Abstract
Recent evidence points to extra-telomeric, noncanonical roles for telomerase in regulating stem cell function. In this study, human embryonic stem cells (hESCs) were cultured in 20% or 2% O2 microenvironments for up to 5 days and evaluated for telomerase reverse transcriptase (TERT) expression and telomerase activity. Results showed increased cell survival and maintenance of the undifferentiated state with elevated levels of nuclear TERT in 2% O2-cultured hESCs despite no significant difference in telomerase activity compared with their high-O2-cultured counterparts. Pharmacological inhibition of telomerase activity using a synthetic tea catechin resulted in spontaneous hESC differentiation, while telomerase inhibition with a phosphorothioate oligonucleotide telomere mimic did not. Reverse transcription polymerase chain reaction (RT-PCR) analysis revealed variations in transcript levels of full-length and alternate splice variants of TERT in hESCs cultured under varying O2 atmospheres. Steric-blocking of Δα and Δβ hTERT splicing using morpholino oligonucleotides altered the hTERT splicing pattern and rapidly induced spontaneous hESC differentiation that appeared biased toward endomesodermal and neuroectodermal cell fates, respectively. Together, these results suggest that post-transcriptional regulation of TERT under varying O2 microenvironments may help regulate hESC survival, self-renewal, and differentiation capabilities through expression of extra-telomeric telomerase isoforms.
Introduction
Embryonic stem cells (ESCs) can be characterized by their ability to self-renew for extended periods in addition to possessing the capacity to give rise to lineage-restricted cell types through differentiation [1]. ESCs undergo long-term self-renewal due, in part, to the maintenance of telomere length/integrity [2]. Human telomeres contain a six-oligonucleotide repeat sequence (TTAGGG)n that is tandemly reiterated up to 15–20 kb at both ends of every chromosome [3]. A conserved set of proteins interact with telomeric DNA to provide protection against chemical modification and nuclease digestion, as well as to regulate telomere length and structure [4]. Maintenance of these telomeric regions results in enhanced chromosomal stability [5] and helps counteract the loss of terminal-coding DNA sequences that occurs during DNA synthesis [6] that leads cells, including stem cells [7], to senesce/apoptose at a dysfunctional (uncapped) telomere length [8].
Telomere shortening can be overcome by de novo synthesis of telomeric repeats, catalyzed by the multisubunit ribonucleoprotein enzyme telomerase [9]. The telomerase reverse transcriptase (TERT) component binds an RNA component (TERC) that aligns telomerase to the chromosomal ends and acts as a template for the addition of telomeric DNA [10]. High telomerase activity is characteristic of germ line and other tissues with high renewal capacity, cancer cells, and stem cells but not somatic cells [11,12]. Tissues with a high cell turnover, such as skin, bone marrow, intestine, and testis, exhibit progressive tissue atrophy, stem cell depletion, organ system failure, and impaired tissue injury responses in telomerase-deficient mice with critically short or uncapped telomeres [13–15]. Aplastic anemia and dyskeratosis congenita patients, who have mutations in hTERT and/or hTERC components of telomerase, display skin abnormalities and bone marrow failure, the latter resulting from defects in maintaining the hematopoietic stem cell pool [16,17]. Conversely, telomerase (TERT) overexpression extends telomeres, reduces DNA damage signaling and associated checkpoint responses, reactivates proliferation in quiescent cultures, and eliminates degenerative phenotypes across multiple organs [18–20]. Telomerase activation by transgenic [19,20] or pharmacological means [21] can reverse tissue degeneration and increase health span in aged mice. Together, these observations support the hypothesis that telomere length and telomerase activity are determinants for tissue homeostasis and regeneration. Interestingly, overexpression of TERT in the epidermal stem cells of transgenic mice promotes stem cell mobilization simultaneously with increased proliferation, enhanced hair growth, and augmented skin hyperplasia in the absence of telomere length alterations [22], indicating that TERT has noncanonical, extra-telomeric functions as well [20,22,23]. Interestingly, alterations in cell function can be achieved by the overexpression of a catalytically inactive TERT mutant lacking reverse transcriptase function [24–28], highlighting potentially novel extra-telomeric roles in stem cell biology.
Naturally occurring TERT isoforms lacking reverse transcriptase function and thus telomerase activity can arise through the generation of splice variants by exon skipping, intron retention, and alternative usage of splice donor and acceptor sites. To date, 22 alternatively spliced hTERT mRNAs have been reported resulting in several in-frame and out-of-frame TERT variants [29–32]. These hTERT splice variants can lack reverse transcriptase (telomeric) function and their expression can modify telomerase activity levels [33]. These hTERT splice forms include the Δβ variant (deletion of exons 7 and 8), resulting in a truncated, enzymatically inactive telomerase. The Δα variant (employing an alternative splice site in exon 6) is also an enzymatically inactive, dominant inhibitor of telomerase activity when overexpressed [34]. Both variants can combine into an hTERT ΔαΔβ variant [31,32]. The Δγ variant is an in-frame deletion of 189 bp, corresponding to the complete loss of exon 11 within the reverse transcriptase domain of hTERT [31]. These hTERT deletion variants are detected in a number of cancers and tumor cell lines and additionally during development displaying tissue-specific and gestational-age-dependent expression patterns that reduce telomerase activity levels and may influence variations in telomere lengths [35–37]. However, additional efforts have revealed that no correlation exists between the levels of TERT splice variants and the decrease in telomerase activity observed during cellular differentiation [38,39]. Therefore, it is necessary to consider other function(s) for alternatively spliced variants of TERT beyond modulating telomerase activity, as these roles would reveal novel mechanisms of controlling stem cell function [40].
The expression profiles of TERT variants indicate that these splicing events are not random and suggest an important physiological role [36]. It is clear that the functions of telomerase in apoptosis, proliferation, and differentiation are independent of its telomeric role [41]. Functional telomerase has been identified in the cytosol or in mitochondria of cells [42–44]. Extra-telomeric TERT alters stem cell function and tissue homeostasis by acting as a transcriptional cofactor to modulate Wnt signaling [24–26,28]. Since several TERT splice variants are abundantly expressed and individually do not exhibit telomeric function, alternative protein isoforms of TERT, especially those without reverse transcriptase activity, are a likely source of these noncanonical, extra-telomeric functions [44–47]. Our previous data [48] and those of others have demonstrated that overexpression of TERT is associated with increased expression of growth-promoting genes and suppression of growth-inhibitory genes [48–50] that promote a progenitor-like stem cell state [48]. TERT overexpression enhances the proliferation and colony-forming ability of human stem cells but suppresses their in vitro differentiation potential [51]. Conversely, knockdown of all TERT transcripts causes reduced ESC proliferation and loss of pluripotency and induces differentiation, indicating an integral role for TERT isoforms in the maintenance of ESC state [51]. Therefore it is critical to investigate the extra-telomeric roles of TERT isoforms to fully understand their contributions to stem cell self-renewal and differentiation.
Herein, we investigated expression of several TERT splice variants (Δα, Δβ, and ΔαΔβ) and their interplay with telomerase activity levels and stem cell function in human embryonic stem cells (hESCs) cultured under varying O2 microenvironments that either promoted or hindered their proliferation in the undifferentiated state. We also utilized two distinct small molecule inhibitors of telomerase activity and morpholino-induced alteration of TERT pre-mRNA splicing to determine the potential roles of these telomerase isoforms in hESC self-renewal. Our findings indicate that extra-telomeric TERT isoforms are essential effectors through which varying O2 microenvironments regulate self-renewal and pluripotency of hESCs.
Materials and Methods
hESC culture
hESCs (H9 line, passage 25; WiCell, Madison, WI) were cultured in six-well tissue culture plates on a monolayer of CF-1-irradiated mouse embryonic fibroblast (MEF) feeder layers (GlobalStem, Rockville, MD). Irradiated MEFs were plated at a density of 250,000 cells/well of a six-well, gelatin-coated culture dish. Prior to plating of hESCs on MEFs, wells were rinsed with hESC medium composed of knockout Dulbecco's modified Eagle's medium (DMEM/F12), 20% knockout serum replacement (KOSR), 1% nonessential amino acids, 2 mM glutamine (CellGro, Manassas, VA), 0.1 mM 2-mercaptoethanol (Fisher, Toronto, Canada), and 4 ng/mL of basic fibroblast growth factor (bFGF). H9 hESCs were grown in hESC medium in all experiments where MEF feeder cells were utilized. Cultures were incubated at 37°C in 5% CO2 and the medium was changed daily. hESCs were passaged mechanically using a glass pick every 5–7 days as required. All media were passed through 0.22-μm filters for sterilization prior to use. All reagents were obtained from Life Technologies (Grand Island, NY) unless noted otherwise. The experiments performed on the human embryonic stem cell lines were approved by the Stem Cell Oversight Committee (SCOC) of the Canadian Institutes of Health Research (CIHR).
Feeder-free hESC cultures
For immunofluorescence, flow cytometry, RNA extraction, telomeric repeat amplification protocol (TRAP) assay, pharmacological inhibition of TERT and TERC, and morpholino oligonucleotide (MO) treatments, H9 hESCs were cultured in feeder-free conditions to limit transmission of MEF cellular material to the analyzed samples. For feeder-free growth, culture dishes were coated with a 1:30 Matrigel™ (Growth Factor Reduced; BD Biosciences, Franklin Lake, NJ) and DMEM mixture and placed at 37°C to promote gelation. hESCs on Matrigel were cultured using hESC medium (as described in hESC culture section, above) that had been preconditioned on MEF feeder layers (MEF-CM) for 48 h and supplemented with a further 8 ng/mL bFGF [52], or in mTeSR1 hESC medium (StemCell Technologies, Vancouver, Canada). For high- and low-O2 culture experiments, H9 hESCs grown in feeder-free conditions were transferred to a 37°C and 5% CO2 environment containing either 20% or 2% O2, respectively, for up to 5 days. All reagents were obtained from Life Technologies unless noted otherwise.
Immunofluorescence microscopy
H9 hESCs grown in feeder-free conditions in either 20% or 2% O2 conditions were rinsed twice in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (in PBS) for 15 min at room temperature. For intracellular staining, fixed cells were rinsed and subsequently permeabilized with 0.1% Triton X-100 in PBS for 15 min. Before antibody staining, fixed cells were blocked using Dako Serum Free Protein Block solution (Dako, Cambridgeshire, United Kingdom). Oct4 (clone 10H11.2), SSEA4 (clone MC-813-70), and SSEA1 (clone MC-480) primary antibodies were obtained from Millipore (Billerica, MA) and used at a 1:500 dilution. The TERT primary antibody (rabbit anti-human polyclonal; 600-401-252) was obtained from Rockland Immunochemicals (Gilbertsville, PA) and used at a 1:100 dilution. Incubation time for all primary antibodies was 1 h, in PBS containing 0.1% bovine serum albumin, at room temperature. After rinsing, the cells were incubated with secondary Alexa-fluor 488 goat anti-mouse IgG (Life Technologies), Alexa-fluor 568 goat anti-mouse IgM (Life Technologies), or goat anti-rabbit IgG (Calbiochem, San Diego, CA) fluorescein-conjugated antibody at 1:2,000 dilution in PBS containing 0.1% bovine serum albumin. Cells were incubated in secondary antibody solutions for 2 h at room temperature and then rinsed three times with PBS. Microscopy images were immediately obtained while immunostained cells remained in PBS using a Leica DMI6000 B inverted microscope (Leica Microsystems, Inc., Concord, ON, Canada) equipped with MetaMorph® image analysis software (Molecular Devices, Sunnyvale, CA). The mean signal intensity of negative control micrographs (without primary antibody incubation) was subtracted from the measured values of the treatment micrographs to eliminate background fluorescence. The adjusted relative signal intensity values, represented in pixels, were plotted graphically with error bars representing the standard error (SE) of the mean.
Western blot analysis
The specificity of rabbit anti-telomerase catalytic subunit Antibody −600-401-252S (Rockland Immunochemicals) was tested by blotting whole-cell lysates of TET-inducible 293T-hTERT-3XFLAG-overexpressing human embryonic kidney (HEK)293T cells in the presence and absence of 1 μg/mL DOX. Following normalization of total protein using the DC Protein Assay (BioRad, Mississauga, Canada), western blotting was performed with the Novex NuPAGE SDS-PAGE Gel System (Life Technologies) using standard protocols. Blots were incubated overnight at 4°C with the antibody of interest, and for 1 h at room temperature with the respective horseradish peroxidase (HRP) secondary antibody. After blotting with Rockland anti-TERT, membranes were stripped and reprobed with anti-FLAG M2 mouse monoclonal antibody (Sigma-Aldrich, St. Louis, MO). Images were merged in Photoshop to show specificity (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd).
In addition, H9 and CA1 hESCs were grown on geltrex under 2% O2 conditions with daily media changes for 48 h. Cells were then placed in 20% O2 for 2, 24, or 48 h. Western blots were carried out on cells lysed in radio-immunoprecipitation assay with protease and phosphatase inhibitors using the Novex NuPAGE SDS-PAGE Gel System (Life Technologies) using standard protocols. Blots were probed with TERT (Y707) and phospho-TERT (Y707) primary antibodies (Abgent, Inc., San Diego, CA) to measure nonphosphorylated and phosphorylated TERT levels at a 1:1,000 dilution overnight. Donkey anti-rabbit HRP was used as a secondary antibody. β-Actin-HRP (Life Technologies) was used as a loading control and human dermal fibroblasts (HDFs) and differentiated CA1 hESCs were used as TERT-low controls. Blots were quantified by densitometric analysis of expected size bands corresponding to ∼127 kDa using ImageLab and the mean of the levels for CA1 and H9 normalized to β-actin. Significance of mean 20% O2 levels from 2% O2 levels was determined using a t-test.
