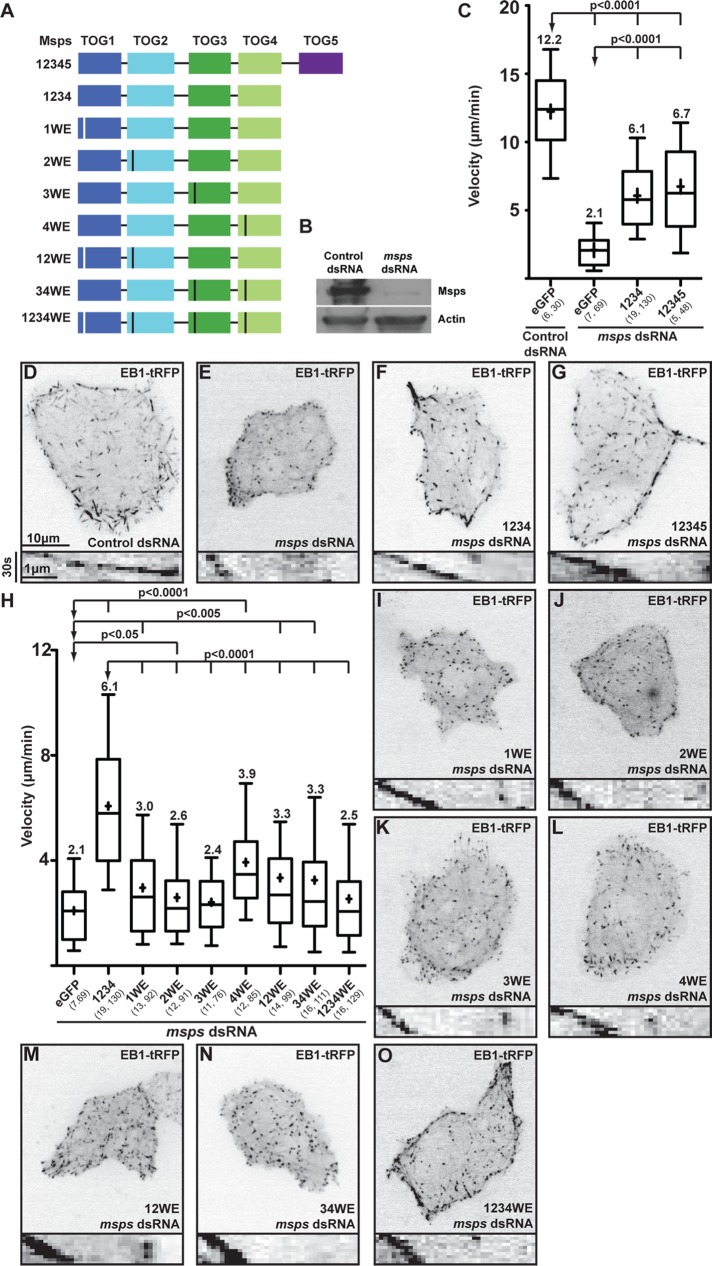
To analyze how the constituent TOG domains in the array contribute to MT polymerization, we performed cellular MT polymerization rescue assays on a minimal Msps construct (Figure 8A; Currie et al., 2011). We treated Drosophila S2 cells with double-stranded RNA (dsRNA) targeting the msps coding region (Msps C-terminal residues 1752–1928). Western blot analysis confirmed RNA interference (RNAi)–mediated Msps depletion (Figure 8B). Msps-depleted cells and control dsRNA-treated cells were transfected with an EB1:EB1-tRFP construct, and time-lapse images were acquired. EB1-tRFP comet velocities were measured as a readout for MT polymerization rates (Figure 8C; Supplemental Movie S1). In control dsRNA-treated cells transfected with a green fluorescent protein (GFP) control, the mean EB1 comet velocity was 12.2 μm/min (Figure 8, C and D). In contrast, cells treated with msps dsRNA had a significantly reduced mean EB1 comet velocity of 2.1 μm/min (Figure 8, C and E; p < 0.0001), consistent with a previous report (Currie et al., 2011). We next attempted to rescue MT polymerization rates in the msps dsRNA-treated cells using a msps construct containing the first four TOG domains (TOG1–4; Figure 8, C and F). In cells transfected with the msps TOG1-4-GFP construct, EB1-tRFP mean comet velocity increased to 6.1 μm/min, largely rescuing MT polymerization rates, as previously observed (Currie et al., 2011; p < 0.0001). An msps TOG1-5-GFP construct showed similar rescue activity, with a mean comet velocity of 6.7 μm/min (Figure 8, C and G; p < 0.0001). Expression of msps-GFP constructs was verified by Western blot (Supplemental Figure S6), and only cells with a GFP intensity that fell within a specified range were scored (see Materials and Methods).