Flow cytometry
H9 hESCs grown in feeder-free conditions in 20% or 2% O2 were harvested using Accutase® (Life Technologies) to obtain a single-cell suspension. After centrifugation, hESC pellets were resuspended in 5% fetal bovine serum (FBS) in PBS. Alexa-488-conjugated SSEA4 (eBioscience, San Diego, CA) and Alexa-647-conjugated SSEA1 (eBioscience) were added to the suspension based on approximate cell number according to the manufacturer's instructions and incubated for 1 h at 4°C. After rinsing, cells were directly analyzed using an Accuri C6 flow cytometer (BD Biosciences). For TERT experiments, harvested cells were fixed in 4% paraformaldehyde in PBS for 10 min and subsequently permeabilized using 0.1% Triton X-100 in PBS. After rinsing, cells were incubated in telomerase-specific primary antibody (mouse anti-human monoclonal, clone 2C4, 1:500 dilution of manufacturer stock; Abcam, Cambridge, MA). Primary antibody incubations were carried out for 1 h in 5% FBS in PBS at 4°C. After rinsing, cells were incubated with an appropriate secondary (goat anti-mouse polyclonal; Abcam). Representative negative control samples contained isotype controls for the primary antibodies used. Secondary antibodies were incubated for 2 h at 4°C. After rinsing, cells were directly analyzed using an Accuri C6 flow cytometer (BD Biosciences). Populations were gated according to forward and side scatter patterns with filtering for viable cells using 7-aminoactinomycin D (7-AAD) stain added prior to analysis. Data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Polyribosome profiling
H9 hESCs were cultured at 5% CO2 in MEF-conditioned DMEM/F12 KO media in 150-mm culture flasks to 70% confluence. Before polysome harvesting, cycloheximide was added to the medium at a concentration of 0.1 mg/mL. The cells were incubated at 37°C for 3 min. Cells were subsequently washed twice with PBS containing 0.1 mg/mL cycloheximide. Lysis buffer [15 mM Tris-Cl (pH 7.4), 15 mM MgCl2, 0.3 M NaCl, 1% Triton X-100, 0.1 mg/mL cycloheximide, and 200U RNase inhibitor] was added directly to the cells. Extracts were incubated on ice for 10 min and then centrifuged to remove nuclei. The supernatant was placed on a 10%–50% sucrose gradient [15 mM Tris-Cl (pH 7.4), 15 mM MgCl2, 0.3 M NaCl, and 0.1 mg/mL cycloheximide]. Polysomes were sedimented by centrifugation at 35,000 rpm in an SW 41 Ti rotor. Ribosome fractions were collected in 200-μL volumes, analyzed for ribosome content at 254 nm, and then pooled into either light- or heavy-molecular-weight fractions with volumes adjusted to 2 mL with H2O. RNA was extracted as previously described [53] by adding 3 mL of 8 M guanidine-HCl followed by 2 min of vortexing. Subsequently, 5 mL of 100% ethanol was added to each fraction and stored overnight at −20°C. The fractions were then centrifuged at 10,000 rpm for 25 min to collect the RNA that was further purified for hydrolysis probe quantitative reverse transcription polymerase chain reaction (RT-qPCR) using a PerfectPure RNA purification kit (5 PRIME, Inc., Gaithersburg, MD).
Hydrolysis probe RT-qPCR
Hydrolysis probe RT-qPCR was carried out on H9 hESC polyribosome fractions to determine whether hTERT isoforms were present in light and heavy polysomal fractions. Oligonucleotide primers and hydrolysis probes, as described by Mavrogiannou et al. [54], directed against all possible alpha/beta hTERT splicing combinations were purchased through Eurofins MWG Operon (Huntsville, AL) and were as follows: TE1/2 (5′-TCAAGGTGGATGTGACGGG-3′); TER3 (5′-CCTGAGCTGTACTTTGTCAAGGA-3′); KAT4b (5′-GGACTTGCCCCTGATGCG-3′); TER2 (5′-GGCACTGGACGTAGGACGTG-3′); and PROBE (5′-[6-FAM]CGTGTTCTGGGGTTTGATGATGCTGGCGA[BHQ1a-6FAM]-3′).
The X-inactive specific transcript (XIST) Taqman assay Hs01079824_m1 was purchased from Life Technologies. RT-qPCR was carried out with QuantiTect Probe PCR Master-Mix (Qiagen, Venlo, Netherlands) on a CFX-384 in quadruplicate. Samples were run for at least 62 cycles to ensure that no amplification could be detected in the negative control. Unfractionated input was used as a positive control and no-template as a negative control. Data were normalized to the input control for a given primer set according to 2−(Cqsample−Cqinput). Amplified products were further run on 2% agarose gels in order to demonstrate that the amplification was specific (data not shown).
Real-time quantitative telomeric repeat amplification protocol
Telomerase activity was measured by the fluorescent telomeric repeat amplification protocol (TRAP) assay with the TRAPeze® RT Telomerase Detection kit (Chemicon International, Temecula, CA) according to the manufacturer's instructions. Briefly, cultured H9 hESCs were lysed in CHAPS buffer for 30 min on ice and centrifuged at 12,000 g for 20 min at 4°C. Protein concentrations were determined using the BCA protein assay (Pierce, Rockford, IL). Telomerase activity was assayed using 2 μg of protein per reaction, and experiments were performed in triplicate. For all experiments, HEK293T cells were used as a positive control and HDF as a negative control. Telomerase, if present in the cell extract, will add telomeric repeat sequences (TTAGGG) to the 3′ end of an oligonucleotide substrate. The number of repeat sequences added by telomerase is quantified using real-time PCR by measuring the increase in SYBR® green fluorescence upon binding to DNA. The parameters for real-time PCR were as follows: 30 min at 30°C, 2 min at 95°C, followed by 45 cycles of 94°C for 15 s, 59°C for 60 s, and 45°C for 10 s.
Pharmacological inhibition of telomerase activity in hESCs
Telomerase inhibitor IX (TI-IX; Calbiochem) is a cell-permeable, bis-catechol containing m-phenylenediamide compound (MST-312) that inhibits telomerase activity [55,56]. Telomerase inhibitor III (TI-III; Calbiochem) is a cell-permeable, hexameric phosphorothioate oligonucleotide (PS-ODN) compound that acts as a telomere mimic resulting in inhibition of telomerase activity [57]. H9 hESCs were grown on six-well plates in feeder-free conditions in the presence of TI-IX (10 and 20 μM), TI-III [3 and 5 μM; 5′-d(TTAGGG)-3′], or 5 μM control PS-ODNs (5′-TGTGAG-3′ and 5′-TGTGAGTGTGAG-3′; Sigma-Aldrich) in MEF-CM at 37°C under a 5% CO2 atmosphere containing either 20% or 2% O2. hESCs were fed daily with MEF-CM that was supplemented with the corresponding inhibitor for 3 days. Treated hESCs were subsequently assayed using real-time quantitative TRAP (RQ-TRAP), immunofluorescence, and flow cytometry (as described in the above sections).
Antisense morpholino oligonucleotide (MO) blocking of TERT splicing events
Antisense morpholino oligonucleotides (MOs; Gene Tools, LLC, Philomath, OR) complementary to specific exon-intron or intron-exon boundaries of the pre-mRNA sequence of TERT were used to specifically block specific alternative splicing events of TERT. The morpholino sequences were selected on the basis of the manufacturer's recommendations (25 nt antisense). We delivered 3′-FITC-conjugated Δα TERT variant (intron 5/exon 6; 5′-CACATCCACCTGTGTGAGTGGAGGC-3′), Δβ TERT variant (exon 8/intron 8; 5′-CACCTGGCCACCTGACTCACTTGCC-3′), or standard control (5′-CCTCTTACCTCAGTTACAATTTATA-3′) antisense morpholinos using a modified scrape method [58]. Briefly, hESCs cultured on an MEF-feeder layer were picked up mechanically into mTeSR1 medium containing 10 μM of each morpholino (Δα MO and Δβ MO) and then transferred to feeder-free conditions under 5% CO2 in air atmosphere. ESCs were fed daily with mTeSRl that contained 20 μM of each morpholino, for 3 days. Treated hESCs were subsequently processed for RNA extraction and quantification, and assayed by real-time RT-qPCR, immunofluorescence, and flow cytometry (as described in the above sections).
Real-time RT-qPCR
RNA was purified from a single well of cells using TriZOL® reagent according to the manufacturer's instructions (Life Technologies). One microgram of RNA based on NanoDrop (Thermo Scientific, Logan, UT) quantification was reverse transcribed to cDNA using the High Capacity Reverse Transcription Kit with RNase inhibitor (Life Technologies). The reaction was carried out in a BioRad Cl000 Thermal Cycler with the following cycle parameters: 25°C for 10 min, 37°C for 2 h, 85°C for 5 min, and 4°C hold. Real-time PCR was performed using the TaqMan® gene expression assay with predesigned FAM-labeled probes and TaqMan Universal PCR Master Mix (Life Technologies). Primers were obtained from Life Technologies and assay information can be found in Supplementary Table S1. Samples were placed in a Cl000™ Thermal Cycler (BioRad) and incubated at 50°C for 2 min followed by 10 min at 95°C. Samples were then amplified at 95°C for 15 s followed by 1 min at 58°C for 46 cycles. All samples were normalized to the large ribosomal protein RPLPO as an internal control, and fold changes were calculated using the ΔΔCT method [59].
The abundance of specific hTERT RNA isoforms was assessed by RT-PCR using specific intron-spanning primers TERT-2164S (5′-GCCTGAGCTGTACTTTGTCAA-3′) and TERT-2620A (5′-CGCAAACAGCTTGTTCTCCATGTC-3′) that contained both the Δα and Δβ TERT splice sites [39]. TERT splice variant amplification used SYBR green detection and the Universal SYBR Green Master Mix. Reactions were initially heated to 94°C for 90 s, followed by 35 cycles of 95°C for 25 s, 68°C for 50 s, and 72°C for 50 s. TERT PCR products were run on a 2% agarose gel and visualized using SYBR Safe (Invitrogen). The amplicon intensities were acquired by ImageJ for the TERT isoform PCR products and were normalized to the band intensity of the β-actin housekeeping gene transcripts (primers: β-actin-774, 5′-GGGAATTCAAAACTGGAACGGTGAAGG-3′, and β-actin-775, 5′-GGAAGCTTATCAAAGTCCTCGGCCACA-3′) [35] and the relative TERT splice variant transcript abundances were subsequently compared between experimental groups. All experiments were carried out according to the manufacturer's instructions.
Statistical analyses
For all experiments, statistical analyses were performed using Microsoft Excel 2010 or Graph Pad Prism software. Results are expressed as SE of biological replicates as indicated in figure legends. Statistical significance was determined using unpaired Student's t-tests. Parametric data were analyzed by a one-way ANOVA followed by a post hoc Tukey's test, while nonparametric data were analyzed by a two-way ANOVA on ranks followed by Bonferroni post hoc test. Statistically significant data were indicated if the P-value was <0.05 (P<0.05).
Results
Low O2 promotes cell viability and maintenance of the undifferentiated state in hESCs
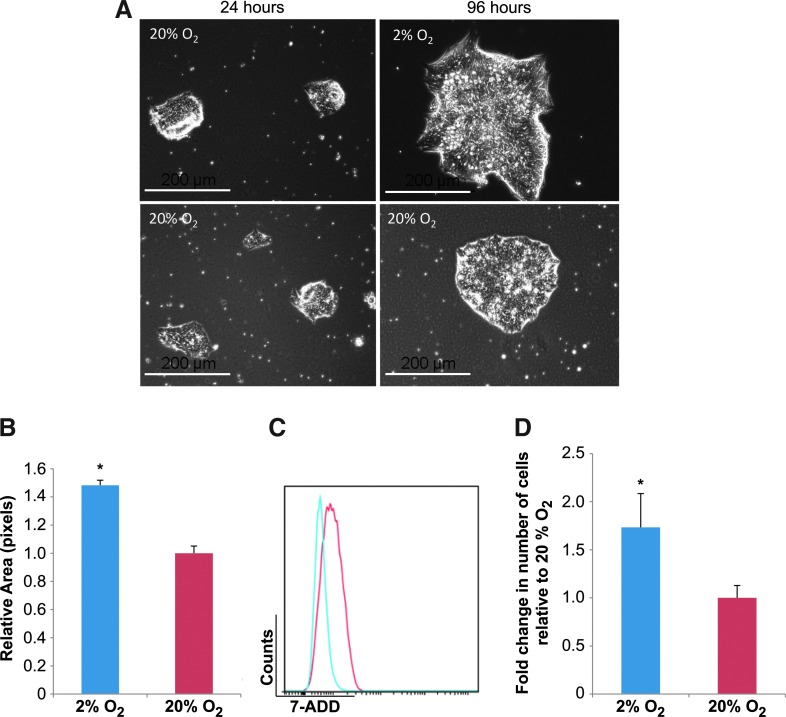
Oxygen availability is one of the most important elements of the microenvironment that has demonstrated roles in mediating both embryonic development and stem cell characteristics [60,61]. To examine the effects of O2 tension on cell survival and maintenance of the undifferentiated cell state, hESCs were cultured for a total of 96 h in feeder-free conditions on Matrigel under an atmosphere that contained either a high (20%) or low (2%) O2 tension. To control for culture variability resulting from colony starting size, cells treated in high- and low-O2 conditions were derived and split from the same population of hESCs. To be consistent in the hESC attachment, they remained in 20% O2 for 24 h after passage to attach under the same conditions (Fig. 1A), and were then either maintained at 20% O2 or moved to 2% O2 culture atmosphere. After 96 h, we observed that hESCs in both O2 conditions displayed uniform colonies of compact cells with sharp edges (Fig. 1A). However, in 20% O2 compared with low-O2 cultures, we observed an increased number of hESC colonies that contained regions of flat cells with elongated morphology consistent with cellular differentiation (Fig. 1A). From measurements of colony area, we observed that hESC colonies in 2% O2 were on average twice as large as those grown in 20% O2 (Fig. 1B). To further investigate the apparent hESC growth rate differences under varying O2 microenvironments, flow cytometry was performed to determine accurate viable cell counts. We observed a decrease in the incidence of cell death, as indicated by 7-AAD staining, in low-O2 cultures compared with their high-O2 counterparts (Fig. 1C, D).