FIGURE 8:
Msps requires a functional TOG array to rescue MT polymerization in cells. (A) Msps GFP constructs analyzed; vertical lines indicate an HR A-loop W → E mutation. (B) Western blot showing RNAi-mediated Msps depletion in Drosophila S2 cells (SK dsRNA control; antibodies: anti-Msps, anti-actin loading control). (C) Distribution of EB1 comet velocities from Msps rescue experiments. (D) Projection image (30 s) of an S2 cell expressing EB1-tRFP treated with control dsRNA (top) and a kymograph of a representative EB1 comet from this cell (bottom). (E–G) EB1 comet velocity is reduced when Msps is depleted (E) but can be largely rescued with Msps TOG1–4 (F) or Msps TOG1-5 (G). (H) Distribution of EB1 comet velocities in Msps dsRNA-treated cells transfected with Msps TOG1–4 mutant rescue constructs. Mutating the conserved HR A-loop tryptophan individually (I–L), in pairs (M–N), or across all four TOG domains (O) fails to rescue EB1 comet velocities. Scale bar, 10 μm (projection images), 30 s and 1 μm (kymographs). Box-and-whisker plots in C and H confer the following information: whiskers, 10th and 90th percentiles; boxes, 25th and 75th percentiles; line, median; cross, mean. Numbers in parentheses are the number of cells analyzed and the total number of EB1 comets tracked. Two-tailed p values were calculated using an unpaired t test.
To determine whether changes in EB1 comet velocities were due to decreased MT polymerization rates or MT sliding, we analyzed EB1 comet length. EB1 binds the MT lattice with highest affinity when tubulin subunits are in a GTP-hydrolytic state mimicked by a GTPγS-bound MT lattice (Maurer et al., 2011, 2012). The on-axis length of tubulin in this transient state should scale linearly with the MT growth rate. We analyzed averaged intensity line scans of EB1-tRFP MT plus-end comets in control dsRNA-treated cells transfected with GFP, msps dsRNA-treated cells transfected with GFP, and msps dsRNA-treated cells transfected with msps TOG1-4-GFP (Supplemental Figure S7A). The EB1 comet lengths measured in these cells scaled linearly with EB1 comet velocity, indicating that EB1 comet velocity correlated with MT polymerization rates under these different treatments (Supplemental Figure S7B).
We then used this system to explore the role of individual TOG domains within the msps TOG1-4 construct and correlate cellular observations with our structural and in vitro results. To probe the role of TOG domains in the TOG1–4 array, we systematically mutated the HR A-loop tryptophan to glutamate in each of the four TOG domains (Figure 8, H–L; Supplemental Movie S2), a mutation previously shown to ablate TOG–tubulin interactions (Slep and Vale, 2007; Leano et al., 2013). When TOG1-4-GFP constructs containing individual mutations in one of the four TOG domains were transfected into msps dsRNA-treated cells, rescued mean EB1 comet velocities were substantially reduced, with rates of 3.0, 2.6, 2.4, and 3.9 μm/min for constructs with TOG domains 1 (1WE), 2 (2WE), 3 (3WE), and 4 (4WE) mutated, respectively (Figure 8, H–L; p < 0.0001). When pairs of TOG domains were mutated (either the TOG1-2 pair [12WE] or the TOG3-4 pair [34WE]), mean EB1 comet velocities were similarly reduced at 3.3 μm/min (Figure 8, H, M, and N; Supplemental Movie S2; p < 0.0001). Mutations in all four TOG domains also disrupted rescue of MT polymerization rates, with a mean EB1 comet velocity of 2.5 μm/min (Figure 8, H and O; Supplemental Movie S2; p < 0.0001). These data suggest that each of the four TOG domains in the TOG1-4 construct is important for Msps’ ability to promote MT polymerization.
DISCUSSION
To better understand the TOG array mechanism, it is important to characterize the structural and functional properties of each domain within the polarized array. We analyzed the TOG domains in the XMAP215-family TOG array and noted TOG class-specific conservation. Our structures of Drosophila Msps TOG4 and human ch-TOG TOG4 revealed that TOG4 is structurally conserved across species, but its architecture differs from structures determined of Stu2 TOG1, Stu2 TOG2, Msps TOG2, and Zyg-9 TOG3 (Al-Bassam et al., 2007; Slep and Vale, 2007; Ayaz et al., 2012). Whereas TOG1, TOG2, and TOG3 are similar, TOG4’s second HR triad is positioned quite differently, and its arrangement predicts a TOG4-specific interaction with α-tubulin. This indicates that the XMAP215 TOG array is structurally polarized, with position-dependent features conserved across species. Why TOG domains in the array would have different tubulin-binding modes may involve the recognition of different tubulin structural states and/or differential interactions with other tubulin/MT-binding proteins.