FIG. 1.
Low-O2 microenvironment promotes cell viability of human embryonic stem cells (hESCs). hESCs were grown in feeder-free conditions in low (2%) and high (20%) O2 for a total of 96 h. (A) Phase-contrast microscopy images of H9 hESCs cultured in 20% O2 for the first 24 h to allow similar cell attachment after cell passage and at 96 h (72 h of additional culture) in 20% and 2% O2 conditions. Scale bars=200 μm. (B) Histogram depicting relative colony area measurements of hESCs in 2% and 20% O2. For each O2 condition, three wells from six-well plates were counted for three different experiments. Colonies were selected in each well by computer software (MetaMorph®) and the average intensity graphed. Error bars represent standard error (SE), n=3, *P<0.05. (C) Representative flow cytometry analysis measuring viable cell counts between hESC cultures grown in 2% and 20% O2. Viability was determined using 7-aminoactinomycin D (7-AAD) staining. Cell populations were gated based on 7-AAD exclusion (7-AAD exclude of nonviable cells in flow cytometric assays); graphs show 7-AAD fluorescence for 2% O2-cultured hESCs (blue) relative to hESCs grown in 20% O2 (red). (D) Mean fluorescence intensity plots derived from flow cytometry data representing number of live cells. Error bars represent SE, n=3, *P<0.05. Color images available online at www.liebertpub.com/scd
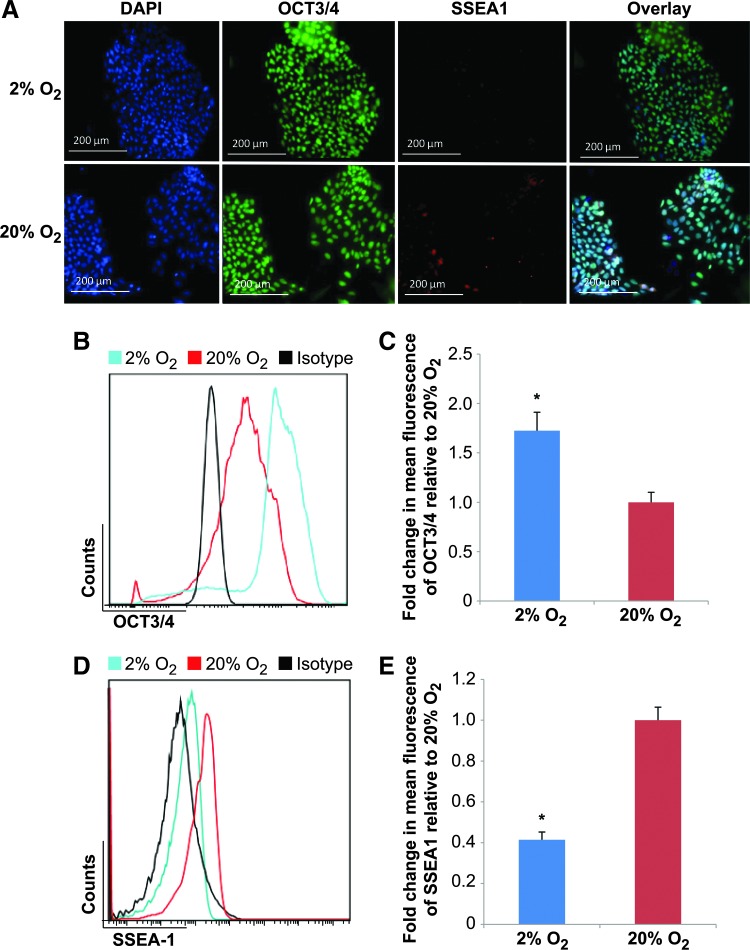
To examine the cellular changes related to pluripotency/differentiation status between hESCs cultured in low- and high-O2 microenvironments, hESCs were examined by immunofluorescence for the levels of pluripotency marker POU5F1 (herein referred to as Oct3/4), and for the cell surface differentiation marker stage-specific embryonic antigen 1 (SSEA1). Oct3/4 is characteristic of undifferentiated hESCs, whereas SSEA1 is typically only found in hESCs that have initiated differentiation [62]. Cultures maintained in both high- and low-O2 conditions contained uniform distributions of Oct3/4 fluorescence within hESC colonies (Fig. 2A). However, hESC colonies grown in 20% O2 conditions contained cells with visible amounts of SSEA1 fluorescence relative to hESCs cultured in 2% O2 (Fig. 2A). Densitometric examination of immunofluorescent signal intensities was performed to quantify the relative fluorescent pixel strength. There was no significant (P>0.05) difference in Oct3/4 fluorescent intensities in hESCs cultured under the two O2 conditions (Supplementary Fig. S2B). However, hESCs grown in 20% O2 contained significantly (P<0.05) higher SSEA1 fluorescent intensities than that of 2% O2 cells (Supplementary Fig. S2A). Flow cytometric analysis verified that the number of SSEA1-expressing cells was increased in high-O2 hESC cultures relative to their low-O2-cultured counterparts (Fig. 2D, E). In addition, flow cytometric data indicated a significant (P<0.05) decrease in Oct3/4 fluorescence in high-O2 hESC cultures compared with those in low O2 (Fig. 2B, C). Taken together, these data indicate that culture of hESCs in a relatively low (2%)–O2 microenvironment promotes cell viability and maintenance of an undifferentiated Oct3/4-positive and SSEA1-negative cellular state.
FIG. 2.
Low-O2 microenvironment promotes maintenance of an undifferentiated hESC state. hESCs were grown in feeder-free conditions in low (2%) and high (20%) O2 for a total of 96 h. (A) Immunocytochemistry detection of Oct3/4 and stage-specific embryonic antigen 1 (SSEA1) in hESCs cultured in 2% or 20% O2 for 96 h. Representative immunofluorecence microscopy images are shown. Scale bars represent 200 μm. Representative flow cytometry data plots representing fluorescence signal for hESC populations harvested from normoxic and hypoxic cultures. Cells were assayed for Oct3/4 and SSEA1. Oct3/4 (B) and SSEA1 (D) fluorescence is shown in 2% (blue) and 20% (red) O2 conditions relative to matched isotype controls (black). Viability was determined using 7-AAD staining. Mean fluorescence intensity plots derived from flow cytometry data representing Oct3/4 (C) and SSEA1 (E) signal intensities of hESCs grown in 2% and 20% O2. Error bars represent SE, n=3, *P<0.05. Color images available online at www.liebertpub.com/scd
Low O2 increases TERT levels and promotes its nuclear localization without significant changes in telomerase activity
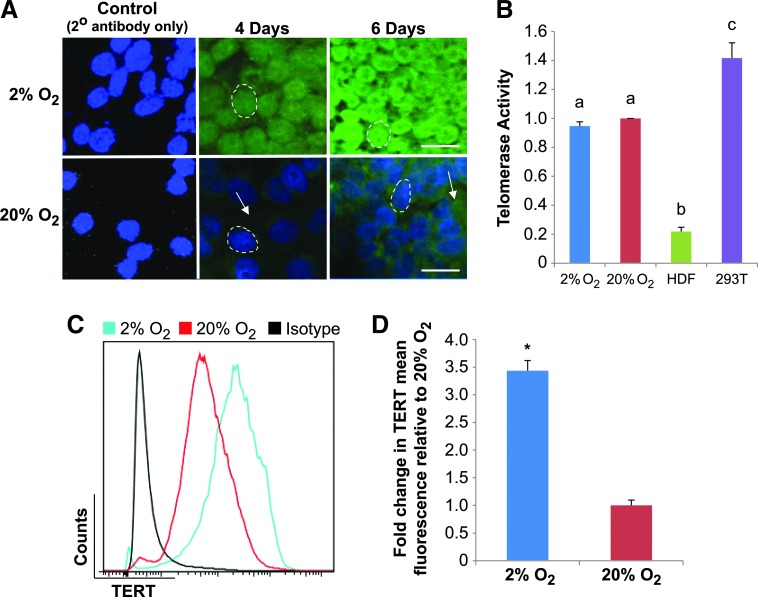
Based on the observation that a low-O2 microenvironment promotes maintenance of an undifferentiated state, we utilized these conditions to investigate the potential molecular mechanisms behind hESC self-renewal and pluripotency. Previous studies have shown that hESCs express TERT and have subsequently high levels of telomerase activity that are reduced upon their spontaneous differentiation [51]. To investigate these observations further, we next sought to determine whether the observed growth and differentiation changes correlated with telomerase activity levels, TERT protein abundance, and TERT localization in hESCs cultured under high and low O2. Immunofluorescence microscopy analyses indicated that TERT protein was elevated and localized predominantly within the nuclei of hESCs when cultured in 2% O2, but displayed a predominantly perinuclear/cytoplasmic localization pattern in cells grown in 20% O2 after both 4 and 6 days of culture (Fig. 3A). Both densitometric analyses of TERT immunofluorescence signals (Supplementary Fig. S3) and flow cytometry indicated a significant (P<0.05) increase in total TERT protein levels in hESCs cultured in 2% O2 compared with those cultured in 20% O2 after 5 days (Fig. 3C, D). In addition, western blot analyses detected elevated levels of total (phosphorylated and nonphosphorylated at Y707) TERT in hESCs grown in 2% O2 that were reduced when the cells were transitioned to 20% O2 conditions (Supplementary Fig. S4). To evaluate whether the observed differences in total TERT protein levels were reflected in the amount of telomerase activity, the RQ-TRAP assay was performed on hESCs cultured in high and low O2 for 5 days. Interestingly, elevated TERT protein in low-O2-cultured hESCs did not result in increased telomerase activity levels, as there was no significant difference (P>0.05) between the hESC groups (Fig. 3B). Overall, these findings indicate that compared with hESCs grown under high-O2 conditions, a low-O2 microenvironment promotes the nuclear localization of elevated TERT levels in hESCs without a significant alteration in telomerase activity levels.
FIG. 3.
Low-O2 microenvironment promotes the nuclear translocation of elevated telomerase reverse transcriptase (TERT) levels in hESCs, despite no significant difference in telomerase activity. hESCs were grown in feeder-free conditions in low (2%) and high (20%) O2 microenvironments to monitor TERT protein abundance and localization, as well as telomerase activity. (A) Representative immunofluorescence images of TERT localization patterns and protein levels in hESCs cultured on Matrigel in 2% and 20% O2. Dashed circles denote single nuclei, and arrows point to areas of extra-nuclear TERT protein abundance within cells. Scale bars=50 μm. (B) Relative telomerase activity in hESCs cultured in 2% or 20% O2 (relative to 20% O2-cultured hESCs). Error bars represent SE, n=3. Letters above histogram bars indicate significant differences (P<0.05). (C) Representative flow cytometric analysis measuring TERT fluorescence levels in H9 hESCs cultured in 20% and 2% O2. TERT fluorescence shown in 2% (blue) and 20% (red) O2 conditions relative to matched isotype controls (black). Viability was determined using 7-AAD staining. (D) Mean fluorescence intensity plots derived from flow cytometry data representing TERT signal of hESCs grown in 2% and 20% O2. Error bars represent SE, n=3, *P<0.05. Color images available online at www.liebertpub.com/scd
Varying effects of pharmacological telomerase activity inhibition on spontaneous hESC differentiation
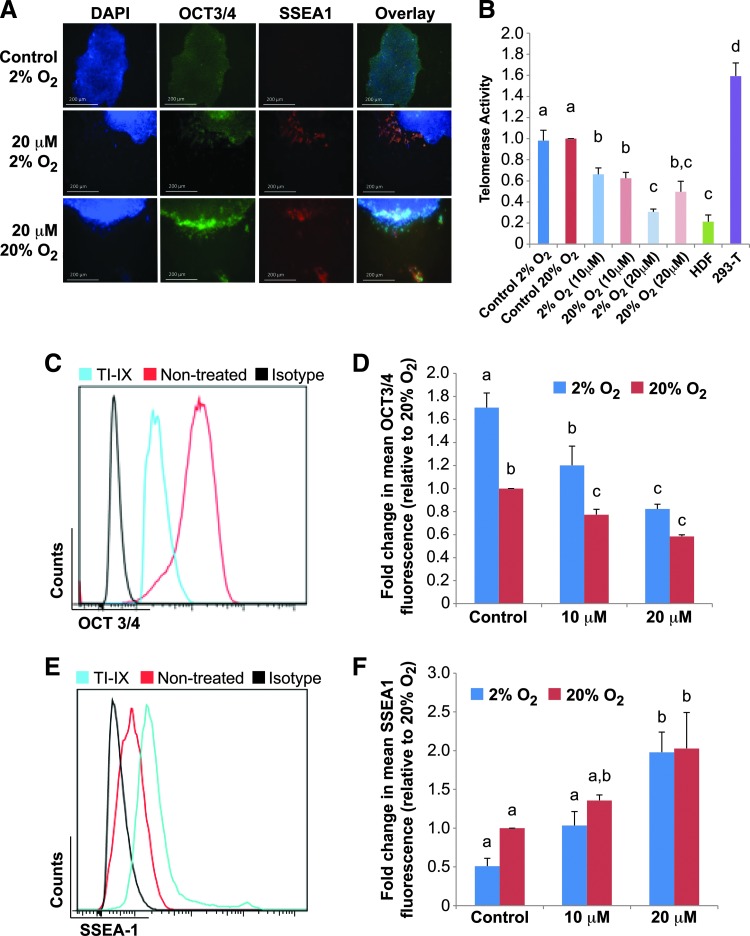
Although previous evidence indicates that hESCs express both TERT and TERC subunit genes [51], recent studies have shown that TERT overexpression in mouse epidermal stem cells activates stem cell division, migration, and self-renewal that is independent of TERC [24]. To build on our data that demonstrate that TERT protein abundance is significantly altered independent of telomerase activity in hESCs grown under different O2 microenvironments, we investigated the potential for additional role(s) of telomerase in hESCs. To undertake this, we utilized the small-molecule telomerase inhibitors TI-IX and TI-III that have different modes of telomerase inhibition [55–57].