Consistent with these structural differences, our in vitro tubulin-binding assays indicate that TOG domains have different affinities for free tubulin. Previous work analyzing Msps TOG1 and TOG2 found that whereas the individual TOG domains do not show detectable tubulin binding over gel filtration, a TOG1-2 construct does bind tubulin. This finding suggests that a multivalent TOG array enhances the TOG–tubulin interaction, potentially by promoting cooperative lateral or longitudinal tubulin–tubulin contacts (Slep and Vale, 2007). Whereas Msps TOG1-2 binds tubulin over gel filtration, Msps TOG3-4 does not display detectable tubulin-binding activity. However, previous work suggests that TOG3 and TOG4 do potentiate tubulin binding in the context of the full TOG array (Widlund et al., 2011). In this study, a full-length XMAP215 construct showed tubulin-binding activity over gel filtration. When the tubulin-binding determinants in TOG1 and TOG2 or TOG3 and TOG4 were mutated, the XMAP215 construct retained some, albeit reduced, tubulin-binding activity. When all five TOG domains were mutated, tubulin-binding activity was nearly abrogated. This suggests that TOG domains have different tubulin-binding activity but use common tubulin-binding determinants in a multivalent array to bind tubulin. Thus, we predict that TOG3 and TOG4 have weak tubulin-binding affinities that preclude detection of tubulin binding by a TOG3-4 construct as assayed by either a gel filtration shift or MT cosedimentation, but that these affinities are tuned to work cooperatively in the context of a larger TOG array with contributions from both the TOG domains and the basic inter-TOG linker regions. The existence of transient interactions between Msps TOG3-4 and tubulin is supported by our in vitro tubulin polymerization assay, in which TOG3-4 induced rapid MT polymerization. This activity required the intra-HEAT A-loop tryptophan that Stu2 TOG1 uses to bind tubulin, thus implicating the same face of TOG3 and TOG4 in tubulin binding. In cells, Msps TOG3-4 only colocalizes with MTs when an N-terminal basic extension implicated in MT binding is included. We found that this linker–TOG3-4 construct's ability to colocalize on MTs was significantly reduced when the intra-HEAT A-loop tryptophan in TOG 3 and/or TOG4 was mutated, again suggesting that TOG3 and TOG4 use canonical intra-HEAT determinants to bind tubulin/MTs but that they require flanking MT/tubulin-binding determinants for enhanced interactions. We hypothesize that along the array, each TOG domain's tubulin-binding affinity is positionally tuned to recognize free tubulin and/or MT lattice features with association and dissociation rates commensurate with the incorporation of 25 tubulin heterodimers into a protofilament per second (calculated based on a MT growth rate of 12 μm/min in Drosophila S2 cells).
The Stu2 TOG1–αβ-tubulin complex shows that TOG1 is polarized on the tubulin heterodimer, with its N- and C-termini oriented toward the MT plus and minus end, respectively (Ayaz et al., 2012). In a simplistic model, XMAP215 members may position TOG1 at the polymerizing plus end with TOG domains 2–5 arranged sequentially, just distal to the MT tip (Supplemental Figure S8). This orientation would position the C-terminal SLAIN2/Sentin-binding elements just behind the polymerizing MT plus end, which would correlate with the localization of EB1. EB1 binds the MT lattice just distal to the polymerizing MT plus end and recruits the SLAIN2/Sentin-XMAP215 complex (Li et al., 2011; van der Vaart et al., 2011; Maurer et al., 2012; Nakamura et al., 2012). This polarized arrangement of TOG domains on the MT lattice would position TOG1 and TOG2 to promote the stable incorporation of free tubulin heterodimers onto the MT plus end, while TOG domains 3–5, the basic linker regions, and the C-terminal SLAIN2/Sentin-binding domain work collectively to associate with the MT lattice.
An XMAP215 mechanism involving a polarized TOG array bound to a polymerizing MT plus end likely engages differential TOG function. These differential TOG functions may range from MT lattice binding and/or free tubulin heterodimer binding to potentiating stable incorporation of tubulin into the MT lattice by templating and/or lowering the tubulin heterodimer's off-rate at the MT plus end. Our cellular MT polymerization rescue experiments indicate that TOG domains work in an array, with each domain mobilizing similar tubulin-binding determinants to promote MT growth. TOG1–4 largely rescued MT polymerization activity, increasing it 2.9-fold above the 2.1 μm/min observed when Msps was depleted. Mutating the predicted tubulin-binding determinants in any single TOG domain in the TOG1–4 construct substantially diminished the rescue activity. The most dramatic effect was evident when TOG1, TOG2, or TOG3 was mutated. In contrast, the TOG4 mutant was able to partially restore polymerization with a 1.9-fold enhancement in the MT growth rate. This suggests that three contiguous N-terminal wild-type TOG domains function with greater efficiency than three wild-type TOG domains interspersed with a mutant TOG domain. This result highlights the importance of a functional, continuous, TOG array, that polymerization activity is polarized from the N-terminal TOG domains, and that larger, contiguous functional TOG arrays enhance MT polymerization rates (TOG1–5 > TOG1–4 > TOG1–4 4WE). Evolutionary support for the idea that polymerization activity is polarized from the N-terminal TOG domains comes from XMAP215 family members that do not have a pentameric TOG array, including S. cerevisiae Stu2 and C. elegans Zyg-9. Stu2 has two TOG domains that are respectively most similar in sequence to TOG1 and TOG2 from XMAP215 pentameric array members (Supplemental Figure S1, D and E). Zyg-9 has three TOG domains, the first two showing greatest sequence similarity to TOG1 from XMAP215 pentameric array members and the third showing greatest sequence similarity to TOG5. This underscores the evolutionary pressure to maintain a functionally polarized TOG array.