Preliminary experimentation and literature sourced concentrations previously used on human cells that effectively inhibited telomerase activity [55–57] indicated that TI-IX and TI-III inhibitors were most effective at nontoxic doses of 20 and 5 μM, respectively, in hESCs cultured in high- and low-O2 conditions (Figs. 4B and 5B and Supplementary Figs. S5 and S6). H9 hESCs were treated with 20 μM TI-IX inhibitor and analyzed by immunofluorescence to detect the levels of pluripotency marker Oct3/4 and differentiation marker SSEA1. In both high- and low-O2 cultures supplemented with TI-IX, we observed noticeable fluorescence for SSEA1 coupled with a decrease in Oct3/4 intensities, indicating that hESCs were spontaneously differentiating (Fig. 4A). These data were further validated through flow cytometry of 20% and 2% O2 hESC cultures in the absence/presence of TI-IX inhibitor at concentrations of 0, 10, and 20 μM. A significant (P<0.05) decrease in Oct3/4 protein abundance was observed at both inhibitor concentrations and O2 concentrations compared with untreated controls (Fig. 4C, D). The significant difference in Oct3/4 protein levels observed between untreated and 10 μM-TI-IX-treated 2% and 20% O2 hESC groups became nonsignificant at the 20 μM TI-IX dose (Fig. 4D). Conversely, SSEA1 levels were significantly (P<0.05) increased in the presence of 20 μM of TI-IX inhibitor compared with untreated controls for both high- and low-O2 hESC cultures (Fig. 4E, F). This data demonstrates that pharmacological inhibition of telomerase activity using a bis-catechol containing m-phenylenediamide compound causes spontaneous hESC differentiation within both high- and low-O2 microenvironments.
FIG. 4.
Pharmacological inhibition of telomerase activity using a synthetic tea catechin induces spontaneous hESC differentiation. H9 hESCs were grown in feeder-free conditions in either 20% or 2% O2 conditions with telomerase inhibitor IX (TI-IX; Calbiochem) for 3 days. (A) Representative immunofluorescence images of Oct3/4 and SSEA1 in hESCs nontreated (control) and treated with 20 μM of TERT inhibitor for 3 days in 2% and 20% O2. Scale bars represent 200 μm. (B) Telomerase activity levels (relative to 20% O2 hESCs) of whole-cell lysates as measured by the real-time quantitative telomeric repeat amplification protocol (RQ-TRAP) assay. Error bars represent SE, n=3. Different letters above the histogram bars indicate significant differences (P<0.05). (C, E) Representative flow cytometry data plots representing fluorescence signal for hESCs treated with 20 μM of TERT inhibitor (TI-IX), and nontreated cells (control) for 3 days in 2% O2. Cells were assayed for Oct3/4 (C) and SSEA1 (E) fluorescence in TI-IX-treated (blue) and nontreated controls (red) relative to matched isotype controls (black). Viability was determined using 7-AAD staining. (D, F) Mean fluorescence intensity plots derived from flow cytometry data representing Oct3/4 and SSEA1 signal of hESCs treated with 10 and 20 μM of TERT inhibitor for 3 days in 2% and 20% O2. Error bars represent SE, n=3. Letters above the histogram bars indicate significant differences (P<0.05). Color images available online at www.liebertpub.com/scd
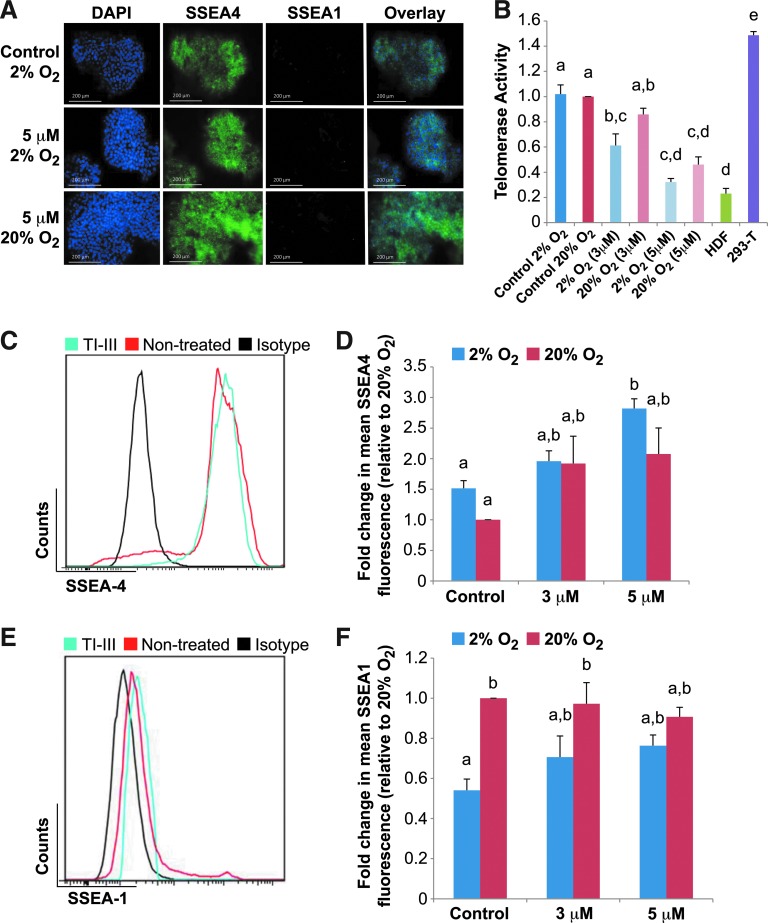
FIG. 5.
Pharmacological inhibition of the telomerase activity using a phosphorothioate oligonucleotide telomere mimic does not cause spontaneous hESC differentiation. H9 hESCs were grown in feeder-free conditions and treated with a small-molecule telomerase inhibitor III (TI-III; Calbiochem). (A) Representative immunofluorescence analysis of SSEA4 and SSEA1 levels in hESCs treated with 5 μM of telomerase RNA component (TERC) inhibitor for 3 days in high (20%) and low (2%) O2. Scale bars=200 μm. (B) Relative telomerase activity levels of whole-cell lysates as measured by the RQ-TRAP assay. Error bars represent SE, n=3. Different letters above the histogram bars indicate significant differences (P<0.05). (C, E) Representative flow cytometry data plots representing fluorescence signal for hESCs treated with 5 μM of TERC inhibitor for 3 days in 2% O2. SSEA4 (C) and SSEA1 (E) fluorescence in TI-III-treated (blue) and nontreated controls (red) relative to isotype control (black). Viability was determined using 7-AAD staining. Mean fluorescence intensity plots derived from flow cytometry data representing SSEA4 (D) and SSEA1 (F) signal intensities, respectively, in hESCs treated with 3 and 5 μM of TERC inhibitor for 3 days in 2% and 20% O2. Error bars represent SE, n=3. Letters above histogram bars indicate significant differences (P<0.05). Color images available online at www.liebertpub.com/scd
We next investigated what effect antagonism of telomerase activity with a small-molecule PS-ODN telomere mimic would have on hESCs cultured under varying O2 conditions. H9 hESCs were treated with 5 μM TI-III and analyzed by immunofluorescence to detect levels of pluripotency marker SSEA4 and the differentiation marker SSEA1 (Fig. 5A). In TI-III-inhibitor-treated hESCs, we observed uniform SSEA4 fluorescence in both high- and low-O2-cultured cells (Fig. 5A). Additionally, we observed no apparent difference in fluorescence signal intensities for SSEA1 in either of the O2 microenvironments after TI-III inhibition (Fig. 5A), although a significant (P<0.05) decrease in telomerase activity levels was observed after a 3-day treatment of hESCs with 5 μM TI-III in both 20% and 2% O2 compared with nontreated controls and compared with PS-ODN-treated hESC controls (Fig. 5B and Supplementary Fig. S7). Similar to the results observed for immunofluorescence, flow cytometry revealed no significant (P>0.05) changes in SSEA4 or SSEA1 abundance in hESCs treated and untreated with the TI-III inhibitor (Fig. 5C, D). Although both pharmacological small-molecule inhibitors can significantly decrease telomerase activity levels, only TI-IX treatment resulted in spontaneous hESC differentiation, whereas TI-III-mediated inhibition did not, suggesting possible extra-telomeric roles for TERT in maintaining the undifferentiated stem cell state.
TERT isoform transcripts are differentially abundant in hESCs cultured under varying O2 atmospheres and are associated with polysomes
Based on the results that demonstrate that a telomere mimic did not induce spontaneous hESC differentiation and that low-O2 conditions elevate total TERT protein levels and promote its nuclear localization without significant changes in telomerase activity levels, we next examined the transcript abundance of several alternatively spliced variants of TERT known not to exhibit telomerase activity [33]. To determine TERT isoform transcript levels in hESCs cultured in 20% and 2% O2 conditions, we performed RT-PCR analysis using intron-spanning primers that will simultaneously detect specific TERT splice variant transcripts [39]. Agarose gel electrophoresis of amplified transcripts revealed that constitutively spliced, full-length (+α+β) TERT as well as the Δα, Δβ, and ΔαΔβ TERT isoforms are expressed in H9 hESCs cultured in both high- and low-O2 conditions (Fig. 6). Interestingly, both full-length TERT and the Δα isoform transcript abundance were significantly (P<0.05) reduced in hESCs cultured in 2% O2 conditions compared with those grown in high 20% O2 (Fig. 6B). The Δβ TERT isoform and the dual-deletion variant (ΔαΔβ TERT isoform) showed no difference in transcript levels between O2 microenvironments; however, the transcript abundance for the ΔαΔβ variant was significantly (P<0.05) less compared with the other TERT splice variants (Fig. 6B).
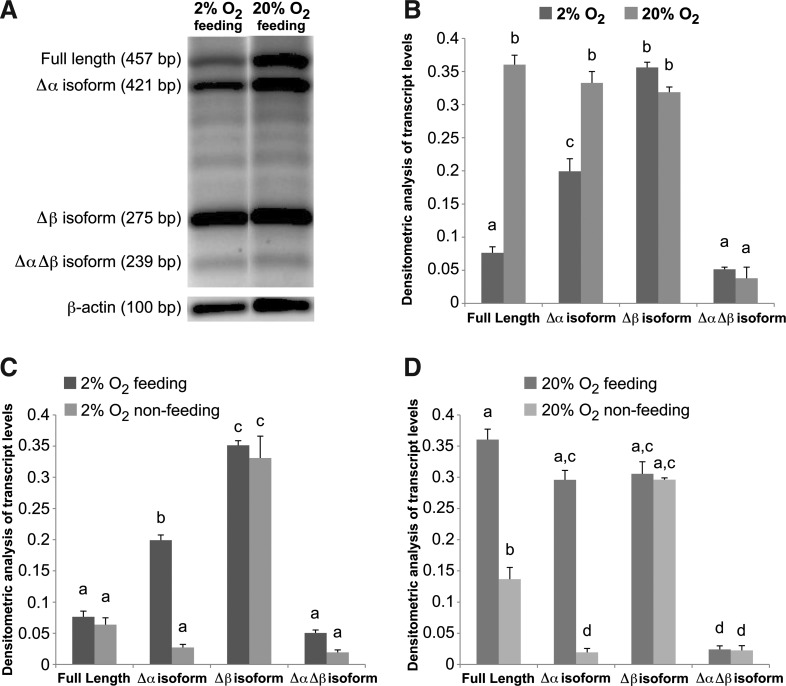
FIG. 6.