Our work highlights key features of the XMAP215 TOG array that further our mechanistic understanding of this critical MT regulator. First, TOG domains function in an array to promote MT polymerization. Second, TOG domains in the array have different structures but are positionally conserved. Third, TOG domains in the array have different affinities for tubulin heterodimers and effects on MT polymerization. Collectively this suggests that TOG domains, in general, mobilize an array-based mechanism to regulate MT dynamics and that differential TOG architecture in the array is mechanistically positioned to perform different regulatory actions on MTs. Our findings correlate with reports in the literature documenting the primary role of TOG1 and TOG2 in XMAP215-family MT polymerase activity in Xenopus, Drosophila, yeast, and Arabidopsis. Mutations in XMAP215 TOG3 and/or TOG4 affected the ability of XMAP215’s pentameric TOG array to bind tubulin, promote MT polymerization, and generate mitotic spindles with proper pole-to-pole length. However, in these studies, the effect of mutations in TOG3 and/or TOG4 were not as dramatic as mutations in TOG1 and/or TOG2. Although this highlights the differential mechanistic role TOG domains in the array play, it also differs slightly from our cellular rescue assays, in which mutations in any single TOG domain across a TOG1–4 array compromised MT polymerization rescue activity. This may be due to the nature of the mutations made in the TOG–tubulin binding determinants in each study. Whereas the studies involving Xenopus XMAP215 used reductive mutations (mutating conserved tryptophan and lysine residues to alanine to reduce binding affinity), our study introduced a negative charge in place of the conserved tryptophan, which may serve to ablate, rather than reduce, tubulin-binding activity.
Recent work examining the structure and function of a cryptic TOG domain in the CLASP family revealed that CLASP comprises a TOG array (Leano et al., 2013). This work showed that CLASP's second TOG domain adopts a unique bent architecture across its tubulin-binding face and that this domain is critical for CLASP's activity. Whereas the XMAP215 TOG array promotes MT polymerization, the CLASP TOG array promotes MT pause in interphase and MT polymerization during mitosis. Collectively a TOG array paradigm is emerging for these two key MT regulators, in which structurally distinct TOG domains in specific arrangements can differentially modulate MT dynamics. Structural insight into how architecturally distinct TOG domain arrays interact with multiple tubulin heterodimers, the role of the linker regions that bridge the TOG domains, and the concerted role their binding partners play in MT dynamics awaits further investigation.
MATERIALS AND METHODS
Multispecies alignment
Alignments were produced using CLUSTALW and adjusted manually based on structural information and secondary structure prediction algorithms (JPred2). Proteins aligned to determine levels of cross-species conservation included ch-TOG (human), Msps (Drosophila melanogaster), XMAP215 (Xenopus laevis), Stu2 (S. cerevisiae), Dis1 (S. pombe), Alp14 (S. pombe), Mor1 (Arabidopsis thaliana), CP224 (Dictyostelium discoideum), TMBP200 (Nicotiana tabacum), CKAP5 (Gallus gallus), CKAP5 (Mus musculus), CKAP5 (Bos taurus), and CKAP5 (Pan troglodytes).
Cloning and expression
Msps and ch-TOG bacterial expression constructs were generated by subcloning single or multiple TOG domains into pET28 (Clontech, Mountain View, CA), which engineered a thrombin-cleavable N-terminal hexahistidine (H6) tag. Fragments subcloned include Msps TOG1 (1–241), Msps TOG2 (267–505), Msps TOG3 (582–825), Msps TOG4 (848–1087), Msps TOG5 (1174–1408), Msps TOG1-2 (1–505), Msps TOG1-2 (1–516), Msps TOG3-4 (581–1080), Msps TOG3-4 (582–1080)–GFP, and ch-TOG TOG4 (846–1081). Mutations were generated using single or multiple rounds of QuikChange mutagenesis (Agilent Technologies, Cary, NC). Constructs were transformed into BL21 DE3 (pLysS) Escherichia coli and grown at 37°C in 6 l of Luria broth under kanamycin selection (50 μg/l) to an optical density of 1.0 (600 nm). Protein expression was induced with 100 μM isopropyl-β-d-thiogalactoside for 16 h at 18°C. Cells were harvested by centrifugation at 2100 × g for 10 min, resuspended in 300 ml buffer A (25 mM Tris, pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.1% β-mercaptoethanol [β-ME]) and stored at −20°C. SeMet-substituted Msps TOG4 and ch-TOG TOG4 were generated using B834 auxotrophic E. coli and minimal media containing l-SeMet (Leahy et al., 1994).
Protein purification
Msps and ch-TOG constructs were purified by sequential Ni2+-nitriloacetic acid (NTA) and ion exchange chromatography as follows. Cell pellets were thawed and lysed by sonication at 4°C. Phenylmethylsulfonyl fluoride (1 mM final) was added during lysis to prevent proteolytic degradation. Cell lysate was clarified by centrifugation at 23,000 × g for 45 min and the supernatant loaded onto a 10 ml Ni2+-NTA column (Qiagen, Venlo, Netherlands). The column was washed with 600 ml of buffer A and protein eluted using a 250-ml linear gradient between buffer A and B (buffer B = buffer A supplemented with 290 mM imidazole). Protein-containing fractions were pooled, and CaCl2 added to 1 mM and 0.1 mg bovine α-thrombin added to proteolytically remove the H6 tag. Thrombin digest proceeded for 16 h at 4°C. Digested protein was filtered over 0.5 ml of benzamadine Sepharose (GE Healthcare, Little Chalfont, UK), concentrated in a Millipore Ultrafree 10,000 MWCO concentrator, and exchanged into buffer C (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.0, 0.1% β-ME). Protein was loaded onto a 10-ml SP-Sepharose fast flow column (GE Healthcare), washed with 200 ml of buffer C, and eluted over a 250-ml linear gradient between buffer C and D (buffer D = buffer C supplemented with 1 M NaCl). Protein fractions were pooled, concentrated, and exchanged into 25 mM HEPES, pH 7.0, in a Millipore Ultrafree 10,000 MWCO concentrator (EMD Millipore, Darmstadt, Germany) to ≥15 mg/ml, frozen in liquid nitrogen, and stored at −80°C. Purification of SeMet-substituted protein proceeded according to the foregoing protocol, except that buffers were supplemented with 5 mM l-methionine.
Crystallization
Msps and ch-TOG TOG4 domains were crystallized using the hanging-drop vapor diffusion method. A 2-μl amount of Msps TOG4 (native and SeMet-substituted protein) at 15 mg/ml was added to an equal volume of a mother liquor containing 21% polyethylene glycol (PEG) 4000, 100 mM Tris, pH 8.5, and 400 mM Li2SO4 and equilibrated against 1 ml of mother liquor at 20°C. Crystals were transferred to a cryoprotection solution containing mother liquor supplemented with 15% PEG 4000 and flash frozen in liquid nitrogen. A 2-μl amount of ch-TOG TOG4 at 10 mg/ml was added to an equal volume of a mother liquor containing 28% PEG 4000, 100 mM Tris, pH 7.5, and 125 mM MgCl2, equilibrated against 0.5 ml of mother liquor at 20°C. SeMet-substituted ch-TOG TOG4 crystallization followed native parameters, except optimal crystals formed using a 0.5-ml well solution of 27.5% PEG 4000, 100 mM Tris, pH 7.5, and 70 mM MgCl2.
Data collection, structure determination, and refinement
Isomorphous Msps TOG4 native and selenium SAD peak data sets were collected on single crystals to a resolution of 1.65 and 1.9 Å, respectively. Diffraction data were collected at the Advanced Light Source 8.3.1 beamline at 100 K. Msps TOG4 crystals belong to the P1 space group with one molecule in the asymmetric unit. ch-TOG TOG4 native and selenium SAD peak data sets were collected on single crystals to resolutions of 1.9 and 2.5 Å, respectively. Diffraction data were collected at the Advanced Photon Source beamlines 22-BM and 22-ID. chTOG native and SeMet-substituted crystals belong to the space groups P212121 and P43212, respectively, with two and one molecules in the asymmetric unit. Data were indexed, integrated, and scaled using HKL2000 (Otwinowski et al., 1997). Selenium sites were identified and used to generate initial density-modified electron density maps using PHENIX AutoSol (Adams et al., 2010). Initial models were built using AutoBuild (PHENIX), followed by reiterative manual building in Coot and refinement using phenix.refine (PHENIX; Adams et al., 2010; Emsley et al., 2010). Selenomethionine-substituted structures were refined against an MLHL target function. The native Msps TOG4 structure was extended to 1.65-Å resolution against an ML target function. ch-TOG SeMet TOG4 coordinates were used to solve the native ch-TOG TOG4 structure by the molecular replacement method (AutoMR, PHENIX) and the structure refined as before using an ML target function. The free R used 10% of the data randomly excluded from refinement. Information regarding data statistics, model building, and refinement is presented in Table 1. Electrostatics was calculated using APBS (Baker et al., 2001). Pairwise alignments and rmsd values were calculated using the Dali pairwise alignment server (Hasegawa and Holm, 2009).