Microenvironmental induced alterations in TERT splice variant expression profiles in hESCs cultured in 2% and 20% O2 conditions. hESCs were cultured in feeder-free conditions in either high (20%) or low (2%) O2 and harvested to monitor TERT isoform transcript abundance. (A) A representative agarose gel electrophoresis of mRNA transcript abundance for constitutively spliced (full length) TERT and alternatively spliced TERT variants (Δα, Δβ, and ΔαΔβ) in hESCs grown under 2% and 20% O2 conditions for 5 days. (B–D) Densitometric analysis of splice variant amplicons normalized to β-actin transcript levels. (B) TERT isoform transcript abundance was compared in hESCs cultured under optimal conditions in 2% versus 20% O2 atmospheres. (C, D) TERT isoform transcript abundance was also compared in hESCs cultured under conditions known to maintain (feeding; daily media changes in 2% and 20% O2 for 72 h) or not maintain (nonfeeding; no media changes in 20% O2 for 72 h) hESC proliferation in the undifferentiated cell state. Error bars represent SE, n=3. Letters above the histogram bars indicate significant differences (P<0.05).
To further delineate the microenvironmental effect on TERT isoform abundance in hESCs, we employed a culture model that reproducibly induces stem cell differentiation at a pinpoint time before overt differentiation when hESCs are making critical cell fate decisions. Studies in our laboratories indicate that hESCs proliferate in all O2 conditions (1%–21%), and that in the absence of media changes (“nonfeeding” conditions), hESCs cultured for 72 h in 21% O2 begin to spontaneously differentiate, whereas hESCs cultured for the same period in 1%–2% O2 remain undifferentiated (Postovit Lab, unpublished data). Using this system we observe that the transcript abundance for both the Δβ and ΔαΔβ TERT isoforms does not change, whereas the Δα TERT isoform transcript levels decrease from feeding to nonfeeding conditions at both O2 tensions (Fig. 6C, D). Interestingly, the transcript abundance for full-length, constitutively spliced TERT, although higher in 20% O2-cultured cells, significantly decreased under nonfeeding conditions, whereas no differences were observed for the lower abundant full-length TERT in hESCs grown under 2% O2 between feeding and nonfeeding regimens (Fig. 6C, D).
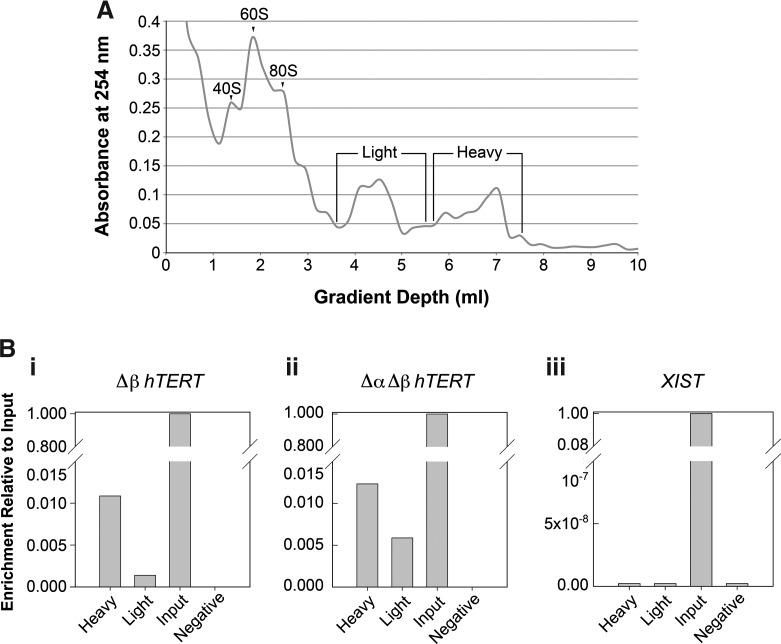
To determine whether these TERT splice variants are translated in hESCs, we assessed their association with polyribosomes in H9 hESCs (Fig. 7). We used a polyribosome profiling approach that separates mRNAs based on the number of ribosomes they bind by sucrose gradient centrifugation [53]. This method allows separation of poorly and efficiently translated mRNAs by their association with light and heavy polyribosome fractions, respectively. Levels of the Δβ and ΔαΔβ hTERT splice variants were evaluated in each fraction by RT-qPCR and compared to the levels of XIST RNA, a well-known nontranslated RNA. In contrast to XIST, which was not associated to either light or heavy polyribosome fractions (Fig. 7Biii), both Δβ (Fig. 7Bi) and Δα Δβ (Fig. 7Bii) hTERT splice variant mRNA transcripts were associated with both light and heavy polyribosome fractions. The Δα hTERT splice variant was not detected in either light or heavy polyribosome fractions isolated from hESCs (data not shown). These results provide strong support that the Δβ and ΔαΔβ hTERT splice variants are translated into proteins within hESCs. Based on the differential expression patterns and evidence of their translation, we next sought to determine isoform-specific roles for the TERT splice variants in hESCs.
FIG. 7.
hTERT splice variant transcripts are associated with polyribosomes in hESCs. (A) Absorbance profile for ribosomes separated on a 10%–50% sucrose gradient. The 40S, 60S, and 80S ribosome subunit peaks and fractions containing light and heavy polysomal peaks are indicated. (B) Relative enrichment of (i) Δβ hTERT isoform, (ii) ΔαΔβ hTERT isoform, and (iii) the nonprotein coding X-inactive specific transcript (XIST) mRNAs relative to input measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR) in the light and heavy polysome fractions and in whole-cell hESC lysates (Input) is plotted (n=2, r=3). Negative, water control samples.
Steric blocking of TERT alternative splicing induces spontaneous hESC differentiation
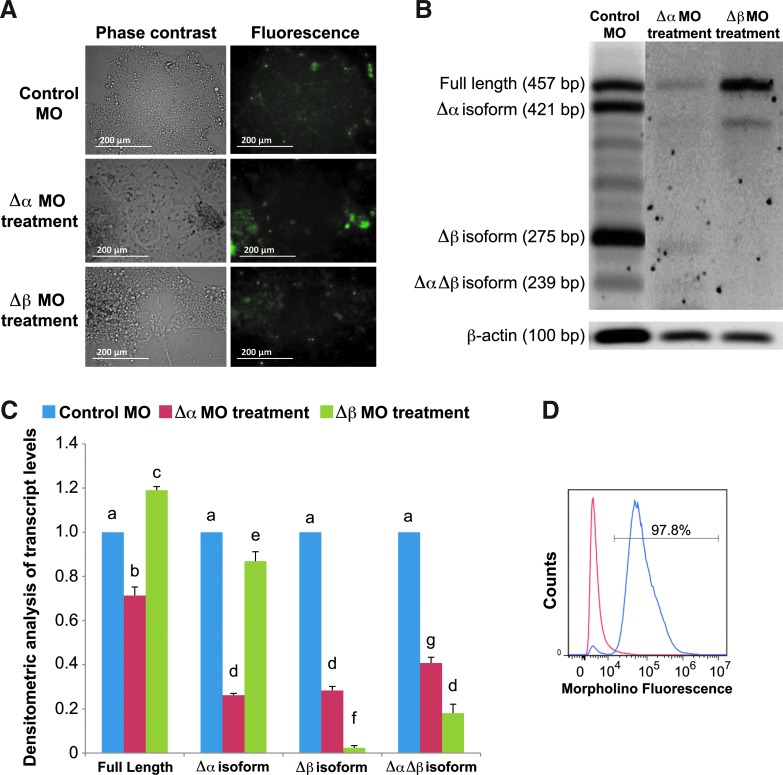
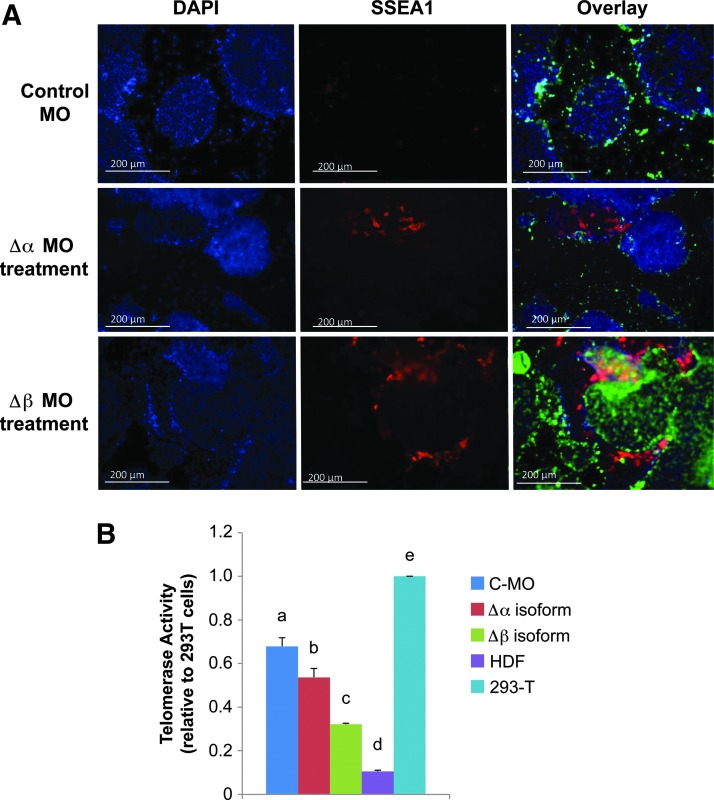
To reveal the potential biological roles for these specific TERT splice variants in hESCs, steric-blocking Morpholino antisense oligonucleotides (MO) were utilized as described previously [63]. In this study MO were designed with a sequence complementary to the Δα (intron 5/exon 6) and Δβ (exon 8/intron 8) boundaries of the TERT pre-mRNA molecule and were 3′-fluorescently tagged to enable visualization and quantification of intracellular uptake. Using immunofluorescence microscopy, we observed that MO specific for Δα and Δβ TERT isoforms, as well as the standard scrambled control MO, were effectively transfected into hESCs (Fig. 8A). Flow cytometric analysis of the control MO in a culture of H9 hESCs showed a very high (97.8%) transfection efficiency that was consistently similar among all the groups (Fig. 8D). To confirm the specificity and effectiveness of the TERT MO, RT-PCR analysis was performed to measure full-length (+α+β) TERT and alternatively spliced TERT isoform transcript levels. cDNA derived from H9 hESCs treated with either Δα or Δβ MOs revealed a significant (P<0.05) decrease in splice variant transcript levels in the presence of each TERT splice variant MO compared with hESCs treated with the standard scrambled MO (Fig. 8B, C). We also observed a significant (P<0.05) decrease of full-length TERT in Δα-MO-treated hESCs but not for Δβ-MO-treated hESCs (Fig. 8B, C), where full-length TERT transcripts were significantly (P<0.05) higher in abundance. Surprisingly, we observed a significant (P<0.05) reduction in telomerase activity levels in the presence of either Δα or Δβ MO, with the Δβ-MO-treated hESCs displaying a much greater decrease in telomerase activity than the Δα-MO-treated cells compared with hESCs exposed to the control MO (Fig. 9B).
FIG. 8.
Steric-blocking morpholino antisense oligonucleotides alter TERT splicing events in hESCs. hESCs were cultured in feeder-free conditions under high (20%) O2 conditions in the presence of 10 μM of antisense morpholino oligonucleotides (MO). hESCs were passaged in the presence of MOs to facilitate uptake. (A) Fluorescently labeled MO as observed by live cell fluorescence microscopy. MO appears as green fluorescence within hESCs. (B) A representative agarose gel electrophoresis of PCR-amplified cDNA samples showing a decrease in transcript abundance of each specific TERT following treatment of either Δα or Δβ TERT splice variant MO compared with standard control-MO-treated hESCs (10 μM MO for 72 h). (C) Densitometry analysis of TERT isoform expression in the presence of each MO. Error bars represent SE, n=3. Letters above histogram bars indicate significant differences (P<0.05). (D) A representative flow cytometry analysis of hESCs carrying scrambled control MO. Percentage within the bar indicates the number of cells positive for MO fluorescence. Color images available online at www.liebertpub.com/scd
FIG. 9.