Circular dichroism
Circular dichroism (CD) spectra of individual Drosophila TOG domains were collected at room temperature using a Chirascan-plus CD spectrometer (Applied Photophysics, Leatherhead, United Kingdom). The TOG domains (TOG1–5) were diluted to 0.1 mg/ml in 10 mM sodium phosphate buffer (pH 7.5) and 50 mM sodium fluoride. Spectra were taken using a 1-mm–path length cuvette from 260 to 185 nm with a step size of 0.5 nm and 1.25 s/point. A baseline CD spectrum was recorded and subtracted from each spectrum. Spectra were smoothed using Chirascan-plus software. Melts were obtained by monitoring CD at 208 and 222 nm in 1°C step size from 20 to 94°C. Melting curve inflection points were obtained by calculating the second derivative of the melt curves.
Gel filtration
A Superdex 200 10/300 gel filtration column (GE Healthcare) was used to perform tubulin-binding gel filtration assays. The column was equilibrated in running buffer (20 mM 1,4-piperazinediethanesulfonic acid [PIPES], pH 6.5, 200 mM KCl, 2 mM MgCl2, 0.1% β-ME, 50 μM GTP) at 4°C. A 120-μl amount of TOG protein (40 μM) and tubulin (20 μM) was incubated for 25 min at 4°C in running buffer supplemented with 150 μM GTP and loaded onto the gel filtration column at a flow rate of 0.5 ml/min.
Microtubule cosedimentation
Taxol-stabilized microtubules were prepared by diluting tubulin to 20 μM in BRB80 (80 mM PIPES, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid [EGTA], pH 6.8) supplemented with 1 mM GTP and 1 mM dithiothreitol (DTT). Taxol was added stepwise as follows: 1/100 volume of 20 μM Taxol in dimethyl sulfoxide (DMSO) was added, and samples were incubated at 37°C for 5 min. A 1/100 volume of 200 μM Taxol in DMSO was added, and samples were incubated at 37°C for 5 min. A 1/100 volume of 2 mM Taxol in DMSO was added, and samples were incubated at 37°C for 15 min. Msps constructs were exchanged into BRB80, and 10 μM protein was mixed with 10 μM Taxol-stabilized microtubules. The sample was incubated for 20 min at 25°C, layered onto a BRB80 Taxol-glycerol cushion, and centrifuged at 100,000 × g for 30 min at 25°C. SDS–PAGE was used to visualize protein in the supernatant and the pellet.
Light scattering tubulin polymerization assay
Tubulin polymerization was monitored using a SPEX Fluorolog-3 spectrofluorometer (Horiba Jobin Yvon, Edison, NJ) in T-format, high-voltage mode with the excitation and detection wavelengths set at 350 nm and the excitation and emission slits set at 0.5 and 0.75 nm, respectively. The cuvette holder was maintained at 37°C. Clarified tubulin samples (20 μM final concentration) were prepared in the presence or absence of Msps TOG constructs (1 μM final concentration) in polymerization buffer (50 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.6, 3.4 M glycerol, 5 mM DTT, 1 mM EGTA, 5 mM MgSO4, 1 mM GTP) and incubated at 4°C for 10 min before the polymerization assay. Samples of 300 μl were injected into a 4-mm–path length quartz cuvette at room temperature, then immediately placed into the 37°C cuvette holder and scattering recorded at 350 nm in 1-s intervals over 1500 s. Spikes in scattering within the first 100 s were the result of samples equilibrating to 37°C.
Microscopy-based analysis of in vitro tubulin polymerization
Tubulin samples (20 μM) doped with 10% rhodamine-labeled tubulin were prepared in the presence or absence of Msps TOG3-4 (1 nM final concentration) in polymerization buffer (BRB80 supplemented with 10% DMSO and 1 mM GTP). After incubation on ice for 20 min, samples were placed at 37°C for 3 min and then diluted twofold into polymerization buffer supplemented with 20 μM Taxol. Samples were then flowed into acid-washed flow chambers precoated with polylysine and DM1α anti–α-tubulin antibody (Sigma-Aldrich) and incubated for 30 min. Samples were then washed with phosphate-buffered saline (PBS) and fixed with PBS supplemented with 4% paraformaldehyde for 10 min. Chambers were washed with PBS, and Dako fluorescent mounting medium was added. Images were taken using an Eclipse Ti inverted microscope with a 40× oil/numerical aperture (NA) 1.0 objective and a CoolSNAP HQ charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) driven by NIS Elements software (Nikon, Tokyo, Japan). Images were processed with ImageJ (National Institutes of Health, Bethesda, MD) software.
Drosophila S2 cell expression constructs
Msps constructs containing TOGs 1–4 (residues 1–1080) were subcloned using Gateway pENTR DTOPO cloning system (Life Technologies, Carlsbad, CA) into a final zeocin-selective pIZ backbone vector that contained a metallothionein promoter, Gateway (Invitrogen) LR recombination sites, and a COOH-terminal GFP tag. Msps 498–1079 was subcloned into a pMT A vector backbone containing a metallothionein promoter, and a COOH-terminal tRFP (Currie et al., 2011). Mutations were generated using single or multiple rounds of QuikChange mutagenesis (Agilent Technologies). EB1:EB1-tRFP was constructed by cloning ∼1.5 kb of genomic DNA containing the EB1 promoter 5′ of EB1-GFP (Rogers et al., 2002). The Msps TOG1–5 GFP construct and Msps 498–1079 construct have been described previously (Currie et al., 2011).
dsRNA generation
The T7 promoter sequence was attached to the 5′ end of primer sequences specific to the Msps COOH-terminal coding region corresponding to residues 1752–1927 (Msps primer annealing sequences used: forward, 5′-GCCGAAGTTTACAGACCTGC-3′; reverse, 5′-TGTACTTGTGAAATGGGGCA-3′). dsRNA was generated using a T7 RiboMAX in vitro transcription kit (Promega, Madison, WI) as previously described (Rogers and Rogers, 2008).
Cell culture and transfection
S2 cells were grown in Sf-900 media and passed every 3–5 d. Transfections were performed using the Amaxa Nucleofector II transfection system (Lonza, Basel, Switzerland) according to the manufacturer's protocol. Cells were treated with msps dsRNA for 7 d as previously described (Rogers and Rogers, 2008) and construct expression induced 12–18 h before imaging with 80 μM copper sulfate.
Fluorescence microscopy
S2 cells were seeded onto concanavalin A–coated glass-bottom dishes (MatTek, Ashland, MA) 30 min before imaging in Schneiders's medium supplemented with 10% fetal bovine serum. Time-lapse images were collected at room temperature using a 100× oil/NA 1.45 Plan Apochromat objective using a VT-Hawk (VisiTech, Sunderland, UK) two-dimensional array scanning confocal system with an Orca-R2 CCD camera (Hamamatsu Photonics, Hamamatsu, Japan) controlled using VisiTech Vox software. Images were acquired at 3-s frame intervals over 4 min.
EB1-tRFP comet tracking
EB1 comet velocities were obtained from time-lapse movies (Supplemental Movies S1 and S2) using the ImageJ plug-in Manual Tracking (Fabrice Cordelières, Institut Curie, Orsay, France). Single EB1 comets were tracked for their lifetime, and the velocity was calculated using 30-s intervals. Mean 30-s velocities were plotted by box-and-whiskers plot (Prism; GraphPad, La Jolla, CA). All TOG1-4–GFP constructs were induced with equivalent levels of copper sulfate. To measure EB1 comet rates in cells expressing Msps GFP constructs within a common expression range, the average GFP fluorescence intensity per area was calculated for each cell. Only cells with an average GFP fluorescence intensity per area that fell within a normalized range of 0.6–1.0 were analyzed, limiting the relative concentration of Msps constructs in cells examined to a range spanning 1.7-fold above the minimal concentration scored. Statistical significance was determined using an unpaired t test to calculate two-tailed p values.
Coordinate deposition
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (www.rcsb.org/), under accession codes 4QMH (Msps TOG4), 4QMJ (ch-TOG SeMet TOG4, space group P43212), and 4QMI (ch-TOG TOG4, space group P212121).
Supplementary Material
Acknowledgments
We thank Mark Peifer for comments and Ashutosh Tripathy for assistance. This work was supported by National Institutes of Health Grant R01GM094415 and March of Dimes Grant FY11-434 to K.C.S. and National Institutes of Health Grant T32GM008570 to the Program in Molecular and Cellular Biophysics, University of North Carolina at Chapel Hill.
Abbreviations used:
- β-ME
β-mercaptoethanol
- HEAT
Huntingtin elongation factor 2
- protein phosphatase 2A
target of rapamycin 1
- HR
HEAT repeat
- MAP
microtubule-associated protein
- MT
microtubule
- PEG
polyethylene glycol
- rmsd
root-mean-square deviation
- SAD
single-wavelength anomalous diffraction
- SeMet
selenomethionine
- TOG
tumor overexpressed gene
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0501) on June 25, 2014.