Morpholino oligonucleotide blocking of TERT splicing induces spontaneous hESC differentiation. hESCs were cultured in feeder-free and 20% O2 conditions in the presence of 10 μM of morpholino oligonucleotides (MO). hESCs were passaged in the presence of MO to facilitate uptake. (A) Representative immunofluorescence images showing the detection of SSEA1 (red fluorescence) in hESCs treated with 3′-fluorescein-labeled morpholino oligonucleotides: standard control morpholino (Control MO) and Δα and Δβ TERT MO (green fluorescence in the overlay). Scale bars represent 200 μm. (B) Relative telomerase activity levels of whole-cell lysates as measured by the RQ-TRAP assay after either control morpholino (C-MO), Δα isoform MO, or Δβ isoform MO. HDFs, human diploid fibroblasts. Error bars represent SE, n=3. Letters above histogram bars indicate significant differences (P<0.05). Color images available online at www.liebertpub.com/scd
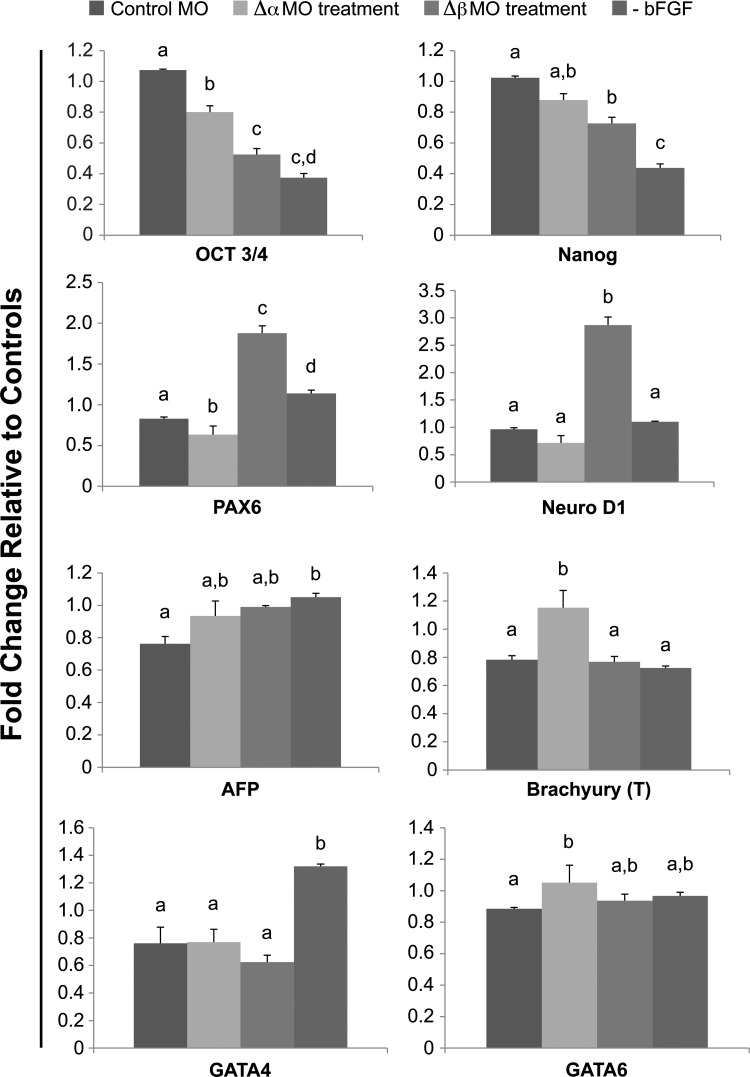
To determine the effect(s) of steric-blocking specific alternative splicing events of TERT on the pluripotent stem cell state, we assayed MO-treated hESCs for SSEA1 expression. There was an apparent increase in SSEA1 fluorescence in the presence of either Δα- or Δβ-isoform-specific MOs but not for hESCs treated with the standard control MO (Fig. 9A). These initial experiments suggested that blocking/altering the alternative splicing events of TERT pre-RNA induces hESC differentiation. hTERT splice variants have been detected during development displaying nonrandom, tissue-specific expression patterns that influence telomerase activity levels during differentiation [36]. To further investigate TERT-splice-variant-dependent regulation of hESC differentiation, MO-treated hESCs were assayed for a panel of lineage-specific markers using real-time quantitative RT-qPCR (Fig. 10). hESCs cultured in the absence of bFGF for 72 h to induce spontaneous differentiation were used as a differentiation control. In the presence of both the Δα- and Δβ-TERT-isoform-specific MO, there was a significant (P<0.05) decrease in Oct3/4 levels, while Nanog transcript levels were significantly decreased in Δβ-MO-treated cells only (Fig. 10). Examining germ-layer-specific markers for ectoderm differentiation, we observed a significant (P<0.05) increase in transcripts for neurogenic differentiation factor 1 (NeuroD1) and paired box 6 (Pax6) only in hESCs treated with Δβ MO (Fig. 10). There were also significant (P<0.05) increases in transcripts for markers representative of endoderm (GATA6) and mesoderm (Brachyury) lineages in the Δα-MO-treated hESCs but not for their Δβ-MO-treated counterparts (Fig. 10). Taken together, these results verify that specific MO inhibition of specific TERT isoforms can promote spontaneous hESC differentiation that appears to be biased toward particular lineages.
FIG. 10.
TERT morpholinos induce spontaneous hESC differentiation that appears to be lineage biased. hESCs grown in feeder-free conditions were assayed for lineage-specific expression after exposure to steric-blocking morpholino oligonucleotides (MOs) specific for alternative splice sites of TERT pre-mRNA. hESCs were passaged in the presence of MO to facilitate uptake. Controls were hESCs grown in medium containing no basic fibroblast growth factor (−bFGF) to induce spontaneous differentiation. Real-time RT-qPCR analysis of hESCs treated with 10 μM of either Δα MO, Δβ MO, or standard control MO for 3 days. Graphs show real-time RT-PCR analysis for various markers: pluripotency (Oct3/4 and Nanog), ectoderm [neurogenic differentiation factor 1 (NeuroD1) and paired box 6 (PAX6)], endoderm [GATA6, GATA4, and alpha-fetoprotein (AFP)], and mesoderm (T and Brachyury). All samples were normalized to the large ribosomal protein (RPLPO) as an internal control, and fold changes calculated using the delta-delta-CT method. Error bars represent SE, n=3. Letters above histogram bars indicate significant differences (P<0.05).
Discussion
The interactions between hESCs and their surrounding microenvironment are critical for the regulation of inter- and intracellular processes, where subtle changes in this niche can induce dramatic alterations in hESC phenotype [64]. Herein, we have shown that a low (2%)–O2 atmosphere induced nuclear localization of elevated TERT levels that correlated with hESC proliferation/cell survival in the undifferentiated state, without significantly increasing telomerase activity levels. Pharmacological perturbation of telomerase activity levels using a synthetic analogue of a major tea catechin promoted spontaneous hESC differentiation, whereas telomerase inhibition by a telomere mimic did not. Expression profiling of a number of alternately spliced variants of TERT that lack reverse transcription function displayed dynamic changes in transcript abundance under varying O2 tensions and in conditions that promote/impede proliferation in undifferentiated cell state. Further, steric blocking of TERT splicing events induced spontaneous hESC differentiation that appears to be lineage biased. Together, our findings suggest a microenvironmental regulation of hESC potency and self-renewal that is modulated, in part, by TERT isoform functions that are beyond telomerase's typical role in telomere synthesis or maintenance.
Our results reveal that a low (2%)–O2 culture atmosphere increases hESC survival and enhances maintenance of an Oct3/4-positive, SSEA1-negative undifferentiated hESC state compared with hESCs cultured under high (20%)–O2 conditions. This is in support of previous studies that have shown that low-O2 tension/hypoxia maintains the undifferentiated states of various stem cell populations and also influences their proliferation and cell-fate commitments [65]. Similar to our findings, constant low-O2/hypoxic culturing allows long-term propagation of hESCs without spontaneous differentiation [66–69]. Likewise, hypoxia induces the return of committed cells back to a pluripotent state [70] and facilitates the generation of induced pluripotent stem cells [71]. These beneficial effects of low O2/hypoxia are due, in part, to the expression/stabilization of hypoxia inducible factors (HIFs) in pluripotent cells grown under low-O2 conditions [67]. However, negative effects of hypoxia on self-renewal have also been observed in mouse ESCs [72,73] and in hESCs cultured with a short splitting interval at low-O2 tensions [74]. Along with inherent genetic variation between individual ESC lines [75], differences in cellular densities, O2 consumption (metabolic) rates, and even medium heights [76] influence the O2 levels encountered by cells at different stages of pluripotency that ultimately determine the distinct proliferation and differentiation capabilities of ESCs under hypoxic conditions [77]. A multifactorial approach that examines hESCs over a range of physiological and nonphysiological O2 tensions with activation/inhibition of various signaling pathways will further define the downstream mechanisms by which O2 tension mediates ESC self-renewal and pluripotency [78].
A number of O2-sensitive transcriptional programs have been identified in hESCs [79,80]. HIFs regulate pluripotency and proliferation in hESCs [67] and HIF-1α in particular has been identified as critical for telomerase function in murine ESCs [81]. Interestingly, both telomerase subunit (TERT and TERC) gene promoter sequences contain hypoxia response element consensus sites, which through HIF-1 transcription factors mediate upregulation of TERT under hypoxic conditions [82,83]. Previous studies have shown that low-O2/hypoxia microenvironments induce telomerase/TERT gene expression [81,82,84,85]. Conversely, oxidant-mediated phosphorylation of TERT (Y707) triggers nuclear export of TERT into the cytoplasm/mitochondria, where it protects mitochondrial function but reduces the anti-apoptotic activity of TERT [86–88]. We have observed TERT phosphorylation and its nuclear export when hESCs are moved from low- to high-O2 microenvironments and after hydrogen peroxide treatments (Teichroeb and Betts, unpublished data). These reports support our observations that low-O2 culture promotes elevation of nuclear TERT levels in hESCs. Although low-O2 culture increased total TERT protein levels, surprisingly no change in telomerase activity was observed. Although this could be due to the poor ability of the CHAPS buffer in the TRAP assay to lyse nuclei, the immunolocalization changes and alterations of total TERT protein levels in hESCs cultured under high- and low-O2 conditions are compelling and warrant additional study. Future subcellular protein fractionation studies will quantify the dynamic movements of TERT localization in hESCs grown under varying culture conditions.
To further investigate the relationship between telomerase function and O2 microenvironment in the context of hESC self-renewal and differentiation, we utilized two small-molecule inhibitors of telomerase activity. Although both molecules significantly decreased telomerase activity in hESCs to similar levels, only the synthetic tea catechin (TI-IX) prompted spontaneous hESC differentiation in both high- and low-O2 microenvironments. In contrast, pharmacological inhibition of telomerase by the hexameric PS-ODN telomere mimic (TI-III) was unable to promote any apparent cellular differentiation of hESCs. Induction of differentiation upon telomerase inhibition by TI-IX occurred within a matter of days, and is therefore not due to critical shortening of telomeres [27,89]. Interestingly, TI-III-mediated telomerase inhibition in hESCs appeared to improve stem-cell-like phenotype compared with nontreated hESC controls. Previously, overexpression of TERT in hESCs increased telomerase activity, enhanced their proliferation and colony forming abilities, and suppressed their in vitro differentiation capabilities [51]. Conversely, knockdown of total TERT levels in hESCs induced their spontaneous differentiation, reduced their proliferation, and caused them to lose pluripotency [51]. However, these modulations of TERT/telomerase levels were carried out in the presence of the TERC [51]. Unlike mTERT−/− mESCs that remain proliferating in the undifferentiated state even with short telomeres [27], inhibition of TERT function by pharmacological means (our study) or siRNAs [51] induces hESCs to undergo rapid and spontaneous differentiation. The species differences in the lack of TERT may be attributed to differing naive and primed pluripotent states; however, previous studies have demonstrated that human and mice cells differ in their telomere signaling pathways [90]. Although the specific mechanism(s) of telomerase inhibition for the synthetic tea catechin (TI-IX) is not known, treatment of cells with TI-IX results in both acute telomere uncapping-mediated cell cycle arrest and long-term telomere erosion [91] that may be mediated by its effects on DNA substrate binding of telomerase [56]. However, many catechols do exhibit antioxidant and/or oxidant properties [92], so TI-IX effects through alteration in cellular redox status cannot be ruled out in our study. In contrast, TI-III is a nuclease-resistant PS-ODN telomere mimic that would sequester telomerase away from the chromosome ends [93] potentially enhancing TERT's ability to act in an extra-telomeric fashion. Our observation of increased levels of the pluripotent marker SSEA4 after TI-III treatment of hESCs supports this hypothesis. Although telomerase activity was inhibited to a similar degree by both small molecules, TI-IX (MST-312) is a reversible inhibitor and thus its effect on inhibiting telomerase levels might have been underestimated by the TRAP assay. This possibility suggests that the different effects observed for each telomerase inhibitor on hESC function could be correlated to varying inhibition levels of total telomerase activity. Although we did not observe significant cell death upon exposure of hESCs to either of the small-molecule inhibitors (Supplementary Fig. S6), hESCs treated with higher doses of TI-III still did not appear to differentiate but seemed to undergo apoptosis and/or senescence (Supplementary Fig. S6). Future studies comparing inducible shRNAi knockdowns of total hTERT and hTERC levels in hESCs and examining telomere dynamics (ie, telomere uncapping) and various extra-telomeric pathways will help decipher the noncanonical function(s) of telomerase in regulating stem cell self-renewal and potency.
Our findings highlight the potential for microenvironmental regulation of hESC function based, in part, on the expression of different TERT isoforms. Increasing evidence points to alternative splicing in controlling regulatory programs for stem cell maintenance and differentiation [94,95]. Alternate splicing provides an important source of protein diversity resulting in alterations or loss of specific function(s), changes in protein localization, or even gain of novel unexpected role(s) [96,97]. Multiple in-frame and out-of-frame alternate splice variants of TERT have been identified [97], many of which lack part of or the entire reverse transcriptase domain, and are expressed in human oocytes and early embryos [37]. Many of these out-of-frame splice variants with a premature termination codon (PTC) are expected to undergo nonsense-mediated decay (NMD) [98]. In this study we investigated characteristics of the most well-known TERT deletion variants (Δα, Δβ, and ΔαΔβ) that lack telomerase activity due to partial or full deletion of the reverse transcriptase domain. The dynamic changes in TERT isoform transcript abundance we observed in hESCs cultured under varying O2 conditions and in microenvironments that maintain or impede proliferation in the undifferentiated state suggest that either the levels of specific TERT isoforms or the relative proportions of the entire TERT isoform expression profile are important for hESC self-renewal and differentiation. Although the mechanism and function of TERT alternative splicing is presently vague [99], the pattern of TERT splice variants correlates with loss of telomerase activity during development [35] and tumor grade [100], suggesting that differential splicing may be a means of regulating TERT gene expression without altering overall transcription levels under certain microenvironments. Interestingly, TERT appears to be regulated by alternative splicing under hypoxic conditions, involving a switch in the splice pattern in favor of the active full-length isoform in ovarian carcinoma cells [83].
Similar to recent studies [46,101], we utilized polyribosome profiling to provide evidence that hTERT splice variants are actively translated within hESCs since commercial antibodies are unreliable in their detection of endogenous levels of specific hTERT isoforms [102]. Blackburn's group recently demonstrated that the Δβ splice variant escapes NMD and is associated with polyribosomes in various human cancer cell lines [46]. We detected both the Δβ and ΔαΔβ hTERT transcripts within light and heavy polyribosome fractions isolated from hESCs. Our inability to detect Δα hTERT spliced transcripts in polyribosomal fractions was probably due to its much lower relative abundance in hESCs. These results further demonstrate that some alternatively spliced TERT mRNAs that contain a PTC (eg, Δβ-containing hTERT isoforms) may not be targeted to NMD and are actually translated [46,101]. The production and application of hTERT-isoform-specific antibodies and the use of polyribosome profiling of hTERT splice variant expression in hESCs grown under varying microenvironments and treatments will help clarify the functional role(s) of TERT isoforms in regulating stem cell function.
As the first step in elucidating their functions we utilized steric-blocking antisense morpholino oligonucleotides (MO) to specifically block hTERT pre-mRNA splicing events and thereby force the expression of altered hTERT transcripts [103–105]. After optimizing MO delivery into hESCs we observed effective steric-blocking of Δα, Δβ, and ΔαΔβ hTERT splice variant transcripts that were associated with their spontaneous differentiation and telomerase downregulation. hTERT splicing has been previously modulated in human prostate carcinoma cells using an oligomer-mediated approach that decreased full-length hTERT transcripts with a concomitant increase in the alternatively spliced transcripts, resulting in significant telomerase activity inhibition that reduced cell growth and induced apoptotic effects that were independent of telomere shortening [106]. Steric-blocking MO in addition to blocking specific splicing events can, at the same time, increase the splicing events (deletions and insertions) at other sites along the same pre-mRNA molecule [103–105]. This would explain the reduction in multiple TERT splice variant transcripts and/or the increase in full-length (+α+β) TERT transcripts after a single MO treatment. These unknown MO-induced alternative-splicing events could confound the interpretation of our results. If the upregulated hTERT splice variants exhibit altered reverse transcriptase function and/or variable TERC binding abilities, then telomerase activity levels would be impacted as well. We are currently conducting hydrolysis-probe-based RT-qPCR to better quantitate the transcript abundance of various hTERT splice variants and to map out the entire hTERT isoform transcriptome of hESCs and rarely find hTERT mRNA containing just a single alternative splice event (Betts, unpublished data). Interestingly, our lineage marker analysis of TERT MO-treated hESCs pointed to a biased differentiation toward endomesodermal and neuroectodermal lineages after MO-induced blocking of the Δα and Δβ hTERT splice variants, respectively. Preliminary studies that examine early lineage commitment in hESCs have observed a nonrandom expression pattern for various hTERT splice variants (Betts, unpublished data). Although our observations could be correlated to total telomerase activity levels as opposed to specific effects on noncanonical TERT activities, a previous study that overexpressed full-length TERT in mESCs showed enhanced differentiation toward the hematopoietic lineage [49] and early work in our lab that overexpresses specific hTERT splice variants as hESCs differentiate display enriched differentiation toward specific cell lineages (Betts, unpublished data). Future studies that knock down specific TERT isoforms and overexpress individual or multiple TERT variants in hESCs during prolonged differentiation regimes in vitro and in vivo will further clarify the precise roles of specific hTERT isoforms during cell fate determination.
Our findings support a mechanistic link between telomerase splice variant expression and stem cell behavior that is regulated by the surrounding O2 microenvironment. Extra-telomeric TERT isoforms, especially those without reverse transcriptase ability, potentially function through Wnt/β-catenin signaling, which is a known O2-regulated pathway in stem cells [107]. Full-length, constitutively spliced TERT modulates Wnt signaling without its telomeric ability [25]. TERT splice variants are expressed in cancer as well during development influencing telomerase activity levels and subsequently telomere length [36,37,108]. However, it is possible that naturally occurring TERT splice variants are not mere bystanders that modulate telomerase activity but are active participants in dictating cellular behavior in an extra-telomeric fashion. Recently, examination of the Δβ deletion has shown that this TERT variant is localized to both the nucleus and mitochondria and confers a growth advantage to cancer cells that is independent of telomere maintenance role [46]. Also, a newly identified human TERT variant containing an in-frame deletion of exons 4–13 (removal of the catalytic domain) stimulates cell proliferation in various telomerase-negative and -positive cell lines [30]. Further examination of alternative splice variants of TERT in various stem and cancer cell populations will help elucidate the various noncanonical roles of TERT isoforms in mediating progenitor/stem cell function and what role they might play in the progression of cancer phenotype under changing microenvironments.
Supplementary Material
Acknowledgments
This research was supported by operating grant to D.H.B. from the Canadian Institutes of Health Research (CIHR; MOP-86453), and grants from the Natural Sciences and Engineering Research Council of Canada (NSERC; 250191–2007), Schulich Gap B funding, and a Children's Health Research Institute (CHRI) Internal Research Fund to D.H.B.
Author Disclosure Statement
The authors declare no commercial associations that might create a conflict of interest in connection with this article. No competing financial interests exist.
References
- 1.Ng HH. and Surani MA. (2011). The transcriptional and signalling networks of pluripotency. Nat Cell Biol 13:490–496 [DOI] [PubMed] [Google Scholar]
- 2.Zeng X. (2007). Human embryonic stem cells: mechanisms to escape replicative senescence?. Stem Cell Rev 3:270–279 [DOI] [PubMed] [Google Scholar]
- 3.Allshire RC, Dempster M. and Hastie ND. (1989). Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res 17:4611–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Steensel B. and de Lange T. (1997). Control of telomere length by the human telomeric protein TRF1. Nature 385:740–743 [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Blasco MA. and Keefe DL. (2002). Requirement of functional telomeres for metaphase chromosome alignments and integrity of meiotic spindles. EMBO Rep 3:230–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhyu MS. (1995). Telomeres, telomerase, and immortality. J Natl Cancer Inst 87:884–894 [DOI] [PubMed] [Google Scholar]
- 7.Flores I, Canela A, Vera E, Tejera A, Cotsarelis G. and Blasco MA. (2008). The longest telomeres: a general signature of adult stem cell compartments. Genes Dev 22:654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harley CB, Vaziri H, Counter CM. and Allsopp RC. (1992). The telomere hypothesis of cellular aging. Exp Gerontol 27:375–382 [DOI] [PubMed] [Google Scholar]
- 9.Greider CW. and Blackburn EH. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405–413 [DOI] [PubMed] [Google Scholar]
- 10.Collins K, Kobayashi R. and Greider CW. (1995). Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell 81:677–686 [DOI] [PubMed] [Google Scholar]
- 11.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL. and Shay JW. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011–2015 [DOI] [PubMed] [Google Scholar]
- 12.Wright WE, Piatyszek MA, Rainey WE, Byrd W. and Shay JW. (1996). Telomerase activity in human germline and embryonic tissues and cells. Dev Genet 18:173–179 [DOI] [PubMed] [Google Scholar]
- 13.Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C. and DePinho RA. (1999). Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96:701–712 [DOI] [PubMed] [Google Scholar]
- 14.Lee HW, Blasco MA, Gottlieb GJ, Horner JW, 2nd, Greider CW. and DePinho RA. (1998). Essential role of mouse telomerase in highly proliferative organs. Nature 392:569–574 [DOI] [PubMed] [Google Scholar]
- 15.Herrera E, Martinez AC. and Blasco MA. (2000). Impaired germinal center reaction in mice with short telomeres. EMBO J 19:472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maciejewski JP. and Risitano A. (2003). Hematopoietic stem cells in aplastic anemia. Arch Med Res 34:520–527 [DOI] [PubMed] [Google Scholar]
- 17.Mason PJ, Wilson DB. and Bessler M. (2005). Dyskeratosis congenita—a disease of dysfunctional telomere maintenance. Curr Mol Med 5:159–170 [DOI] [PubMed] [Google Scholar]
- 18.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S. and Wright WE. (1998). Extension of life-span by introduction of telomerase into normal human cells. Science 279:349–352 [DOI] [PubMed] [Google Scholar]
- 19.Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, et al. (2011). Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469:102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegl-Cachedenier I, Flores I, Klatt P. and Blasco MA. (2007). Telomerase reverses epidermal hair follicle stem cell defects and loss of long-term survival associated with critically short telomeres. J Cell Biol 179:277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jesus BB, Schneeberger K, Vera E, Tejera A, Harley CB. and Blasco MA. (2011). The telomerase activator TA-65 elongates short telomeres and increases health span of adult/old mice without increasing cancer incidence. Aging Cell 10:604–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flores I, Cayuela ML. and Blasco MA. (2005). Effects of telomerase and telomere length on epidermal stem cell behavior. Science 309:1253–1256 [DOI] [PubMed] [Google Scholar]
- 23.Cong Y. and Shay JW. (2008). Actions of human telomerase beyond telomeres. Cell Res 18:725–732 [DOI] [PubMed] [Google Scholar]
- 24.Choi J, Southworth LK, Sarin KY, Venteicher AS, Ma W, Chang W, Cheung P, Jun S, Artandi MK, et al. (2008). TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet 4:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, et al. (2009). Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE. and Artandi SE. (2005). Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436:1048–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pucci F, Gardano L. and Harrington L. (2013). Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell 12:479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shkreli M, Sarin KY, Pech MF, Papeta N, Chang W, Brockman SA, Cheung P, Lee E, Kuhnert F, et al. (2012). Reversible cell-cycle entry in adult kidney podocytes through regulated control of telomerase and Wnt signaling. Nat Med 18:111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR. and Jefferson RA. (1997). Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet 6:2011–2019 [DOI] [PubMed] [Google Scholar]
- 30.Hrdlickova R, Nehyba J. and Bose HR., Jr (2012). Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol Cell Biol 32:4283–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hisatomi H, Ohyashiki K, Ohyashiki JH, Nagao K, Kanamaru T, Hirata H, Hibi N. and Tsukada Y. (2003). Expression profile of a gamma-deletion variant of the human telomerase reverse transcriptase gene. Neoplasia 5:193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saeboe-Larssen S, Fossberg E. and Gaudernack G. (2006). Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol Biol 7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi X, White DM, Aisner DL, Baur JA, Wright WE. and Shay JW. (2000). An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia 2:433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colgin LM, Wilkinson C, Englezou A, Kilian A, Robinson MO. and Reddel RR. (2000). The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia 2:426–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulaner GA, Hu JF, Vu TH, Giudice LC. and Hoffman AR. (1998). Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res 58:4168–4172 [PubMed] [Google Scholar]
- 36.Ulaner GA, Hu JF, Vu TH, Giudice LC. and Hoffman AR. (2001). Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int J Cancer 91:644–649 [PubMed] [Google Scholar]
- 37.Brenner CA, Wolny YM, Adler RR. and Cohen J. (1999). Alternative splicing of the telomerase catalytic subunit in human oocytes and embryos. Mol Hum Reprod 5:845–850 [DOI] [PubMed] [Google Scholar]
- 38.Liu WJ, Zhang YW, Zhang ZX. and Ding J. (2004). Alternative splicing of human telomerase reverse transcriptase may not be involved in telomerase regulation during all-trans-retinoic acid-induced HL-60 cell differentiation. J Pharmacol Sci 96:106–114 [DOI] [PubMed] [Google Scholar]
- 39.Yi X, Shay JW. and Wright WE. (2001). Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res 29:4818–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majerska J, Sykorova E. and Fajkus J. (2011). Non-telomeric activities of telomerase. Mol Biosyst 7:1013–1023 [DOI] [PubMed] [Google Scholar]
- 41.Chung HK, Cheong C, Song J. and Lee HW. (2005). Extratelomeric functions of telomerase. Curr Mol Med 5:233–241 [DOI] [PubMed] [Google Scholar]
- 42.Santos JH, Meyer JN. and Van Houten B. (2006). Mitochondrial localization of telomerase as a determinant for hydrogen peroxide-induced mitochondrial DNA damage and apoptosis. Hum Mol Genet 15:1757–1768 [DOI] [PubMed] [Google Scholar]
- 43.Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, et al. (2007). Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol 5:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, Birch-Machin MA, von Zglinicki T. and Saretzki G. (2008). Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J Cell Sci 121:1046–1053 [DOI] [PubMed] [Google Scholar]