REFERENCES
- Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Flor-Parra I, Lal N, Velji H, Chang F. Fission yeast Alp14 is a dose-dependent plus end-tracking microtubule polymerase. Mol Biol Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–362. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, van Breugel M, Harrison SC, Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittle AL, Ohkura H. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 2005;24:1387–1396. doi: 10.1038/sj.emboj.7600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Slep KC. αβ-Tubulin and microtubule-binding assays. Methods Mol Biol. 2011;777:87–97. doi: 10.1007/978-1-61779-252-6_6. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Becker B, Carney B. TOGp regulates microtubule assembly and density during mitosis and contributes to chromosome directional instability. Cell Motil Cytoskeleton. 2009;66:535–545. doi: 10.1002/cm.20359. [DOI] [PubMed] [Google Scholar]
- Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci. 1998;111:1371–1383. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- Cullen CF, Deak P, Glover DM, Ohkura H. Mini spindles: a gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie JD, Stewman S, Schimizzi G, Slep KC, Ma A, Rogers SL. The microtubule lattice and plus-end association of Drosophila Mini spindles is spatially regulated to fine-tune microtubule dynamics. Mol Biol Cell. 2011;22:4343–4361. doi: 10.1091/mbc.E11-06-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Becker BE, Josh Romney S. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int Rev Cytol. 2004;239:179–272. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr Opin Struct Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Hsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E, Wasteneys GO. MOR1, the Arabidopsis thaliana homologue of Xenopus MAP215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. J Cell Sci. 2008;121:4114–4123. doi: 10.1242/jcs.039065. [DOI] [PubMed] [Google Scholar]
- Kerssemakers JW, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Kosco KA, Pearson CG, Maddox PS, Wang PJ, Adams IR, Salmon ED, Bloom K, Huffaker TC. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol Biol Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Kruljac-Letunic A, Caudron-Herger M, Bieling P, Karsenti E. XMAP215-EB1 interaction is required for proper spindle assembly and chromosome segregation in Xenopus egg extract. Mol Biol Cell. 2009;20:2684–2696. doi: 10.1091/mbc.E08-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy DJ, Erickson HP, Aukhil I, Joshi P, Hendrickson WA. Crystallization of a fragment of human fibronectin: introduction of methionine by site-directed mutagenesis to allow phasing via selenomethionine. Proteins. 1994;19:48–54. doi: 10.1002/prot.340190107. [DOI] [PubMed] [Google Scholar]
- Leano JB, Rogers SL, Slep KC. A cryptic TOG domain with a distinct architecture underlies CLASP-dependent bipolar spindle formation. Structure 21, 939–950. 2013 doi: 10.1016/j.str.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner B, Rashbrooke MC, Collings DA, Eng RC, Kawamura E, Whittington AT, Wasteneys GO. The N-terminal TOG domain of Arabidopsis MOR1 modulates affinity for microtubule polymers. J Cell Sci. 2012;125:4812–4821. doi: 10.1242/jcs.107045. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- Li W, Miki T, Watanabe T, Kakeno M, Sugiyama I, Kaibuchi K, Goshima G. EB1 promotes microtubule dynamics by recruiting Sentin in Drosophila cells. J Cell Biol. 2011;193:973–983. doi: 10.1083/jcb.201101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T. GTPgammaS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proc Natl Acad Sci USA. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Grigoriev I, Nogi T, Hamaji T, Cassimeris L, Mimori-Kiyosue Y. Dissecting the nanoscale distributions and functions of microtubule-end-binding proteins EB1 and ch-TOG in interphase HeLa cells. PLoS One. 2012;7:e51442. doi: 10.1371/journal.pone.0051442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology. Charlottesville: University of Virginia; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pecqueur L, Duellberg C, Dreier B, Jiang QY, Wang CG, Pluckthun A, Surrey T, Gigant B, Knossow M. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc Natl Acad Sci USA. 2012;109:12011–12016. doi: 10.1073/pnas.1204129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov AV, Pozniakovsky A, Arnal I, Antony C, Ashford AJ, Kinoshita K, Tournebize R, Hyman AA, Karsenti E. XMAP215 regulates microtubule dynamics through two distinct domains. EMBO J. 2001;20:397–410. doi: 10.1093/emboj/20.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, Jülicher F. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat Cell Biol. 2013;15:1116–1122. doi: 10.1038/ncb2834. [DOI] [PubMed] [Google Scholar]
- Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: Structural studies of alpha beta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci USA. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC. The role of TOG domains in microtubule plus end dynamics. Biochem Soc Trans. 2009;37:1002–1006. doi: 10.1042/BST0371002. [DOI] [PubMed] [Google Scholar]
- Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittle C, Charrasse S, Larroque C, Cassimeris L. The interaction of TOGp with microtubules and tubulin. J Biol Chem. 2000;275:20748–20753. doi: 10.1074/jbc.M002597200. [DOI] [PubMed] [Google Scholar]
- Tournebize R, Popov A, Kinoshita K, Ashford AJ, Rybina S, Pozniakovsky A, Mayer TU, Walczak CE, Karsenti E, Hyman AA. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- van der Vaart B, et al. SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J Cell Biol. 2011;193:1083–1099. doi: 10.1083/jcb.201012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez RJ, Gard DL, Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO. MOR1 is essential for organizing cortical microtubules in plants. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, Howard J. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci USA. 2011;108:2741–2746. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M, Widlund PO, Hyman AA, Howard J. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat Cell Biol. 2013;15:688–693. doi: 10.1038/ncb2744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.