- 45.Zhu H, Fu W. and Mattson MP. (2000). The catalytic subunit of telomerase protects neurons against amyloid beta-peptide-induced apoptosis. J Neurochem 75:117–124 [DOI] [PubMed] [Google Scholar]
- 46.Listerman I, Sun J, Gazzaniga FS, Lukas JL. and Blackburn EH. (2013). The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res 73:2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y, Li H, Deb S. and Liu JP. (2002). TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene 21:3130–3138 [DOI] [PubMed] [Google Scholar]
- 48.Perrault SD, Hornsby PJ. and Betts DH. (2005). Global gene expression response to telomerase in bovine adrenocortical cells. Biochem Biophys Res Commun 335:925–936 [DOI] [PubMed] [Google Scholar]
- 49.Armstrong L, Saretzki G, Peters H, Wappler I, Evans J, Hole N, von Zglinicki T. and Lako M. (2005). Overexpression of telomerase confers growth advantage, stress resistance, and enhanced differentiation of ESCs toward the hematopoietic lineage. Stem Cells 23:516–529 [DOI] [PubMed] [Google Scholar]
- 50.Lee MK, Hande MP. and Sabapathy K. (2005). Ectopic mTERT expression in mouse embryonic stem cells does not affect differentiation but confers resistance to differentiation- and stress-induced p53-dependent apoptosis. J Cell Sci 118:819–829 [DOI] [PubMed] [Google Scholar]
- 51.Yang C, Przyborski S, Cooke MJ, Zhang X, Stewart R, Anyfantis G, Atkinson SP, Saretzki G, Armstrong L. and Lako M. (2008). A key role for telomerase reverse transcriptase unit in modulating human embryonic stem cell proliferation, cell cycle dynamics, and in vitro differentiation. Stem Cells 26:850–863 [DOI] [PubMed] [Google Scholar]
- 52.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD. and Carpenter MK. (2001). Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol 19:971–974 [DOI] [PubMed] [Google Scholar]
- 53.Johannes G. and Sarnow P. (1998). Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4:1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mavrogiannou E, Strati A, Stathopoulou A, Tsaroucha EG, Kaklamanis L. and Lianidou ES. (2007). Real-time RT-PCR quantification of human telomerase reverse transcriptase splice variants in tumor cell lines and non-small cell lung cancer. Clin Chem 53:53–61 [DOI] [PubMed] [Google Scholar]
- 55.Seimiya H, Oh-hara T, Suzuki T, Naasani I, Shimazaki T, Tsuchiya K. and Tsuruo T. (2002). Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-1991. Mol Cancer Ther 1:657–665 [PubMed] [Google Scholar]
- 56.Cohn EP, Wu KL, Pettus TR. and Reich NO. (2012). A new strategy for detection and development of tractable telomerase inhibitors. J Med Chem 55:3678–3686 [DOI] [PubMed] [Google Scholar]
- 57.Pitts AE. and Corey DR. (1998). Inhibition of human telomerase by 2′-O-methyl-RNA. Proc Natl Acad Sci U S A 95:11549–11554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J. and Hendrix MJ. (2006). Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat Med 12:925–932 [DOI] [PubMed] [Google Scholar]
- 59.Pfaffl MW. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon MC. and Keith B. (2008). The role of oxygen availability in embryonic development and stem cell function. Nat Rev Mol Cell Biol 9:285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunwoodie SL. (2009). The role of hypoxia in development of the mammalian embryo. Dev Cell 17:755–773 [DOI] [PubMed] [Google Scholar]
- 62.Lin CY, Peng CY, Huang TT, Wu ML, Lai YL, Peng DH, Chen PF, Chen HF, Yen BL, Wu KK. and Yet SF. (2012). Exacerbation of oxidative stress-induced cell death and differentiation in induced pluripotent stem cells lacking heme oxygenase-1. Stem Cells Dev 21:1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Postovit LM, Margaryan NV, Seftor EA, Kirschmann DA, Lipavsky A, Wheaton WW, Abbott DE, Seftor RE. and Hendrix MJ. (2008). Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc Natl Acad Sci U S A 105:4329–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hazeltine LB, Selekman JA. and Palecek SP. (2013). Engineering the human pluripotent stem cell microenvironment to direct cell fate. Biotechnol Adv 31:1002–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohyeldin A, Garzon-Muvdi T. and Quinones-Hinojosa A. (2010). Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell 7:150–161 [DOI] [PubMed] [Google Scholar]
- 66.Ezashi T, Das P. and Roberts RM. (2005). Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci U S A 102:4783–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forristal CE, Wright KL, Hanley NA, Oreffo RO. and Houghton FD. (2010). Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction 139: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zachar V, Prasad SM, Weli SC, Gabrielsen A, Petersen K, Petersen MB. and Fink T. (2010). The effect of human embryonic stem cells (hESCs) long-term normoxic and hypoxic cultures on the maintenance of pluripotency. In Vitro Cell Dev Biol Anim 46:276–283 [DOI] [PubMed] [Google Scholar]
- 69.Chen HF, Kuo HC, Lin SP, Chien CL, Chiang MS. and Ho HN. (2010). Hypoxic culture maintains self-renewal and enhances embryoid body formation of human embryonic stem cells. Tissue Eng Part A 16:2901–2913 [DOI] [PubMed] [Google Scholar]
- 70.Mathieu J, Zhang Z, Nelson A, Lamba DA, Reh TA, Ware C. and Ruohola-Baker H. (2013). Hypoxia induces re-entry of committed cells into pluripotency. Stem Cells 31:1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida Y, Takahashi K, Okita K, Ichisaka T. and Yamanaka S. (2009). Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 5:237–241 [DOI] [PubMed] [Google Scholar]
- 72.Powers DE, Millman JR, Huang RB. and Colton CK. (2008). Effects of oxygen on mouse embryonic stem cell growth, phenotype retention, and cellular energetics. Biotechnol Bioeng 101:241–254 [DOI] [PubMed] [Google Scholar]
- 73.Ramirez MA, Pericuesta E, Yanez-Mo M, Palasz A. and Gutierrez-Adan A. (2011). Effect of long-term culture of mouse embryonic stem cells under low oxygen concentration as well as on glycosaminoglycan hyaluronan on cell proliferation and differentiation. Cell Prolif 44:75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen HF, Kuo HC, Chen W, Wu FC, Yang YS. and Ho HN. (2009). A reduced oxygen tension (5%) is not beneficial for maintaining human embryonic stem cells in the undifferentiated state with short splitting intervals. Hum Reprod 24:71–80 [DOI] [PubMed] [Google Scholar]
- 75.Allegrucci C. and Young LE. (2007). Differences between human embryonic stem cell lines. Hum Reprod Update 13:103–120 [DOI] [PubMed] [Google Scholar]
- 76.Millman JR, Tan JH. and Colton CK. (2009). The effects of low oxygen on self-renewal and differentiation of embryonic stem cells. Curr Opin Organ Transplant 14:694–700 [DOI] [PubMed] [Google Scholar]
- 77.Fernandes TG, Diogo MM, Fernandes-Platzgummer A, da Silva CL. and Cabral JM. (2010). Different stages of pluripotency determine distinct patterns of proliferation, metabolism, and lineage commitment of embryonic stem cells under hypoxia. Stem Cell Res 5:76–89 [DOI] [PubMed] [Google Scholar]
- 78.Barbosa HS, Fernandes TG, Dias TP, Diogo MM. and Cabral JM. (2012). New insights into the mechanisms of embryonic stem cell self-renewal under hypoxia: a multifactorial analysis approach. PLoS One 7:e38963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Westfall SD, Sachdev S, Das P, Hearne LB, Hannink M, Roberts RM. and Ezashi T. (2008). Identification of oxygen-sensitive transcriptional programs in human embryonic stem cells. Stem Cells Dev 17:869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forsyth NR, Kay A, Hampson K, Downing A, Talbot R. and McWhir J. (2008). Transcriptome alterations due to physiological normoxic (2% O2) culture of human embryonic stem cells. Regen Med 3:817–833 [DOI] [PubMed] [Google Scholar]
- 81.Coussens M, Davy P, Brown L, Foster C, Andrews WH, Nagata M. and Allsopp R. (2010). RNAi screen for telomerase reverse transcriptase transcriptional regulators identifies HIF1alpha as critical for telomerase function in murine embryonic stem cells. Proc Natl Acad Sci U S A 107:13842–13847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishi H, Nakada T, Kyo S, Inoue M, Shay JW. and Isaka K. (2004). Hypoxia-inducible factor 1 mediates upregulation of telomerase (hTERT). Mol Cell Biol 24:6076–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anderson CJ, Hoare SF, Ashcroft M, Bilsland AE. and Keith WN. (2006). Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene 25:61–69 [DOI] [PubMed] [Google Scholar]
- 84.Yu RM, Chen EX, Kong RY, Ng PK, Mok HO. and Au DW. (2006). Hypoxia induces telomerase reverse transcriptase (TERT) gene expression in non-tumor fish tissues in vivo: the marine medaka (Oryzias melastigma) model. BMC Mol Biol 7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Minamino T, Mitsialis SA. and Kourembanas S. (2001). Hypoxia extends the life span of vascular smooth muscle cells through telomerase activation. Mol Cell Biol 21:3336–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haendeler J, Hoffmann J, Diehl JF, Vasa M, Spyridopoulos I, Zeiher AM. and Dimmeler S. (2004). Antioxidants inhibit nuclear export of telomerase reverse transcriptase and delay replicative senescence of endothelial cells. Circ Res 94:768–775 [DOI] [PubMed] [Google Scholar]
- 87.Haendeler J, Hoffmann J, Brandes RP, Zeiher AM. and Dimmeler S. (2003). Hydrogen peroxide triggers nuclear export of telomerase reverse transcriptase via Src kinase family-dependent phosphorylation of tyrosine 707. Mol Cell Biol 23:4598–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haendeler J, Drose S, Buchner N, Jakob S, Altschmied J, Goy C, Spyridopoulos I, Zeiher AM, Brandt U. and Dimmeler S. (2009). Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arterioscler Thromb Vasc Biol 29:929–935 [DOI] [PubMed] [Google Scholar]
- 89.Hirashima K, Migita T, Sato S, Muramatsu Y, Ishikawa Y. and Seimiya H. (2013). Telomere length influences cancer cell differentiation in vivo. Mol Cell Biol 33:2988–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smogorzewska A. and de Lange T. (2004). Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73:177–208 [DOI] [PubMed] [Google Scholar]
- 91.Wong VC, Ma J. and Hawkins CE. (2009). Telomerase inhibition induces acute ATM-dependent growth arrest in human astrocytomas. Cancer Lett 274:151–159 [DOI] [PubMed] [Google Scholar]
- 92.Kim HS, Quon MJ. and Kim JA. (2014). New insights into the mechanisms of polyphenols beyond antioxidant properties; lessons from the green tea polyphenol, epigallocatechin 3-gallate. Redox Biol 2:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mata JE, Joshi SS, Palen B, Pirruccello SJ, Jackson JD, Elias N, Page TJ, Medlin KL. and Iversen PL. (1997). A hexameric phosphorothioate oligonucleotide telomerase inhibitor arrests growth of Burkitt's lymphoma cells in vitro and in vivo. Toxicol Appl Pharmacol 144:189–197 [DOI] [PubMed] [Google Scholar]
- 94.Chepelev I. and Chen X. (2013). Alternative splicing switching in stem cell lineages. Front Biol (Beijing) 8:50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu H, He L. and Tang L. (2012). Alternative splicing regulation and cell lineage differentiation. Curr Stem Cell Res Ther 7:400–406 [DOI] [PubMed] [Google Scholar]
- 96.Venables JP. (2006). Unbalanced alternative splicing and its significance in cancer. Bioessays 28:378–386 [DOI] [PubMed] [Google Scholar]
- 97.Sykorova E. and Fajkus J. (2009). Structure-function relationships in telomerase genes. Biol Cell 101:375–392; 1 p following 392. [DOI] [PubMed] [Google Scholar]
- 98.Amor S, Remy S, Dambrine G, Le Vern Y, Rasschaert D. and Laurent S. (2010). Alternative splicing and nonsense-mediated decay regulate telomerase reverse transcriptase (TERT) expression during virus-induced lymphomagenesis in vivo. BMC Cancer 10:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wong MS, Chen L, Foster C, Kainthla R, Shay JW. and Wright WE. (2013). Regulation of telomerase alternative splicing: a target for chemotherapy. Cell Rep 3:1028–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y, Meeker AK, Kowalski J, Tsai HL, Somervell H, Heaphy C, Sangenario LE, Prasad N, Westra WH, Zeiger MA. and Umbricht CB. (2011). Telomere length is related to alternative splice patterns of telomerase in thyroid tumors. Am J Pathol 179:1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu S, Rousseau P, Lauzon C, Gandin V, Topisirovic I. and Autexier C. (2014). Inactive C-terminal telomerase reverse transcriptase insertion splicing variants are dominant-negative inhibitors of telomerase. Biochimie 101:93–103 [DOI] [PubMed] [Google Scholar]
- 102.Wu YL, Dudognon C, Nguyen E, Hillion J, Pendino F, Tarkanyi I, Aradi J, Lanotte M, Tong JH, Chen GQ. and Segal-Bendirdjian E. (2006). Immunodetection of human telomerase reverse-transcriptase (hTERT) re-appraised: nucleolin and telomerase cross paths. J Cell Sci 119:2797–2806 [DOI] [PubMed] [Google Scholar]
- 103.Morcos PA. (2007). Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos. Biochem Biophys Res Commun 358:521–527 [DOI] [PubMed] [Google Scholar]
- 104.Schmajuk G, Sierakowska H. and Kole R. (1999). Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J Biol Chem 274:21783–21789 [DOI] [PubMed] [Google Scholar]
- 105.Gebski BL, Mann CJ, Fletcher S. and Wilton SD. (2003). Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet 12:1801–1811 [DOI] [PubMed] [Google Scholar]
- 106.Brambilla C, Folini M, Gandellini P, Daprai L, Daidone MG. and Zaffaroni N. (2004). Oligomer-mediated modulation of hTERT alternative splicing induces telomerase inhibition and cell growth decline in human prostate cancer cells. Cell Mol Life Sci 61:1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mazumdar J, O'Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS. and Simon MC. (2010). O2 regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol 12:1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, et al. (2013). Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet 45:371–384, 384e1–384e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.